Abstract
Lipoma in patients with the multiple endocrine neoplasia type 1 (MEN1) syndrome is a type of benign fat-cell tumor that has biallelic inactivation of MEN1 that encodes menin and could serve as a model to investigate normal and pathologic fat-cell (adipocyte) proliferation and function. The role of menin and its target genes in adipocytes is not known. We used in vitro differentiation to derive matched normal and menin-deficient adipocytes from wild type (WT) and menin-null (Men1-KO) mouse embryonic stem cells (mESCs), respectively, or 3T3-L1 cells without or with menin knockdown to investigate cell size, lipid content, and gene expression changes. Adipocytes derived from Men1-KO mESCs or after menin knockdown in 3T3-L1 cells showed a 1.5–1.7-fold increase in fat-cell size. Global gene expression analysis of mESC-derived adipocytes showed that lack of menin downregulated the expression of many differentially methylated genes including the tumor suppressor long noncoding RNA Meg3 but upregulated gene expression from the prolactin gene family locus. Our results show that menin deficiency leads to fat-cell hypertrophy and provide model systems that could be used to study the regulation of fat-cell size.
1. Introduction
Patients with the multiple endocrine neoplasia type 1 (MEN1) syndrome carry a heterozygous germline inactivating mutation in MEN1 that predisposes to tumors of multiple endocrine and nonendocrine tissues (OMIM ID 131100 and 613733) [1–3]. Lipoma in patients with MEN1 occurs as an overgrowth of fat tissue, frequently multiple, and usually in the subcutaneous area of the back, neck, shoulder, arm, thigh, or abdomen [4, 5]. The MEN1-associated lipoma is a type of benign fat-cell tumor occurring in about 30% of MEN1 patients and similar to the other MEN1-associated endocrine and nonendocrine tumors has biallelic inactivation of MEN1 (1st hit in the germline, 2nd hit tissue-specific) [5–9]. Lipoma tissue samples are rarely available for analysis because the tumor is harmless and removed only for cosmetic reasons by surgery or liposuction, and the tumor does not recur after removal. In the mouse models of Men1 loss generated in different laboratories, the endocrine tumors of MEN1 are frequently observed [10, 11]. However, lipoma was observed only in one mouse [12]. Therefore, there is a dearth of molecular studies exploring the pathogenesis of MEN1-associated lipoma.
The MEN1 encoded menin is a widely expressed, predominantly nuclear protein involved in transcriptional regulation and other vital cellular functions [13, 14]. Comparing gene expression changes in normal and menin-deficient fat-cells (adipocytes) could provide insights into the pathogenesis of MEN1-associated lipomas; at the same time such studies could also provide insights about normal adipocyte growth and function. No such studies have been performed perhaps due to the limitation of procuring normal adipocytes that can serve as a matched control for the abnormal adipocytes in the lipoma. Specific cell types produced by in vitro differentiation of embryonic stem cells (ESCs) or induced pluripotent stem cells (iPSCs) that are normal or mutant for a disease-causing gene can serve as useful models to study molecular processes that are altered in disease [15, 16]. This is highly desirable for affected cell types that are not easily available, are difficult to isolate, and are not abundant or that lie among other normal cell types. Menin-null (Men1-KO) mouse ESCs (mESCs) can differentiate in vitro into pancreatic islet-like endocrine cells (PILECs); however, they are defective in hematopoietic differentiation [17, 18]. It is not known whether Men1-KO mESCs can differentiate into adipocytes.
Gene expression analyses of wild type (WT) and Men1-KO mESCs, and mESC-derived WT and Men1-KO PILECs have shown a role of the menin-dependent histone H3 lysine-4 trimethyl mark (H3K4me3) in the regulation of genes clustered at the Meg3 locus and the four Hox gene clusters [18]. Most gene clusters are highly conserved, such as the four Hox gene clusters [19]. However, the prolactin family gene cluster is present in rodents and not in humans [20]. In humans, the prolactin gene resides alone in a gene poor region of the genome on chromosome 6. In rodents, gene duplication has generated a large prolactin gene family of 26 genes near the prolactin gene (mouse chromosome 13) with varying expression and functions in pregnancy and lactation [21, 22].
In this study, we show that adipocytes could be generated by in vitro differentiation of Men1-KO mESCs and upon menin knockdown from 3T3-L1 preadipocyte cells. These menin-deficient adipocytes and their corresponding normal adipocytes were analyzed for cell size, lipid content, and gene expression changes. Menin-deficient adipocytes from both cell types showed increased cell size with a modest increase in lipid content; and adipocytes derived from Men1-KO mESCs showed reduced expression of a few differentially methylated genes including the long noncoding RNA Meg3 and increased expression of prolactin family genes. These cells could enable further investigations by genomic and proteomic approaches to understand physiological and pathological states of adipocytes.
2. Materials and Methods
2.1. Primers
All the primers used in this study are provided in Supplementary Table 1 in Supplementary Material available online at http://dx.doi.org/10.1155/2015/149826.
2.2. Differentiation of Mouse Embryonic Stem Cells (mESCs) into Adipocytes
Adipogenesis was performed by using the reagents and protocol from a mESC adipogenesis kit (Millipore). Wild type TC-1 and Men1-KO-3.2N mESCs [17] were cultured on a feeder layer of primary mouse embryonic fibroblasts (MEFs) (ATCC) in ESC maintenance medium containing leukemia inhibitory factor (LIF). The mESCs were processed to obtain feeder-free cells by culturing on tissue culture treated dishes few times for an hour each to get rid of the attached feeder layer cells, and the unattached feeder-free mESCs were subsequently cultured on gelatin-coated dishes in ESC maintenance medium containing LIF. Cells were detached with accutase and plated in embryoid body (EB) formation medium (EB medium) (ESC maintenance medium without LIF) in nonadherent dishes for 2 days and then in medium containing inducer-A solution (all-trans-retinoic acid) for 3 days with a medium change every day. On day 5, the retinoic acid induced EBs were plated in gelatin-coated 24-well plates in adipocyte differentiation medium (EB medium + 3,3′,5-triiodo-L-thyronine (T3) + insulin). The adipocyte differentiation medium was changed every 2 days for a total of 21 days to allow adipocyte formation.
2.3. Transfection and Differentiation of 3T3-L1 Cells into Adipocytes
3T3-L1 cells (ATCC) were cultured in high glucose Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% Fetal Bovine Serum (FBS) and 1X antibiotic/antimycotic (3T3-L1 cell propagation medium). For menin knockdown, 1 × 106 cells were electroporated with 10 μg of control shRNA plasmid or Men1-shRNA plasmid [23] by using the Nepa Gene Super Electroporator NEPA21 Type II (Bulldog-Bio) (Porting Pulse: volts (V) = 175, length milliseconds (ms) = 5, interval (ms) = 50, number = 2, and decay rate (%) = 10; transfer pulse: V = 20, length (ms) = 50, interval (ms) = 50, number = 5, decay rate (%) = 40, and polarity = +/−). Electroporated cells were plated in 3T3-L1 cell propagation medium overnight and induced to differentiate into adipocytes using an established protocol [24]. On the next day medium was changed to adipocyte differentiation medium (3T3-L1 cell propagation medium with isobutylmethylxanthine, dexamethasone, and insulin) and incubated for 2 days. Thereafter the medium was changed to maintenance medium (3T3-L1 cell propagation medium with insulin) for the next 4 days with a change of medium every 2 days to allow adipocyte formation.
2.4. Oil Red O Staining and Lipid Content
Adipocytes obtained after 21 days of mESC differentiation were processed for Oil Red O staining by using the reagents provided in the mESC adipogenesis kit (Millipore). Oil Red O specifically stains lipids such as triglycerides and cholesteryl oleate [25]. For adipocytes derived from 3T3-L1 cells, Oil Red O (Sigma) reconstituted in isopropanol was diluted 3 : 2 with distilled water (dH2O). Cells in the culture dish were rinsed with 1X Dulbecco's Phosphate-Buffered Saline (DPBS) and fixed with 4% paraformaldehyde for 20 minutes and then rinsed with 1X DPBS. Oil Red O staining solution was added and the cells were allowed to stain for 20 minutes. Cells were rinsed 4 times with 1X DPBS. To determine lipid accumulation in the adipocytes, after aspirating the last 1X DPBS rinse, 0.25 mL dye extraction solution was added to each well to extract the Oil Red O dye from the stained lipids. The plate was allowed to shake on an orbital shaker for 30 minutes and the extracted dye was transferred to a 96-well plate, and optical density (OD) was measured at 520 nm in a plate reader (Molecular Devices).
2.5. Microscopy and Cell Size Measurement
Images of Oil Red O stained cells were captured by bright-field microscopy on the Axiovert 40 CFL (Zeiss) or DM5000B (Leica Microsystems) at 400x magnification. The size of Oil Red O stained cells was determined from cells contained in 3–5 microscopy fields per experiment. As an index of cell size, the cell diameter was measured from microscopy images. Microscopic morphological images of the adipocytes were printed, circles were drawn at the periphery of the individual cells around the circular shape formed by the orange lipid droplets accumulated inside the cell, and the diameter of the circles was noted using a ruler [26–28]. The diameter mean and SD were plotted for each experiment.
2.6. RNA Isolation, Conventional Reverse-Transcriptase PCR (RT-PCR), Microarray, and Real-Time Quantitative PCR (QPCR)
Total RNA was isolated by using the RNeasy mini kit (Qiagen) and DNase treated by using the Turbo DNA-free kit (Ambion/Life Technologies).
Expression of adipogenesis marker genes (Adiponectin, Pgc1α, and Pparγ), Men1, and an internal control gene (Gapdh) was determined by conventional RT-PCR using RNA isolated from cells before and after differentiation. OligodT-primed first strand cDNA was used for PCR using the GoTaq Green Master Mix (Promega). Equal amount of PCR products was analyzed by agarose gel electrophoresis.
RNA from WT and Men1-KO mESC-derived adipocytes was analyzed for global gene expression changes at the NIDDK genomics core facility (Affymetrix microarray platform). RNA samples (n = 3 of each) were labeled and hybridized to Genechip mouse genome 430 arrays. Microarray data were normalized and analyzed using Affymetrix Genechip software, Microarray analysis Suite 5.0 (Affymetrix). For the validation and further analysis of the target genes identified by microarray, OligodT-primed first strand cDNA was used for QPCR using the Brilliant II SYBR Green QPCR Kit (Agilent Technologies) and Mx3000P PCR machine (Stratagene). To calculate fold change, 2−ΔΔCt values were calculated from the raw threshold values generated by the Mx3000P software. Gapdh was used as the internal control.
2.7. Western Blot Analysis
Whole cell extracts (WCE) were prepared in cell culture lysis reagent (Promega), and protein quantitation was performed with the DC protein assay kit (Bio-Rad). Equal amounts of protein were separated on 10% Tris-Glycine gels, transferred to nitrocellulose membrane, and probed with appropriate dilution of primary and secondary antibodies, and the blots were developed using enhanced chemiluminescence (ECL) (Millipore). Antibodies used were rabbit antimenin (Bethyl, A300-105A) and mouse antitubulin (Calbiochem, CP06).
2.8. Statistics
Data from at least 3 independent experiments or measurements were plotted as mean and standard deviation (SD). Differences between groups were compared by Student's t-tests. p < 0.05 was considered significant.
3. Results
3.1. Men1-KO mESCs Can Undergo In Vitro Differentiation into Adipocytes and Show Increased Adipocyte Cell Size
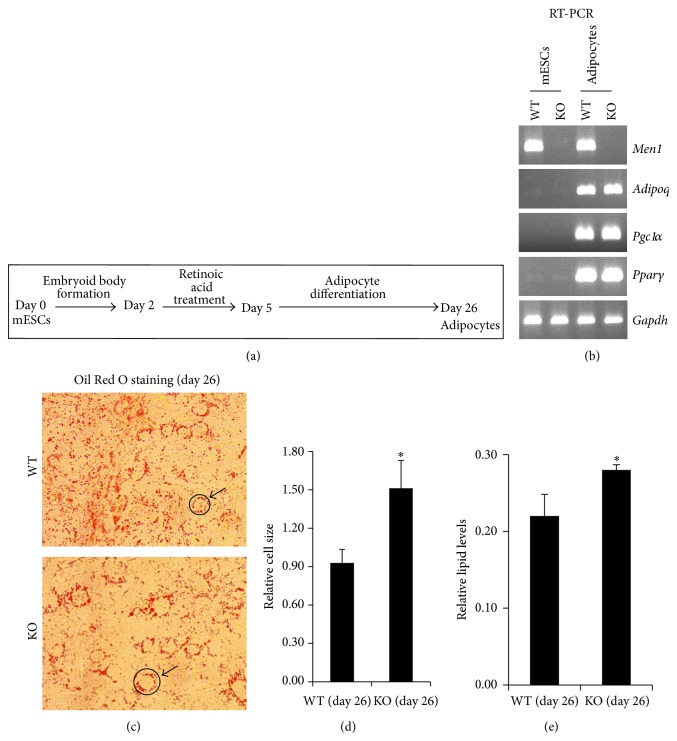
To determine the ability of Men1-KO mESCs to differentiate into adipocytes, in vitro differentiation of wild type (WT) and Men1-KO mESCs was performed using an established protocol (Figure 1(a)) [29]. Both cell types were equally capable of differentiating into adipocytes with similar expression of adipocyte-specific marker genes (Adiponectin, Pgc1α, and Pparγ) and similar level of Oil Red O staining in lipid droplets (Figures 1(b) and 1(c)). Adipocytes derived from Men1-KO mESCs were 1.5- to 1.7-fold larger than WT adipocytes accompanied with a slight increase in lipid content (Figures 1(d) and 1(e)). Thus, in vitro adipogenesis of menin-null mESCs is possible and the menin-null adipocytes are larger than normal, suggesting a role for menin in determining fat-cell size.
Figure 1.
Menin-null (Men1-KO) mouse embryonic stem cells (mESCs) can undergo in vitro differentiation into adipocytes and show increased adipocyte cell size. (a) In vitro adipogenesis. Feeder-free mESCs (WT or Men1-KO) were allowed to form embryoid bodies (EBs) in medium without LIF for 2 days, followed by retinoic acid treatment for 3 days. On day 5, retinoic acid induced EBs were cultured in gelatin-coated plates with adipocyte differentiation medium for 21 days to obtain adipocytes, and the medium was replaced every 2 days. (b) Expression of adipocyte marker genes. Conventional RT-PCR followed by agarose gel electrophoresis of the adipocyte marker genes (Adiponectin, Pgc1α, and Pparγ), Men1, and an internal control gene (Gapdh) using RNA samples before and after in vitro differentiation of WT or Men1-KO (KO) mESCs into adipocytes. (c) Oil Red O staining. Bright-field microscopy images of WT and Men1-KO mESCs stained for lipids/triglycerides after adipogenesis by Oil Red O staining show round adipocyte cells with lipid droplets (orange colored spots). Magnification = 400x. (d) Relative cell size. After adipogenesis, Oil Red O staining for lipids/triglycerides was performed, and images were captured by bright-field microscopy shown in (c). Cell diameter was measured as an index of cell size from 3 to 5 microscopy fields using the outline formed by the circle of orange lipid droplets accumulated inside the round adipocytes (an example marked with a black circle and arrow is shown in (c)). Diameter mean and SD are shown (∗ p < 0.05). Note: the units on the y-axis do not reflect the actual diameter of the cells but they reflect the measurement of the cell diameter from images captured at 400x magnification. (e) Relative lipid content. After adipogenesis, Oil Red O staining for lipids/triglycerides was performed, dye was extracted, and the OD was measured at 520 nm. Relative lipid level (OD at 520 nm) is shown for adipocytes. Error bar = SD. ∗ p < 0.05.
3.2. Gene Expression Profile of mESC-Derived Menin-Null Adipocytes Shows Reduced Expression of Several Differentially Methylated Genes
To examine the consequence of menin loss in lipoma cells, WT and Men1-KO mESC-derived adipocytes were used as a surrogate for gene expression microarray analysis. The top 40 genes differentially expressed in the Men1-KO adipocytes (at least 3-fold change compared with WT adipocytes; p < 0.001) were validated by QPCR. Unknown/predicted genes were not considered in this validation set of 40 genes (Table 1). More than 2-fold change was confirmed for 38 genes in Men1-KO versus WT adipocytes (Table 1).
Table 1.
Differentially regulated genes in Men1-KO versus WT adipocytes derived from mESCs.
| Affymetrix probe set ID | Ref seq. transcript ID | Gene symbol | Gene title | Microarray fold change | QPCR fold change |
|---|---|---|---|---|---|
| 1451634_at | NR_002853 | Airn | Antisense Igf2r RNA | −9.24 | −18.5 |
| 1457030_at | NR_028265 | Mirg | miRNA containing gene | −8.36 | −6.54 |
| 1438588_at | NM_009538 | Plagl1 | Pleiomorphic adenoma gene-like 1 | −7.04 | −7.46 |
| 1427580_a_at | NR_028261 | Rian | RNA imprinted and accumulated in nucleus | −6.96 | −10.7 |
| 1436713_s_at | NR_003633 | Meg3 | Maternally expressed gene 3 | −6.72 | −7.35 |
| 1417836_at | NM_024198 | Gpx7 | Glutathione peroxidase 7 | −5.91 | −6.49 |
| 1421461_at | NM_001122949 | Mpl | Myeloproliferative leukemia virus oncogene | −5.68 | −1.74 |
| 1418061_at | NM_013589 | Ltbp2 | Latent transforming growth factor beta binding protein 2 | −5.51 | −2.28 |
| 1428896_at | NM_026840 | Pdgfrl | Platelet-derived growth factor receptor-like | −4.52 | −2.9 |
| 1419411_at | NM_009312 | Tac2 | Tachykinin 2 | −4.34 | −3.29 |
| 1423506_a_at | NM_010923 | Nnat | Neuronatin | −3.92 | −2.86 |
| 1460412_at | NM_024237 | Fbln7 | Fibulin 7 | −3.91 | −7.51 |
| 1448788_at | NM_010818 | Cd200 | CD200 antigen | −3.76 | −4.5 |
| 1421074_at | NM_007825 | Cyp7b1 | Cytochrome P450, family 7, subfamily b, and polypeptide 1 | −3.73 | −3.09 |
| 1419197_x_at | NM_032541 | Hamp | Hepcidin antimicrobial peptide | −3.73 | −2.84 |
| 1417979_at | NM_022322 | Tnmd | Tenomodulin | −3.63 | −3.36 |
| 1425040_at | NM_028593 | Cybrd1 | Cytochrome b reductase 1 | −3.57 | −4.69 |
| 1419292_at | NM_001042615 | Htra3 | HtrA serine peptidase 3 | −3.49 | −5.69 |
| 1435476_a_at | NM_001077189 | Fcgr2b | Fc receptor, IgG, low affinity IIb | −3.27 | −8.81 |
| 1416347_at | NM_001168488 | Men1 | Multiple endocrine neoplasia 1 | −3.18 | −714.1 |
| 1450285_at | NM_011667 | Ube1y1 | Ubiquitin-activating enzyme E1, Chr Y 1 | 4.34 | 9.38 |
| 1435064_a_at | NM_020626 | Tmem27 | Transmembrane protein 27 | 4.39 | 9.44 |
| 1448783_at | NM_021291 | Slc7a9 | Solute carrier family 7 (cationic amino acid transporter, y+ system), member 9 | 4.43 | 17.26 |
| 1456601_x_at | NM_007503 | Fxyd2 | FXYD domain-containing ion transport regulator 2 | 4.51 | 19.42 |
| 1424279_at | NM_001111048 | Fga | Fibrinogen alpha chain | 4.74 | 7.31 |
| 1418916_a_at | NM_029269 | Spp2 | Secreted phosphoprotein 2 | 4.86 | 19.56 |
| 1449907_at | NM_001163028 | Bcmo1 | Beta-carotene 15,15′-monooxygenase | 4.92 | 2.84 |
| 1422289_a_at | NM_029636 | Ctsq | Cathepsin Q | 4.94 | 6.32 |
| 1418766_s_at | NM_001161355 | Timd2 | T-cell immunoglobulin and mucin domain-containing 2 | 5.2 | 13.92 |
| 1443824_s_at | NM_053070 | Car7 | Carbonic anhydrase 7 | 5.39 | 5.89 |
| 1417761_at | NM_007468 | Apoa4 | Apolipoprotein A-IV | 5.59 | 7.67 |
| 1418724_at | NM_007686 | Cfi | Complement component factor i | 5.86 | 9.12 |
| 1440218_at | NM_001033364 | Cdhr2 | Cadherin-related family member 2 | 6.02 | −1.07 |
| 1452165_at | NM_025532 | Prl2b1 | Prolactin family 2, subfamily b, member 1 | 6.06 | 19.69 |
| 1451513_x_at | NM_009243 | Serpina1a | Serine (or cysteine) peptidase inhibitor, clade A, member 1A | 6.28 | 12.46 |
| 1418282_x_at | NM_009244 | Serpina1b | Serine (or cysteine) peptidase inhibitor, clade A, member 1B | 6.34 | 6.02 |
| 1424673_at | NM_053165 | Clec2h | C-type lectin domain family 2, member h | 7.19 | 7.11 |
| 1429826_at | NM_029355 | Prl7b1 | Prolactin family 7, subfamily b, member 1 | 7.58 | 29.44 |
| 1448572_at | NM_011165 | Prl4a1 | Prolactin family 4, subfamily a, member 1 | 15.49 | 29.65 |
| 1436717_x_at | NM_008221.4 | Hbb-y | Hemoglobin Y, beta-like embryonic chain | 19.31 | 2.8 |
Validation of top 20 downregulated or upregulated genes (p < 0.001) identified in the gene expression microarray analysis. QPCR fold change was normalized to Gapdh. Bold, genes regulated by differential DNA methylation. Italic, genes from the prolactin gene family.
Men1-KO adipocytes showed more than 3-fold downregulation of 12 differentially methylated genes including 3 genes from the Meg3 locus (Meg3, Mirg, and Rian) and showed more than 6-fold upregulation of 3 genes from the prolactin locus (Prl2b1, Prl7b1, and Prl4a1) (Table 1). Differential expression of genes that regulate the cell cycle has been observed in pancreatic islet β-cell tumors of mice with tissue-specific conditional loss of menin [30]. Analysis of gene expression microarray data showed that the expression of cell cycle genes was unaffected in Men1-KO adipocytes (not more than 2-fold change and p < 0.001). Also, menin-binding protein partners [13] were not present among the differentially expressed genes.
DNA methylation inversely correlates with a gene activating chromatin modification, the histone H3 lysine-4 trimethyl mark (H3K4me3) [31]. Menin partners with the MLL-protein-complex to place this active mark in chromatin, and loss of menin leads to loss of H3K4me3 on specific genes [18, 32]. Among the 12 genes found to have differential DNA methylation, a comparison with our previously published data [18] showed that only one gene (Meg3) downregulated in mESC-derived menin-null adipocytes also showed significantly reduced histone methylation (H3K4me3) in menin-null mESCs. We have previously shown that menin loss in mESCs resulted in reduced expression of the long noncoding RNA Meg3 by facilitating DNA methylation in the Meg3 promoter region [33]. These observations suggest a role for menin in regulating H3K4me3 and consequently DNA methylation at specific genes in adipocytes and other MEN1-associated target tissues.
3.3. Prolactin Family Genes Are Upregulated in mESC-Derived Menin-Null Adipocytes
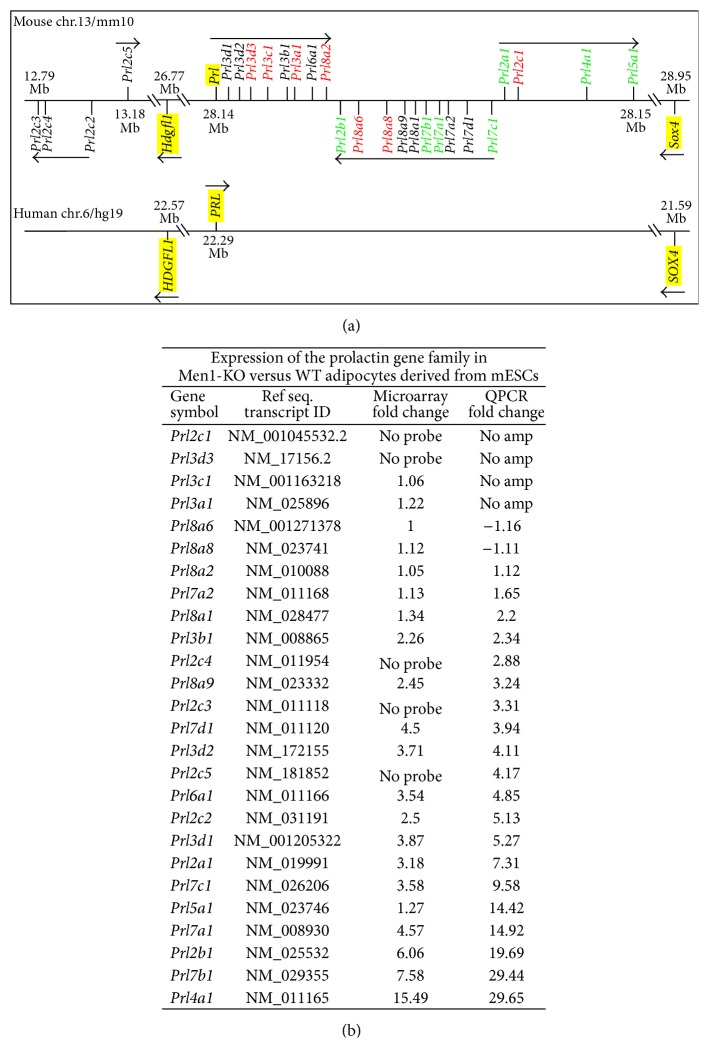
The prolactin gene family consists of 26 members clustered in a region of mouse chromosome 13 that includes Hgdfl1, Prl, and Sox4 (Figure 2(a)). The syntenic region on human chromosome 6 only contains HGDF1L, PRL, and SOX4 (Figure 2(a)). Therefore, the prolactin gene family is unique to the rodent genome. Gene expression microarray analysis showed upregulation of 3 genes from this family in adipocytes derived from Men1-KO mESCs (Table 1). QPCR analysis of all the prolactin family genes (n = 26) using RNA isolated from mESC-derived adipocytes showed 1.6- to 29-fold upregulation of 19 genes in menin-null adipocytes; the expression of 3 genes was unchanged, and 4 genes did not yield any PCR product (Figure 2(b)). These data show that loss of menin regulates gene expression at the mouse prolactin gene family locus.
Figure 2.
Prolactin family genes are upregulated in adipocytes derived from Men1-KO mESCs. (a) Schematic diagram comparing the mouse prolactin gene family locus with the syntenic human locus. The mouse prolactin gene family locus on chromosome 13 is shown on top and the syntenic human locus on chromosome 6 is shown at the bottom. Red, genes not differentially expressed in Men1-KO versus WT adipocytes or that did not yield a PCR product. Green, genes with the highest fold change (>5-fold) in Men1-KO versus WT adipocytes. Yellow highlight, genes present in both mouse and human. Arrows indicate the orientation of the coding strand. Chromosomal location in megabases (Mb) is from the UCSC genome browser using mouse genome version mm9 and human genome version hg19. (b) Relative expression of prolactin family genes. Prolactin family genes (n = 26) were analyzed by QPCR using RNA isolated from adipocytes derived by in vitro differentiation of WT or Men1-KO mESCs. Microarray data and QPCR relative fold change (normalized to Gapdh) are shown. “No probe” indicates genes not present on the microarray, and “no amp” indicates genes that did not yield a PCR product.
3.4. 3T3-L1 Cells with Menin Knockdown Can Undergo In Vitro Differentiation into Adipocytes and Also Show Increased Adipocyte Cell Size
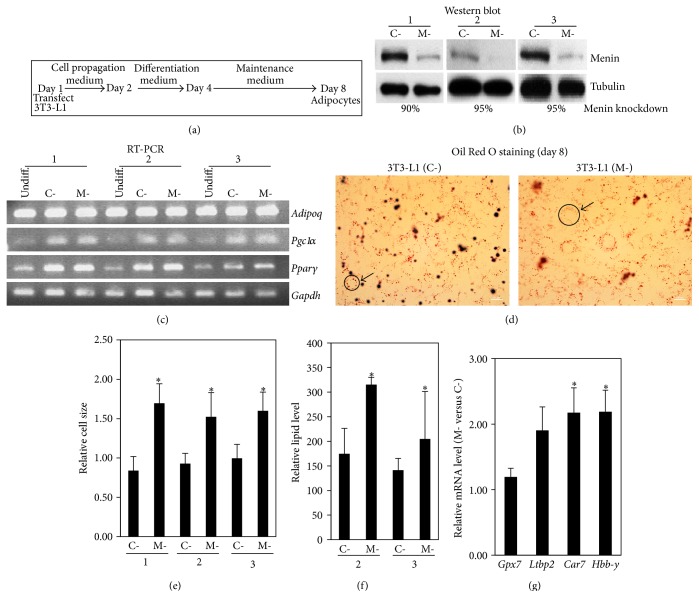
To further investigate the results obtained from mESC-derived adipocytes, an independent experimental model of in vitro adipogenesis was employed. The consequence of menin deficiency on adipocytes was studied in 3T3-L1 cells, a well-characterized preadipocyte cell line that is used for in vitro adipogenesis [24] (Figure 3(a)). Menin knockdown in differentiated 3T3-L1 cells was confirmed by western blot (Figure 3(b)). Induction of adipocyte marker gene expression (Pgc1α and Pparγ) and Oil Red O staining was also observed in adipocytes derived after menin knockdown (Figures 3(c) and 3(d)). Similar to the observation in the Men1-KO mESC-derived adipocytes, the 3T3-L1 adipocytes with menin knockdown showed 1.5- to 1.7-fold increase in cell size with a modest increase in lipid content (Figures 3(e) and 3(f)). Genes with more than 5-fold change in mESC-derived Men1-KO adipocytes (Table 1) were analyzed for expression changes in the 3T3-L1 adipogenesis model. Among the 20 genes analyzed (8 downregulated and 12 upregulated in Men1-KO mESC-derived adipocytes), only 4 were expressed in 3T3-L1-derived adipocytes with or without menin knockdown. Modest expression change (about 2-fold) was observed for 2 of these 4 genes in the menin knockdown 3T3-L1-derived adipocytes (Car7 and Hbb-y) (Figure 3(g)). The expression of 7 prolactin family genes with the highest fold change between mESC-derived Men1-KO and WT adipocytes was also analyzed by QPCR (Prl4a1, Prl7b1, Prl2b1, Prl7a1, Prl5a1, Prl7d1, and Prl2a1). Three genes (Prl4a1, Prl5a1, and Prl7a1) gave no product, and no significant differences were observed for the other 4 genes. Therefore, using an independent model of in vitro adipogenesis, we could confirm that menin deficiency did not affect adipocyte differentiation and produced adipocytes with increased cell size. However, the gene expression changes from mESC-derived adipocytes were not observed or could not be investigated in the 3T3-L1 adipogenesis model.
Figure 3.
3T3-L1 cells with menin knockdown can also undergo in vitro differentiation into adipocytes and show increased adipocyte cell size. (a) In vitro adipogenesis. Control shRNA plasmid (C-) or Men1 shRNA plasmid (M-) transfected 3T3-L1 cells were plated on day 1 in 3T3-L1 cell propagation medium overnight. The medium was replaced with differentiation medium for 2 days, followed by maintenance medium for 4 days to produce adipocytes, with a medium change (maintenance medium) every two days. (b) Menin knockdown. Whole cell extracts prepared from 3T3-L1 cells differentiated as shown in (a) were analyzed for the level of menin knockdown by western blot (day 8 adipocytes). Antitubulin was used as the loading control. Percent knockdown of menin was calculated after normalization to tubulin. Data are shown for 3 independent experiments. (c) Expression of adipocyte marker genes. RNA isolated from the 3 independent experiments shown in (b) was analyzed for the indicated adipocyte marker genes (Adiponectin, Pgc1α, and Pparγ) and an internal control gene (Gapdh) by conventional RT-PCR followed by agarose gel electrophoresis. Lanes marked “undiff” are 3T3-L1 cells before in vitro differentiation. (d) Oil Red O staining. Bright-field microscopy images of C- and M- differentiated 3T3-L1 cells stained for lipids/triglycerides after adipogenesis by Oil Red O staining show round adipocyte cells with lipid droplets (orange colored spots). Images are shown for experiment 2. Magnification = 400x. (e) Relative cell size. Cell diameter was measured as an index of cell size from 3 to 5 microscopy fields using the cell outline formed by the circle of orange lipid droplets accumulated inside the round adipocytes from the 3 independent 3T3-L1 differentiation experiments performed in (b) (an example marked with a black circle and arrow is shown in (d)). Diameter mean and SD are shown (∗ p < 0.05). Note: the units on the Y-axis do not reflect the actual diameter of the cells but they reflect the measurement of the cell diameter from images captured at 400x magnification. (f) Relative lipid content. After adipogenesis, Oil Red O staining for lipids/triglycerides was performed, dye was extracted, and the OD was measured at 520 nm. Relative lipid level is shown for 2 independent experiments performed in (b) (OD at 520 nm). Error bar = SD. ∗ p < 0.05. (g) Relative expression of differentially expressed genes identified by microarray analysis of mESC-derived adipocytes. Genes more than 5-fold downregulated (n = 8) or upregulated (n = 12) in mESC-derived menin-null adipocytes were analyzed by QPCR using RNA isolated from adipocytes derived by in vitro differentiation of 3T3-L1 cells shown in (b) (experiment 2). QPCR relative fold change (normalized to Gapdh) is shown for 4 genes. The other 16 genes did not yield a PCR product in either cell type (C- or M-). ∗ p < 0.05 (>2-fold change in M- versus C-).
4. Discussion
The availability of lipoma specimens from human MEN1 patients or from the mouse models of MEN1 and the availability of normal cells matched to lipoma are a challenge in the study of MEN1-associated lipoma. Here, we demonstrate the potential of using in vitro differentiation to derive menin-deficient and matched normal fat-cells (adipocytes) for such investigations. Using 2 different models of in vitro adipogenesis (mESCs and 3T3-L1 cells), we demonstrate that lack of menin or reduced level of menin did not affect the derivation of adipocytes. Interestingly, we observed that in both models the menin-deficient adipocytes were 1.5- to 1.7-fold larger compared with their WT/menin-containing control adipocytes and with a modest increase in lipid content which perhaps results from the increased cell size and not due to increased production of lipids. Gene expression analysis did not reveal any obviously relevant downstream effectors that could explain fat-cell hypertrophy. Analysis of these cells by posttranscriptional and proteomic approaches will be of interest.
Pparγ-induced in vitro adipogenesis was inefficient in producing adipocytes from immortalized Men1-KO mouse embryo fibroblasts (MEFs) or upon menin knockdown in 3T3-L1 cells [34]. The different methods used for in vitro differentiation in the previous study [34, 35] and in the present study [24, 29] could account for our successful derivation of adipocytes from Men1-KO mESC or from 3T3-L1 with menin knockdown. Similarly, menin inactivation by antisense oligonucleotides did not affect BMP-2-induced commitment of a mouse multipotential mesenchymal stem cell line (10T1/2) and mouse bone marrow stromal cell lines (ST2 and PA6) into the adipocyte lineage [36].
Men1-KO mESC-derived adipocytes showed downregulation of a few differentially methylated genes including the long noncoding RNA Meg3 and upregulation of most of the prolactin family genes. This observation could not be confirmed in the 3T3-L1 adipogenesis model. Meg3 was not expressed in 3T3-L1 or 3T3-L1-derived adipocytes and therefore further decrease from menin knockdown could not be assessed. Similarly, the prolactin family genes analyzed (n = 7) were either not expressed in WT or menin-deficient 3T3-L1-derived adipocytes or unaffected upon menin knockdown. The difference in gene expression in mESC or 3T3-L1-derived adipocytes could be attributed to their differentiation ability. Embryonic stem cells are multipotent and able to commit to different cell lineages, and 3T3-L1 cells are already committed to the adipocyte lineage [15, 24]. The reduced expression of several genes that are affected by differential DNA methylation highlights the involvement of menin in the regulation of DNA methylation at specific genes in the mESC-derived adipocytes.
We previously reported that menin loss in mESCs impaired H3K4me3 and gene expression at the Meg3 locus; and in mESC-derived pancreatic islet-like endocrine cells, menin loss impaired H3K4me3 and gene expression at the four Hox loci [18]. Also, in Men1-KO MEFs gene expression and H3K4me3 are significantly reduced at the four Hox loci [32]. Consistent with these data, in the present study, we found that, in mESC-derived adipocytes, menin loss increased gene expression from the prolactin gene family locus and decreased the expression of genes from the Meg3 locus. These observations suggest that the structural organization at loci with gene clusters is a target of menin to up- or downregulate gene expression. Gene rich chromatin domains that contain coordinately expressed genes in clusters such as the Hox genes are highly conserved in evolution [19]. However, the prolactin family genes are present in rodents and not in humans [22]. Therefore, the effect from menin loss on the expression of prolactin gene family is not relevant in the context of MEN1-associated human lipoma.
About 30% of MEN1 patients develop lipomas as one of the cutaneous manifestations of the MEN1 syndrome. These patients carry a heterozygous germline mutation in MEN1, and loss of the remaining normal copy of MEN1 has been documented in such MEN1-associated lipomas [5–9]. Our findings from in vitro differentiation studies in mouse cells that adipocytes can form despite menin deficiency are in agreement with the observation that lipomas resemble normal fat. However, how menin loss leads to localized increased fatty mass and tumor formation remains to be determined. Further studies of these benign fatty lesions could provide insights into the pathogenesis of MEN1-related lipomas and the growth of normal fat tissue.
5. Conclusion
In vitro adipogenesis showed that menin deficiency could result in fat-cell hypertrophy and differential gene expression from the methylated Meg3 locus and the prolactin gene family locus. Our data highlight the potential of two different in vitro experimental systems to study MEN1-associated lipoma biology and to investigate the mechanisms that govern fat-cell size, proliferation, and function.
Supplementary Material
Supplementary Table 1 shows the sequences of all the primers used in this study.
Acknowledgments
The authors are grateful to Settara Chandrasekarappa of the NHGRI for kindly providing the WT and Men1-KO mESCs. They thank the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) genomics core facility (Chithra Keembiyehetty and Weiping Chen) for their advice and help in generating microarray data. This work was supported by the Intramural Research Program of the NIH, NIDDK (SKA; Project no. 1ZIADK075035-03). All the gene expression microarray data generated for this study are deposited in the NCBI GEO repository under accession no. GSE65859.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Chandrasekharappa S. C., Guru S. C., Manickam P., et al. Positional cloning of the gene for multiple endocrine neoplasia-type 1. Science. 1997;276(5311):404–406. doi: 10.1126/science.276.5311.404. [DOI] [PubMed] [Google Scholar]
- 2.Lemmens I., van de ven Wim J. M., Kas K., et al. Identification of the multiple endocrine neoplasia type 1 (MEN1) gene. Human Molecular Genetics. 1997;6(7):1177–1183. doi: 10.1093/hmg/6.7.1177. [DOI] [PubMed] [Google Scholar]
- 3.Thakker R. V., Newey P. J., Walls G. V., et al. Clinical practice guidelines for multiple endocrine neoplasia type 1 (MEN1) The Journal of Clinical Endocrinology and Metabolism. 2012;97(9):2990–3011. doi: 10.1210/jc.2012-1230. [DOI] [PubMed] [Google Scholar]
- 4.Darling T. N., Skarulis M. C., Steinberg S. M., Marx S. J., Spiegel A. M., Turner M. Multiple facial angiofibromas and collagenomas in patients with multiple endocrine neoplasia type 1. Archives of Dermatology. 1997;133(7):853–857. doi: 10.1001/archderm.1997.03890430067009. [DOI] [PubMed] [Google Scholar]
- 5.Vidal A., Iglesias M. J., Fernández B., Fonseca E., Cordido F. Cutaneous lesions associated to multiple endocrine neoplasia syndrome type 1. Journal of the European Academy of Dermatology and Venereology. 2008;22(7):835–838. doi: 10.1111/j.1468-3083.2008.02578.x. [DOI] [PubMed] [Google Scholar]
- 6.Dong Q., Debelenko L. V., Chandrasekharappa S. C., et al. Loss of heterozygosity at 11q13: analysis of pituitary tumors, lung carcinoids, lipomas, and other uncommon tumors in subjects with familial multiple endocrine neoplasia type 1. The Journal of Clinical Endocrinology and Metabolism. 1997;82(5):1416–1420. doi: 10.1210/jcem.82.5.3944. [DOI] [PubMed] [Google Scholar]
- 7.Pack S., Turner M. L., Zhuang Z., et al. Cutaneous tumors in patients with multiple endocrine neoplasia type 1 show allelic deletion of the MEN1 gene. The Journal of Investigative Dermatology. 1998;110(4):438–440. doi: 10.1046/j.1523-1747.1998.00140.x. [DOI] [PubMed] [Google Scholar]
- 8.Vortmeyer A. O., Böni R., Pak E., Pack S., Zhuang Z. Multiple endocrine neoplasia 1 gene alterations in MEN1-associated and sporadic lipomas. Journal of the National Cancer Institute. 1998;90(5):398–399. doi: 10.1093/jnci/90.5.398. [DOI] [PubMed] [Google Scholar]
- 9.Rusconi D., Valtorta E., Rodeschini O., et al. Combined characterization of a pituitary adenoma and a subcutaneous lipoma in a MEN1 patient with a whole gene deletion. Cancer Genetics. 2011;204(6):309–315. doi: 10.1016/j.cancergen.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 10.Agarwal S. K. Exploring the tumors of multiple endocrine neoplasia type 1 in mouse models for basic and preclinical studies. International Journal of Endocrine Oncology. 2014;1(2):153–161. doi: 10.2217/ije.14.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Piret S. E., Thakker R. V. Mouse models for inherited endocrine and metabolic disorders. The Journal of Endocrinology. 2011;211(3):211–230. doi: 10.1530/joe-11-0193. [DOI] [PubMed] [Google Scholar]
- 12.Harding B., Lemos M. C., Reed A. A. C., et al. Multiple endocrine neoplasia type 1 knockout mice develop parathyroid, pancreatic, pituitary and adrenal tumours with hypercalcaemia, hypophosphataemia and hypercorticosteronaemia. Endocrine-Related Cancer. 2009;16(4):1313–1327. doi: 10.1677/erc-09-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Agarwal S. K. Multiple endocrine neoplasia type 1. Frontiers of Hormone Research. 2013;41:1–15. doi: 10.1159/000345666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matkar S., Thiel A., Hua X. Menin: a scaffold protein that controls gene expression and cell signaling. Trends in Biochemical Sciences. 2013;38(8):394–402. doi: 10.1016/j.tibs.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keller G. Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes & Development. 2005;19(10):1129–1155. doi: 10.1101/gad.1303605. [DOI] [PubMed] [Google Scholar]
- 16.Sterneckert J. L., Reinhardt P., Schöler H. R. Investigating human disease using stem cell models. Nature Reviews Genetics. 2014;15(9):625–639. doi: 10.1038/nrg3764. [DOI] [PubMed] [Google Scholar]
- 17.Novotny E., Compton S., Liu P. P., Collins F. S., Chandrasekharappa S. C. In vitro hematopoietic differentiation of mouse embryonic stem cells requires the tumor suppressor menin and is mediated by Hoxa9. Mechanisms of Development. 2009;126(7):517–522. doi: 10.1016/j.mod.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Agarwal S. K., Jothi R. Genome-wide characterization of menin-dependent H3K4me3 reveals a specific role for menin in the regulation of genes implicated in men1-like tumors. PLoS ONE. 2012;7(5) doi: 10.1371/journal.pone.0037952.e37952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee A. P., Koh E. G. L., Tay A., Brenner S., Venkatesh B. Highly conserved syntenic blocks at the vertebrate Hox loci and conserved regulatory elements within and outside Hox gene clusters. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(18):6994–6999. doi: 10.1073/pnas.0601492103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emes R. D., Goodstadt L., Winter E. E., Ponting C. P. Comparison of the genomes of human and mouse lays the foundation of genome zoology. Human Molecular Genetics. 2003;12(7):701–709. doi: 10.1093/hmg/ddg078. [DOI] [PubMed] [Google Scholar]
- 21.Alam S. M. K., Ain R., Konno T., Ho-Chen J. K., Soares M. J. The rat prolactin gene family locus: species-specific gene family expansion. Mammalian Genome. 2006;17(8):858–877. doi: 10.1007/s00335-006-0010-1. [DOI] [PubMed] [Google Scholar]
- 22.Soares M. J., Konno T., Alam S. M. K. The prolactin family: effectors of pregnancy-dependent adaptations. Trends in Endocrinology and Metabolism. 2007;18(3):114–121. doi: 10.1016/j.tem.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 23.Ang Y.-S., Tsai S.-Y., Lee D.-F., et al. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell. 2011;145(2):183–187. doi: 10.1016/j.cell.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Green H., Kehinde O. An established preadipose cell line and its differentiation in culture II. Factors affecting the adipose conversion. Cell. 1975;5(1):19–27. doi: 10.1016/0092-8674(75)90087-2. [DOI] [PubMed] [Google Scholar]
- 25.Ramírez-Zacarías J. L., Castro-Muñozledo F., Kuri-Harcuch W. Quantitation of adipose conversion and triglycerides by staining intracytoplasmic lipids with oil red O. Histochemistry. 1992;97(6):493–497. doi: 10.1007/BF00316069. [DOI] [PubMed] [Google Scholar]
- 26.Yamaguchi M., Nishimura F., Naruishi H., Soga Y., Kokeguchi S., Takashiba S. Thiazolidinedione (pioglitazone) blocks P. gingivalis- and F. nucleatum, but not E. coli, lipopolysaccharide (LPS)-induced interleukin-6 (IL-6) production in adipocytes. Journal of Dental Research. 2005;84(3):240–244. doi: 10.1177/154405910508400306. [DOI] [PubMed] [Google Scholar]
- 27.Sakuma S., Nishioka Y., Imanishi R., et al. cis9, trans11-conjugated linoleic acid differentiates mouse 3T3-L1 preadipocytes into mature small adipocytes through induction of peroxisome proliferator-activated receptor gamma. Journal of Clinical Biochemistry and Nutrition. 2010;47(2):167–173. doi: 10.3164/jcbn.10-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Almendro V., Fuster G., Ametller E., et al. Interleukin-15 increases calcineurin expression in 3T3-L1 cells: possible involvement on in vivo adipocyte differentiation. International Journal of Molecular Medicine. 2009;24(4):453–458. doi: 10.3892/ijmm_00000252. [DOI] [PubMed] [Google Scholar]
- 29.Dani C., Smith A. G., Dessolin S., et al. Differentiation of embryonic stem cells into adipocytes in vitro. Journal of Cell Science. 1997;110(11):1279–1285. doi: 10.1242/jcs.110.11.1279. [DOI] [PubMed] [Google Scholar]
- 30.Karnik S. K., Hughes C. M., Gu X., et al. Menin regulates pancreatic islet growth by promoting histone methylation and expression of genes encoding p27Kip1 and p18INK4c. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(41):14659–14664. doi: 10.1073/pnas.0503484102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Balasubramanian D., Akhtar-Zaidi B., Song L., et al. H3K4me3 inversely correlates with DNA methylation at a large class of non-CpG-island-containing start sites. Genome Medicine. 2012;4(5, article 47) doi: 10.1186/gm346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang P., Lin C., Smith E. R., et al. Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Molecular and Cellular Biology. 2009;29(22):6074–6085. doi: 10.1128/MCB.00924-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Modali S. D., Parekh V. I., Kebebew E., Agarwal S. K. Epigenetic regulation of the lncRNA MEG3 and its target c-MET in pancreatic neuroendocrine tumors. Molecular Endocrinology. 2015;29(2):224–237. doi: 10.1210/me.2014-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dreijerink K. M. A., Varier R. A., van Beekum O., et al. The multiple endocrine neoplasia type 1 (MEN1) tumor suppressor regulates peroxisome proliferator-activated receptor gamma-dependent adipocyte differentiation. Molecular and Cellular Biology. 2009;29(18):5060–5069. doi: 10.1128/mcb.01001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeninga E. H., van Beekum O., van Dijk A. D. J., et al. Impaired peroxisome proliferator-activated receptor γ function through mutation of a conserved salt bridge (R425C) in familial partial lipodystrophy. Molecular Endocrinology. 2007;21(5):1049–1065. doi: 10.1210/me.2006-0485. [DOI] [PubMed] [Google Scholar]
- 36.Sowa H., Kaji H., Canaff L., et al. Inactivation of menin, the product of the multiple endocrine neoplasia type 1 gene, inhibits the commitment of multipotential mesenchymal stem cells into the osteoblast lineage. The Journal of Biological Chemistry. 2003;278(23):21058–21069. doi: 10.1074/jbc.m302044200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1 shows the sequences of all the primers used in this study.