Abstract
Parent-of-origin imprints have been implicated in the regulation of neural differentiation and brain development. Previously we have shown that, despite the lack of a paternal genome, human parthenogenetic (PG) embryonic stem cells (hESCs) can form proliferating neural stem cells (NSCs) that are capable of differentiation into physiologically functional neurons while maintaining allele-specific expression of imprinted genes. Since biparental (“normal”) hESC–derived NSCs (N NSCs) are targeted by immune cells, we characterized the immunogenicity of PG NSCs. Flow cytometry and immunocytochemistry revealed that both N NSCs and PG NSCs exhibited surface expression of human leukocyte antigen (HLA) class I but not HLA-DR molecules. Functional analyses using an in vitro mixed lymphocyte reaction assay resulted in less proliferation of peripheral blood mononuclear cells (PBMC) with PG compared with N NSCs. In addition, natural killer (NK) cells cytolyzed PG less than N NSCs. At a molecular level, expression analyses of immune regulatory factors revealed higher HLA-G levels in PG compared with N NSCs. In line with this finding, MIR152, which represses HLA-G expression, is less transcribed in PG compared with N cells. Blockage of HLA-G receptors ILT2 and KIR2DL4 on natural killer cell leukemia (NKL) cells increased cytolysis of PG NSCs. Together this indicates that PG NSCs have unique immunological properties due to elevated HLA-G expression.
INTRODUCTION
Mammalian parthenogenetic (PG) embryos undergo early developmental demise and do not develop to full term. However, PG human embryonic stem cells (hESCs) can be established from blastocysts originating from unfertilized oocytes that have been activated artificially (1–5). PG hESCs contain genetic material exclusively from the oocyte donor. Due to meiotic recombination, PG hESC derived from human leukocyte antigen (HLA) heterozygous donors can be both HLA heterozygous and homozygous; and the induction of haploidy during oocyte activation protocols can be utilized to generate HLA homozygous PG hESC (6,7). Homozygous PG hESCs may serve as an alternative for immunomatched therapies for a large population of patients (8).
There is increasing evidence that paternally and maternally inherited alleles influence brain development, function and behavior (9). Therefore, PG hESCs are a unique model system to study the distinct roles of paternal and maternal genomes during neural development. Chimera studies in the mouse have shown that neural development requires tight control of imprinted gene expression: when combined with normal embryos to form chimeras, murine PG cells contributed preferentially to the cortex, striatum and hippocampus, but not to the hypothalamic structures (10). Conversely, androgenetic cells, with two copies of a paternal genome, were found in hypothalamic structures but not in the cortex. These experiments suggested that both parental genomes play nonredundant roles during brain development. However, uniparental murine and human ESCs resemble biparental (“normal”) ESCs (N ESCs) in their capacity to proliferate and undergo multilineage in vitro differentiation with similar functional neurogenesis and in vivo neural engraftment (1,11–14).
In vitro and in vivo studies revealed that N hESCs and hESC-derived progeny are not immune-privileged (15). N hESCs and their differentiated derivatives express low levels of HLA class I (HLA-I), which can be induced by interferon-γ (IFN-γ), but they do not express costimulatory or HLA class II (HLA-II) molecules (16,17). Whether or not N hESCs stimulate allogeneic T cell proliferation remains contradictory (17,18). However, N hESC–derived neural stem cells (N NSCs) stimulate the proliferation of peripheral blood mononuclear cells (PBMCs) in vitro, particularly of CD4+ T cells; N NSCs also are susceptible to natural killer (NK) cell–mediated lysis (19,20). In vivo analyses further showed that xeno-rejection of hESCs and of hESC-derived cells is mainly T cell–mediated and that NK cells also are involved (21,22).
The nonclassical HLA-Ib molecule HLA-G has been identified as a ligand that can induce tolerance. HLA-G has properties that differ from classical HLA-I molecules. Classical HLA molecules are highly polymorphic and, therefore, can present a wide range of antigenic peptides, whereas HLA-G displays only very limited polymorphism. The expression of HLA-G is restricted mainly to extravillous cytotrophoblast cells of the placenta with a role in maternal-fetal immunological tolerance during pregnancy (23,24). HLA-G mRNA also is present in human oocytes and preimplantation embryos, in tumor and in virus-infected cells, in the adult human brain and in mesenchymal stem cells (25–28). Inflammatory conditions induce HLA-G expression in microglia, macrophages and neurons to counteract inflammatory responses (29). A small number of HLA-G-expressing cells is sufficient to maintain an antiinflammatory milieu in the central nervous system (CNS) (29). Whether N hESCs express HLA-G remains unclear, as disparate results have been reported (16,30,31).
HLA-G inhibits T and NK cell proliferation, the cytolytic function of NK cells and alloproliferative responses of CD4+ T cells (24). HLA-G exerts its tolerogenic functions through direct binding to its inhibitory receptors ILT2 (on B, T and NK cells), ILT4 (on myeloid cells) and KIR2DL4 (on the CD56+ subset of NK cells), even though the latter interaction has become controversial (24,32).
To validate that PG hESC–derived NSCs (PG NSCs) have no deficits in HLA biology it is critical to characterize their immunological properties in more detail. We therefore assessed HLA expression and function in PG and N NSCs using in vitro assays. We show that, in contrast to N-derived cells, PG NSCs exhibit elevated expression of HLA-G and thereby inhibit both the proliferation of PBMCs and cytolytic activity of NKL cells.
MATERIALS AND METHODS
Cell Lines and Culture
PG hESC lines LLC6P and LLC9P (previously phESC-3 and phESC-6) were obtained from the International Stem Cell Corporation (Carlsbad, CA, USA) (1). N hESC lines H9 (WiCell, Madison, WI, USA) (33) and HS401 (Karolinska Institute, Stockholm, Sweden) (34) were used as controls. PG and N hESC lines were cultured on feeders (human foreskin fibroblasts [ATCC-LGC Standards, Wesel, Germany] or murine embryonic fibroblasts) treated with mitomycin C (Sigma-Aldrich, St. Louis, MO, USA). Cells were differentiated into the neural lineage as described (12). Briefly, for embryoid body (EB) formation, detached hESCs were cultured for 4 d under floating conditions in KnockOut DMEM medium (Life Technologies [Thermo Fisher Scientific Inc., Waltham, MA, USA]) supplemented with 15% KnockOut Serum Replacement (Life Technologies [Thermo Fisher Scientific]), 10 mL/L penicillin/streptomycin, 2 mmol/L L-glutamate, 1% nonessential amino acids, 10 mmol/L HEPES (all from PAA Laboratories, Cölbe, Germany) and FGF2 (R&D Systems, Abingdon, UK). To generate attached EBs (attEBs), EBs were plated onto polyornithin/laminin-coated plates (polyornithin/laminin from Sigma-Aldrich) in N2 medium consisting of DMEM/F-12 medium (Life Technologies [Thermo Fisher Scientific]), 10 mL/L penicillin/streptomycin and N2 supplement (1:100) (Life Technologies [Thermo Fisher Scientific]). 20 ng/mL fibronectin (Sigma-Aldrich) and 10 ng/mL FGF2 were added freshly. For expansion of attEBs of line LLC6P, 20 ng/mL laminin was added. After 10 d of culturing attEBs, rosette structures were isolated mechanically with a needle and isolates were propagated for 4 d as free-floating neurospheres in N2 medium supplemented with 10 ng/mL FGF2. Neurospheres were dissociated with trypsin/EDTA (GE Healthcare, Munich, Germany) and cells were seeded onto polyornithin/laminin-coated plates in N2 medium supplemented with 0.16 g/mL glucose (AppliChem, Darmstadt, Germany) to generate NSCs. Media with freshly added 10 ng/mL FGF, 10 ng/mL EGF (R&D Systems) and B27 supplement (1:100, Life Technologies [Thermo Fisher Scientific]) was changed every second day. For passaging, NSCs were dissociated with trypsin/EDTA. Trypsin inhibitor (Sigma-Aldrich) was added to stop the reaction. Cells were replated onto polyornithin/laminin-coated plates. IFN-γ (Millipore, Schwalbach, Germany) treatment was done using 25 ng/mL for 48 h. EDTA (AppliChem) treatment was done using 1 mmol/L for 24 h.
The human CML cell line K562 and the human choriocarcinoma cell line JEG-3 were cultured in RPMI-1640 medium (Sigma-Aldrich) supplemented with 10% FCS (Life Technologies [Thermo Fisher Scientific]), 10 mL/L penicillin/streptomycin and 2 mmol/L L-glutamate. PBMCs, isolated by Ficoll-Paque (GE Healthcare) gradient centrifugation, were cultured in RPMI-1640 medium as detailed above.
HLA Fine Typing
Feeder cells were depleted by repeated passages of PG and N hESCs onto Matrigel-coated plates (Matrigel from BD Biosciences, Heidelberg, Germany) using feeder-conditioned medium. For HLA fine typing, genomic DNAs of PBMCs, feeder-depleted PG and N hESCs were isolated by phenol/chloroform extraction. HLA typing of PG and N hESCs required testing for HLA-A, -B, -C, -DRB1, and -DQB1 at high resolution as described previously (35). Exons 2 and 3 were analyzed for HLA class I typing, and exon 2 only for HLA class II typing. High resolution HLA typing was performed by sequencing analysis (sequence-based typing) for HLA class I and by polymerase chain reaction (PCR)-sequence–specific oligonucleotide probes method or sequence-based typing for HLA class II (35).
Immunocytochemistry
Cells grown on coverslips or cytospun cells were fixed in 3.7% formaldehyde/PBS (formaldehyde from AppliChem) and were either permeabilized and blocked in PBS containing 0.1% Triton X-100 and 0.2% gelatin (both AppliChem) or in 1% gelatin/PBS. Staining was performed overnight at 4°C with the following primary antibodies: mouse anti-OCT3/4 (Santa-Cruz Biotechnology Inc., Dallas, TX, USA), mouse anti-nestin (Abcam, Cambridge, UK), rabbit anti-SOX1 (Millipore), mouse anti-HLA class I (clone W6/32, Abcam), mouse anti-HLA-DR (clone TAL14.1, Abcam). Cy3- or Cy5- labeled anti-mouse or Cy3-labeled anti-rabbit (Millipore) secondary antibodies were diluted in blocking solution and incubated (1 h, room temperature) with samples. DAPI (Sigma-Aldrich) was used to counterstain nuclei. Samples were mounted in ProLong Gold antifade reagent (Life Technologies [Thermo Fisher Scientific]) and analyzed using SP5 Confocal (Leica, Wetzlar, Germany) or AxioVision Microscopes (Carl Zeiss, Oberkochen, Germany). Images were processed using the ImageJ (NIH, Bethesda, MD, USA) or AxioVision software. To assess the pluripotent status of the cells, fixed hESCs were stained with the alkaline phosphatase kit from Sigma-Aldrich.
Flow Cytometry
PG, N NSCs and control cells were trypsinized, resuspended in staining buffer (phosphate buffered saline [PBS] with 0.4% BSA) and passed through a cell strainer (70 μm) (BD Biosciences). Two × 105 PG, N NSCs or control cells were incubated for 1 h at 4°C with mouse anti-HLA class I (Abcam), mouse anti-HLA-DR (Abcam), mouse anti-HLA-DQ (clone TÜ169, Imusyn, Göttingen, Germany), DyLight 680 conjugated mouse anti-HLA-DP (clone B7/21, Leinco Technologies, St. Louis, MO, USA), APC-conjugated mouse anti-HLA-DR (clone L243, BioLegend, London, UK), APC-conjugated mouse anti-HLA-G (clone MEM-G/9, Life Technologies [Thermo Fisher Scientific]) or appropriate isotype control antibodies. Cells were washed and incubated (1 h, 4°C) with anti-mouse Cy3-labeled secondary antibody (Cy3 from Millipore). After staining, cells were washed and resuspended in staining buffer. Samples were analyzed on a FACSCanto flow cytometer (BD Biosciences). A live cell FSC/SSC gate was used. Data were analyzed using FlowJo software (Treestar, Ashland, OR, USA). Specific fluorescence indexes (SFIs) were calculated by dividing median fluorescence values of each specific antibody by the median fluorescence values of the respective isotype control reaction.
Mixed Lymphocyte Reaction (MLR)
NSCs and control cells (PBMCs, JEG-3 cells) were irradiated (20 Gy; Faxitron CP-160 X-ray radiation cabinet [160 kV, 6.3 mA, 0.4 Gy/min, filter: 0.5 mm Cu]) and used as stimulator cells in a colorimetric MLR (Roche, Mannheim, Germany). In a 96-well flat bottom plate, 1 × 105 PBMCs and 1 × 105 irradiated stimulator cells were mixed and incubated. As a positive control, PBMCs in coculture with irradiated autologous PBMCs were treated with 2.5 μg/mL concanavalin A (ConA) (Sigma-Aldrich) and 1% (v/v) phyto-hemagglutinin (PHA-M) (Life Technologies [Thermo Fisher Scientific]). Each experimental group was plated in triplicates. After 4 d, cocultures were incubated for 24 h with 10 μmol/L bromodeoxyuridine (BrdU). Cells were fixed, incubated with blocking solution (Roche) and anti-BrdU-peroxidase, then washed before 3,3′,5,5′-tetramethylbenzidine (TMB), the visualizing reagent, was added. 1 mol/L H2SO4 was added to stop the reaction. Enzymatic reactions were quantified in a SPECTRA Thermo enzyme-linked immunosorbent assay (ELISA) plate reader (Tecan Group Ltd., Männedorf, Switzerland) at 450 nm (reference wavelength 620 nm). Levels of background signals in unstimulated PBMCs and stimulator cells were subtracted.
Cytotoxicity Assay
To assess the NSC-lysis ability of NKL cells, a nonradioactive cytotoxicity assay was used (Promega, Mannheim, Germany). NKL effector cells were cultured in RPMI-1640 medium supplemented with 10% FCS, 10% horse serum (Life Technologies [Thermo Fisher Scientific]), 10 mL/L penicillin/streptomycin, 2 mmol/L L-glutamate and 200 U/mL IL-2 (PeproTech, Hamburg, Germany). NKL cells were added to 5 × 104 target cells to achieve 10:1 NKL-to-target cell ratios in a final volume of 100 μL. Target cells (PG, N NSCs, or control cells [K562 and JEG-3]) and NKL cells were cultivated either alone or in coculture for 4 h at 37°C in polyornithin/laminin-coated plates in triplicates. Cells were centrifuged, supernatants were transferred into new plates and the substrate mix was added. After 30 min, the reaction was stopped and absorbance values that represent lactate dehydrogenase (LDH) activity were determined by SPECTRA Thermo ELISA plate reader at 490 nm. Background signals were subtracted and percentages of cytotoxicity were calculated with the following formula:
where “experimental” is absorbance values from cocultures of effector and target cells, “effector spontaneous” or “target spontaneous” is spontaneous LDH release of cultured effector or target cells, and where. “target maximum” is maximum LDH release in cultures of target cells after chemically-induced cell lysis.
Detection of Active Caspase-3/7
For quantification of apoptotic cells, the CellEvent Caspase-3/7 Green Flow Cytometry Assay (Life Technologies [Thermo Fisher Scientific]) was used. Target cells (PG or N NSCs) were labeled with 5 μmol/L Cell Proliferation Dye eFluor670 (eBioScience, Frankfurt, Germany) to distinguish target and effector cells. To assess specific binding of HLA-G to NKL receptors, NKL cells were preincubated for 30 min with 0.1 μg/mL of functional blocking monoclonal antibodies raised against the inhibitory NKL receptors ILT2 (HP-F1, eBioscience) or KIR2DL4 (clone mAb 33, BioLegend). Next, 5 × 104 labeled target and 5 × 105 effector NKL cells were cocultured in RPMI-1640 medium supplemented with 10 mL/L penicillin/streptomycin and 2 mmol/L L-glutamate for 4 h at 37°C, 5% CO2. Cells were trypsinized and stained (30 min, 37°C) with 0.5 μmol/L CellEvent Caspase-3/7 green detection reagent. Samples were measured in a FACSCanto. Data were analyzed using FlowJo software. A live cell gate on APC/FSC channel was used to select for highly cell proliferation dye-positive target cells.
Semiquantitative Reverse Transcriptase (RT)-PCR
Total RNAs were isolated from feeder-depleted PG and N hESCs and from differentiated progeny using peqGOLD RNAPure (Peqlabs, Göttingen, Germany). RNAs were treated with DNase I (Applied Biosystems [Thermo Fisher Scientific]) and 1 μg RNA was reverse transcribed using M-MLV reverse transcriptase (Life Technologies [Thermo Fisher Scientific]). As controls, cDNAs were generated from human fetal brain total RNA (19 wks, male, single donor) (Agilent Technologies, Santa Clara, CA, USA) and from PBMCs, JEG-3 and K562 cells. For amplification, Taq Polymerase (H T Biotechnology, Cambridge, UK) was used. PCR conditions were: 94°C, 30 s, 56.5°C to 62°C, 30 s according to the primers, 72°C, 30 s (28 to 38 cycles). RPL30 was used as housekeeper control. Primers (Eurofins MWG Operon, Ebersberg, Germany) used were: OCT4 forward (frw): 5′-AGCCCTCATTTCACCAGGCC-3′, reverse (rev): 5′-CAAAACCCGGAGGAG TCCCA-3′, 62°C; REX1 frw: 5′-TAGAA TGCGTCATAAGGGGTGA-3′, rev: 5′-TCTTGCCTGTCATGTACTCAGAA-3′, 57°C; CD40 frw: 5′-ACCCTTGGAC AAGCTGTGAG-3′, rev: 5′-TGGCA AACAGGATCCCGAAG-3′, 57.5°C; CD80 frw: 5′-TGCTGGCTGGTCTTT CTCAC-3′, rev: 5′-GGTTCTTGTA CTCGGGCCAT-3′, 56.5°C; CD84 frw: 5′-ATCCAAGAACCACGGCAAGT-3′, rev: 5′-CTCCTGCGTCTTCCATCCTC-3′, 57.5°C; CD86 frw: 5′-CTGCTCATCTATACA CGGTTACC-3′, rev: 5′-GGAAACGTCG TACAGTTCTGTG-3′, 60.6°C; B7H1 frw: 5′-TTTGCTGAACGCCCCATACA-3′, rev: 5′-TTGGTGGT GGTGGTCTTACC-3′, 57.5°C; 2B4 frw: 5′-TCGTGATTCTAAGCG CACTGT-3′, rev: 5′-CAGGTTCTTG TGACGTGGGAG-3′, 60.6°C; HLA-DP frw: 5′-AGGAATGCTACGCGTTTAAT-3′, rev: 5′-CAGCTCGTAGTTGTGTCTGC-3′, 56.6°C; HLA-DQ frw: 5′-CTTGTGACCA GATACATCTA-3′, rev: 5′-GGTCGTGCGG AGCTCCAACT-3′, 57.8°C; HLA-DR frw: 5′-GCGGTTGCTGGAAAGATGCA-3′, rev: 5′-GTGAAGCTCTCACCAACCCC-3′, 57.8°C; RPL30 frw: 5′-ACAGCATGCG GAAAATACTAC-3′, rev: 5′-AAAGG AAAATTTTGCAGGTTT-3′, 60°C.
Real-Time RT-PCR
As described above, 1 μg RNA was reverse transcribed into cDNA. For mature miRNA analysis, 4 μg RNA were reverse transcribed. For the formation of stem-loop structures, a gene-specific RT primer was added during cDNA synthesis (36). RT-PCR reactions were performed and quantified using ABsolute QPCR SYBR Green Mix (ABgene, Hamburg, Germany) and Rotor-Gene 3000 (Corbett Life Science, LTF Labortechnologie, Wasserburg, Germany). Real-time RT-PCR conditions were: 95°C, 45 s, 60°C, 40 s and 72°C, 60 s (35 cycles). Relative gene expression levels were calculated with the 2−ΔΔCt method. The housekeeping gene RPL30 was used as reference, whereas for mature miRNA detection small nuclear RNA U6 was used. Primer sequences were: HLA-G frw: 5′-TGCTGAGATGGAAGCAGTCTTC-3′, rev: 5′-ACTACAGCTGCAAGGACAAC CA-3′; MIR152 frw: 5′-GTCGTCAGTG CATGACAGAACTT-3′, rev: 5′-GTGCA GGGTCCGAGGT-3′; U6 frw: 5′-CTCGC TTCGGCAGCACA-3′, rev: 5′-AACGC TTCACGAATTTGCGT-3′.
Western Blotting
For Western blotting, 1 × 106 cells were lysed in 120 μL 6 × SDS loading buffer (0.5 mol/L Tris pH 6.8 buffer containing 0.4% SDS, 33% glycerol, 0.4 mol/L β-mercaptoethanol and bromphenolblue) (all from Sigma-Aldrich). Extracts were heated (95°C, 5 min). Discontinuous SDS-PAGE was performed using 12% acrylamide resolving and 4% to 5% stacking gel. Following electrophoresis, proteins were electroblotted onto Nitrocellulose membranes (GE Healthcare). Membranes were washed three times with PBS + 0.1% Tween-20 (PBST) (AppliChem) and blocked with 5% non-fat dry milk/PBST solution (milk from AppliChem) for 1 h at room temperature. Staining with primary anti-HLA-G MEM-G/1 (1:200, AbD Serotec by Bio-Rad, Puchheim, Germany), anti-GAPDH (Merck Millipore, Darmstadt, Germany); or anti-actin (1:1000, Santa Cruz Biotechnology) antibodies was performed at 4°C overnight. Membranes were washed three times in PBST and incubated with stabilized peroxidase-conjugated goat anti-mouse (H + L) secondary antibody (1:1000; Thermo Scientific [Thermo Fisher Scientific]) or goat anti-rabbit secondary antibodies (1:1250; Thermo Scientific [Thermo Fisher Scientific]) for 1 h at room temperature. Membranes were again washed three times before incubation with Immobilon Western HRP Substrate (Millipore) for 1 min. Immunoreactive protein bands were visualized by autoradiography (Bio-Rad, München, Germany). HLA-G blots were stripped and reprobed for staining with an anti-GAPDH or anti-actin antibody as loading control.
Transfection with MIR152 Mimic
For transfection, 2 × 105 PG NSCs were plated onto polyornithin/laminin-coated 6-well plates. On the next day, cells were transfected with 5 μmol/L MIR152 synthetic RNA duplex (MIR152 mimic) or with 5 μmol/L anti-GAPDH RNA duplex (control) using Dharmafect transfection medium (Thermo Scientific [Thermo Fisher Scientific]). Fifty-six hours after transfection, cells were stained for FACS analysis, or RNA and protein were isolated.
Bisulfite Sequencing
Bisulfite-modified DNA from PG and N NSCs was prepared using the EZ DNA Methylation-Direct kit (Zymo Research, Irvine, TX, USA). DNA fragments covering a region of 26 CpGs within the MIR152 promoter were amplified by PCR using frw 5′-GGGTTAGGGGGAGTAGTTAATT-3′ and rev 5′-TTCTTCCAACTAATCCCTAA CC-3′ primers as described previously (37). PCR fragments were subcloned into pJET 2.1 (Thermo Scientific [Thermo Fisher Scientific]) for sequencing. Analysis of sequences and diagram generation was performed using BISMA (38).
Statistical Analysis
Data are presented as mean ± SD for the indicated number of biological replica (n). Data plotting and statistical analysis were done using Student t test (two-tailed, paired). Differences were considered significant at p < 0.05.
All supplementary materials are available online at www.molmed.org.
RESULTS
PG hESC–Derived NSCs Express HLA-I but Not HLA-DR Antigens
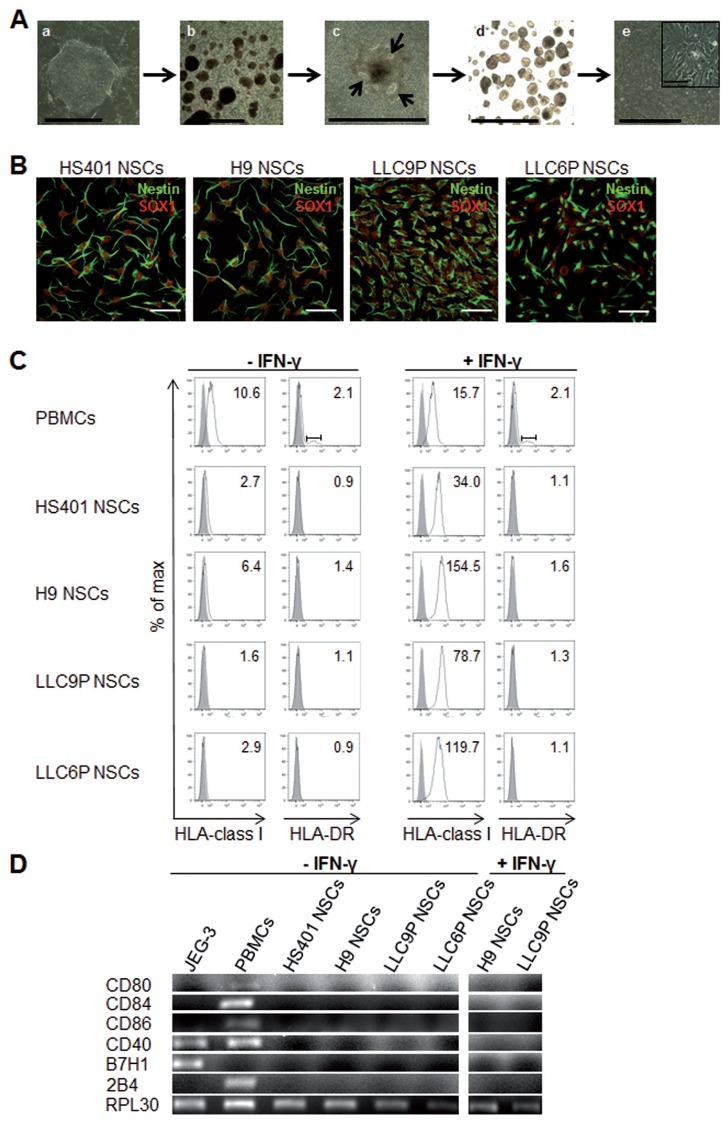
PG hESCs exhibited typical hESC morphology, expressed pluripotency- related markers and formed neural rosettes and NSCs when subjected to a multistep differentiation protocol (Figures 1A, B, Supplementary Figure 1) as described previously (12). HLA fine typing revealed that the PG hESC lines used in this study were fully heterozygous in the HLA region (Table 1). To assess the expression of HLA-I and HLA-II molecules in PG in comparison to N NSCs, we used the W6/32 monoclonal antibody (mAb) directed against HLA class I and the TAL14.1 mAb recognizing HLA-DR beta. Flow cytometry and immunofluorescence staining of NSCs derived from 2 PG and 2 N control hESC lines revealed relatively low expression of HLA-I and absence of HLA-DR (Figure 1C, Supplementary Figures 2, 3). In addition, HLA-DP and -DQ were not expressed in PG or N NSCs (see Supplementary Figure 3). To assess the induction of HLA-I protein expression in NSCs, we treated the cells with IFN-γ. Following treatment with IFN-γ, both PG and N NSCs exhibited 12- to 50-fold elevated HLA-I expression (see Figure 1C, Supplementary Figure 2) but remained negative for HLA class II (see Figure 1C, Supplementary Figure 3). Expression of costimulatory molecules including CD80, CD84, CD86, CD40, B7H1 and 2B4 that are required for T cell activation in addition to HLA binding was not detectable by RT-PCR in PG or N NSCs prior to or following IFN-γ stimulation (Figure 1D).
Figure 1.
Differentiation of PG hESCs into NSCs and expression of HLA and costimulatory molecules. (A) Phase contrast images of different stages during in vitro neural differentiation: (a) PG hESCs on feeders, (b) floating EBs, (c) attEBs (arrows indicate rosette-like structures), (d) floating neurospheres and (e) NSCs. Scale bar: 0.15 nm. Magnification showed NSCs. Scale bar: 0.1 mm. (B) Confocal images of NSC cultures immunostained with NSC-specific markers SOX1 (red) and Nestin (green) in PG and control N NSCs. Scale bar: 50 μm. (C) Flow cytometry of untreated (left) or 48 h IFN-γ-treated (25 ng/mL) (right) NSC cultures. Cells were stained with monoclonal antibodies specific for HLA class I or HLA-DR. Histograms show specific fluorescence signal for HLA class I or -DR (black lines) compared with isotype control (filled gray). Specific fluorescence indexes (SFI: specific geometric median/geometric median of unspecific control) are indicated in the upper right. (D) RT-PCR analysis for expression of costimulatory molecules in untreated PG and N NSCs and in IFN-γ treated NSCs. JEG-3 cells and PBMCs were shown as controls and RPL30 as housekeeper gene.
Table 1.
| HLA-I | HLA-II | ||||
|---|---|---|---|---|---|
|
|
|
||||
| HLA-A | HLA-B | HLA-C | HLA-DRB1 | HLA-DQB1 | |
| HS401 hESCs | *01:01 | *07:02 | *07:01 | *07:01 | *03:03 |
| *24:02 | *57:01 | *07:02 | *15:01 | *06:02 | |
| H9 hESCs | *02:01 | *35:03 | *04:01 | *15:01 | *05:02 |
| *03:01 | *44:02 | *07:04 | *16:01 | *06:02 | |
| LLC9P hESCs | *02:01 | *07:02 | *02:02 | *04:04 | *06:03 |
| *03:01 | *27:05 | *07:02 | *15:01 | *03:02 | |
| LLC6P hESCs | *02:01 | *52:01 | *03:03 | *01:01 | *02:01 |
| *03:01 | *55:01 | *04:01 | *03:01 | *05:01 | |
| PBMCs | *02:01 | *40:01 | *03:04 | *11:01 | *03:01 |
| *03:01 | *44:03 | *16:01 | *13:01 | *06:01 | |
HLA fine-typing of PG and N hESCs and PBMCs was performed by PCR.
The alleles examined were in HLA-I and -II loci (HLA-A, -B, -C, -DRB1 and DQB1).
PG NSCs Are Less Immunogenic than N NSCs
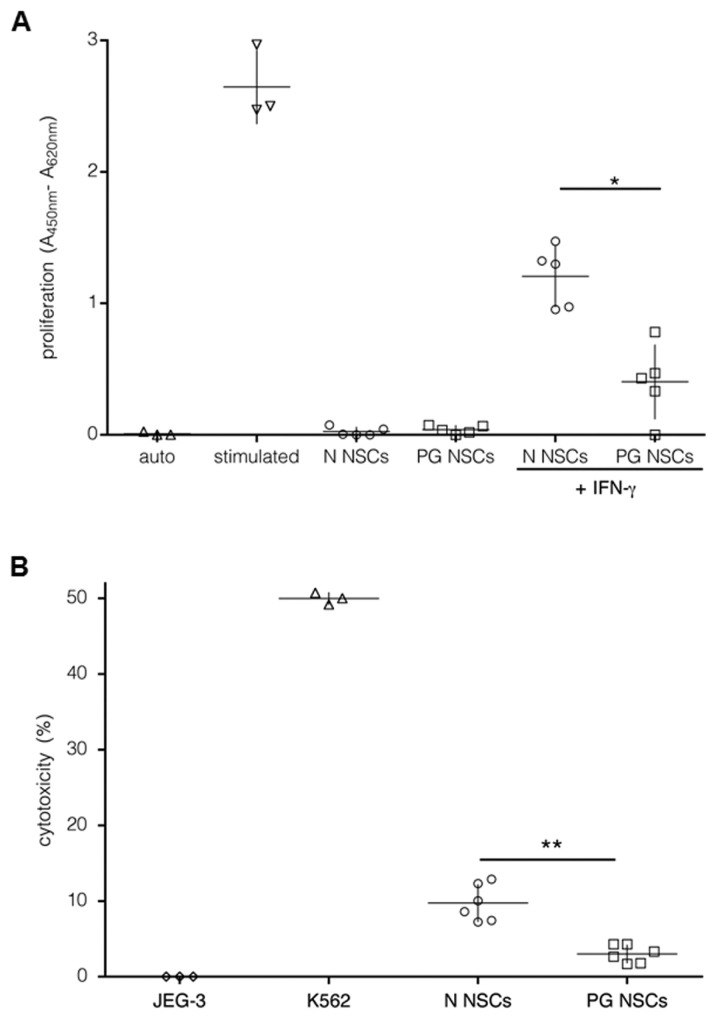
N NSCs induce proliferation of allogeneic T cells in a mixed lymphocyte reaction (MLR) despite the absence of costimulatory molecules (19,20). To address this property for PG NSCs, we recorded the response of healthy donor PMBCs to irradiated PG NSCs in allogeneic MLR reactions. Stimulated PBMCs were included as a positive control and JEG-3 cells, which suppress proliferative responses by surface HLA-G, as a negative control (39). As shown in Figure 2A, unstimulated PG and N NSCs only moderately induced proliferation of PBMCs, probably due to their low HLA-I and absent HLA-II expression. As IFN-γ treatment induced HLA-I protein levels on NSCs (see Figure 1C), we next treated PG and N NSCs with IFN-γ prior to PBMC coculture. IFN-γ-treated PG NSCs stimulated the proliferation of PBMCs, but less strongly than N NSCs (see Figure 2A). We observed similarly low levels of stimulation with PBMCs from all blood donors tested; therefore, these co-culture assays suggested either reduced immunogenicity or stronger immunosuppressive properties of PG NSCs. We further investigated NK cell–mediated target cell killing, which, depending on the balance of stimulatory and inhibitory ligands on target cells, can lead to rejection of allogeneic cells. The lack of HLA-I protein expression can render NSCs a target for NK cell–mediated cytolysis (40). To test whether NK cells target PG NSCs, we performed cytotoxicity assays using NKL cells as effector cells, and JEG-3 and NKL-sensitive K562 cells as negative and positive controls, respectively. At an NKL-to-target ratio of 10:1, N NSCs were more than three-fold more susceptible to NKL cell lysis than PG NSCs (Figure 2B).
Figure 2.
Immunostimulatory properties of PG NSCs. (A) Cell proliferation in mixed lymphocyte reaction with irradiated autologous PBMCs (△), PG (□) or N (○) NSCs as stimulator cells and PBMCs as responder cells at a 1:1 stimulator-to-responder ratio. Proliferation was measured by BrdU incorporation and ELISA. For maximum stimulation, a PBMC sample was treated with ConA and PHA (stimulated auto, ▽). *p < 0.05 by Student t test. We analyzed three different donor samples (each sample was analyzed two to three times). Shown are the median for each condition. (B) Cytotoxicity assay. NKL effector cells were mixed with JEG-3 cells (⋄), K562 (▵), N (○) or PG NSCs (□) at 10:1 NKL-to-target ratios. NKL cell–mediated lysis was determined by lactate dehydrogenase release into the medium quantified by ELISA. **p < 0.001. n = 5.
HLA-G in PG NSCs Protects against NKL-Induced Apoptosis
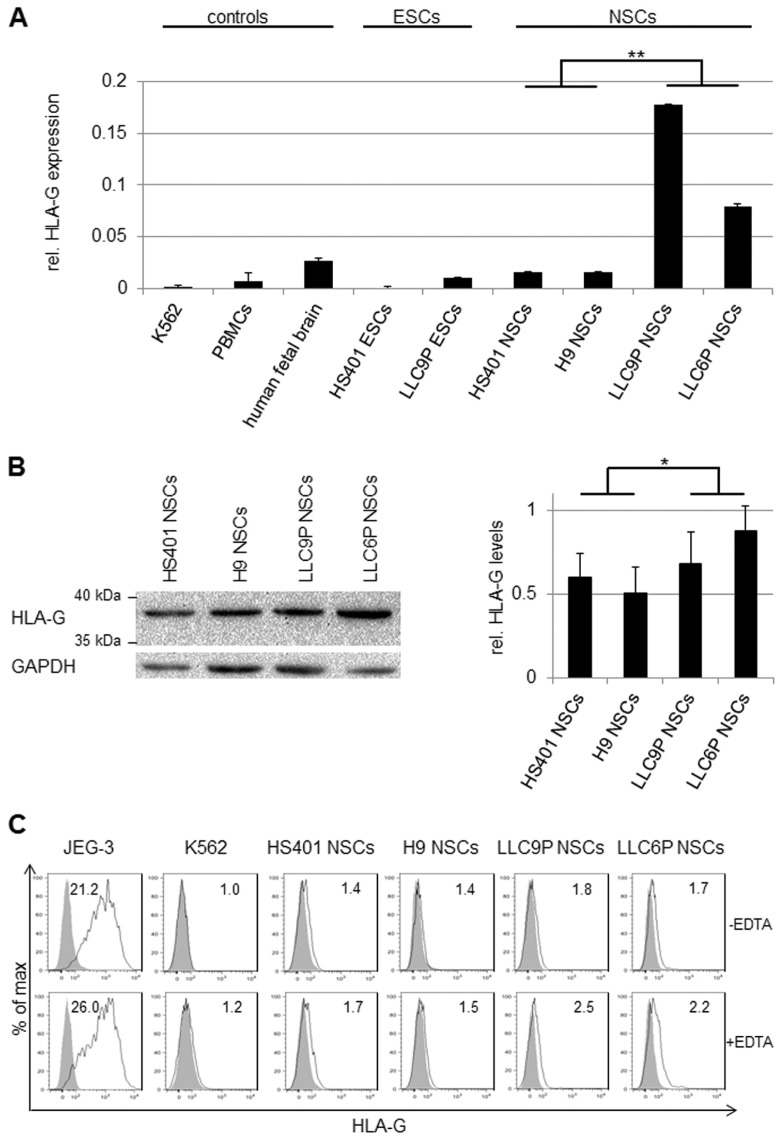
HLA-I and HLA-G are known ligands for inhibitory NK cell receptors such that higher expression of HLA-I or HLA-G ligands on PG NSCs could cause reduced NK cell–mediated cytolysis. Because PG and N NSCs express similar levels of HLA-I (see Figure 1C), we focused on HLA-G, which has been shown to reduce immune responses when expressed constitutively in hESC-derived dopaminergic neurons or in mesenchymal stem cells (28,41). To test whether PG in comparison to N NSCs have altered HLA-G expression leading to reduced immunogenicity, we first performed HLA-G- specific gene expression analysis. As shown in Figure 3A, HLA-G transcript levels were low in human PBMCs, fetal brain, N, PG hESCs and in N NSCs, but elevated in PG NSCs (see Figure 3A). HLA-G mRNA in PG NSCs was expressed at 10% to 20% of JEG-3 levels, a cell line known for its extremely high HLA-G expression, and at about 10-fold lower amounts in N NSCs (1% to 3%; see Figure 3A). Western blotting and flow cytometry revealed minor differences in HLA-G protein levels between PG and N NSCs (Figures 3B, C). Western analysis was consistent with the size of HLA-G1 (39 kDa) (see Figure 3B).
Figure 3.
HLA-G expression in PG NSCs. (A) HLA-G-specific RT-PCR analysis of human K562 CML cells, PBMCs, fetal brain (19 wks of gestation), N and PG hESCs and NSCs. Samples were normalized to the housekeeper gene RPL30. Relative gene expression levels were determined using the 2−ΔΔCt method; HLA-G gene expression in human JEG-3 cells was set to 1. **p < 0.001; n = 3. (B) Western blot analysis to determine HLA-G protein levels in PG and N NSCs (antibody: MEM-G/1). GAPDH was used as loading control (left panel). Densitometric analysis of three independent Western blots was performed using ImageJ. HLA-G protein levels were normalized to GAPDH. *p < 0.05 (right panel). (C) Flow cytometry of untreated (upper panel) or 24-h EDTA-treated (1 mmol/L) (lower panel) NSC cultures. Cells were stained with the HLA-G-specific antibody MEM-G/9. Histograms show specific fluorescence signal for HLA-G (black lines) compared with isotype control (filled gray). Specific fluorescence indexes (SFI: specific geometric median/geometric median of unspecific control) are indicated in the upper right.
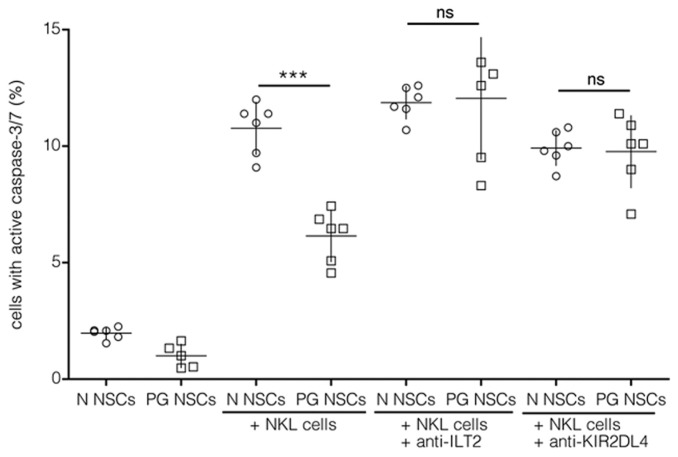
Next, to determine the function of HLA-G during the interaction of immune cells with allogeneic, hESC-derived NSCs, we analyzed apoptotic mechanisms. Flow cytometric detection of activated caspase-3 and -7 in PG and N NSCs following coculture in a 10:1 ratio was performed. Consistent with reduced NKL cell–mediated lysis of PG NSCs (see Figure 2B), PG cells exhibited reduced caspase-3/7 activity compared with N NSCs (Figure 4). Interaction of HLA-G with the inhibitory NK receptors ILT2, ILT4 or KIR2DL4 can inhibit the cytolytic function of NK cells (24). Therefore, we asked whether different HLA-G levels in PG and N NSCs caused the observed differences in susceptibility to NKL cell lysis. As shown in Figure 4, masking the inhibitory NKL receptors ILT2 or KIR2DL4 with functional blocking antibodies during coculture of NKL and target cells increased caspase-3/7 activity in PG NSC cultures. In N NSC cultures, blockage of ILT2 or KIR2DL4 had, however, no effect on NKL-induced apoptosis. We refrained from also blocking ILT4 because this receptor is not expressed by NKL cells. Thus, masking of inhibitory NKL receptors leads to increased apoptosis of PG but not N NSCs after NKL exposure, suggesting a specific role of the ILT2 and KIR2DL4 ligand HLA-G in protecting PG NSCs against NKL cell lysis.
Figure 4.
Increased apoptosis in PG NSCs after blockage of inhibitory NKL receptors. Flow cytometric analysis of activated caspase-3/7 in the target cells PG (□) or N (○) NSCs, co-cultures of effector NKL cells and target cells (E:T) at 10:1 NKL-to-target ratios, E:T in the presence of blocking ILT2 antibody (E:T anti-ILT2) or KIR2DL4 antibody (E:T anti-KIR). ***p < 0.0001, not significant (ns); n ≥ 5.
MIR152 Regulates HLA-G Expression in PG NSCs
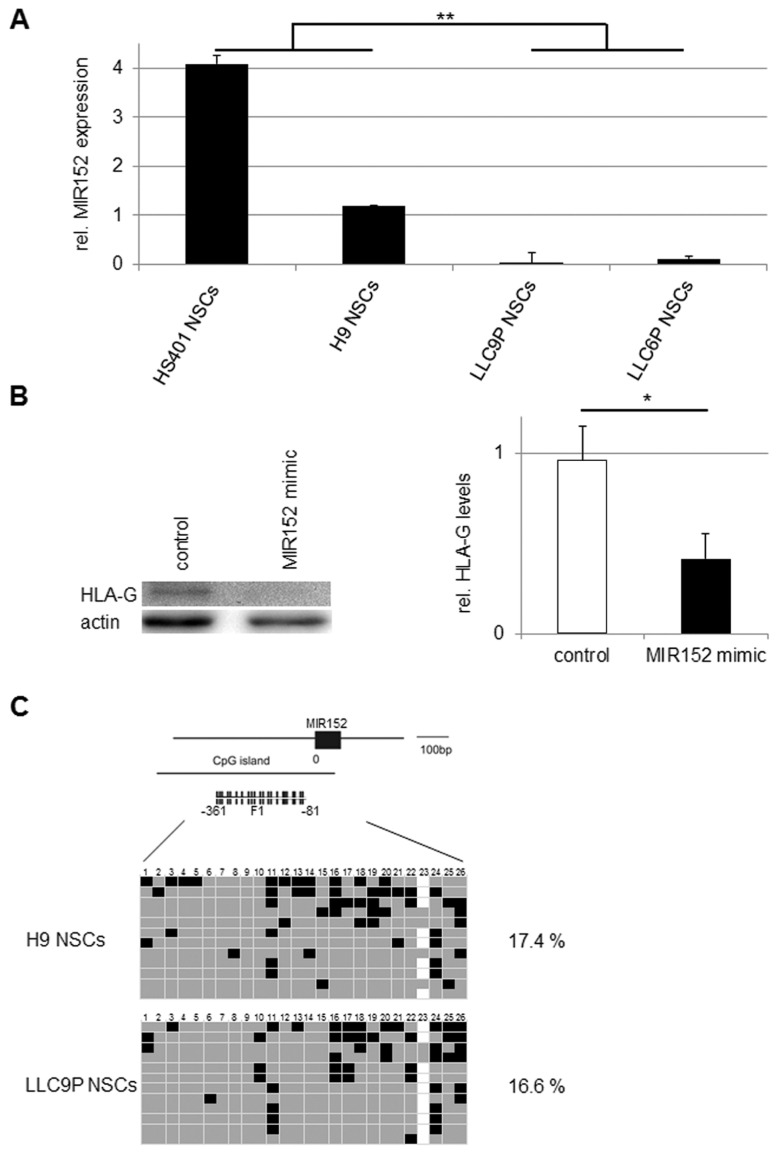
As the promoter region for HLA-G with its variable sites has not yet been characterized in detail, a potential regulation of HLA-G by promoter methylation could not be assessed. HLA-G is, however, a known target of MIR152, a microRNA which downregulates HLA-G expression and decreases ILT2-mediated inhibition of NK cell–mediated cytolysis (42). Real-time RT-PCR analysis to determine whether differences in HLA-G expression in PG and N NSCs correlated with the respective MIR152 levels confirmed lower expression levels of MIR152 in PG compared with N NSCs (Figure 5A). To perform gain-of-function analyses, we overexpressed MIR152 in PG NSCs. As shown in Figure 5B, over-expression of MIR152 in PG NSCs decreased HLA-G expression, while transfection of an unrelated RNA mimic did not affect HLA-G levels. This indicates that HLA-G expression is regulated by MIR152 in NSCs. Due to the different expression levels of MIR152 in PG and N NSCs, we hypothesized that MIR152 expression may be subject to parent-of-origin–specific regulation. Bisulfite sequencing of the MIR152 promoter region, however, did not reveal differences in CpG methylation between PG and N NSCs (Figure 5C). The observed differential expression of MIR152 in PG and N NSCs is therefore independent of promoter DNA methylation.
Figure 5.
MIR152 expression in PG NSCs. (A) Real-time RT-PCR analyses of MIR152 expression in PG and N NSCs normalized to the housekeeper gene U6. Gene expression was calculated with the 2−ΔΔCt method; MIR152 gene expression in PBMCs was set to 1. **p < 0.001; n = 3. (B) Overexpression of MIR152 in LLC9P NSCs. Western blot analysis of HLA-G (MEM-G/1) in PG (LLC9P) NSCs transfected with anti-GAPDH (control) or MIR152 mimic double-stranded oligonucleotides. Actin was used as loading control (left panel). Densitometric analysis of three independent western blots was performed using ImageJ. HLA-G protein levels were normalized to actin. *p < 0.05 (right panel). (C) Analysis of CpG methylation of the MIR152 promoter in PG and N NSCs. F1 represents the genomic region selected for bisulfite sequencing. Each row represents a separate DNA molecule (clone); CpG numbers are indicated on top. Gray and black boxes represent unmethylated and methylated CpG dinucleotides, respectively; white boxes: no data. Percentages of CpG methylation are indicated. n = 2.
DISCUSSION
Our objective was to characterize HLA expression and function in PG NSCs. In summary, we observed that PG NSCs, which contain only maternally derived genomes, and N NSCs both express HLA-I proteins. Surprisingly, however, PG NSCs express high levels of HLA-G, a nonclassical HLA molecule that is expressed in a limited range of cell types. Functional immunological analyses revealed that, compared with N NSCs, PG NSCs stimulated less proliferation of allogeneic PBMCs. It is highly plausible that the impaired T cell proliferation is linked to the presence of HLA-G, though this link was not ultimately proven by direct blocking experiments. Blockage of HLA-G receptors ILT2 and KIR2DL4 confirmed that high HLA-G expression levels decreased the susceptibility of PG NSCs toward NKL cell lysis.
Similar to earlier reports on N hESCs and their differentiated progeny, we observed that PG NSCs expressed low levels of HLA-I proteins but no HLA-II or costimulatory molecules (16,18,19). HLA-I expression in PG NSCs was increased by IFN-γ treatment and reached PBMC levels as reported for N hESCs (16). Thus, HLA-I molecules are expressed on the surface of PG NSCs. The absence of HLA-II and costimulatory molecules on PG and N NSCs predicts that these cells do not deliver appropriate stimulatory signals for robust immune responses by direct allorecognition (16,17).
The status of HLA-G expression in N hESCs and their progeny remains unclear as the literature reports conflicting results. One report concluded that HLA-G is not expressed on N hESCs or differentiated progeny (16). Others showed that functionally active HLA-G is expressed (30,31). However, these discrepant results were generated using different hESC lines and different HLA-G antibodies. Despite substantial variation among pluripotent cell lines, the cell lines used in the present study showed similar results (43). In particular, this relates to HLA-G being expressed at higher levels in PG compared with N cells. Our analysis did not discriminate between the differentially spliced membrane-bound and soluble HLA-G isoforms, as all of these forms are immunosuppressive (30). Likewise, we did not test to which extent HLA-G on PG NSCs was bound to β2-microglobulin (β2M). Of note, free HLA-G also can engage the inhibitory receptor ILT4. Moreover, there is no neutralizing antibody for free HLA-G available (compare patent application WO2014072534A1/EP 2730588 A1, published May 14, 2014). Western blotting revealed a predominance of HLA-G1 in both N and PG NSCs as reported previously for N hESCs (31). The elevated HLA-G levels in PG compared with N NSCs may be caused by parent-of-origin–specific mechanisms. Although a small number of genes are imprinted, these genes have important effects on development and physiology, and many are expressed in the brain (9). Earlier analyses revealed that, in first trimester trophoblast and placental cells, HLA-G is expressed from both alleles (44,45). However, for most tissues, allele specificity of expression has not yet been determined.
HLA-G expression underlies post-transcriptional regulation. Overexpression of MIR152 revealed that MIR152 targets and downregulates HLA-G expression and thereby sensitizes cells to NK cell–mediated killing (36,42). Low levels of MIR152 were found in tumors with elevated HLA-G expression, such as glioblastoma (46,47). We show that MIR152 expression levels were lower in PG compared with N NSCs and that overexpression of MIR152 in PG NSCs reduced HLA-G protein levels. Although bisulfite analyses of the MIR152 promoter did not reveal differential methylation patterns between PG and N cells, differential DNA methylation may occur at distant regions controlling MIR152 expression. Therefore, our data support a model that includes MIR152 as a regulator of HLA-G expression.
HLA-G expression in fetal cells of the placenta has been associated with inhibition of the maternal immune system. Its presence in other fetal tissues indicates additional roles during development (30); for example, HLA-G expression in primitive erythroid cells throughout prenatal life suggests a possible role in erythroid differentiation and angiogenesis (48).
In agreement with elevated HLA-G levels, PG NSCs were recognized by allogeneic immune cells but at reduced levels compared with N NSCs. At the maternal-fetal interface, HLA-G suppresses a wide range of immune responses by binding to the inhibitory receptors ILT2, ILT4 and KIR2DL4 (24). Antibody-mediated masking of either ILT2 or KIR2DL4 prior to HLA-G binding resulted in increased apoptosis of PG NSCs, further supporting the notion that HLA-G molecules on PG NSCs can functionally activate inhibitory NK cell receptors. Blockade of ILT4 was not investigated since this receptor is not expressed on NKL cells.
Parent-of-origin effects influence the etiology of multiple sclerosis as the paternally expressed DLK1 was identified as a novel disease-predisposing gene in an autoimmunity encephalomyelitis model in rat (49). Our data show that the presence of maternal-only genomes in neural cells is associated with more pronounced HLA-G expression, suggesting higher HLA-G-mediated immune tolerance. The function of HLA-I proteins is, however, not limited to the immune system. HLA-I proteins also have been found to participate in brain development and plasticity and to regulate synaptic development and function of cortical connections in the developing mouse brain (50). Our observation that HLA-G is transcribed in the human fetal brain and that HLA-G expression is upregulated during neural hESC differentiation indicates that HLA-G may also play a role during early developmental stages in the human brain, possibly via interaction with its receptor ILT2, which is expressed on microglial cells (51).
The brain is often considered an immune-privileged tissue. However, potent immune reactions occur in the central nervous system (52). HLA-G as a tolerogenic molecule could be employed in therapeutic applications. Along this line, hESC-derived dopaminergic neurons with stable HLA-G expression elicited a far lower immune response than non-transgenic hESC derivatives (41). HLA-G is also a key immunomodulatory molecule expressed by hESC-derived mesenchymal progenitors (53). In addition to the immunomodulatory functions of HLA-G in respect to NK cells, other aspects determine the fate of transplanted stem cells. For example, NSCs secrete paracrine factors including transforming growth factor (TGF)-β1, prostaglandins (PGE2), nitric oxide (NO) and heme oxygenases (HOs) (54). TGF-β1 maintains immune tolerance, T-cell homeostasis and balances the immunogenicity of NSCs (20,55,56). In addition, HOs alone or in combination with NO production mediate immunosuppressive effects of NSCs (57,58), and growth factors and neurotrophins influence stem cell–immune cell interactions (54). Also immune-independent parameters, such as the ability to cope with oxidative stress, are critical for stem cell survival in vivo (59). Thus, the survival of transplanted NSCs is influenced by multiple factors, and a better understanding of the mechanisms of, for example, inflammatory cascades induced by transplanted NSCs is essential for the development of stem cell-based therapies for the brain (60).
Obviously, the best test for the immunogenicity of hESC-derived cells would require in vivo analyses. However, due to the lack of a murine HLA-G homologue and the lack of a suitable transplant model with a functional human immune system, in vivo testing of HLA-G function is difficult to perform and not always conclusive. Considering the difficulties with humanized animal models, we decided not to develop in vivo approaches and performed in vitro assays exclusively involving human cells. We conclude that in vitro, PG NSCs were found to express HLA-G and to be protected against NK cell–mediated lysis in an HLA-G dependent manner. Therefore, PG cells may be an alternative to transgenic HLA-G cells to alleviate or even avoid immune rejection in allogeneic therapies.
CONCLUSION
In summary, PG hESC–derived NSCs with a maternally derived genome exhibit reduced immunogenicity due to elevated HLA-G expression, further enhancing the application of PG hESCs as a possible source of transplantation-based stem cell therapies. Furthermore, PG hESCs are a unique tool to investigate parent-of-origin–specific expression of HLA encoding genes.
Supplemental Data
ACKNOWLEDGMENTS
We thank Ruslan Semechkin for kindly providing the PG hESCs LLC6P and LLC9P (International Stem Cell Corporation), Outi Hovatta and Liselotte Antonsson (Division of Obstetrics and Gynecology, Karolinska Institutet, Stockholm, Sweden) for the HS401 hESCs, WiCell Research Institute Wisconsin (Madison, USA) for the H9 hESCs, Ulrike Kämmerer (Department of Obstetrics and Gynecology, University of Würzburg, Germany) for the JEG-3 cells and Winfried S Wels (Georg-Speyer-Haus, Frankfurt am Main, Germany) for the NKL cell line. We are grateful to Andrea Reusch and Doris Heim (MSZ, Würzburg, Germany) for technical assistance, and to Andrea Niklaus and Katharina Mattenheimer (ZEMM, Würzburg, Germany) for help with sample collection. We thank Joannis Mytilineos (Institute of Clinical Transfusion Medicine and Immunogenetics Ulm, Germany) for high-resolution HLA typing. Funding for this work was provided by the Interdisciplinary Center for Clinical Research (IZKF), University of Würzburg (TP D103), DFG-funded SPP 1738 and by a fellowship, Chancengleichheit für Frauen in Forschung und Lehre, from the University of Würzburg (Würzburg, Germany).
Footnotes
Online address: http://www.molmed.org
DISCLOSURE
The authors declare they have no competing interests as defined by Molecular Medicine, or other interests that might be perceived to influence the results and discussion reported in this paper.
Cite this article as: Schmitt J, et al. (2015) Human parthenogenetic embryonic stem cell–derived neural stem cells express HLA-G and show unique resistance to NK cell–mediated killing. Mol. Med. 21:185–96.
REFERENCES
- 1.Revazova ES, et al. Patient-specific stem cell lines derived from human parthenogenetic blastocysts. Cloning Stem Cells. 2007;9:432–49. doi: 10.1089/clo.2007.0033. [DOI] [PubMed] [Google Scholar]
- 2.Mai Q, et al. Derivation of human embryonic stem cell lines from parthenogenetic blastocysts. Cell Res. 2007;17:1008–19. doi: 10.1038/cr.2007.102. [DOI] [PubMed] [Google Scholar]
- 3.Lin G, et al. A highly homozygous and parthenogenetic human embryonic stem cell line derived from a one-pronuclear oocyte following in vitro fertilization procedure. Cell Res. 2007;17:999–1007. doi: 10.1038/cr.2007.97. [DOI] [PubMed] [Google Scholar]
- 4.Kim K, et al. Recombination signatures distinguish embryonic stem cells derived by parthenogenesis and somatic cell nuclear transfer. Cell Stem Cell. 2007;1:346–52. doi: 10.1016/j.stem.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 5.Brevini TA, et al. Cell lines derived from human parthenogenetic embryos can display aberrant centriole distribution and altered expression levels of mitotic spindle check-point transcripts. Stem Cell Rev. 2009;5:340–52. doi: 10.1007/s12015-009-9086-9. [DOI] [PubMed] [Google Scholar]
- 6.Revazova ES, et al. HLA homozygous stem cell lines derived from human parthenogenetic blastocysts. Cloning Stem Cells. 2008;10:11–24. doi: 10.1089/clo.2007.0063. [DOI] [PubMed] [Google Scholar]
- 7.Turovets N, et al. Derivation of human parthenogenetic stem cell lines. Methods Mol Biol. 2011;767:37–54. doi: 10.1007/978-1-61779-201-4_4. [DOI] [PubMed] [Google Scholar]
- 8.Daughtry B, Mitalipov S. Concise review: parthenote stem cells for regenerative medicine: genetic, epigenetic, and developmental features. Stem Cells Transl Med. 2014;3:290–8. doi: 10.5966/sctm.2013-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilkinson LS, Davies W, Isles AR. Genomic imprinting effects on brain development and function. Nat Rev Neurosci. 2007;8:832–43. doi: 10.1038/nrn2235. [DOI] [PubMed] [Google Scholar]
- 10.Keverne EB, Fundele R, Narasimha M, Barton SC, Surani MA. Genomic imprinting and the differential roles of parental genomes in brain development. Brain Res Dev Brain Res. 1996;92:91–100. doi: 10.1016/0165-3806(95)00209-x. [DOI] [PubMed] [Google Scholar]
- 11.Dinger TC, et al. Androgenetic embryonic stem cells form neural progenitor cells in vivo and in vitro. Stem Cells. 2008;26:1474–83. doi: 10.1634/stemcells.2007-0877. [DOI] [PubMed] [Google Scholar]
- 12.Ahmad R, et al. Functional neuronal cells generated by human parthenogenetic stem cells. PLoS One. 2012;7:e42800. doi: 10.1371/journal.pone.0042800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi SW, et al. Two paternal genomes are compatible with dopaminergic in vitro and in vivo differentiation. Int J Dev Biol. 2010;54:1755–62. doi: 10.1387/ijdb.103188sc. [DOI] [PubMed] [Google Scholar]
- 14.Wolber W, et al. Phenotype and stability of neural differentiation of androgenetic murine ES cell-derived neural progenitor cells. Cell Medicine. 2013;5:29–42. doi: 10.3727/215517913X666468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pearl JI, Kean LS, Davis MM, Wu JC. Pluripotent stem cells: immune to the immune system? Sci Transl Med. 2012;4:164ps125. doi: 10.1126/scitranslmed.3005090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drukker M, et al. Characterization of the expression of MHC proteins in human embryonic stem cells. Proc Natl Acad Sci U S A. 2002;99:9864–9. doi: 10.1073/pnas.142298299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grinnemo KH, et al. Human embryonic stem cells are immunogenic in allogeneic and xenogeneic settings. Reprod Biomed Online. 2006;13:712–24. doi: 10.1016/s1472-6483(10)60663-3. [DOI] [PubMed] [Google Scholar]
- 18.Li L, et al. Human embryonic stem cells possess immune-privileged properties. Stem Cells. 2004;22:448–56. doi: 10.1634/stemcells.22-4-448. [DOI] [PubMed] [Google Scholar]
- 19.Preynat-Seauve O, et al. Neural progenitors derived from human embryonic stem cells are targeted by allogeneic T and natural killer cells. J Cell Mol Med. 2009;13:3556–69. doi: 10.1111/j.1582-4934.2009.00746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J, et al. Human neural stem/progenitor cells derived from embryonic stem cells and fetal nervous system present differences in immunogenicity and immunomodulatory potentials in vitro. Stem Cell Res. 2013;10:325–37. doi: 10.1016/j.scr.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 21.Drukker M, et al. Human embryonic stem cells and their differentiated derivatives are less susceptible to immune rejection than adult cells. Stem Cells. 2006;24:221–9. doi: 10.1634/stemcells.2005-0188. [DOI] [PubMed] [Google Scholar]
- 22.Tian X, Woll PS, Morris JK, Linehan JL, Kaufman DS. Hematopoietic engraftment of human embryonic stem cell-derived cells is regulated by recipient innate immunity. Stem Cells. 2006;24:1370–80. doi: 10.1634/stemcells.2005-0340. [DOI] [PubMed] [Google Scholar]
- 23.Rouas-Freiss N, Goncalves RM, Menier C, Dausset J, Carosella ED. Direct evidence to support the role of HLA-G in protecting the fetus from maternal uterine natural killer cytolysis. Proc Natl Acad Sci U S A. 1997;94:11520–5. doi: 10.1073/pnas.94.21.11520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carosella ED, Favier B, Rouas-Freiss N, Moreau P, Lemaoult J. Beyond the increasing complexity of the immunomodulatory HLA-G molecule. Blood. 2008;111:4862–70. doi: 10.1182/blood-2007-12-127662. [DOI] [PubMed] [Google Scholar]
- 25.Jurisicova A, Casper RF, MacLusky NJ, Mills GB, Librach CL. HLA-G expression during preimplantation human embryo development. Proc Natl Acad Sci U S A. 1996;93:161–5. doi: 10.1073/pnas.93.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maier S, Geraghty DE, Weiss EH. Expression and regulation of HLA-G in human glioma cell lines. Transplant Proc. 1999;31:1849–53. doi: 10.1016/s0041-1345(99)00186-4. [DOI] [PubMed] [Google Scholar]
- 27.Lafon M, et al. Modulation of HLA-G expression in human neural cells after neurotropic viral infections. J Virol. 2005;79:15226–37. doi: 10.1128/JVI.79.24.15226-15237.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nasef A, et al. Immunosuppressive effects of mesenchymal stem cells: involvement of HLA-G. Transplantation. 2007;84:231–7. doi: 10.1097/01.tp.0000267918.07906.08. [DOI] [PubMed] [Google Scholar]
- 29.Wiendl H. HLA-G in the nervous system. Hum Immunol. 2007;68:286–93. doi: 10.1016/j.humimm.2006.10.021. [DOI] [PubMed] [Google Scholar]
- 30.LeMaoult J, et al. Biology and functions of human leukocyte antigen-G in health and sickness. Tissue Antigens. 2003;62:273–84. doi: 10.1034/j.1399-0039.2003.00143.x. [DOI] [PubMed] [Google Scholar]
- 31.Verloes A, et al. HLA-G expression in human embryonic stem cells and preimplantation embryos. J Immunol. 2011;186:2663–71. doi: 10.4049/jimmunol.1001081. [DOI] [PubMed] [Google Scholar]
- 32.Le Page ME, Goodridge JP, John E, Christiansen FT, Witt CS. Killer Ig-like receptor 2DL4 does not mediate NK cell IFN-gamma responses to soluble HLA-G preparations. J Immunol. 2014;192:732–40. doi: 10.4049/jimmunol.1301748. [DOI] [PubMed] [Google Scholar]
- 33.Thomson JA, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–7. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 34.Ström S, Holm F, Bergstrom R, Stromberg AM, Hovatta O. Derivation of 30 human embryonic stem cell lines-improving the quality. In Vitro Cell Dev Biol Anim. 2010;46:337–44. doi: 10.1007/s11626-010-9308-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fürst D, et al. High-resolution HLA matching in hematopoietic stem cell transplantation: a retrospective collaborative analysis. Blood. 2013;122:3220–9. doi: 10.1182/blood-2013-02-482547. [DOI] [PubMed] [Google Scholar]
- 36.Zhu XM, et al. Overexpression of miR-152 leads to reduced expression of human leukocyte antigen-G and increased natural killer cell mediated cytolysis in JEG-3 cells. Am J Obstet Gynecol. 2010;202:592.e1–7. doi: 10.1016/j.ajog.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 37.Ji W, et al. MicroRNA-152 targets DNA methyltransferase 1 in NiS-transformed cells via a feedback mechanism. Carcinogenesis. 2013;34:446–53. doi: 10.1093/carcin/bgs343. [DOI] [PubMed] [Google Scholar]
- 38.Rohde C, Zhang Y, Reinhardt R, Jeltsch A. BISMA-fast and accurate bisulfite sequencing data analysis of individual clones from unique and repetitive sequences. BMC Bioinformatics. 2010;11:230. doi: 10.1186/1471-2105-11-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burt D, Johnston D, Rinke de Wit T, Van den Elsen P, Stern PL. Cellular immune recognition of HLA-G-expressing choriocarcinoma cell line Jeg-3. Int J Cancer Suppl. 1991;6:117–22. doi: 10.1002/ijc.2910470722. [DOI] [PubMed] [Google Scholar]
- 40.Ljunggren HGKK. In search of the ‘missing self’: MHC molecules and NK cell recognition. Immunol Today. 1990;11:237–44. doi: 10.1016/0167-5699(90)90097-s. [DOI] [PubMed] [Google Scholar]
- 41.Zhu Y, et al. DA neurons derived from hES cells that express HLA-G1 are capable of im-munosuppression. Brain Res. 2012;1437:134–42. doi: 10.1016/j.brainres.2011.12.033. [DOI] [PubMed] [Google Scholar]
- 42.Manaster I, et al. MiRNA-mediated control of HLA-G expression and function. PLoS One. 2012;7:e33395. doi: 10.1371/journal.pone.0033395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bock C, et al. Reference maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144:439–52. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hiby SE, King A, Sharkey A, Loke YW. Molecular studies of trophoblast HLA-G: polymorphism, isoforms, imprinting and expression in preimplantation embryo. Tissue Antigens. 1999;53:1–13. doi: 10.1034/j.1399-0039.1999.530101.x. [DOI] [PubMed] [Google Scholar]
- 45.Hviid TVF, Møller C, Sørensen S, Morling N. Co-dominant expression of the HLA-G gene and various forms of alternatively spliced HLA-G mRNA in human first trimester trophoblast. Hum Immunology. 1998;59:87–98. doi: 10.1016/s0198-8859(97)00259-0. [DOI] [PubMed] [Google Scholar]
- 46.Zheng X, Chopp M, Lu Y, Buller B, Jiang F. MiR-15b and miR-152 reduce glioma cell invasion and angiogenesis via NRP-2 and MMP-3. Cancer Lett. 2013;329:146–54. doi: 10.1016/j.canlet.2012.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Veit TD, Chies JA. Tolerance versus immune response — microRNAs as important elements in the regulation of the HLA-G gene expression. Transpl Immunol. 2009;20:229–31. doi: 10.1016/j.trim.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 48.Menier C, et al. Erythroblasts secrete the nonclassical HLA-G molecule from primitive to definitive hematopoiesis. Blood. 2004;104:3153–60. doi: 10.1182/blood-2004-03-0809. [DOI] [PubMed] [Google Scholar]
- 49.Stridh P, et al. Parent-of-origin effects implicate epigenetic regulation of experimental autoimmune encephalomyelitis and identify imprinted Dlk1 as a novel risk gene. PLoS Genet. 2014;10:e1004265. doi: 10.1371/journal.pgen.1004265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elmer BM, McAllister AK. Major histocompatibility complex class I proteins in brain development and plasticity. Trends Neurosci. 2012;35:660–70. doi: 10.1016/j.tins.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wiendl H, et al. Expression of the immune-tolerogenic major histocompatibility molecule HLA-G in multiple sclerosis: implications for CNS immunity. Brain. 2005;128:2689–704. doi: 10.1093/brain/awh609. [DOI] [PubMed] [Google Scholar]
- 52.Hickey WF. Basic principles of immunological surveillance of the normal central nervous system. Glia. 2001;36:118–24. doi: 10.1002/glia.1101. [DOI] [PubMed] [Google Scholar]
- 53.Yen BL, et al. Brief report—human embryonic stem cell-derived mesenchymal progenitors possess strong immunosuppressive effects toward natural killer cells as well as T lymphocytes. Stem Cells. 2009;27:451–6. doi: 10.1634/stemcells.2008-0390. [DOI] [PubMed] [Google Scholar]
- 54.Pluchino S, Cossetti C. How stem cells speak with host immune cells in inflammatory brain diseases. Glia. 2013;61:1379–401. doi: 10.1002/glia.22500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bommireddy R, et al. Calcineurin deficiency decreases inflammatory lesions in transforming growth factor beta1-deficient mice. Clin Exp Immunol. 2009;158:317–24. doi: 10.1111/j.1365-2249.2009.04015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ubiali F, et al. Allorecognition of human neural stem cells by peripheral blood lymphocytes despite low expression of MHC molecules: role of TGF-beta in modulating proliferation. Int Immunol. 2007;19:1063–74. doi: 10.1093/intimm/dxm079. [DOI] [PubMed] [Google Scholar]
- 57.Bonnamain V, et al. Expression of heme oxygenase-1 in neural stem/progenitor cells as a potential mechanism to evade host immune response. Stem Cells. 2012;30:2342–53. doi: 10.1002/stem.1199. [DOI] [PubMed] [Google Scholar]
- 58.Wang L, et al. Neural stem/progenitor cells modulate immune responses by suppressing T lymphocytes with nitric oxide and prostaglandin E2. Exp Neurol. 2009;216:177–83. doi: 10.1016/j.expneurol.2008.11.017. [DOI] [PubMed] [Google Scholar]
- 59.Carletti B, Piemonte F, Rossi F. Neuroprotection: the emerging concept of restorative neural stem cell biology for the treatment of neurodegenerative diseases. Curr Neuropharmacol. 2011;9:313–7. doi: 10.2174/157015911795596603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lindvall O, Barker RA, Brustle O, Isacson O, Svendsen CN. Clinical translation of stem cells in neurodegenerative disorders. Cell Stem Cell. 2012;10:151–5. doi: 10.1016/j.stem.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.