Abstract
Phosphorylation of myosin regulatory light chain (RLC) N-terminal extension (NTE) activates myosin in thick filaments. RLC phosphorylation plays a primary regulatory role in smooth muscle and a secondary (modulatory) role in striated muscle, which is regulated by Ca2+ via TnC/TM on the thin filament. Tarantula striated muscle exhibits both regulatory systems: one switches on/off contraction through thin filament regulation, and another through PKC constitutively Ser35 phosphorylated swaying free heads in the thick filaments that produces quick force on twitches regulated from 0 to 50% and modulation is accomplished recruiting additional force-potentiating free and blocked heads via Ca2+4-CaM-MLCK Ser45 phosphorylation. We have used microsecond molecular dynamics (MD) simulations of tarantula RLC NTE to understand the structural basis for phosphorylation-based regulation in tarantula thick filament activation. Trajectories analysis revealed that an inter-domain salt bridges network (R39/E58,E61) facilitates formation of a stable helix-coil-helix (HCH) motif made up by helices P and A in the unphosphorylated NTE of both myosin heads. Phosphorylation of blocked head on Ser45 does not induce any substantial structural change. However, phosphorylation of free head on Ser35 disrupts this salt bridge network and induces a partial extension of helix P along RLC helix A. While not directly participating in the HCH inter-domain folding, phosphorylation of Ser35 unlocks compact structure and allows the NTE to spontaneously undergo coil-helix transitions. The modest structural change induced by subsequent Ser45 diphosphorylation monophosphorylated Ser35 free head, facilitates full helix P extension into a single structurally stable α-helix through a network of intra-domain salt bridges (pS35/R38,R39,R42). We conclude that tarantula thick filament activation is controlled by sequential Ser35-Ser45 phosphorylation via a conserved disorder-to-order transition.
Introduction
Modulation of Ca2+ concentration regulates the actin-myosin ATPase, myosin crossbridge cycling on actin and hence contraction in all muscles. This control mechanism is linked to molecular switches located either on thin (actin-containing) or thick (myosin-containing) filaments that assemble to form the sarcomere.1 In the first case (actin-linked regulation), troponintropomyosin (TN/TM) regulates access of myosin heads to the thin filament. In the second case (myosin-linked regulation), the myosin head activity itself is regulated, either by Ca2+ binding to the essential light chains (ELC) as in molluscan muscles,2 or to calmodulin (CaM), resulting in activation of myosin light chain kinase (MLCK) and phosphorylation of the myosin regulatory light chain (RLC). In vertebrate smooth muscle, this phosphorylation-based regulatory scheme constitutes the primary regulatory mechanism for muscle contraction.3 In contrast, actin-linked regulation constitute the primary regulatory mechanism in arthropod (Limulus,4 tarantula,5-11 scorpion12) and vertebrate13 striated muscle, whereas RLC phosphorylation plays a secondary role. This secondary (or modulatory) role seem to be controlled by two phosphorylation sites in Limulus,4 tarantula5-7 and scorpion.12
Tarantula striated muscle exhibits a dual regulation mechanism:5-11 (A) a primary regulatory mechanism in which contraction is switched on/off via TN/TM in the thin filaments8 and (B) a secondary regulatory mechanism in the thick filament producing 0 to 50% of the total force through a fixed number of Ser35 monophosphorylated swaying free heads, predetermined constitutively by a protein kinase C (PKC)9-11 where force can be potentiated via modulation of the net available heads by MLCK Ser45 phosphorylation.10, 11 Therefore in tarantula striated muscle contraction is switched on by the increase of Ca2+ concentration through the TN/TM switch at thin filaments. The force produced depends on the available swaying Ser35 monophosphorylated free heads.10 Ser45 phosphorylation of heads that are not constitutively Ser35 monophosphorylated allows recruiting additional heads, potentiating the force, i.e. phosphorylation modulates the produced force.10
Myosin II, the myosin isoform in muscles, contains two heavy chains each with a pair (ELC and RLC) of light chains.14 The heavy chains form a long coiled-coil tail (light meromyosin) and subfragment 2 (S2), while the remaining N-terminal part forms the subfragment 1 (S1) globular head. This S1 has an N-terminal catalytic motor domain, which binds actin and ATP, and a C-terminal regulatory domain, which binds the light chains and acts as a lever arm, to move thick filaments along thin filaments.
Electron microscopy (EM) studies of 2D crystals have shown that the two heads of isolated vertebrate smooth muscle myosin molecules form an asymmetric interacting-heads structure. This structural model showed that the actin-binding interface of one head (i.e., the blocked head) interacts with the converter and catalytic domains of the other head (i.e., the free head). As a result, the blocked head is unable to bind to thin filaments.15-17 Furthermore, the actin-binding activity of the free head is not blocked, but its ATPase activity, however, is inhibited. Myosin filament structure has been most extensively studied in tarantula muscle. Cryo-EM and single particle analysis have shown that the interacting-heads motif is present in native thick filaments,9,18 demonstrating that it is not an artifact of myosin isolation or 2D-crystallization techniques. The myosin “interacting-heads” motif (Fig. 1Aa) has been observed in thick filaments isolated from striated (tarantula,18 Limulus,19 scorpion,12 scallop20), cardiac (mouse,21 human,22 zebra fish23) or smooth (Schistosome)24 muscle, as well as on isolated myosin molecules from striated, cardiac, smooth and nonmuscle tissue.25-27 This motif is highly conserved, underlying the relaxed state of thick filaments in both smooth and striated muscles over a wide range of species since vertebrates and invertebrates diverged through evolution.26
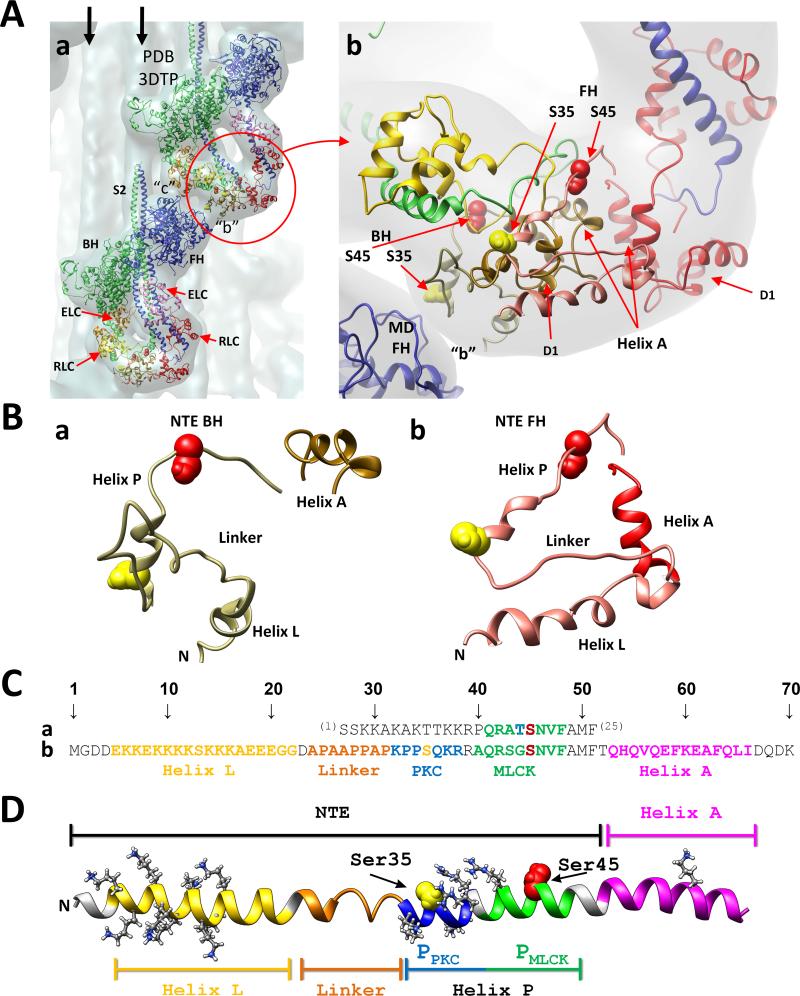
Fig. 1.
(A) Two tarantula myosin interacting-heads motifs (PDB 3DTP) are shown in (a) fitted to the tarantula thick filament 3D-map (EMD 1950, in gray).9 Both motifs interact in “b” and “c” forming part of a helix on the thick filament. The free (FH, blue) and blocked (BH, green) heads and their essential (ELC, magenta and orange) and regulatory (RLC, red/pink and yellow/tan) light chains are shown as ribbons models, and are labelled in the bottom motif. Vertical arrows: neighbour myosin tails. Bare zone: top. A zoom of the RLCs region (red circle) of (a) is shown on (b) with the blocked and free head RLCs respectively in yellow and red, with their NTEs in tan and pink. Phosphorylatable serines: Ser35 (yellow spheres), Ser45 (red spheres). Both serines of BH are located below the BH domain 1 (D1), which is located below the FH NTE9 while both FH serines are located above, freely exposed. (B) Enlarged view of the isolated blocked (a) and free (b) NTEs formed by helix L, linker and helix P, adjacent to the RLC helix A. (C) Sequence alignment of the residues 1-2529 of chicken smooth muscle RLC NTE (a) and residues 1-709 of tarantula striated muscle which included the 52-aa RLC NTE and the adjacent 14-aa helix A (b). (D) Extended model of the tarantula NTE formed by helix L (yellow), flexible coil Pro/Ala linker (orange) and helix P (blue-green), adjacent to RLC EF helix A (magenta). Helix P is formed by the target consensus sequences of PCK (PPKC, blue) and MLCK (PMLCK, green) with its phosphorylatable serines Ser35 (yellow) and Ser45 (red). Helices L and P have respectively 9 and 5 positively charged residues (white).
The study of the tarantula myosin interacting-heads structure9, 18 has opened the way to understand the role of the myosin RLC phosphorylation on the sequential release of the free and blocked heads on activation. The tarantula RLC N-terminal extension (NTE) is very long (52-aa long) and possesses target consensus sequences for PKC and MLCK kinases for the phosphorylatable serines Ser35 and Ser459 (Fig. 1Cb). The free head RLC has been found to be constitutively monophosphorylated at Ser35.9 This constitutive phosphorylation makes this head capable of swaying away (i.e. swaying heads) to quickly interact with the thin filament to produce force in twitches.10 After a longer exposure on high Ca2+ concentration the MLCK becomes activated enabling it to diphosphorylate the Ser35 monophosphorylated swaying free head at Ser45, inducing its active release.10 This release then makes possible the phosphorylation at Ser45 of its partner blocked head, which becomes a swaying head.10 Therefore phosphorylation is the key on the sequential release of heads on activation.
The myosin RLC has two domains (connected by a linker helix) and a NTE.28 The length of this NTE varies depending on the species, being short in vertebrates and long in invertebrates.9
The most detailed information on the structure and dynamics of the NTE comes from smooth muscle RLC,29 which possesses a 25 residue-long NTE, so called phosphorylation domain (PD, residues 1-24)29 and features a single phosphorylation site (Ser-19) (Fig. 1Ca). Electronic paramagnetic resonance (EPR) spectroscopy has shown that, upon phosphorylation, the smooth muscle NTE becomes more helically ordered and solvent accessible.29 Molecular dynamics (MD) simulations of the isolated smooth muscle NTE further predicted that this domain undergoes a disorder-to-order transition upon phosphorylation.30 Furthermore, MD simulations and free energy calculations showed that phosphorylation acts as a molecular switch that allows reversibility of the phosphorylation-induced conformational transition.31 Complementary time-resolved fluorescence resonance energy transfer (TR-FRET) experiments and MD simulations revealed the coexistence of two RLC structural states, where phosphorylation switches the system from a closed state (with the NTE contacting the C-terminal lobe of the RLC) to a more dynamic (but helically ordered) open structural state.32 EM evidences have showed that in phosphorylated smooth muscle, the heavy meromyosin shows an open conformation linked to activation.33 Site directed mutagenesis results show that interactions between the phosphorylated smooth muscle RLC NTE and helix-A of the ELC are required for phosphorylation to activate smooth muscle myosin.34
NTE structure is unknown because it is absent in myosin head crystal structures.14 Nevertheless, using secondary structure prediction a RLC structure was obtained and flexible fitted to the frozen-hydrated thick filament 2.0 nm 3D-reconstruction deposited in the Electron Microscopy Data Bank (EMDB)35 as EMDB-19509 obtaining the myosin interacting-heads motif PDB 3DTP9 (Fig. 1A) deposited in the Protein Data Bank (PDB).36 The final 3D-map revealed that the blocked NTE region is more compact than the free NTE region (Fig. 1Ab,B).9 Furthermore, the free head is above but separated from the backbone, interacting only with its own subfragment 2 (S2) (Fig. 1Aa), while the blocked head reside completely above the backbone, interacting with it and with the motor domain of the neighbour free head (Fig. 1Aa).9 Based on these observations, it was concluded that the phosphorylation sites of the free head RLC are exposed to the surrounding solvent, allowing the free NTE to be phosphorylated both at Ser35 and Ser45 (Fig. 1Ab).9,10 In contrast Ser35 and Ser45 of the blocked heads are not exposed and cannot be phosphorylated (Fig. 1Ab).9,10 This is so because these two serines are located between the backbone and domain 1 of the blocked head RLC, which covers them completely, hindering their phosphorylation by PKC or MLCK (Fig. 1Ab).9-11 Only when the free head is released, these two blocked head serines become exposed so the Ser45 can be phosphorylated by the activated MLCK. Since Ser35 is located on a different consensus sequence, it cannot be phosphorylated by activated MLCK neither can it be phosphorylated by inactivated PKC.9-11
These structural differences between the free and blocked heads have important functional consequences: the free head NTE is involved in the sway away and active release of the swaying free heads, whereas the blocked head NTE is involved in the swaying away of blocked heads on potentiation.9-11 Therefore, studying these differences at atomic detail is essential to understand the functional implications of NTE phosphorylation on each case. In this study, we performed unbiased MD simulations to determine the mechanisms by which sequential phosphorylation induces structural changes in the NTE. These studies are important because we simulated the effects of phosphorylation on NTE starting from a largely unstructured segment as opposed to previous studies of Espinoza-Fonseca et al.30, 31 where the NTE was initially modelled as a straight helix. The phosphorylation-induced structural changes of the NTE observed in our simulations are supported by previous structural and functional studies performed by our groups9-11,30,31,37 thus validating our simulations.
Methods
Setup for MD simulations
We followed methods described in Espinoza-Fonseca et al.30 We performed MD simulations starting from relaxed NTE conformations either of the PDB 3DTP free or blocked head RLCs (Fig. 1B, Fig. 2A) plus the adjacent 14-aa RLC helix A (Fig. 1Cb,D) to obtain the blocked and free head equilibrated conformations (Fig. 2B). To investigate the changes in the NTE structure upon phosphorylation, these equilibrated structures were used as starting conformations for two phosphorylated peptides: blocked head Ser45 monophosphorylated NTE and free head Ser35 monophosphorylated NTE. This last peptide was used for the free head NTE diphosphorylated at Ser35 and Ser45. Peptides were capped N-methylamide at the C-terminus. All peptides were solvated using TIP3P water molecules; the size of the water box was large enough to prevent the peptide from interacting with its periodic image. Na+ and Cl- ions were added to the system to neutralize the charge of the system and to produce a NaCl concentration of approximately 150 mM. CHARMM22 force field topologies and parameters38 with CMAP corrections39 were used for the protein, water and ions.
Fig. 2.
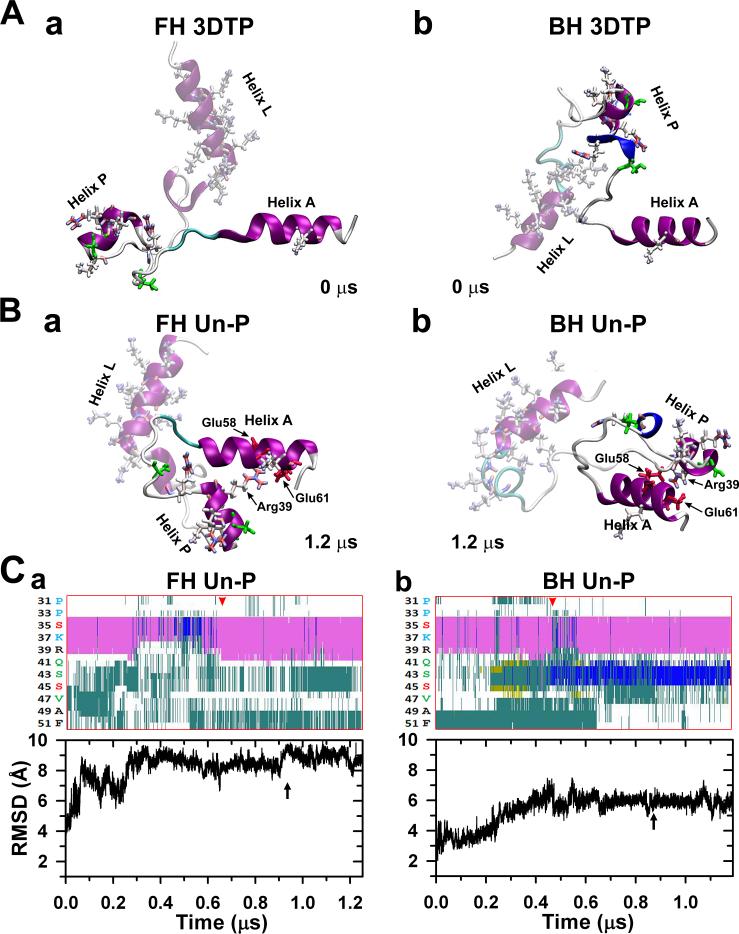
(A) Interacting heads motif (PDB 3DTP) free (a) and blocked (b) heads RLCs NTE peptides starting structures used for the MD simulations. The helices A of both myosin RLCs are at the right. Both unphosphorylated (un-P) compact peptides stabilized after ~1.2 νs in the two conformations shown in (B), remaining compact, with helix P inter-domain folded or bent close to helix A making a helix-coil-helix (HCH) motif. A network of salt bridges was established between R39 of helix P and residues E58 and E61 of helix A (Table 1). The evolution of the secondary structure of both NTE helices PPKC (Ser32-Arg38) and PMLCK (Ala40-Phe48) regions (cf. Fig. 1Cb) along each peptide trajectories is shown at top of (C): the α-helix content (pink) of helix PPKC sequence of the free head (a) does not change substantially after ~ 0.65 μs (red arrowhead) while the helix PMLCK is mostly turn (cyan) and coil (white). The blocked head helix content (b) increase by the presence of 3-10 helix (blue) after ~ 0.45 μs (red arrowhead), and the turn (cyan) and coil (white) content stabilizes. In both heads the root mean square deviation (RMSD) shows little changes after ~ 0.9 μs (C, bottom, black arrows). After stabilizing to the final snapshots shown in (B), the free and blocked head peptides were then phosphorylated at Ser35 (Fig. 5a) or Ser45 (Fig. 4a) respectively. Secondary structure colour key: 3-10 helix (blue), α-helix (pink), turn (cyan), coil (white), β-sheet (yellow). In this figure and in figures 4-6 the helix L is shadowed to emphasize the changes in the helix P conformation.
It is worth noting that in vivo, the diphosphorylated free head NTE can only be reached from a previously stabilized Ser35 monophosphorylated free head structure.10,11 Therefore there will be only one final true structure and chemical model for the diphosphorylated previously Ser35 monophosphorylated free head NTE that would reach a single local minimum in its folding pathway. Also, in vivo, the NTE -as part of the RLC and the myosin interacting-heads motif- may not fold as we have studied it in this work with isolated NTEs. Instead the NTE conformation would probably evolve in the context of the rest of the RLC structure to which it belongs and of its myosin heavy chain partner. This might completely change its stabilized structure from the ones we observed here. Also, the trajectories may be exploring a space unavailable in vivo due to the space occupied by the rest of the RLC. The highly positively charged and flexible helix L could make salt bridges with the charged surface of the calmodulin-like part of its own RLC or other adjacent parts of the thick filament, especially given the eventual flexibility contributed by the NTE linker. This last point was observed in the current simulations, in which helix L docks back on the rest of the negatively charged end of the helix A. This, however, does not seem to interfere with the local changes occurring in the helix-P. In this paper we focused only on the structural changes occurring on the NTE, specifically on helix P where both critical phosphorylatable serines (Ser35 and Ser45) are located. For that reason we did not take into account the changes on helix L and the NTE coil linker.
Molecular dynamics simulation protocol
MD simulations were performed using NAMD version 2.5.30,40 Periodic boundary conditions,41 particle mesh Ewald method,42, 43 a nonbonded cutoff of 9 Å and a 2 fs time step were used. The NPT assemblage was maintained with a Langevin thermostat (310K) and a Langevin piston barostat (1 atm). Energy minimizations were attained with initial 1000 steps of conjugate-gradient algorithm with restraints to the protein backbone, followed by additional 1000 steps, but without restraints. Systems were warmed up for 20 ps, then equilibrated for 60 ps with lower restraints, finishing at 310 K with no restraints. A description of the systems and conditions used for each simulation are shown in Supplementary Table 1.
Molecular graphics images for the Fig. 1 were produced using the UCSF Chimera package,44 Figs. 2, 4, 5 and 6, Supplementary Fig. 1 and 2 and movies S1 and S2 were produced using the Visual Molecular Dynamics (VMD) program.45
Fig. 4.
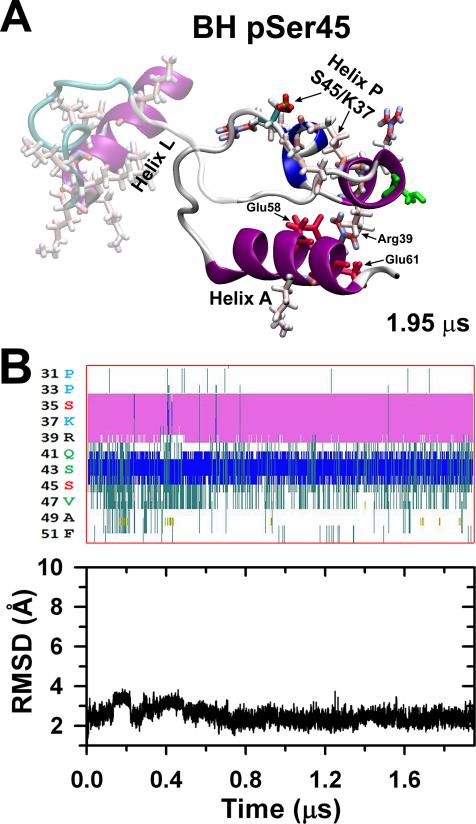
The stabilized BH un-P compact peptide (Fig. 2Bb) was phosphorylated at Ser45, stabilizing after 1.95 μs to a compact conformation (A) in which a salt bridge is formed between the pSer45 and the Lys37 (Table 1) but maintaining the R39/E58,E61 salt bridge network. The α-helix (pink), 3-10 helix (blue) and coil (white) contents of helix P do not change much (B, top), and the RMSD is low remaining stable over all simulation (C, bottom).
Fig. 5.
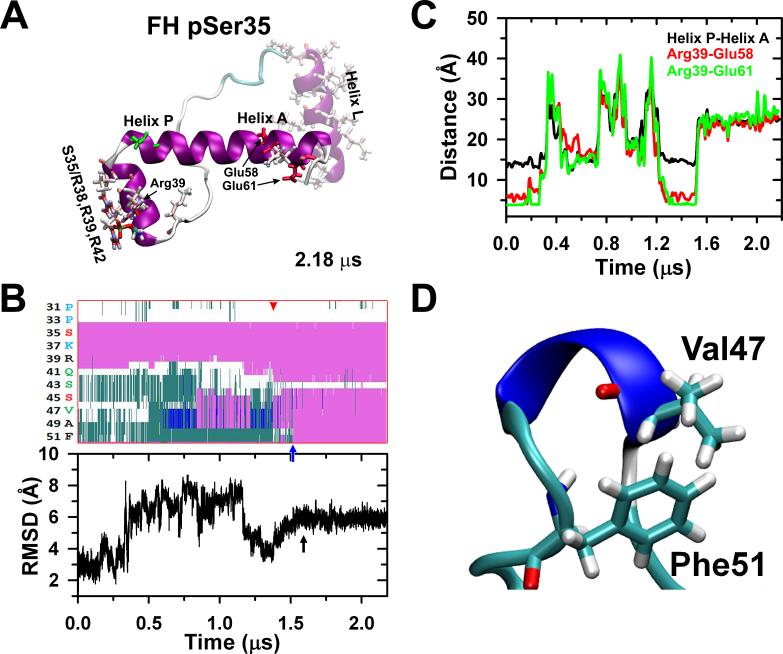
The stabilized FH unphosphorylated compact peptide (Fig. 2Ba) was phosphorylated at Ser35, stabilizing after 2.18 μs to a less compact conformation (A) in which part of helix P become an extension of helix A (HCH motif), partially elongating it partially as a consequence of the formation of another salt bridge network between the pSer35 and Arg38, Arg39 and Arg42 (Table 1) that breaks the R39/E58,E61 salt bridge network. The secondary structure evolution of the unphosphorylated free head after monophosphorylation in Ser35 is shown in (B, top). After ~1.5 μs (red arrowhead) there is an increase of the α-helix (pink) content of the helix P (PMLCK and PPKC) becoming an extension of the helix A (blue arrow) leaving a coil gap around Ser43. The stabilization of the structure is also shown by the RMSD (B, bottom, black arrow). The monophosphorylated Ser35 forms hydrogen bonds with Arg39 destabilizing the inter-domain salt bridges Arg39-Glu58 and Arg39-Glu61 (Table 1), inducing the separation of helices P and A (C) and removing the structural restraints in Val47-Phe48-Ala49 allowing a main chain reorganization which facilitates interaction between the Val47 methyl group and Phe51 aromatic ring (D).
Fig. 6.
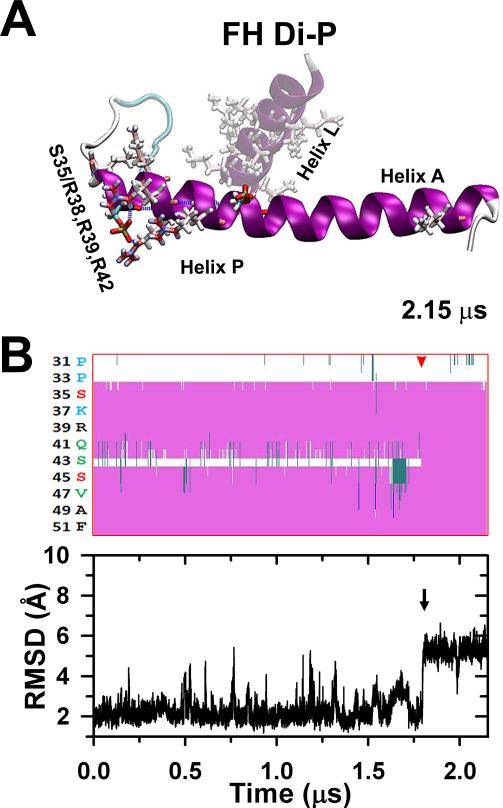
The stabilized FH pSer35 less compact peptide (Fig. 5A) was phosphorylated at Ser45, stabilized after 2.15 μs to a full elongated conformation in which the rest of helix P gets aligned along helix A culminating the disorder-to-order transition.30 The diphosphorylation does not establish new salt bridges (A). The secondary structure evolution of the stabilized Ser35 monophosphorylated peptide after Ser45 diphosphorylation is shown in (B, top) indicating that after ~1.8 μs (red arrowhead) the Ser35 and Ser45 parts of helix P become a continuous helix along helix A. The black arrow corresponds to the last conformational change that stabilizes the peptide correlated to the RMSD evolution (B, bottom).
Results
Structure of blocked and free head unphosphorylated NTE
The NTE is formed by two helices connected by a short loop linker: the N-terminal helix L and the helix P (with phosphorylation sites), and is appended to the RLC helix A (Fig. 1B, D, Fig. 2A). RLC NTE's in 3DTP9 (Fig. 1A) from both the free and blocked heads were used as starting structures for MD simulations (Fig. 2A). Initial performed tests suggested that simulations on the nanosecond range was not enough to achieve a stable state, thus indicating that microsecond-long MD trajectories are needed to capture the structural dynamics of the NTE. We performed 1.2 μs long MD simulations to determine whether free and blocked heads unphosphorylated NTE segments undergoes structural changes in solution. The secondary structure of the NTE is preserved in both simulations (Fig. 2B, C). Helices L, P and A do not fold themselves in our simulations (i.e. HCH inter-domain folding), indicating that these segments are intrinsically structured in solution (Fig. 2B). In the absence of phosphorylation, the regions of NTE adjacent to sites Ser35 and Ser45 do not undergo intra-domain folding in this time scale (Fig. 2B).
Structures extracted at the end of the 1.2 μs trajectories showed that both NTE peptides from the free and blocked head converged to two different structural states (Fig. 2B). A RMSD value of ~10 Å between the two structures at the end of the trajectories shows a substantial difference on the spatial arrangement of the helical segments between the NTE from the free and blocked heads (Fig. 2B). In addition, we observed some minor structural differences in the regions that contain phosphorylation sites Ser35 (PPKC, residues Ser32-Arg-38) and Ser45 (PMLCK, residues Ala40-Phe48). Although the α-helix content of helix PPKC of the free and blocked head remains largely unchanged in the trajectories (Fig. 2B), the helix PMLCK is structurally different between peptides for the free and blocked heads. PMLCK of the free head conform to mostly turn and coil structures (Fig. 2Ca), whereas residues 42-44 of PMLCK in the blocked head folds into a 3-10 helix (Fig. 2Cb).
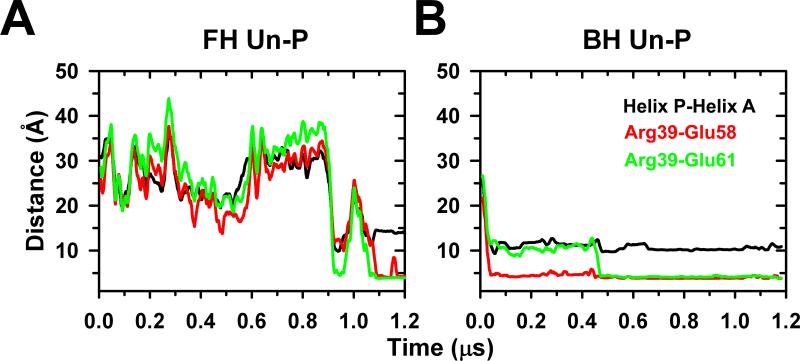
Despite these structural differences, quantitative analysis showed structural similarities between the NTE from the free and blocked heads. Analysis of the distances between P and A helices of the free head shows that it fluctuates between 20 and 45 Å during the first 0.9 μs of simulations, and then converges to an inter-domain distance of 14 Å after 1 μs (Fig. 3A). The inter-domain distance between P and A helices from the blocked head showed that although these segments are initially separated by a distance of ~25 Å, this distance rapidly decreases and settles at a plateau around 10 Å (Fig. 3B), showing that helices interact with each other to form a helix-coil-helix (HCH) structure in both NTE peptides from the free and blocked heads (Fig. 2B). We observed that the interaction between helices P and A facilitates the formation of a compact structure of NTE C-terminus and helix A of the RLC in both heads. The HCH structure formed between the P and A helices is not stabilized by residues between hydrophobic residues, but by a stable salt bridge network between residues R39 of helix P and E58/E61 of helix A (“R39/E58,E61”, Table 1). The formation of salt bridges R39-E58 or R39-E61 correlates with inter-domain distance decrease between P and A helices (Fig. 3). These observations suggest that this network of salt bridges is important for the formation and stability of the HCH structure of P and A helices in the unphosphorylated peptides of both free and blocked heads. The stable equilibrated compact HCH structure of the blocked head NTEs (cf. Fig. 2Ab vs. 2Bb) matches with the compact shape of this region in the EM density maps (Fig. 1Ab, Table 1).9
Fig. 3.
Free (A) and blocked (B) heads inter-domain distance between the P and A helices (black) of the NTE HCH motif. In the free head (A) both helices are initially separated by ~35 Å, decreasing in ~0.9 μs to ~ 10 Å, settling finally to ~14 Å after 1 μs. In contrast the P-A helix separation in the blocked head (B) shortens from ~26 Å in ~0.05 μs and finally settling to ~10 Å. The distances between Arg39 and Glu58 (red) and Arg39 and Glu61 (green) diminished to less than 5 Å after 1.1 μs for the free head and 0.46 μs for the blocked head settling down the salt bridge network.
Table 1.
Comparison between the un-, mono- and diphosphorylated tarantula RLC NTE peptides charge, conformation, secondary structure and shape
| Head | Phosphorylation level |
Net charge | RLC shape according to the 3D-Map9 |
HCH inter-domain salt bridges |
HCH inter-domain folding |
Helix P intra-domain salt bridges |
Helix P extension |
Disorder-to-order transition**** |
|---|---|---|---|---|---|---|---|---|
| BH | Un-P | 0 | Compact | R39/E58-E61 | Folded | none | None | No |
| BH* | S45 Mono-P | −2 | N/A | R39/E58-E61 | Folded | pS45/K37 | None | No |
| FH | Un-P | 0 | N/A | R39/E58-E61 | Folded | none | None | No |
| FH** | S35 Mono-P | −2 | Less compact | none | Disrupted | pS35/R38-R39-R42 | Partial | Partial |
| FH*** | S35 & S45 di-P | −4 | N/A | none | Disrupted | pS35/R38-R39-R42 | Full | Full |
Only the free head is constitutively monophosphorylated at Ser35.10, 11 There is no constitutive Ser35 monophosphorylation in the blocked head.10, 11
Only the blocked head can be monophosphorylated at Ser45 as the free head is already constitutively monophosphorylated in Ser35. 10, 11
Only the free head can be biphosphorylated as it is constitutively monophosphorylated at Ser35 while the blocked head, which is not constitutively monophosphorylated, cannot be diphosphorylated.10, 11
Disorder-to-order is used here describe a coil-to-helix transition as originally proposed by Espinoza-Fonseca et al.30, 31
N/A: 3D-map not available, as thick filaments are disordered when activated.5
Effects of Ser45 phosphorylation on the structure of the blocked head NTE
According to the cooperative phosphorylation mechanism previously described by our group, MLCK can only phosphorylate Ser45 of the blocked head upon release of the free head.10,11 Therefore, we performed a 1.95 μs long MD simulation of the blocked head NTE to determine the effects of Ser45 phosphorylation (pSer45) on the structure of this peptide. We used the equilibrated conformation of unphosphorylated blocked head NTE at the end of the 1.2 μs as a starting structure for this simulation (Fig. 2Bb). To determine the global effects of pSer45 on the structure of blocked head NTE, we calculated the average RMSD differences between the structures generated in the trajectory pSer45 and the structure of unphosphorylated head NTE at 1.2 μs. The average RMSD difference between the two peptides was 3.9±0.7 Å, indicating that the structure of blocked head Ser45 NTE phosphorylated is compact and nearly identical to the unphosphorylated one in solution (cf. Fig. 4A vs. 2Bb). Analysis of the secondary structure showed that pSer45 does not induce changes in the secondary structure of the blocked head peptide (cf. Supplementary Fig. S1Ab vs. S1Bb).
We also analyzed the effect of pSer45 on the structure of the HCH motif formed between helices P and A. The secondary structure of helices P and A of unphosphorylated and Ser45 phosphorylated blocked head is identical (Fig. 2Cb, 4B, Supplementary Fig. S1Bb). Furthermore, analysis of the time-dependent changes in the RMSD of this segment showed that the structural arrangement of the HCH structure observed in unphosphorylated blocked head remains intact upon Ser45 monophosphorylation (cd. Fig. 2Cb vs. 4B). In fact, pSer45 does not interfere with the stabilizing interactions in this motif since inter-domain salt bridges between residues R39 of helix P and E58 and E61 are present in >97% of the simulation time. The α-helical and the 3-10 helical contents of PPKC and PMLCK helices remain mostly unchanged upon Ser45 phosphorylation (Fig. 4B top). So, compared to unphosphorylated blocked head NTE, pSer45 does not induce any structural changes on this peptide under physiological conditions.
The lack of a 3D-map for an activated tarantula thick filament, as activation produces the disordering of the myosin interacting-heads motifs helices,5,6 precludes the fitting of the monophosphorylated at Ser45 or diphosphorylated NTE conformations (cf.).17
We also performed a 2.4 μs MD simulation of the free head NTE phosphorylated on Ser45 (Fig. S2, Aa). We found that phosphorylation of Ser45 induces the formation of a β-sheet between Ser43 and Val47. However, phosphorylation of this residue does not have any effect on the helical order of the NTE. In fact, we found that pSer45 disrupts the helical structure of the helix P (Fig. S2, Ba). Furthermore, phosphorylation of Ser45 does not alter the compact structure of the NTE, as revealed by calculation of the time-dependent changes in RMSD (Fig. S2, Ba bottom plot). These findings confirm that phosphorylation of Ser45 alone does not induce substantial structural changes in either free or blocked head NTE.
Structural changes in free head NTE induced by Ser35 phosphorylation
We have previously shown that the free heads are constitutively monophosphorylated at Ser35.10,11 Therefore, we performed a 2.18 μs MD simulation of the free head NTE phosphorylated at Ser35 (pSer35). To this aim, we used the compact conformation of unphosphorylated free head NTE obtained at the end of the 1.2 μs trajectory (Fig. 2Ba); this starting structure was monophosphorylated at pSer35 and equilibrated as described in Methods. Analysis of the RMSD differences between unphosphorylated and free head pSer35 NTE showed that average RMSD difference between the two peptides is >10 Å. This reveals that Ser35 phosphorylation induces large global changes in the structure of the free head NTE. In addition, we observed that the free head Ser35 phosphorylated NTE conforms to a less compact structure as compared to the free head unphosphorylated NTE (Fig. 5A vs. 2Ba, Table 1, Movie S1).
Analysis of the secondary structure of the helix P showed that phosphorylation of Ser35 induces important changes in the structure of the HCH segment formed by helices P and A. In particular, pSer35 induces a partial extension of the helix A and induces the formation of a continuous single helix with a segment of helix P (Fig. 5B, top). Unlike free head unphosphorylated NTE, phosphorylation at Ser35 induces the formation of a 3-10 helix centered on residues Val47-Ala49 at ~0.6 μs (blue helix in Fig. 5D); this 3-10 helix remains fairly stable between 0.6 and 0.85 μs (Fig. 5B). After this period of time, this segment undergoes transitions between 3-10 helix, α-helix and β-turn until ~1.55 μs (Fig. 5B, top). Time-dependent evolution of both the secondary structure show that residues 43-51 form a single helix, and that this segment settles a plateau at ~1.6 μs (Fig. 5B, bottom). These results show that upon Ser35 phosphorylation, the HCH motif formed helices P and A is structurally disrupted and undergoes intra-domain folding into a single, structurally stable α-helix in solution. This structural change is responsible for the diminished compactness of the free head NTE Ser35 phosphorylated, in agreement with the EM density map of the RLCs region, which is less compact in the free head NTE than in the blocked head NTE (Fig. 1Ab, Table 1).9
According to our results the intra-domain folding NTE starting region is located ~12 residues away from pSer35 (Val47 near helix A). Therefore, we set out to determine, in our simulation, the mechanisms by which Ser35 phosphorylation allosterically induce intra-domain folding of the free head NTE. First, we analyzed the local changes in residue-residue interactions at the HCH motif formed by helices P and A. We noted that pSer35 forms hydrogen bonds with residues Arg38, Arg39 and Arg42; this interaction results in inter-domain salt bridges network Arg39/Glu58, Glu61 destabilization (Table 1). The disruption of this salt bridge network induces the spatial separation of P and A helices upon Ser35 phosphorylation (Fig. 5C). Nevertheless, pSer35 only forms a local network of salt bridges with Arg38 and Arg39 (>90 % of the time) upon separation of the P and A helices and does not form direct stabilizing/ordering interactions in the region of NTE that undergoes intra-domain folding (residues Gly44-Phe51).
It seems then, that Ser35 phosphorylation only disrupts the compact structure of the free head NTE, but does not form stabilizing interactions that directly induce inter-domain folding of this domain. By which mechanisms pSer35 facilitates the inter-domain folding of the HCH into a stable helix in solution? A total of 45% of the residues in the loop connecting helices P and A are hydrophobic (Val47-Phe51), and the formation of the 3-10 helix upon phosphorylation on Ser35 begins around residues Val47, Phe48 and Ala49. Analysis of the RMSD showed that Val47, Phe48 and Ala49 have low mobility (RMSD <1.5 Å) in the HCH motif formed between helices P and A. Upon Ser35 phosphorylation, the RMSD values of this three-residue segment fluctuate between 4 and 13 Å, indicating that separation of helices P and A releases the structural restraints granting a structural reorganization of the main chain. Indeed, removal of the structural restraints in this segment seems to facilitate the interaction between the methyl group of Val47 and the aromatic ring of Phe51 (Fig. 5D). In the unphosphorylated free head NTE, these residues are separated by a distance of ~12 Å, upon Ser35 phosphorylation this distance decreases to ~7 Å to form direct side chain-side chain interactions. The interaction between these residues stabilizes the 3-10 helix between 0.6 and 0.85 μs (Fig. 5B top). This helix eventually propagates toward the C-terminus and extends toward helix A to form a kinked helix at position Ser43 (Fig. 5A, B). This intra-domain folding mechanism supports previous findings showing that 3-10 helix plays an important role in helix nucleation and that helix propagation occurs preferentially from the N- to C-terminus direction.46 So it seems that Ser35 phosphorylation does not directly participate in the intra-domain folding of the NTE; instead, it unlocks – via the inter-domain unfolding - the compact structure of this peptide, allowing its spontaneous coil-helix transitions by altering its intra-domain folding.
We performed an additional 1.3 μs MD simulation of the blocked head NTE phosphorylated on Ser35 to determine if structural changes observed in our simulations are phosphorylation- or free/blocked head-dependent. Trajectory analysis of secondary structure and RMSD (Fig. S2, Bb) showed that phosphorylation of Ser35 does not induce folding of the blocked head. We found that pSer35 forms a salt bridge with Arg38, which probably stabilizes the compact structure of the NTE (Fig. S2, Ba). These results indicate that the effect of phosphorylation on Ser35 is dependent on the structural arrangement of the free and blocked heads, thus providing a structural explanation as to why only the free head needs to be constitutively phosphorylated for function.10, 11
Taking together these findings shows that the constitutive Ser35 phosphorylation initially disrupt the inter-domain salt bridges network (R39/E58,E61) and consequently the HCH structure, enabling the establishment of a new intra-domain salt bridges network (pSer35/R38,R39,R42) (Table 1). Therefore the phosphorylation at Ser35 induces a constitutive structural change in the free head in contrast to what happen with phosphorylation at Ser45 in the blocked head, which due to the action of MLCK is not constitutive and does not induce a constitutive structural change.
Structural consequences of sequential free head NTE Ser45 phosphorylation
We also performed a 2.15 μs MD simulation of the free head NTE diphosphorylated at Ser45 after Ser35 to determine the structural changes associated with sequential phosphorylation of the free head NTE. For this, we used the structure of free head NTE phosphorylated at Ser35 extracted at the end of the 2.18 μs trajectory (Fig. 5A). Trajectory analysis showed that during the first 1.75 μs of simulation, the secondary structure content of free head diphosphorylated NTE is almost identical to that observed in the last 0.5 μs of the Ser35 monophosphorylated simulation (Supplementary Fig. S1Ba vs. Ca). The kinked helix formed upon Ser35 phosphorylation is stable in the trajectory of free head diphosphorylated NTE, showing that the ordering effect of pSer35 monophosphorylation is preserved in the microsecond time scale (Fig. 6A).
The most important structural effect of sequential phosphorylation was observed after t=1.75 μs: Ser43 undergoes a disorder-order transition, allowing the kinked helix to extend completely and form a long helix by spanning the chain from residue Pro33 to Gln64 (Supplementary Fig. S1Ca, Table 1, movie S2). This extended helix remains completely folded throughout the remaining trajectory. Additional analysis showed that pSer45 does not form salt bridge interactions with helix P, which is rich in basic residues arginine and lysine. This data suggests that the mechanism by which the free head NTE undergoes a modest disorder-order transition upon diphosphoryaltion occurs via long-range electrostatic interactions in a similar fashion to that previously observed in the NTE of smooth muscle RLC.30,32
Discussion
Functional consequences of the asymmetric disposition of the free and blocked heads
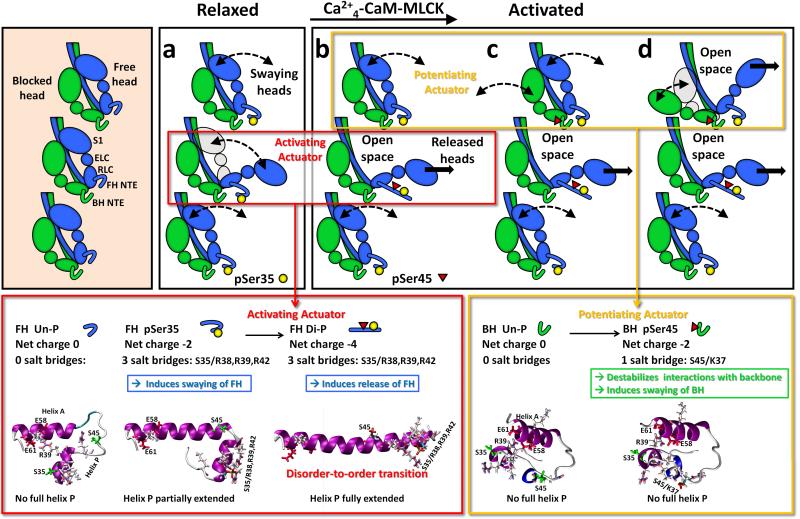
Studies on the tarantula thick filament structure have shown that the free and blocked head molecular environments are very different (Fig. 1Aa).9-11 Based on in vitro motility assays, sequence analysis, and mass spectrometry,10 Western blots, immunofluorescence, MLCK inhibition,11 and O18-MS (Biasutto et al, in preparation) a molecular cooperative phosphorylation mechanism for muscle activation (Fig. 7) was proposed.10, 11 In this mechanism heads are phosphorylated by PKC (Fig. 7a) and MLCK (Fig. 7b-d) in a way that can explain tarantula's (and possible other arthropods) striated muscle force development, force potentiation and post-tetanic potentiation: the Ser35 of the free heads can be constitutively phosphorylated by PKC (Fig. 7a), while the blocked head are unphosphorylated; on activation (Fig. 7b-d), MLCK phosphorylates the free heads (Fig. 7b) and blocked heads, both at Ser45 (Fig. 7c). In this mechanism, the sequential control release of free and blocked heads was hypothesized to reside on two phosphorylation-induced disorder-to-order transition actuators (Fig. 7a,b red box, Fig. 7b-d, yellow box),9,10 similarly to the one reported for the smooth myosin RLC NTE.30
Fig. 7.
The cooperative phosphorylation-controlled mechanism for recruiting active heads in tarantula thick filament activation (a-d)10, 11 showing the two actuators (red and yellow boxes) that control the sequential release of free (red box) and blocked (yellow box) myosin heads on tarantula thick filament activation. On the left three myosin interacting-heads motifs are shown along one thick filament helix with RLC NTEs unphosphorylated (bare zone at the top). In the “activating actuator” (red boxes) the Ser45 phosphorylation of the constitutively Ser35 monophosphorylated swaying free heads in (a) induces a complete disorder-to-order transition. This transition fully elongates the helix P along the helix A (depicted as a blue cylinder) by establishing three salt bridges pSer35/Arg38,Arg39,Arg42 (b). This elongation suggests that the diphosphorylated NTE would possibly stiffen, altering the free head regulatory domain, producing the release of the swaying free heads (i.e. released heads) in (b). In the “potentiating actuator” (yellow boxes), the Ser45 monophosphorylation of the unphosphorylated blocked head in (b), does not produce any conformational change on the NTE, but only a decrease of its charge by −2. This could weaken the interaction of its NTE with a loop on the motor domain of the neighbour free head, allowing it to sway away (c, curved arrows), producing the release of its partner constitutively Ser35 monophosphorylated free head (d, arrow).
In tarantula striated muscle the blocked and free heads have very different functional and structural roles: the free head NTE is mainly involved in setting the initial level of activation of the thick filament by the swaying free heads through Ser35 monophosphorylation (Fig. 7a) while its release through Ser35-Ser45 diphosphorylation (Fig. 7b center), is part of a cooperative mechanism that allows phosphorylation at Ser45 of the blocked head (Fig. 7c top), whereas the blocked head NTE, through Ser45 monophosphorylation, is only involved in the swaying away (Fig. 7c,d top) during potentiation.9-11 These different roles are a consequence of the presence of two key structural features in the myosin RLC NTE of myosin: the free head local secondary structure can be regulated by activated PKC Ser35 phosphorylation and activated Ca2+4-CaM-MLCK Ser45 phosphorylation (Fig. 1Cb); in contrast, blocked head regulation occurs only at Ser45 by activated Ca2+4-CaM-MLCK (Fig. 7b-d). We hypothesize that phosphorylation-induced secondary structure changes in the helix P of free and blocked head RLC NTEs which allows them to act as “molecular actuators” (Fig. 7, red and yellow boxes), i.e. actively responsible for these heads flexibility and for altering some of the interactions between them, with their neighbour heads and with the backbone, as well as its own. Our aim here was, by using predictions from all-atom μs MD simulations, to disclose which secondary structure changes are induced by Ser35 and/or Ser45 phosphorylation that could provide to these two molecular actuators the faculty to assign very different Ser45 functional roles to each head.
The consequences of Ser45 mono- and diphosphorylation could produce, along the filament and toward the bare zone, a cooperative unzipping of adjacent myosin interacting-heads motifs, starting with the diphosphorylation of one free head and followed by the Ser45 monophosphorylation of the next blocked head above, and continuing along this helix without the requirement that each free head diphosphorylation have to occur to facilitate phosphorylation of the neighbouring blocked head (Fig. 7a-d) 10. In other words the diphosphorylation produces just a “seed hole” on one of the myosin heads helices, unzipping the myosin interacting-heads motifs towards the bare zone in a cooperative way.10 The mechanism also suggests that, upon relaxation, when both free and blocked heads become unphosphorylated at Ser45, the blocked head would dock back first onto the backbone avoiding so steric clashes. Then its partner free head would do the same on top of it, locking the blocked head with its NTE (Fig. 7b) and recovering the initial structural relaxed state (Fig. 7a).10
Structural basis of phosphorylation-based tarantula thick filament activation
Our results (Figs. 2-6) reveal that, upon mono- and diphosphorylation, different conformational changes occur on both free and blocked heads RLC NTEs. It is through these different changes that Ca2+ might control the differential release of these heads via RLC phosphorylation. These two changes - driven by two phosphorylation-dependent molecular actuators30,47 and ascribed to the blocked or free heads provides, as shown below, a plausible structural basis to explain the steps a-b of the molecular mechanism for tarantula thick filament activation (Fig. 7a,b). Our results identify one of these two molecular actuators (Fig. 7, red box) as residing in the helix P of the free head: the Ser45 diphosphorylation of a constitutively Ser35 monophosphorylated free head. We have observed, by spectroscopy and MD simulations30-32,37 that this phosphorylation induces a modest but crucial structural change through long-range electrostatic interactions that facilitates the full helix P extension as in smooth muscle RLC NTE.32 In contrast, the blocked head Ser45 phosphorylation does not induce any structural changes on this peptide under physiological conditions, leaving unclear how the mechanism of the other molecular actuator (Fig. 7, yellow box) promotes the swaying away of the pSer45 blocked heads. Since at physiological activating conditions only Ser45 phosphorylation and unphosphorylation might occur,9-11 we concentrate below on the functional role of Ser45 phosphorylation on the activation of tarantula thick filaments.
The mechanism for phosphorylation-based activation in tarantula depends on simple steric/accessibility factors that control which head would be phosphorylated and so enabled to interact with actin to generate filament sliding.10,11 Our MD simulations (Figs. 2-6, Table 1) of the 3DTP NTE structures provide additional structural information, suggesting the structural basis of how the constitutive free head Ser35 monophosphorylation allows them to sway away. We also obtained additional structural information on how the sequential phosphorylation on Ser45 of the constitutively Ser35 monophosphorylated free head release it and making it mobile, prompted us to include these new features in the improved cooperative phosphorylation mechanism, as shown in Fig. 7.
Blocked and free heads unphosphorylated NTEs are locked in a compact conformation by the inter-domain folding of its HCH motifs
Our results show structural similarities between the free and blocked heads unphosphorylated NTE. In both heads a compact NTE structure was facilitated by the interaction between helices P and A, forming a HCH structure induced by a stable salt bridge network between residues R39 of helix P and E58/E61 of helix A (Table 1). The regions of NTE adjacent to unphosphorylated sites Ser35 and Ser45 do not undergo intra-domain folding. The inter-domain network of salt bridges is important for the formation and stability of the HCH structure of helices P and A on both free and blocked heads unphosphorylated NTEs. It is important to notice that in the relaxed state (Fig. 7a) only blocked heads are unphosphorylated, while free heads are constitutively Ser35 monophosphorylated. Therefore only the conformational result on the unphosphorylated blocked head NTE peptide is relevant in relaxed physiological conditions (Fig. 7a), in agreement with the 3D map densities.9 Our previous work9-11 showed that there are not thick filaments in which all the RLC NTE of either free and blocked heads are unphosphorylated as depicted in the left panel of Fig. 7 (shadowed box).
Ser45 phosphorylation of the blocked head does not change the NTE secondary structure
Ser45 phosphorylation does not induce changes in the structural arrangement of the HCH structure nor the secondary structure of the blocked head NTE, which remains compact and nearly identical to the unphosphorylated one in solution (Fig. 4A, Fig. 7c top). Ser45 phosphorylation does not interfere with the stabilized interactions since inter-domain salt bridges between residues R39 of helix P and E58/E61 (Table 1) that are present and the α-helical and the 3-10 helical contents of PPKC and PMLCK helices remain mostly unchanged. In spite of the lack of conformational changes on the blocked head after Ser45 phosphorylation, the blocked heads do sway away (Fig. 7c, top motif),10,11 supporting the detected force potentiation. It is very striking that blocked head Ser45 monophosphorylation does not stimulate the partial helix P elongation that Ser35 monophosphorylation promotes on the free head as discussed below (cf. Fig. 4 vs. 5). This apparent disparity is probably a direct consequence of the actual locations of the Ser45 and Ser35 consensus sequences on PMLCK and PPKC helices of the RLC NTE (Fig. 1D). Neither the intra-domain salt bridge pS45/K37 detected upon Ser45 phosphorylation nor the other intra-domain salt bridges shown in Table 1 generate by themselves any functional structural changes. On view of this lack of structural changes on the blocked head NTE, we suggest that the striking functional effect caused by Ser45 monophosphorylation of the blocked head, i.e. its swaying away (Fig. 7c),10,11 could be induced by the actual NTE electrical charge reduction (Table 1) after Ser45 monophosphorylation. This charge reduction (−2) could impair the interaction “b” 9 between the blocked head RLC NTE and the adjacent free head motor domain (Fig. 1Aa red circle, Fig. 1Ab), allowing a blocked head to sway away and back (Fig. 7c). Additionally, the Ser45 monophosphorylation would induce a blocked head regulatory domain IQ motif flexibilization,48 favouring its swaying away.
Constitutive Ser35 phosphorylation of the free head NTE unlocks its secondary structure making it to sway away
Free head NTE Ser35 phosphorylation induces a partial extension of the helix A hence the formation of a continuous single helix with a segment of helix P (Fig. 5A). This structural change is responsible for loosen compactness of the free head NTE Ser35 phosphorylated in agreement with the EM density map in the RLCs region, which shows a less compact shape in the free head NTE than in the blocked head NTE (Table 1).9 pSer35 forms an exclusive local network of salt bridges with Arg38, Arg39 and Arg42 upon separation of the helices P and A (Table 1), disrupting its compact structure and does not form direct stabilizing/ordering interactions in the region of NTE that undergoes intra-domain folding (residues Gly44-Phe51). Consequently it just unlocks the compact structure of this peptide, which allows it to spontaneously undergo a disorder-to-order transition. As the free head Ser35 monophosphorylation is constitutive, it could imply an enforced presetting in the relaxed state, that only the free heads are swaying heads, to take advantage of their favourable location to sway away (Fig. 1A, Fig. 7a) that can eventually be released by diphosphorylation (Fig. 7b). Functionally free heads are completely different to blocked heads: they are completely exposed to the solvent and their RLC NTEs are not involved in any intermolecular interaction (Fig. 1Aa, red circle). The formation on the free head of three salt bridges (pS35/R38,R39,R42) upon Ser35 monophosphorylation, disrupts first the inter-domain network of salt bridges (Fig. 5A, Table 1), and induces then a partial helix P elongation along helix A (Fig. 2Ba vs. 5A, Table 1). This elongation induces changes in the free head regulatory domain, such that, in relaxing conditions, these heads may sway away (Fig. 7a), supporting quick twitch responses when neighbouring thin filaments are Ca2+-activated via TnC/TM (Fig. 7a).10
Ser45 phosphorylation of the constitutively Ser35 monophosphorylated free head NTEs endows its secondary structure to trigger activation
Diphosphorylation only induces a modest but functionally important structural change that facilitates full extension of helix A (Fig. 6A, Table 1). The conformational transition induced by diphosphorylation on the free head is similar to the disorder-to-order transition reported for vertebrate smooth muscle upon phosphorylation.30 The three salt bridges (pS35/R38,R39,R42) present on the constitutive Ser35 monophosphorylated free head remain present after Ser45 diphosphorylation, and does not directly produce any functional structural changes as well. We suggest that the free head diphosphorylation induces a complete disorder-to-order transition through an increase of α-helicity as quantified by the rigidification and straightening of the NTE49 (Fig. 6) releasing and making it mobile (Fig. 7b). Our predictions here allows us to extent our inquiry on the structural changes on the NTE structure with a different approach as done in the companion paper49 in which we report an analysis of the static and dynamic persistence lengths of the tarantula RLC NTE peptides showing that diphosphorylation produces an important 24-fold straightening and a 16-fold rigidification of the RLC NTE, while monophosphorylation at Ser35 (or Ser45) has a less profound effect that induces the release of the constitutively Ser35 monophosphorylated free head because the stiffened NTE would hinder the proper re-docking of the free head after swaying away, being unable to recovering its original interacting position so becoming released and mobile until it is Ser45 unphosphorylated by the endogenous MLCP. In the case of the Ser35 monophosphorylated free head this effect does not occur as there is not such increase in the flexural rigidity of the NTE, so the Ser35 monophosphorylated free head can sway away and back without problem. On the Ser45 monophosphorylated blocked head, there is not such increase in flexural rigidity either, and a similar effect cannot occur as the blocked head need to dock back first than the free head.49
Summarizing the free head phosphorylation condition clearly makes the difference between simply enabling it for swaying away (when it is only Ser35 monophosphorylated) or to become released and mobile (after diphosphorylation) (Fig. 7a,b).
In conclusion, MD simulations showed that in contrast to the vertebrate smooth muscle NTE disorder-to-order transition which is induced by Ser19 monophosphorylation by the smooth muscle activated Ca2+4-CaM-MLCK,30-32 in tarantula striated muscle NTE Ser35 monophosphorylation induces only a partial disorder-to-order transition, while Ser45 diphosphorylation induces a complete disorder-to-order transition through an increase of α-helicity, suggesting it is a functional improvement on striated muscle. Therefore, molecular actuators, located in the PKC/MLCK consensus sequences of the RLC NTE, allow the sequential release of the free and blocked heads as proposed by the cooperative phosphorylation mechanism for activation and potentiation of tarantula striated muscle. Direct structural information about the conformation of the tarantula RLC NTE, as well as concomitant changes on helix P associated to salt bridges upon mono- and diphosphorylation could be achieved by future EPR or FRET studies. Similarly, the possible elucidation of the atomic structure of the myosin interacting-heads motif in the relaxed state by X-ray crystallography50,51 would allow building a homologous tarantula atomic model to further complement the frozen-hydrated 3D-map, as well as the present MD simulations.
Supplementary Material
Acknowledgements
We thank Dr. Gustavo Márquez for his help with the manuscript. Molecular graphics images (Fig. 1) were produced using the UCSF Chimera package44 from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH P41 RR-01081). Figs. 2, 4, 5 and 6, Supplementary Fig. 1 and Movies S1 and S2 were produced using the Visual Molecular Dynamics (VMD) program45. Computational resources were also provided by the Minnesota Supercomputing Institute. This work was supported by grants from the National Institutes of Health (AR32961 to D.D.T., HL86655), the American Heart Association (10POST4350076 to L.M.E.F.) and the Howard Hughes Medical Institute (HHMI, to R.P.).
References
- 1.Lehman W, Szent-Györgyi AG. J. Gen. Physiol. 1975;66:1– 30. doi: 10.1085/jgp.66.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Szent-Gyorgyi AG. Adv. Exp. Med. Biol. 2007;592:253–264. doi: 10.1007/978-4-431-38453-3_21. [DOI] [PubMed] [Google Scholar]
- 3.Stull JT, K Blumenthal D, Cooke R. Biochem. Pharmacol. 1980;29:2537–2543. doi: 10.1016/0006-2952(80)90063-5. [DOI] [PubMed] [Google Scholar]
- 4.Sellers JR. J. Biol. Chem. 1981;256:9274–9278. [PubMed] [Google Scholar]
- 5.Craig R, Padrón R, Kendrick-Jones J. J. Cell Biol. 1987;105:1319–1327. doi: 10.1083/jcb.105.3.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Padrón R, Pante N, Sosa H, Kendrick-Jones J. J. Muscle Res. Cell Motil. 1991;12:235–241. doi: 10.1007/BF01745112. [DOI] [PubMed] [Google Scholar]
- 7.Hidalgo C, Craig R, Ikebe M, Padrón R. J. Muscle Res. Cell Motil. 2001;22:51–59. doi: 10.1023/a:1010388103354. [DOI] [PubMed] [Google Scholar]
- 8.Craig R, Lehman W. J. Mol. Biol. 2001;311:1027–1036. doi: 10.1006/jmbi.2001.4897. [DOI] [PubMed] [Google Scholar]
- 9.Alamo L, Wriggers W, Pinto A, Bartoli F, Salazar L, Zhao FQ, Craig R, Padrón R. J. Mol. Biol. 2008;384:780–797. doi: 10.1016/j.jmb.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brito R, Alamo L, Lundberg U, Guerrero JR, Pinto A, Sulbaran G, Gawinowicz MA, Craig R, Padrón R. J. Mol. Biol. 2011;414:44–61. doi: 10.1016/j.jmb.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sulbarán G, Biasutto A, Alamo L, Riggs C, Pinto A, Mendéz F, Craig R, Padrón R. Biophys. J. 2013;105:2114–2122. doi: 10.1016/j.bpj.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pinto A, Sanchez F, Alamo L, Padrón R. J. Struct. Biol. 2012;180:469–478. doi: 10.1016/j.jsb.2012.08.010. [DOI] [PubMed] [Google Scholar]
- 13.Sweeney HL, Bowman BF, Stull JT. Am. J. Physiol. 1993;264:C1085–C1095. doi: 10.1152/ajpcell.1993.264.5.C1085. [DOI] [PubMed] [Google Scholar]
- 14.Rayment I, Rypniewski WR, Schmidt-Base K, Smith R, Tomchick DR, Benning MM, Winkelmann DA, Wesenberg G, Holden HM. Science. 1993;261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- 15.Wendt T, Taylor D, Messier T, Trybus KM, Taylor KA. J. Cell Biol. 1999;147:1385–1390. doi: 10.1083/jcb.147.7.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wendt T, Taylor D, Trybus KM, Taylor K. Proc. Natl. Acad. Sci. U. S. A. 2001;98:4361–4366. doi: 10.1073/pnas.071051098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J, Wendt T, Taylor D, Taylor K. J. Mol. Biol. 2003;329:963–972. doi: 10.1016/s0022-2836(03)00516-3. [DOI] [PubMed] [Google Scholar]
- 18.Woodhead JL, Zhao FQ, Craig R, Egelman EH, Alamo L, Padrón R. Nature. 2005;436:1195–1199. doi: 10.1038/nature03920. [DOI] [PubMed] [Google Scholar]
- 19.Zhao FQ, Craig R, Woodhead JL. J. Mol. Biol. 2009;385:423–431. doi: 10.1016/j.jmb.2008.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woodhead JL, Zhao FQ, Craig R. Proc. Natl. Acad. Sci. U. S. A. 2013;110:8561–8566. doi: 10.1073/pnas.1218462110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zoghbi ME, Woodhead JL, Moss RL, Craig R. Proc. Natl. Acad. Sci. U. S. A. 2008;105:2386–2390. doi: 10.1073/pnas.0708912105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.AL-Khayat HA, Kensler RW, Squire JM, Marston SB, Morris EP. Proc. Natl. Acad. Sci. U. S. A. 2013;110:318–323. doi: 10.1073/pnas.1212708110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonzalez-Sola M, AL-Khayat HA, Behra M, Kensler RW. Biophys. J. 2014;106:1671–1680. doi: 10.1016/j.bpj.2014.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sulbarán G, Alamo L, Pinto A, Marquéz G, Méndez F, Padrón R, Craig R. Biophys. J. 2015;106:159a. [Google Scholar]
- 25.Burgess SA, Yu S, Walker ML, Hawkins RJ, Chalovich JM, Knight PJ. J. Mol. Biol. 2007;372:1165–1178. doi: 10.1016/j.jmb.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 26.Jung HS, Burgess SA, Billington N, Colegrave M, Patel H, Chalovich JM, Chantler PD, Knight PJ. Proc. Natl. Acad. Sci. U.S.A. 2008;105:6022–6026. doi: 10.1073/pnas.0707846105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jung HS, Komatsu S, Ikebe M, Craig R. Mol. Biol. Cell. 2008;19:3234–3242. doi: 10.1091/mbc.E08-02-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chacko S, Conti MA, Adelstein RS. Proc. Natl. Acad. Sci. U.S.A. 1977;74:129–133. doi: 10.1073/pnas.74.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nelson WD, Blakely SE, Nesmelov YE, Thomas DD. Proc. Natl. Acad. Sci. U.S.A. 2005;102:4000–4005. doi: 10.1073/pnas.0401664102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Espinoza-Fonseca LM, Kast D, Thomas DD. Biophys. J. 2007;93:2083–2090. doi: 10.1529/biophysj.106.095802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Espinoza-Fonseca LM, Kast D, Thomas DD. J. Am. Chem. Soc. 2008;130:12208–12209. doi: 10.1021/ja803143g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kast D, Espinoza-Fonseca LM, Yi C, Thomas DD. Proc. Natl. Acad. Sci. U.S.A. 2010;107:8207–8212. doi: 10.1073/pnas.1001941107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baumann BAJ, Taylor DW, Huang Z, Tama F, Fagnant PM, Trybus KM, Taylor KA. J. Mol. Biol. 2012;415:274–287. doi: 10.1016/j.jmb.2011.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor KA, Feig M, Brooks CL, Fagnant PM, Lowey S, Trybus KM. J. Struct. Biol. 2014;185:375–382. doi: 10.1016/j.jsb.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lawson CL, Baker ML, Best C, Bi C, Dougherty M, Feng P, van Ginkel G, Devkota B, Lagerstedt I, Ludtke SJ, Newman RH, Oldfield TJ, Rees I, Sahni G, Sala R, Velankar S, Warren J, Westbrook JD, Henrick K, Kleywegt GJ, Berman HM, Chiu W. Nucleic Acids Res. 2011;39:D456–D464. doi: 10.1093/nar/gkq880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berman HM, Bhat T, Bourne PE, Feng Z, Gilliland G, Weissig H, Westbrook J. Nat. Struct. Biol. 2000;11:957–959. doi: 10.1038/80734. [DOI] [PubMed] [Google Scholar]
- 37.Espinoza-Fonseca LM, Colson BA, Thomas DD. Mol. BioSyst. 2014;10:2693–2698. doi: 10.1039/c4mb00364k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiórkiewicz-Kuczera J, Yin D, Karplus M. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 39.Buck M, Bouguet-Bonnet S, Pastor RW, MacKerell AD., Jr Biophys. J. 2006;90:L36–L38. doi: 10.1529/biophysj.105.078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kalé L, Skeel R, Bhandarkar M, Brunner R, Gursoy A, Krawetz N, Phillips J, Shinozaki A, Varadarajan K, Schulten K. J. Comput. Phys. 1999;151:283–312. [Google Scholar]
- 41.Weber W, Hünenberger PH, McCammon JA. J. Phys. Chem. B. 2000;104:3668–3675. [Google Scholar]
- 42.Darden T, York D, Pedersen L. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 43.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 44.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 45.Humphrey W, Dalke A, Schulten K. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 46.Monticelli L, Tieleman DP, Colombo G. J. Phys. Chem. B. 2005;109:20064–20067. doi: 10.1021/jp054729b. [DOI] [PubMed] [Google Scholar]
- 47.Colson B, Gruber S, Thomas D. J. Muscle Res. Cell Motil. 2012;33:419–429. doi: 10.1007/s10974-012-9317-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Himmel DM, Mui S, O'Neall-Hennessey E, Szent-Györgyi AG, Cohen C. J. Mol. Biol. 2009;394:496–505. doi: 10.1016/j.jmb.2009.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alamo L, Li X, Espinoza-Fonseca LM, Pinto A, Thomas DD, Lehman W, Padrón R. Mol. Biosyst. 2015 doi: 10.1039/c5mb00163c. 2015, (submitted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O'Neall-Hennessey E, Reshetnikova L, Senthil Kumar VS, Robinson H, Szent-Györgyi AG, Cohen C. Acta Crystallogr., Sect. F: Struct. Biol. Cryst. Commun. 2013;69:248–252. doi: 10.1107/S1744309112049925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gillilan RE, Kumar VSS, O'Neall-Hennessey E, Cohen C, Brown JH. PLoS One. 2013;8:e81994. doi: 10.1371/journal.pone.0081994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.