Abstract
On the basis of their characteristics, we presume that developmental stage-specific hepatocytes should have the ability to induce maturation of hepatoma cells. A regulatory circuit formed by hepatocyte nuclear factor (HNF)-4α, HNF-1α, HNF-6 and the upstream stimulatory factor (USF-1) play a key role in the maturation of embryonic hepatocytes; however, it is unclear whether the regulatory circuit mediates the embryonic induction of hepatoma cell maturation. In this study, 12.5-d to 15.5-d mouse embryonic hepatocytes or their medium were used to coculture or treat HepG2 cells, and the induced maturation was evaluated in vitro and in vivo. In the induced HepG2 cells, the components of the regulatory circuit were detected, their cross-regulation was evaluated and HNF-4α RNA interference was performed. We found that 13.5-d to 14.5-d embryonic hepatocytes could induce HepG2 cell maturation, demonstrated by morphological changes, increased maturation markers and decreased c-Myc and α-fetoprotein (AFP) in vitro. The majority of HepG2 tumors were eliminated by 13.5-d embryonic induction in vivo. All components of the regulatory circuit were upregulated and the binding of HNF-4α, HNF-1α, HNF-6 and USF-1 to their target sites was promoted to rebuild the regulatory circuit in the induced HepG2 cells. Moreover, RNA interference targeting HNF-4α, which is the core of the regulatory circuit, attenuated the induced maturation of HepG2 cells with downregulation of the regulatory circuit. These results revealed that developmental stage-specific hepatocytes could induce the maturation of HepG2 cells by rebuilding the regulatory circuit.
INTRODUCTION
Hepatocellular carcinoma (HCC), which is a major liver malignancy, is the third leading cause of cancer death (1–3). Although the current treatments are able to effectively kill tumor cells, they fail to completely cure HCC and inhibit its recurrence (4,5). Therefore, the search for a specific therapy remains a struggle in medical research.
HCC cells own common characteristics that are often found to be hepatic stem/progenitor cell biomarkers (2,6–10), although multiple environmental and genetic risk factors exist. HCC progression and embryonic liver development share many same properties, and a tumor may originate from progenitor/stem cells that receive abnormal proliferation and maturation signals (11–13). Thus, knowledge about cancer stem cells may reveal the exact biology of tumor cells and will offer new insights on HCC therapy (5,14–16).
A series of studies focused on the embryo (including embryonic stem cells, embryonary sac and parts of fetus) to induce tumor cell maturation (17,18). However, not all the induction experiments succeed, and the intrinsic explanation is not fully clarified (19,20). On the basis of their characteristics, we presumed that developmental stage-specific hepatocytes, in theory, should have the ability to direct and induce maturation of tumor cells (21). The question of whether developmental stage-specific hepatocytes could induce the maturation of HepG2 cells has not been explored.
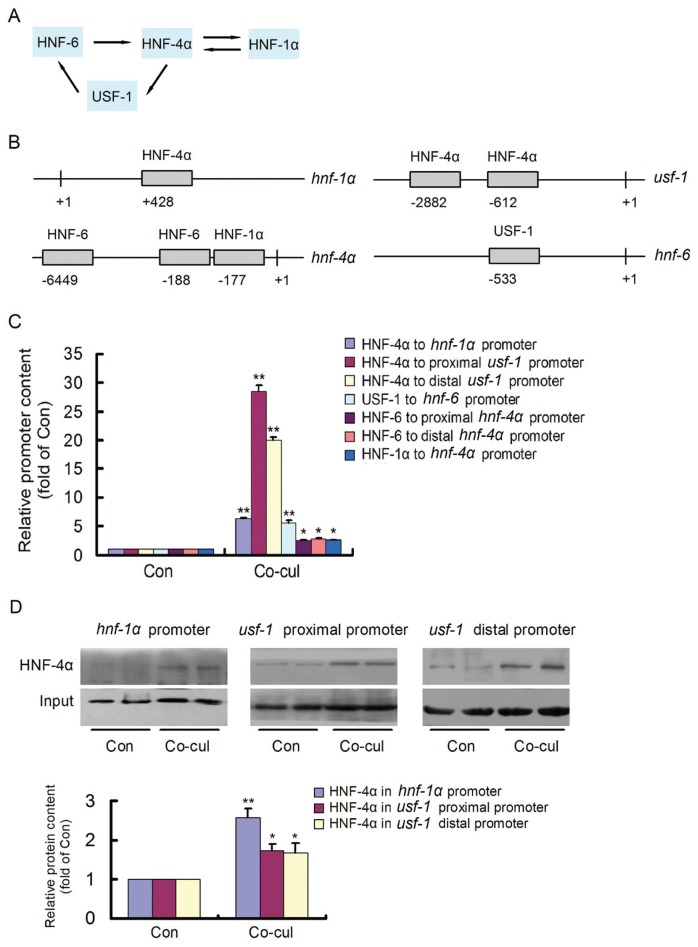
During liver development, complex regulatory cascades of hepatic transcription factors control hepatocyte maturation (22,23), and the regulatory circuit formed by cross-regulation of hepatocyte nuclear factor (HNF)-4α, HNF-1α, HNF-6 and upstream stimulatory factor (USF)-1 plays a pivotal role in hepatocyte maturation and functional formation (24–26). HNF-4α, the core of the regulatory circuit, is believed to be a controller and marker of mature hepatocytes (27,28). As a downstream target of HNF-4α, HNF-1α is also essential for hepatocyte maturation (29,30). USF-1, which competes with the oncogene c-Myc for the E-box regulatory element, is another downstream target of HNF-4α that plays crucial roles in the maturation and formation of functional hepatocytes (31,32). USF-1 can activate HNF-6, which is another important transcription factor for hepatocyte maturation that has been found to regulate HNF-4α (23,33). Therefore, HNF-4α, HNF-1α, HNF-6 and USF-1 form a positive feedback loop that drives hepatocyte maturation in liver development (Figure 1A). Because the transcription factor binding sites are generally believed to scatter within 10 kb upstream of the transcriptional start site (34), the 10-kb gene promoters were all analyzed. On the basis of previous reports (25), the predicted binding sites of HNF-4α HNF-1α, HNF-6 and USF-1 were identified, respectively (Figure 1B). It remains unknown whether developmental stage-specific hepatocytes rebuild the regulatory circuit to induce the maturation of HepG2 cells.
Figure 1.
The cross-regulation of the regulatory circuit in induced HepG2 cells. (A) As illustrated, HNF-4α, HNF-1α, HNF-6 and USF-1 were cross-regulated to form a positive feedback circuit. (B) The predicted binding sites of HNF-4α, HNF-1α, HNF-6 and USF-1 within their target promoters were identified. (C) Binding of HNF-4α, HNF-1α, HNF-6 and USF-1 to their target sites was evaluated by ChIP. After HepG2 cells were cocultured with 13.5-d mouse embryonic hepatocytes for 48 h, the binding of HNF-4α, HNF-1α, HNF-6 and USF-1 to their target elements was significantly promoted. (D) The contents of HNF-4α bound in hnf-1α and usf-1 promoters were detected by DNA pull-down assay. After HepG2 cells were treated with 13.5-d mouse embryonic hepatocyte medium for 48 h, the bound HNF-4α increased significantly. *P < 0.05 and **P < 0.01 versus control. The experiments were repeated three times. Con, control.
In this study, HepG2 cells were cocultured with mouse embryonic hepatocytes at gestation of 12.5–15.5 d, because the mouse hepatocytes differentiated at 12.5 d, and the liver structure became firmly established between 12 and 15 d of gestation (35–37). The induced maturation of HepG2 cells was evaluated on the basis of their morphological changes, HNF-4α and c-Myc expression and α-fetoprotein (AFP) content. In addition, expressions of cytochrome P450, family 3, subfamily A, polypeptide 4 (CYP3A4), CYP1B1, ornithine carbamoyltransferase (OTC), arginase 1 (ARG1) and alcohol dehydrogenase 1C (class I), γ polypeptide (ADH1C), in 13.5-d embryonic hepatocyte medium–treated HepG2 cells were evaluated. Moreover, the effects of the 13.5-d embryonic hepatocyte medium on the HepG2 tumor cells that were inoculated into nude mice were observed to confirm their induction effects in vivo. All components of the regulatory circuit (HNF-4α, HNF-1α, HNF-6 and USF-1) were detected, and their cross-regulation was measured to investigate whether the regulatory circuit was rebuilt in the induced HepG2 cells. In addition, RNA interference of HNF-4α was performed to confirm the role of the regulatory circuit in the induced maturation of HepG2 cells. In parallel, 10.5-d to 13.5-d embryonic hepatocyte medium was used to treat another human hepatoma cell line SMMC-7721 for 48 h, and its proliferation and maturation markers CYP1B1 and ADH1C were evaluated.
MATERIALS AND METHODS
Cells and Treatment
HepG2 (human hepatoma cell line) and SMMC-7721 cells were a product of the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and were cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% (v/v) fetal bovine serum (Gibco [Thermo Fisher Scientific Inc., Waltham, MA, USA]).
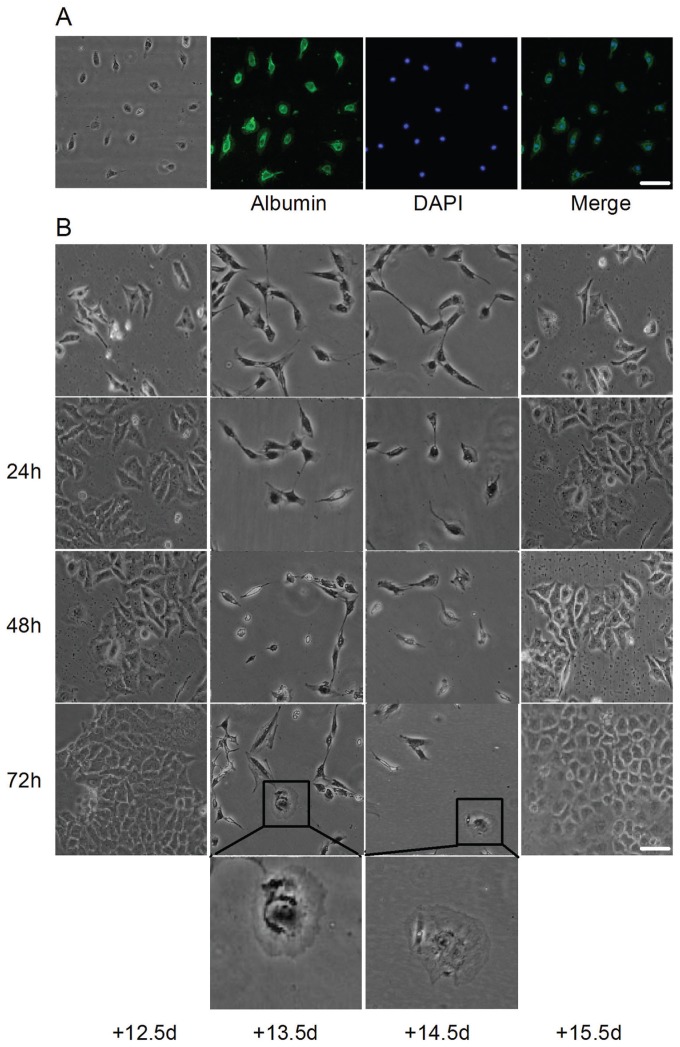
Six- to eight-week-old Swiss mice (25–30 g) from the Department of Experimental Animals of Hebei Medical University (1203166) were housed in a controlled environment with free access to food and water and maintained on a 12-h light–dark cycle. The animal experimental protocol was approved by the institutional animal care committee. Estrus female and male mice were placed together at night, and the following morning, female mice with a vaginal plug were designated as 0.5 d gestation. The female mice at 12.5-, 13.5-, 14.5- and 15.5-d gestation were sacrificed, and their embryonic livers were isolated. After each embryonic liver was sheared into tissue pieces, 0.25% collagenase IV in D-Hanks was used for digestion. Embryonic hepatocytes were collected by centrifugation at 1000g and filtration and purified by differential adhesion. The embryonic hepatocytes were identified by albumin immunofluorescence (Figure 2A). The freshly isolated embryonic hepatocytes were directly used for coculture. HepG2 cells (105) were seeded in six-well plates, and embryonic hepatocytes (107) were cocultured in inserts of Transwell chambers (EMD Millipore, Billerica, MA, USA), which were divided into 12.5-d, 13.5-d, 14.5-d and 15.5-d groups. After coculturing for different times, HepG2 cells were observed, photographed and used for further detection. On the other hand, after the freshly isolated embryonic hepatocytes were cultured with DMEM (1 mL per embryonic liver) for 48 h, the supernatant was collected and used to treat HepG2 cells in 1:1 volume. After being treated for 48 h, the cells were photographed, counted and used for MTT and other detections. In addition, expressions of cyp3a4, cyp1b1, otc, arg1 and adh1c were detected. Meanwhile, 10.5-d, 11.5-d, 12.5-d and 13.5-d hepatocyte medium was used to treat SMMC-7721 cells, and its morphological changes, proliferation rate by MTT and expressions of cyp1b1 and adh1c were detected.
Figure 2.
Morphological changes in HepG2 cells cocultured with mouse embryonic hepatocytes. (A) Mouse primary embryonic hepatocytes were identified by detecting albumin. (B) After coculturing with mouse embryonic hepatocytes at 12.5-, 13.5-, 14.5- and 15.5-d gestation, HepG2 cells were observed. No proliferation was observed, and most HepG2 cells became round and shrunken in shape and detached and drifted in the 13.5-d and 14.5-d groups. Moreover, only a few cells were observed, and they appeared mononuclear and hexagonal, similar to hepatocytes. However, in 12.5-d and 15.5-d groups, apparent proliferation was observed with no obvious morphological changes. Scale bar = 20 μm.
Quantitative Real-Time Reverse Transcription–Polymerase Chain Reaction
Total RNA of HepG2 cells was isolated by using Trizol reagent (Takara, Japan) and reverse-transcribed into cDNA by using the RevertAid First-Strand cDNA Synthesis Kit (Fermentas, Canada) followed by real-time polymerase chain reaction (PCR) amplification with specific primers (Supplementary Table S1). Actin was used as a normalization gene.
Western Blotting
HNF-4α, HNF-1α, HNF-6, USF-1 and c-Myc protein content was measured by Western blotting by using a previously described protocol (38,39). Anti–HNF-4α, anti-HNF-1α, anti–HNF-6, anti–USF-1, anti–c-Myc and anti-actin antibodies (Santa Cruz Biotechnology, Santa Cruz, CA, USA) were used, and band intensities were quantified and calculated.
In Vivo Experiment
The 13.5-d embryonic hepatocytes were cultured for 48 h, and then the medium was collected. After having been centrifuged at 8000g, the supernatant was collected, filtered and used to treat HepG2 tumors.
Four-week-old nude mice were a product of Beijing HFK Bioscience (11401300001123; Beijing HFK Biotechnology Co. Ltd., Beijing, P.R. China), and the animal experimental protocol was approved by the institutional animal care committee. Sixteen mice were inoculated subcutaneously in armpit with 1 × 107 HepG2 cells in 1 mL phosphate-buffered saline (PBS) and divided equally into control and treated groups. When tumors became apparent, the mice in the treated group were locally injected with 0.1 mL of the 13.5-d embryonic hepatocyte medium, and the same volume of medium was injected into the control group once daily. The mice were treated for 17 d, and the changes in the tumors were observed for 25 d.
Chromatin Immunoprecipitation
The binding of HNF-4α, HNF-1α, HNF-6 and USF-1 to their target promoters was analyzed by chromatin immunoprecipitation (ChIP), which was performed according to a previously published protocol (39,40). Briefly, the cells were cross-linked, collected and washed. Then, the cell suspension was sonicated, and an aliquot of the sonicated DNA was precipitated with ethanol and analyzed by electrophoresis. The average fragment sizes were 0.5–1 kb. Anti–HNF-4α, anti–HNF-1α, anti–HNF-6, anti–USF-1 (Santa Cruz Biotechnology) or control rabbit IgG (Santa Cruz Biotechnology) was added to an aliquot of 200 μL sonicated lysate, and 20 μL washed protein G-agarose beads (Santa Cruz Biotechnology) was then added. The mixture was rotated and subsequently centrifuged at 500g to wash the beads. The washed beads were resuspended, vortexed and boiled, and the sample and the sonicated lysate were treated with proteinase K. After centrifugation at 12,000g, the digested DNA was used in real-time PCR assays. The primers for the binding sites are listed in Supplementary Table S2. Additionally, controls of the nuclear factor (NF)-κB binding site in inducible nitric oxide synthase (inos) gene promoter and controls with only the antibody or beads were performed.
DNA Pull-down
The protein contents of HNF-4α bound in hnf-1α and usf-1 promoters were measured by DNA pull-down assay. The cells were collected and nuclear proteins were extracted. After protein concentration was determined, DNA affinity precipitation assay was performed. The oligonucleotides containing biotin on the 5′-end of the each strand were used. The sequences of oligonucleotides for the predicted HNF-4α binding sites were in Supplementary Table S3. Each pair of oligonucleotides was annealed following standard protocols. Nuclear protein extracts (200 μg) were precleared with ImmunoPure streptavidin-agarose beads (20 μL/sample, Thermo Fisher Scientific). After centrifugation at 12,000g, the supernatant was incubated with 4 μg biotinylated double-stranded oligonucleotides overnight. A total of 20 μL streptavidin-agarose beads was added and incubated, and the protein-DNA-streptavidin-agarose complex was subjected to Western blotting with anti–HNF-4α antibody.
RNA Interference
A small interfering RNA (siRNA) targeting HNF-4α (si-HNF-4α: 5′-CCACA UGUACUCCUGCAGATT-3′ and 5′-UCUGCAGGAGUACAUGUGGTT-3′), a siRNA control (si-control: 5′-UUCUC CGAACGUGUCACGUTT-3′ and 5′-ACGUGACACGUUCGGAGAATT-3′) and fluorescein-labeled (FAM)-siRNA were products of Integrated Biotech Solutions (Shanghai, China). Transfections were performed by using the Lipofectamine Reagent (Invitrogen [Thermo Fisher Scientific]) following the manufacturer’s instructions. First, after transfection with the FAM-siRNA, the cells were observed by using a fluorescence microscope (Olympus IX51, Olympus, Tokyo, Japan) and were then subjected to flow cytometry (Guava EasyCyte Mini, EMD Millipore) to evaluate the transfection efficiency. Then, 24 h after transfection, HepG2 cells were cocultured with 13.5-d embryonic hepatocytes for 48 h, and the HNF-4α mRNA and protein contents were examined to confirm the interfering effect. After the cells were successfully treated, the AFP content was determined by immunofluorescence, and the mRNA and protein contents of HNF-1α, HNF-6, USF-1 and c-Myc were measured.
Immunofluorescence
Albumin in primary embryonic hepatocytes and AFP in the cocultured HepG2 cells or with si-HNF-4α were visualized by immunofluorescence detection. After fixation, the cells were incubated with 1% Triton X-100 for 30 min, blocked with 10% goat serum for 1 h and incubated with primary antibody (albumin or AFP; ZSGB-BIO, Beijing, China) at 37°C for 2 h. After washing with PBS, the cells were incubated with the secondary antibody (fluorescein isothiocyanate [FITC]; ZSGB-BIO). Subsequently, the cells were exposed to 0.5 μg/mL 4′,6-diamidino-2-phenylindole (DAPI) for 15 min at room temperature and sealed with 50% glycerol. Images were taken by using a fluorescence microscope (Olympus IX51). Controls were also performed with PBS in place of the first or second antibody.
Statistical Analysis
Data are presented as the means ± standard deviation (SD), and all statistical tests were two-sided. Student t test was used to assess statistical significance, and a P value <0.05 was considered significant.
All supplementary materials are available online at www.molmed.org.
RESULTS
Developmental Stage-Specific Hepatocytes Induced Maturation of HepG2 Cells
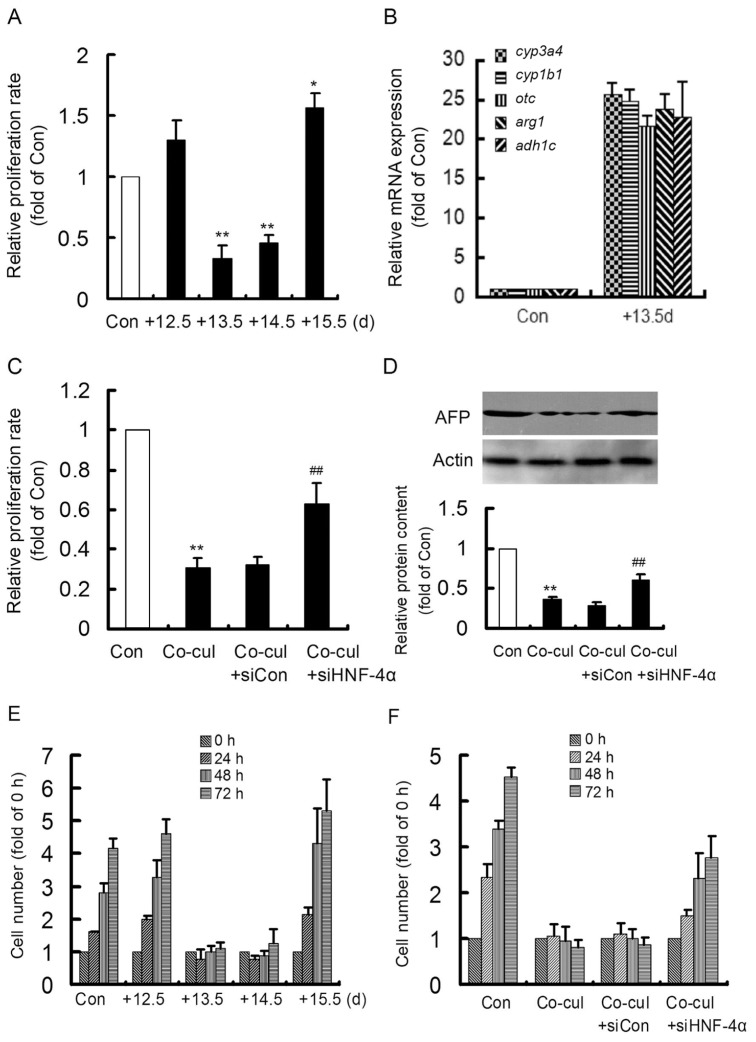
Morphological changes in HepG2 cells were observed after coculturing with 12.5-d to 15.5-d embryonic hepatocytes for different times (Figure 2B). Although HepG2 cells cocultured with 12.5-d and 15.5-d embryonic hepatocytes proliferated and did not show conspicuous morphological changes, when HepG2 cells were cocultured with 13.5-d and 14.5-d embryonic hepatocytes, their proliferation was completely inhibited, and they showed distinct morphological changes. Most HepG2 cells were round and shrunken in shape and had detached and drifted; however, the existing cells were mononuclear and hexagonal, similar to hepatocytes. And the proliferation rate of HepG2 cells treated with 13.5-d and 14.5-d embryonic hepatocyte medium decreased significantly, which showed an increase in the 15.5-d group (Figures 3A, E).
Figure 3.
The proliferation, maturation marker gene expression and AFP content in treated HepG2 cells. After HepG2 cells were treated with 12.5-, 13.5-, 14.5- and 15.5-d mouse embryonic hepatocyte medium, their proliferation rates were detected by MTT, which was significantly inhibited in the 13.5-d and 14.5-d groups (A), and meanwhile, the cell number along time was counted (E). (B) The expression of cyp3a4, cyp1b1, otc, arg1 and adh1c in HepG2 cells treated with 13.5-d mouse embryonic hepatocyte medium was dramatically induced. After HepG2 cells were cocultured with 13.5-d embryonic hepatocytes and HNF-4α RNA interference, their proliferation rate (C), AFP content (D) and cell number (F) were detected. *P < 0.05 and **P < 0.01 versus Con. ##P < 0.01 versus Co-cul. Con, control; Co-cul, coculture. The experiment was repeated three times.
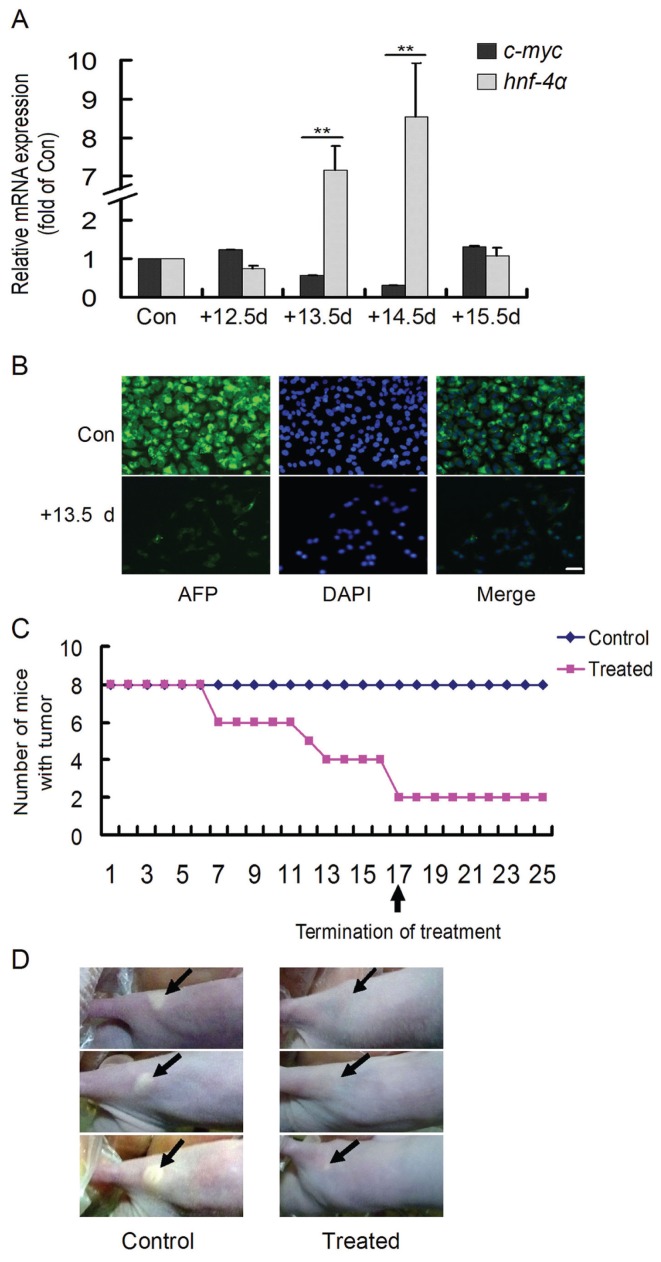
Subsequently, after HepG2 cells were cocultured with mouse embryonic hepatocytes at 12.5-, 13.5-, 14.5- and 15.5-d gestation for 48 h, expressions of the oncogene c-myc and the maturation marker hnf-4α were detected (Figure 4A). In the 13.5-d and 14.5-d groups, the expression of c-myc decreased, whereas that of hnf-4α increased significantly, consistent with the morphological changes in HepG2 cells. The AFP content in the HepG2 cells that were cocultured with 13.5-d embryonic hepatocytes for 48 h was visualized by immunofluorescence. As shown, AFP was apparent in HepG2 cells but nearly disappeared in the cocultured cells (Figure 4B). Besides, cyp3a4, cyp1b1, otc, arg1 and adh1c in HepG2 cells treated with 13.5-d embryonic hepatocyte medium increased dramatically (Figure 3B). These results suggest that developmental stage-specific hepatocytes have the ability to induce the maturation of HepG2 cells.
Figure 4.
Induced maturation of HepG2 cells by mouse embryonic hepatocytes. (A) In the 13.5-d and 14.5-d groups, c-myc decreased and hnf-4α increased significantly. **P < 0.01 versus control. The experiments were repeated three times. Con, control. (B) In contrast to the apparent AFP in the control (Con) group, AFP was nearly absent in the 13.5-d coculture group for 48 h treatment. Scale bar = 20 μm. (C) Changes in mice inoculated with HepG2 tumors. (D) At the end of the experiment, in contrast to the control, tumor masses were eliminated in the majority of treated mice.
In addition, this embryonic induction effect was evaluated in vivo. When inoculated HepG2 tumors became apparent, the mice were treated with 13.5-d embryonic hepatocyte medium, and the tumor masses were observed once daily (Figure 4C). Over the course of the study, tumor masses shrunk and gradually disappeared in the treated mice, whereas no changes were observed in the control mice. Importantly, after termination of the treatment, no recurrence of the tumors was observed in the treated mice. At the end of the experiment, in contrast to the control mice, no tumor masses were observed in the majority of treated mice (Figure 4D). The results provided in vivo evidence that HepG2 cells that were inoculated into nude mice could be eliminated by 13.5-d embryonic hepatocyte medium.
Developmental Stage-Specific Hepatocytes Rebuilt the Regulatory Circuit in HepG2 Cells
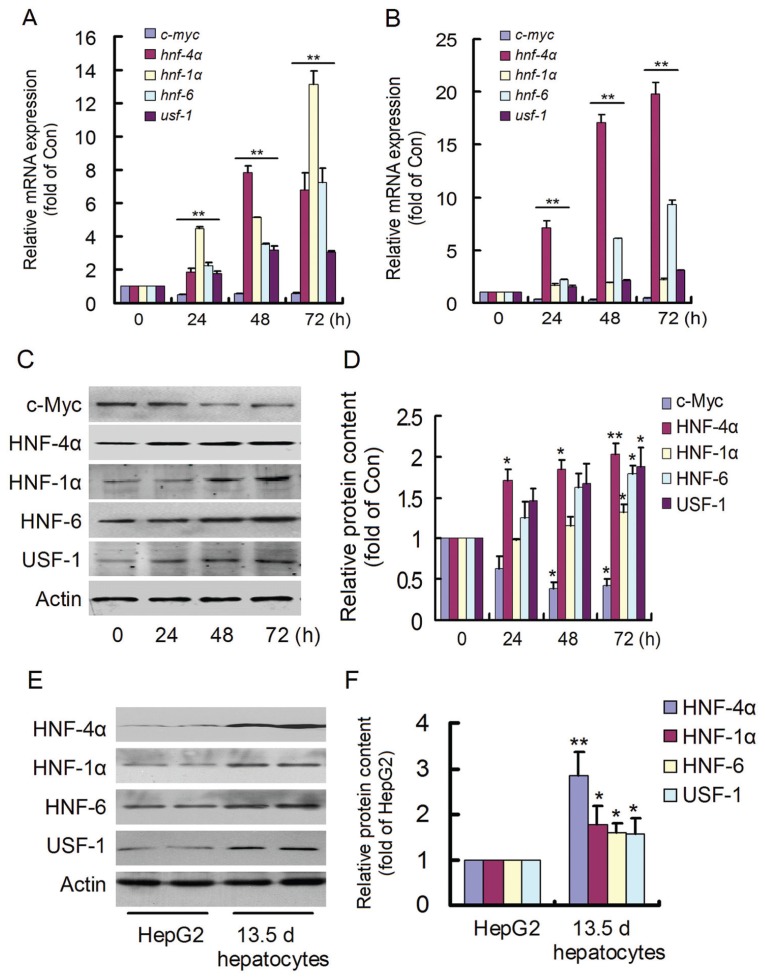
HepG2 cells were cocultured with 13.5-d and 14.5-d embryonic hepatocytes for different times, and the expression levels of the circuit components and c-myc were detected (Figures 5A, B). The expression of hnf-4α, hnf-1α, hnf-6 and usf-1 increased significantly with time, whereas the expression of the oncogene c-myc was decreased at all time points. In addition, the protein contents of the circuit components and c-Myc were determined in 13.5-d embryonic hepatocyte cocultures (Figure 5C). The protein contents of HNF-4α, HNF-1α, HNF-6 and USF-1 increased significantly; however, that of the oncogene c-Myc decreased (Figure 5D). These results were consistent with the gene expression changes. In addition, the circuit components, which were high in 13.5-d embryonic hepatocytes, decreased in HepG2 cells (Figures 5E, F). The results confirmed that developmental stage- specific hepatocytes could upregulate the circuit components with suppression of c-Myc in HepG2 cells.
Figure 5.
Changes of the regulatory circuit components in cocultured HepG2 cells. After HepG2, cells were cocultured with 13.5-d (A) and 14.5-d (B) mouse embryonic hepato-cytes for different times and the expression of the circuit components, and c-myc was evaluated. The expression of hnf-4α, hnf-1α, hnf-6 and usf-1 increased, whereas that of c-myc decreased significantly. (C) Their protein contents were measured for 13.5-d embryonic hepatocyte coculture. (D) The increases in HNF-4α, HNF-1α, HNF-6 and USF-1 levels and decreases in c-Myc levels were verified. The regulatory circuit components were detected (E), which were low in HepG2 cells and significantly high in 13.5-d mouse embryonic hepatocytes (F). *P < 0.05 and **P < 0.01 versus 0 h or HepG2 group. The experiments were repeated three times. Con, control.
After HepG2 cells were cocultured with 13.5-d embryonic hepatocytes for 48 h, the cross-regulation of the circuit components was evaluated. The binding of HNF-4α, HNF-1α, HNF-6 and USF-1 to their predicted sites was detected by ChIP (Figure 1C). The 13.5-d embryonic hepatocyte coculture promoted binding of the factors to their sites, which might result in their elevated expression and rebuilding of the regulatory circuit. The NF-κB binding site in inos gene promoter control showed no meaningful signal and was not shown. The binding of HNF-4α to hnf-1α and HNF-4α to usf-1 was promoted, verifying the core position of HNF-4α. Meanwhile, the content of HNF-4α bound in hnf-1α and usf-1 promoters was detected by DNA pull-down assay, and the results showed that 13.5-d embryonic hepatocyte medium could significantly increase the content of bound HNF-4α in HepG2 cells. All the data indicated that the developmental stage-specific hepatocytes could rebuild the regulatory circuit that drives the maturation of HepG2 cells.
Inhibition of the Regulatory Circuit Attenuated the Maturation of HepG2 Cells
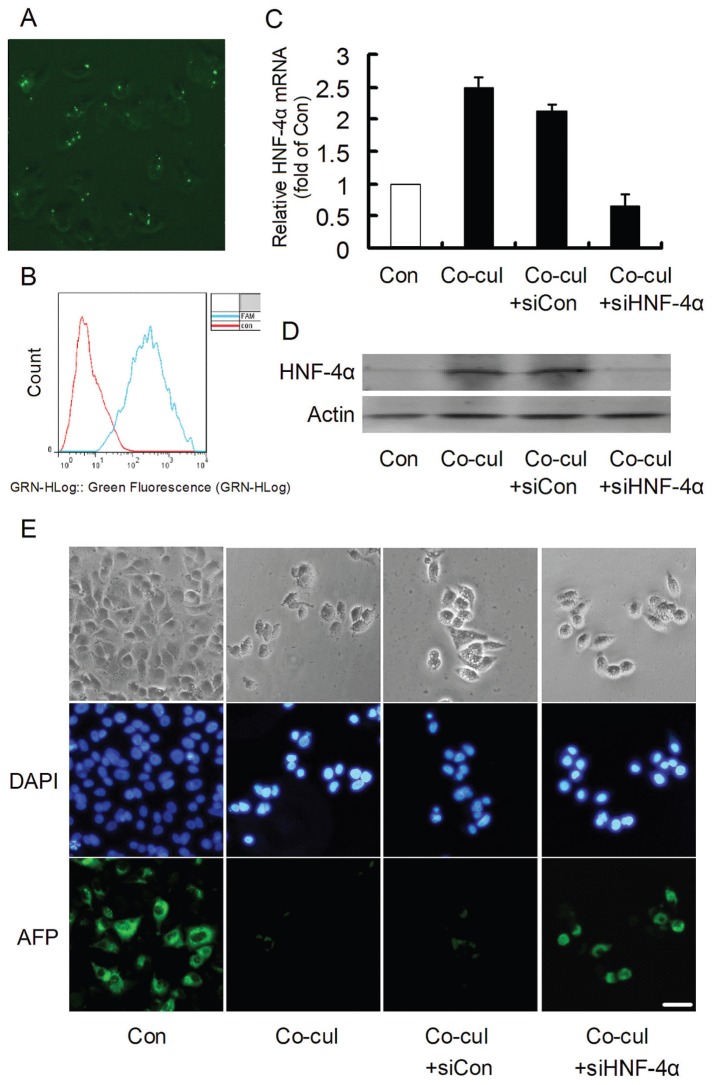
RNA interference targeting HNF-4α, which is the core of the regulatory circuit, was adopted to confirm the role of the circuit in the induced maturation of HepG2 cells. Successful transfection was indicated by FAM-siRNA (Figures 6A, B). The HepG2 cells were then treated with si-HNF-4α and cocultured with 13.5-d embryonic hepatocytes for 48 h. The mRNA and protein contents of HNF-4α were evaluated to analyze the effects of RNA interference (Figures 6C, D), and the effective inhibition of HNF-4α was confirmed by the dramatic inhibition of its mRNA and protein content in cocultured HepG2 cells. Then, the AFP content was examined by immunofluorescence (Figure 6E) and Western blotting (Figure 3D). Although AFP was apparently abolished in HepG2 cells cocultured with 13.5-d embryonic hepatocytes, si-HNF-4α treatment reversed the near disappearance of AFP in the cocultured HepG2 cells. Meanwhile, the inhibited proliferation rate of cocultured HepG2 cells could be recovered significantly by si-HNF-4α treatment (Figures 3C, F). These results indicated that inhibition of the regulatory circuit could attenuate the induced maturation of HepG2 cells.
Figure 6.
HNF-4α RNA interference. Successful transfection was determined by fluorescence (A) and flow cytometric (B) detection of FAM-siRNA. The HNF-4α mRNA (C) and protein content (D) were evaluated to analyze the interfering effect. Treatment with si-HNF-4α dramatically inhibited the increase in its mRNA and protein content in cocultured HepG2 cells. The experiment was repeated twice. (E) The apparent AFP that was observed in the control group nearly disappeared in the cocultured group. However, the AFP level was increased in cocultured HepG2 cells treated with si-HNF-4α, whereas treatment with si-control had no effect. Con, control; Co-cul, coculture. Scale bar = 20 μm.
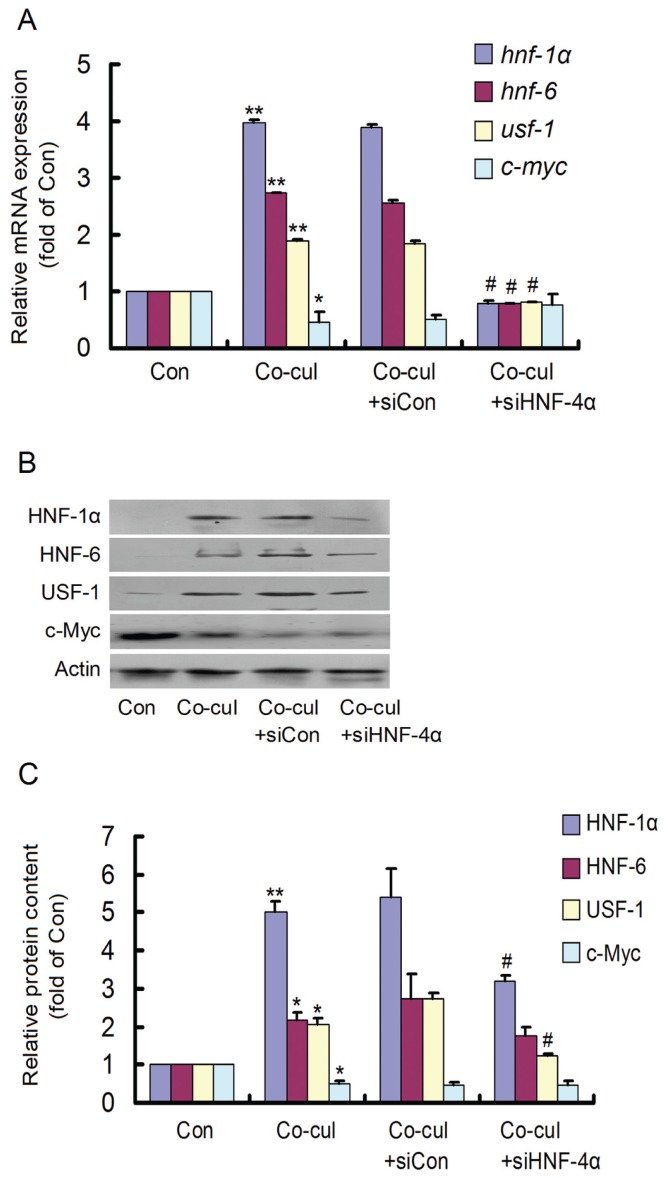
In addition, the mRNA and protein contents of other elements of the regulatory circuit as well as c-Myc were measured after HNF-4α interference in the cocultured HepG2 cells (Figures 7A, B). The si-HNF-4α treatment significantly inhibited the induced expression of hnf-1α, hnf-6 and usf-1 in HepG2 cells that were cocultured with 13.5-d embryonic hepatocytes. However, no obvious change in c-myc expression was observed after si-HNF-4α treatment, but HNF-1α and USF-1 protein expression was significantly inhibited (Figure 7C). These data revealed that inhibition of HNF-4α may result in suppression of the circuit components HNF-1α and USF-1.
Figure 7.
Changes in the regulatory circuit components after HNF-4α RNA interference in cocultured HepG2 cells. (A) After HepG2 cells were treated with si-HNF-4α and cocul-tured with 13.5-d embryonic hepatocytes for 48 h, the expression of the circuit components and c-myc were detected. HNF-4α RNA interference dramatically inhibited the increased expression of hnf-1α, hnf-6 and usf-1 in the cocultured HepG2 cells but did not significantly affect that of c-myc. (B) The HNF-1α, HNF-6, USF-1 and c-Myc protein content was reexamined. (C) HNF-4α RNA interference significantly decreased the protein content of HNF-1α and USF-1. *P < 0.05 and **P < 0.01 versus Con. #P < 0.05 versus Co-cul. Con, control; Co-cul, coculture. The experiment was repeated twice.
DISCUSSION
Although current treatments can effectively kill tumor cells, they fail to completely cure cancer and inhibit its recurrence. The treatment strategy should transit to the induction of tumor cell normalization (41–44). Because embryonic development process has the ability to coordinate the synchronism of cell maturation, we questioned whether developmental stage-specific hepatocytes induce the maturation of HCC cells. If so, we hypothesized that this method would provide an effective and harmless strategy for HCC therapy.
On the basis of their characteristics, we presumed that only developmental stage-specific hepatocytes should be able to direct and induce their maturation. In this study, HepG2 cells were cocultured with 12.5-d to 15.5-d mouse embryonic hepatocytes because of hepatocyte development (35–37). Embryonic hepatocytes at different developmental stages had distinct effects on the HepG2 cells, and the 13.5-d to 14.5-d embryonic hepatocytes could completely inhibit the proliferation and apparently induce the maturation of HepG2 cells.
The induced maturation of HepG2 cells was confirmed by increased HNF-4α and decreased c-Myc and AFP levels. Meanwhile, cyp3a4, cyp1b1, otc, arg1 and adh1c increased dramatically for the 13.5-d embryonic hepatocyte medium treatment. Furthermore, 13.5-d embryonic hepatocyte medium could completely eliminate most of the HepG2 tumors that were inoculated into nude mice. Most importantly, no tumor recurrence was observed in the treated mice after termination of the treatment. In parallel, it was found that 11.5-d embryonic hepatocyte medium could inhibit proliferation and induce maturation of SMMC-7721 cells. These results demonstrated that only the developmental stage-specific embryonic hepatocytes could induce maturation of liver tumor cells. Further clarifying the corresponding effective developmental stage for every type of liver tumor cells may provide an alternative promising approach for HCC therapy. Even every person’s tumor cell can find its unique developmental stage, and therefore, individualized treatment should and must be applied for the cure of cancer.
The molecular mechanism underlying the induced maturation of HepG2 cells is not clarified. During normal liver development, the transcription factors HNF-4α, HNF-1α, HNF-6 and USF-1 cross-regulate each other to form a positive feedback regulatory circuit, which drives hepatocyte maturation (24–26). And we revealed that compared with that in 13.5-d embryonic hepatocytes, the circuit components HNF-4α, HNF-1α, HNF-6 and USF-1 were decreased in HepG2 cells. However, it is unclear whether the developmental stage-specific hepatocytes induce the maturation of HepG2 cells by rebuilding the regulatory circuit.
First, the components of the regulatory circuit were evaluated in the cocultured HepG2 cells. After coculture, HNF-4α, HNF-1α, HNF-6 and USF-1 increased; however, c-Myc expression decreased with time, showing that developmental stage-specific hepatocytes could upregulate the regulatory circuit components and inhibit the oncogene c-Myc. Then, the cross-regulation of HNF-4α, HNF-1α, HNF-6 and USF-1 was measured by ChIP. The 13.5-d embryonic hepatocyte cocultures promoted the binding of the factors to their target sites and rebuilt the regulatory circuit in HepG2 cells. Promotion of the binding of these factors to their promoters might result in their elevated expression, thus forming a positive feedback circuit that drives the maturation of HepG2 cells. The binding of HNF-4α to hnf-1α and HNF-4α to usf-1 was especially promoted and the accumulation of HNF-4α bound in hnf-1α and usf-1 promoters was proved again by DNA pull-down assay, verifying the core position of HNF-4α.
RNA interference of HNF-4α was performed to confirm the role of the regulatory circuit in the induced maturation of HepG2 cells. The results suggested that HNF-4α RNA interference could reverse the inhibited proliferation rate and the near disappearance of AFP protein in HepG2 cells that were cocultured with 13.5-d embryonic hepatocytes, indicating that inhibition of the regulatory circuit may attenuate the induced HepG2 cell maturation. In addition, si-HNF-4α treatment significantly inhibited the induction of hnf-1α, hnf-6 and usf-1 expression and the increased HNF-1α and USF-1 protein contents in HepG2 cells that were cocultured with 13.5-d embryonic hepatocytes. These data suggested that inhibition of the regulatory circuit might result in the attenuation of the induced maturation of HepG2 cells.
CONCLUSION
This study revealed that developmental stage-specific hepatocytes could induce the maturation of HepG2 cells in vitro and in vivo, and this induced maturation was mediated by rebuilding the regulatory circuit. It was confirmed that differences in the expression of genes in embryonic stem cells at different developmental stages results in the production of diverse types and intensity of signaling molecules (45,46). And several signals and pathways of regulation mechanism of induced maturation of tumor cells have been identified (20). Therefore, clarification of the molecules expressed in tumor cells and the signaling pathways that mediate their inductive effects may reveal the intrinsic mechanism of induced maturation of tumor cells and then allow for the clinical application of this embryonic-induced maturation therapy.
Supplemental Data
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (81470595) and Hebei Natural Science Foundation (H2012206005).
Footnotes
Online address: http://www.molmed.org
DISCLOSURE
The authors declare that they have no competing interests as defined by Molecular Medicine, or other interests that might be perceived to influence the results and discussion reported in this paper.
Cite this article as: Li Y, et al. (2015) Developmental stage-specific hepatocytes induce maturation of HepG2 cells by rebuilding the regulatory circuit. Mol. Med. 21:285–95.
REFERENCES
- 1.Yong KJ, et al. Oncofetal gene SALL4 in aggressive hepatocellular carcinoma. N Engl J Med. 2013;368:2266–76. doi: 10.1056/NEJMoa1300297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Han ZG. Functional genomic studies: insights into the pathogenesis of liver cancer. Annu Rev Genomics Hum Genet. 2012;13:171–205. doi: 10.1146/annurev-genom-090711-163752. [DOI] [PubMed] [Google Scholar]
- 3.Viatour P, et al. Notch signaling inhibits hepatocellular carcinoma following inactivation of the RB pathway. J Exp Med. 2011;208:1963–76. doi: 10.1084/jem.20110198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gou L, et al. Proteomic identification of RhoA as a potential biomarker for proliferation and metastasis in hepatocellular carcinoma. J Mol Med. 2011;89:817–27. doi: 10.1007/s00109-011-0753-3. [DOI] [PubMed] [Google Scholar]
- 5.Lee TK, Cheung VC, Ng IO. Liver tumor-initiating cells as a therapeutic target for hepatocellular carcinoma. Cancer Lett. 2013;338:101–9. doi: 10.1016/j.canlet.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 6.Dragani TA. Risk of HCC: genetic heterogeneity and complex genetics. J Hepatol. 2010;52:252–7. doi: 10.1016/j.jhep.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 7.Wang K, et al. Genomic landscape of copy number aberrations enables the identification of oncogenic drivers in hepatocellular carcinoma. Hepatology. 2013;58:706–17. doi: 10.1002/hep.26402. [DOI] [PubMed] [Google Scholar]
- 8.Yang XR, et al. High expression levels of putative hepatic stem/progenitor cell biomarkers related to tumour angiogenesis and poor prognosis of hepatocellular carcinoma. Gut. 2010;59:953–62. doi: 10.1136/gut.2008.176271. [DOI] [PubMed] [Google Scholar]
- 9.Oikawa T, et al. Sal-like protein 4 (SALL4), a stem cell biomarker in liver cancers. Hepatology. 2013;57:1469–83. doi: 10.1002/hep.26159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muramatsu S, et al. Visualization of stem cell features in human hepatocellular carcinoma reveals in vivo significance of tumor-host interaction and clinical course. Hepatology. 2013;58:218–28. doi: 10.1002/hep.26345. [DOI] [PubMed] [Google Scholar]
- 11.Yamashita T, et al. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology. 2009;136:1012–24. doi: 10.1053/j.gastro.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang Y, et al. Progenitor/stem cells give rise to liver cancer due to aberrant TGF-beta and IL-6 signaling. Proc Natl Acad Sci U S A. 2008;105:2445–50. doi: 10.1073/pnas.0705395105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Majumdar A, et al. Hepatic stem cells and transforming growth factor β in hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2012;9:530–8. doi: 10.1038/nrgastro.2012.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marquardt JU, Galle PR, Teufel A. Molecular diagnosis and therapy of hepatocellular carcinoma (HCC): an emerging field for advanced technologies. J Hepatol. 2012;56:267–75. doi: 10.1016/j.jhep.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 15.Yamashita T, Wang XW. Cancer stem cells in the development of liver cancer. J Clin Invest. 2013;123:1911–8. doi: 10.1172/JCI66024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rountree CB, Mishra L, Willenbring H. Stem cells in liver diseases and cancer: recent advances on the path to new therapies. Hepatology. 2012;55:298–306. doi: 10.1002/hep.24762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hendrix MJ, et al. Reprogramming metastatic tumour cells with embryonic microenvironments. Nat Rev Cancer. 2007;7:246–55. doi: 10.1038/nrc2108. [DOI] [PubMed] [Google Scholar]
- 18.Postovit LM, et al. Human embryonic stem cell microenvironment suppresses the tumorigenic phenotype of aggressive cancer cells. Proc Natl Acad Sci U S A. 2008;105:4329–34. doi: 10.1073/pnas.0800467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu JM, Jun ES, Bae YC, Jung JS. Mesenchymal stem cells derived from human adipose tissues favor tumor cell growth in vivo. Stem Cells Dev. 2008;17:463–73. doi: 10.1089/scd.2007.0181. [DOI] [PubMed] [Google Scholar]
- 20.Biava PM, Nicolini A, Ferrari P, Carpi A, Sell S. A systemic approach to cancer treatment: tumor cell reprogramming focused on endocrine-related cancers. Curr Med Chem. 2014;21:1072–81. doi: 10.2174/0929867321666131201143124. [DOI] [PubMed] [Google Scholar]
- 21.Li T, et al. Multi-stage analysis of gene expression and transcription regulation in C57/B6 mouse liver development. Genomics. 2009;93:235–42. doi: 10.1016/j.ygeno.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kyrmizi I, et al. Plasticity and expanding complexity of the hepatic transcription factor network during liver development. Genes Dev. 2006;20:2293–305. doi: 10.1101/gad.390906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laudadio I, et al. A feedback loop between the liver-enriched transcription factor network and miR-122 controls hepatocyte differentiation. Gastroenterology. 2012;142:119–29. doi: 10.1053/j.gastro.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 24.Odom DT, et al. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378–81. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Odom DT, et al. Core transcriptional regulatory circuitry in human hepatocytes. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100059. 2006.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagaki M, Moriwaki H. Transcription factor HNF and hepatocyte differentiation. Hepatol Res. 2008;38:961–9. doi: 10.1111/j.1872-034X.2008.00367.x. [DOI] [PubMed] [Google Scholar]
- 27.Dankel SN, Hoang T, Flågeng MH, Sagen JV, Mellgren G. cAMP-mediated regulation of HNF-4alpha depends on the level of coactivator PGC-1alpha. Biochim Biophys Acta. 20101803:1013–9. doi: 10.1016/j.bbamcr.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 28.Hatziapostolou M, et al. An HNF4α-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell. 2011;147:1233–47. doi: 10.1016/j.cell.2011.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pastoret A, et al. Inhibition of hepatocyte nuclear factor 1 and 4 alpha (HNF1α and HNF4α) as a mechanism of arsenic carcinogenesis. Arch Toxicol. 2013;87:1001–12. doi: 10.1007/s00204-012-0948-6. [DOI] [PubMed] [Google Scholar]
- 30.Zeng X, et al. Recombinant adenovirus carrying the hepatocyte nuclear factor-1alpha gene inhibits hepatocellular carcinoma xenograft growth in mice. Hepatology. 2011;54:2036–47. doi: 10.1002/hep.24647. [DOI] [PubMed] [Google Scholar]
- 31.van Deursen D, van Leeuwen M, Vaulont S, Jansen H, Verhoeven AJ. Upstream stimulatory factors 1 and 2 activate the human hepatic lipase promoter via E-box dependent and independent mechanisms. Biochim Biophys Acta. 2009;1791:229–37. doi: 10.1016/j.bbalip.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 32.Wu S, et al. Upstream transcription factor 1 influences plasma lipid and metabolic traits in mice. Hum Mol Genet. 2010;19:597–608. doi: 10.1093/hmg/ddp526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beaudry JB, et al. Threshold levels of hepatocyte nuclear factor 6 (HNF-6) acting in synergy with HNF-4 and PGC-1alpha are required for time-specific gene expression during liver development. Mol Cell Biol. 2006;26:6037–46. doi: 10.1128/MCB.02445-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teng X, Zhang H, Snead C, Catravas JD. Molecular mechanisms of iNOS induction by IL-1 beta and IFN-gamma in rat aortic smooth muscle cells. Am J Physiol Cell Physiol. 2002;282:C144–52. doi: 10.1152/ajpcell.2002.282.1.C144. [DOI] [PubMed] [Google Scholar]
- 35.Duncan SA. Mechanisms controlling early development of the liver. Mech Dev. 2003;120:19–33. doi: 10.1016/s0925-4773(02)00328-3. [DOI] [PubMed] [Google Scholar]
- 36.Lemaigre F, Zaret KS. Liver development update: new embryo models, cell lineage control, and morphogenesis. Curr Opin Genet Dev. 2004;14:582–90. doi: 10.1016/j.gde.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 37.Zaret KS, Grompe M. Generation and regeneration of cells of the liver and pancreas. Science. 2008;322:1490–4. doi: 10.1126/science.1161431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y, et al. Peroxynitrite-induced nitration of cyclooxygenase-2 and inducible nitric oxide synthase promotes their binding in diabetic angiopathy. Mol Med. 2010;16:335–42. doi: 10.2119/molmed.2010.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y, et al. Protein nitration promotes inducible nitric oxide synthase transcription mediated by NF-κB in high glucose-stimulated human lens epithelial cells. Mol Cell Endocrinol. 2013;370:78–86. doi: 10.1016/j.mce.2013.02.015. [DOI] [PubMed] [Google Scholar]
- 40.Nelson JD, Denisenko O, Bomsztyk K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat Protoc. 2006;1:179–85. doi: 10.1038/nprot.2006.27. [DOI] [PubMed] [Google Scholar]
- 41.Campos B, et al. Differentiation therapy exerts antitumor effects on stem-like glioma cells. Clin Cancer Res. 2010;16:2715–28. doi: 10.1158/1078-0432.CCR-09-1800. [DOI] [PubMed] [Google Scholar]
- 42.Negrotto S, et al. Noncytotoxic differentiation treatment of renal cell cancer. Cancer Res. 2011;71:1431–41. doi: 10.1158/0008-5472.CAN-10-2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Azzi S, et al. Differentiation therapy: targeting human renal cancer stem cells with interleukin 15. J Natl Cancer Inst. 2011;103:1884–98. doi: 10.1093/jnci/djr451. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y, et al. All-trans retinoic acid potentiates the chemotherapeutic effect of Cisplatin by inducing differentiation of tumor initiating cells in liver cancer. J Hepatol. 2013;59:1255–63. doi: 10.1016/j.jhep.2013.07.009. [DOI] [PubMed] [Google Scholar]
- 45.Biava PM, et al. Cancer cell reprogramming: stem cell differentiation stage factors and an agent based model to optimize cancer treatment. Curr Pharm Biotechnol. 2011;12:231–42. doi: 10.2174/138920111794295783. [DOI] [PubMed] [Google Scholar]
- 46.McKinney-Freeman S, et al. The transcriptional landscape of hematopoietic stem cell ontogeny. Cell Stem Cell. 2012;11:701–14. doi: 10.1016/j.stem.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.