Abstract
Background
Reference diagnostic tests for leptospirosis include nucleic acid amplification tests, bacterial culture, and microscopic agglutination testing (MAT) of acute and convalescent serum. However, clinical laboratories often do not receive paired specimens. In the current study, we tested serum samples using a highly sensitive real-time nucleic acid amplification test for Leptospira and compared results to MAT performed on the same specimens.
Methods/Principal Findings
478 serum samples from suspected leptospirosis cases in Rio de Janeiro were tested using a real-time RT-PCR for the diagnosis of leptospirosis, malaria and dengue (the Lepto-MD assay). The Lepto-MD assay detects all species of Leptospira (saprophytic, intermediate, and pathogenic), and in the current study, we demonstrate that this assay amplifies both Leptospira RNA and DNA. Dengue virus RNA was identified in 10 patients, and no cases of malaria were detected. A total of 65 samples (13.6%) were positive for Leptospira: 35 samples (7.3%) in the Lepto-MD assay, 33 samples (6.9%) by MAT, and 3 samples tested positive by both (kappa statistic 0.02). Poor agreement between methods was consistent regardless of the titer used to define positive MAT results or the day of disease at sample collection. Leptospira nucleic acids were detected in the Lepto-MD assay as late as day 22, and cycle threshold values did not differ based on the day of disease. When Lepto-MD assay results were added to the MAT results for all patients in 2008 (n=818), the number of detected leptospirosis cases increased by 30.4%, from 102 (12.5%) to 133 (16.3%).
Conclusions/Significance
This study demonstrates a lack of agreement between nucleic acid detection of Leptospira and single-specimen MAT, which may result from the clearance of bacteremia coinciding with the appearance of agglutinating antibodies. A combined testing strategy for acute leptospirosis, including molecular and serologic testing, appears necessary to maximize case detection.
Introduction
Leptospirosis is a potentially fatal zoonotic disease that occurs throughout the world, but reported incidence rates likely underestimate the true burden of disease [1,2]. This situation results from many factors, but it is due, in part, to limitations of available diagnostics to accurately detect leptospirosis in the acute setting [3]. Laboratory confirmation of leptospirosis requires one of the following: a four-fold increase in antibody titer between acute and convalescent serum samples, as detected by microscopic agglutination testing (MAT); a high MAT titer (≥ 1:400 to 1:800) in single or paired serum samples; isolation of pathogenic Leptospira species from a normally sterile site; or the detection of DNA from pathogenic Leptospira species by PCR [2,4]. Outside of a structured research setting, it can be difficult to obtain paired specimens for serologic testing [5]. Laboratories often receive only one sample from a suspected case, which leaves single-specimen MAT and molecular diagnostics as the principal means of confirming leptospirosis.
The Centro de Referência Nacional para Leptospirose (CRNL) at the Instituto Oswaldo Cruz, Fiocruz, in Rio de Janeiro receives 500 to 1,000 serum samples from patients in the Rio de Janeiro State annually for reference Leptospira testing. In the majority of cases (~90%), only a single specimen is received for testing by MAT, conventional PCR, and/or bacterial culture. Recently, our group reported the development of two molecular diagnostics for Leptospira: 1) an internally-controlled, multiplex assay for detection of all species of Leptospira, Plasmodium species with a specific call-out for P. falciparum, and the four dengue virus (DENV) serotypes (referred to here as the Lepto-MD assay, previously described as the UFI assay) [6]; and 2) an assay for the detection of pathogenic Leptospira species [7]. These assays target a region of the 16S rrs gene, and using a set of 65 serum samples from CRNL, both proved more sensitive than a reference Leptospira 16S PCR [7]. Of these samples, 55 had been tested by MAT, including only 6 positives, which limited our ability to compare test results from these two methods. Also, given the design of the Lepto-MD and pathogenic Leptospira assays, it was possible that improved sensitivity resulted from RNA amplification with or without concomitant DNA amplification.
In the current study, we tested a larger set of serum samples, collected in Rio de Janeiro in 2008, to compare results of screening with the Lepto-MD assay and single-specimen MAT for the detection of leptospirosis cases. The improved clinical sensitivity of the Lepto-MD assay, compared to other molecular tests, was utilized to identify missed cases of leptospirosis in this population and generate a new estimate of leptospirosis incidence for the study period. Finally, we evaluated the contribution of Leptospira RNA detection to the performance of the Lepto-MD and pathogenic Leptospira assays.
Methods
Ethics Statement
The study protocol was reviewed and approved by the Stanford Institutional Review Board and the Scientific Review Board of Instituto Oswaldo Cruz, Fiocruz, Rio de Janeiro, Brazil (CAAE:32200914.2.00005248).
Clinical Samples and MAT
Archived, de-identified serum samples were selected for RT-PCR from specimens that had been sent from patients in Rio de Janeiro to CRNL for reference Leptospira testing between January 1 and December 31, 2008. During this period, CRNL received 894 serum samples for MAT from 818 patients in Rio de Janeiro: 749 patients had one sample, 62 patients had two samples, and 7 patients had three samples sent for testing. Samples were obtained as part of routine care from suspected leptospirosis cases. Day of disease at sample collection, when provided, was recorded by the requesting provider. All samples were tested upon receipt at CRNL by MAT using a reference panel of 19 Leptospira serovars, as described [7,8]. A positive MAT result was defined as any one of the following: a negative acute-phase titer and a follow-up sample with a titer ≥1:100; a 4-fold rise in titer by MAT between paired samples; or a single acute-phase MAT titer of ≥1:800. For analyses involving a comparison of test results for individual samples, samples were considered positive by MAT if the agglutination titer was ≥ 1:800 and indeterminate if the titers were 1:100–1:400. Up to 55 samples per month with at least 200μL of volume were selected for molecular testing. If sufficient numbers were available from a given month, only MAT-negative samples were selected. MAT-positive samples were included from months with <55 MAT-negative samples.
Molecular Testing
Serum was stored at -20°C at CRNL until shipment to Stanford. Total nucleic acids were extracted from 200μL of serum using an easyMAG instrument (Biomerieux) with a 60μL elution volume. Two eluate aliquots were stored at -80°C. All samples were screened using the Lepto-MD assay, which was performed and interpreted as previously described [6]. Samples positive for Leptospira in the Lepto-MD assay were tested using three additional assays: 1) a 16S RT-PCR for the detection of pathogenic Leptospira species [7]; 2) the Leptospira 16S PCR, originally reported by Smythe, et al. [9]; and 3) a PCR for the detection of lipL32 [10]. The latter two assays were performed as described elsewhere [11]. Results were analyzed on the linear scale, threshold was set at 0.05, and samples were considered positive in either assay if an exponential curve crossed the threshold prior to cycle 45. All molecular tests were performed on a Rotor-Gene Q instrument (Qiagen) using 5μL of eluate.
Results of MAT and molecular testing were compared for individual samples based on the timing of sample collection and cutoff used to define a positive MAT result. Samples obtained at ≤7 days of disease, 8–14 days, and >14 days were categorized as acute, late-acute, and convalescent, respectively. Results were organized by month of the year, and the proportion of positive tests was evaluated in relation to recorded rainfall. Data regarding precipitation in Rio de Janeiro during the study period was obtained from the Armazém de Dados, Portal da Prefeitura da Cidade do Rio de Janeiro (armazemdedados.rio.rj.gov.br, accessed 5 March 2015).
PCR Evaluation and RNase A Treatment
The sensitivity of Leptospira DNA detection was evaluated using two real-time PCR kits: 1) the TaqMan Universal PCR Master Mix and 2) Platinum Taq DNA Polymerase (both from Life Technologies). The Leptospira and RNase P primers and probes from the Lepto-MD assay were used at the same final reaction concentrations with both real-time PCR kits. Real-time PCRs were evaluated by testing extracted nucleic acids from cultured isolates of L. interrogans serovar Australis (reference strain Ballico), L. biflexa serovars Patoc (reference strain Patoc I) and Andamana (reference strain CH 11), L. borgpetersenii serovar Castellonis (reference strain Castellon 3), L. kirschneri serovar Grippotyphosa (reference strain Moskva V), and L. weilii serovar Vughia (reference strain LT 89–68) [6]. Nucleic acids were extracted from cultured isolates using the DNeasy Blood & Tissue Kit (Qiagen) according to manufacturer recommendations. Real-time Leptospira PCR performance was compared to the Lepto-MD assay [6].
RNase A treatment was performed on extracted nucleic acids from the cultured Leptospira isolates, cultured strains of DENV-1 (Hawaii 1944) and DENV-2 (New Guinea C), plasmid DNA containing the Leptospira target sequence, and synthetic ssDNA containing the target sequence for DENV-1. To each 25uL of nucleic acid, RNase A (Thermo Scientific) was added to a final concentration of 100ug/mL. Samples were incubated at 37°C for 1hr and then placed on ice. Water was added to control aliquots of each sample. Treated samples and controls were immediately run side-by-side in the Lepto-MD assay.
Statistics
A confirmed case of leptospirosis required nucleic acid detection by RT-PCR and/or a positive result by MAT. Basic statistics were calculated using Excel software (Microsoft). GraphPad software (GraphPad) was used for the calculation of kappa-statistics, two-tailed Fisher’s exact tests, and t-tests. Kappa-statistics were calculated to evaluate the agreement between MAT and the Lepto-MD assay for individual samples. Fisher’s exact tests were used in comparisons of categorical variables, and t-tests were performed for comparisons of continuous variables. Significance was defined as a p-value ≤ 0.05.
Results
Lepto-MD Assay Testing
A total of 479 serum samples from suspected leptospirosis cases were extracted and tested in the Lepto-MD assay. An assay for RNase P detection serves as the internal control in the Lepto-MD assay [6]. A single internal-control failure occurred in a sample from November; this was excluded from further analysis. Results are shown in Table 1 for the remaining 478 samples (99.8%) based on the month in which the specimen was obtained. No samples tested positive for Plasmodium species. DENV was detected in 10 samples (2.1%). Compared to leptospirosis cases (Table 2), patients with DENV were younger (p = 0.028), more likely to be female (p≤0.001), and presented earlier in the course of their illness [mean day of disease, 4.6 (standard deviation 2.4) versus 12.3 (standard deviation 7.9), respectively; p = 0.015). This latter association remained significant if leptospirosis cases that were only detected by MAT were excluded from the analysis (p = 0.028). No DENV and Leptospira co-infected patients were detected.
Table 1. Total number of samples tested from 2008 and positive results in the Lepto-MD assay by month.
| Positive Samples | ||||
|---|---|---|---|---|
| Study Month | Samples Tested | MAT Positive | Dengue virus | Leptospira |
| n | n | n (%) | n (%) | |
| January | 55 | 0 | 1 (1.8) | 3 (5.5) |
| February | 23 | 2 | 2 (8.7) | 2 (8.7) |
| March | 55 | 0 | 5 (9.1) | 5 (9.1) |
| April | 50 | 4 | 0 (0) | 2 (4.0) |
| May | 18 | 3 | 0 (0) | 2 (11.1) |
| June | 55 | 0 | 2 (3.6) | 4 (7.3) |
| July | 54 | 0 | 0 (0) | 6 (11.1) |
| August | 36 | 1 | 0 (0) | 2 (5.6) |
| September | 8 | 1 | 0 (0) | 1 (12.5) |
| October | 49 | 8 | 0 (0) | 4 (8.2) |
| November 1 | 40 | 8 | 0 (0) | 1 (2.5) |
| December | 35 | 6 | 0 (0) | 3 (8.6) |
| Total | 478 | 33 (6.9) | 10 (2.1) | 35 (7.3) |
1 Forty-one samples were extracted in November. One sample had a failed internal control reaction and was not included in further analyses.
Table 2. Patient characteristics associated with 478 samples that tested positive and negative for Leptospira.
| Leptospira Positive | Leptospira Negative | ||||
|---|---|---|---|---|---|
| Total 1 | Lepto-MD Assay | MAT | Total | DENV-positive | |
| Number, n | 65 | 35 | 33 | 413 | 10 |
| Male gender, n (%) | 55 (84.6) | 28 (80.0) | 27 (90.0) | 253 (61.3) | 3 (30.0) |
| Age, years, mean (SD) | 36.7 (15.3) | 35.6 (17.0) | 38.0 (13.4) | 32.8 (18.4) | 23.2 (20.2) |
| Day of Disease, n (%) | |||||
| Acute | 12 (18.5) | 8 (22.9) | 4 (12.1) | 115 (27.8) | 6 (60.0) |
| Late Acute | 11 (16.9) | 6 (17.1) | 5 (15.2) | 58 (14.0) | 1 (10.0) |
| Convalescent | 13 (20.0) | 4 (11.4) | 10 (30.3) | 73 (17.7) | 0 (0.0) |
| Not provided | 29 (44.6) | 17 (48.6) | 14 (42.4) | 167 (40.4) | 3 (3.0) |
Abbreviations: SD, standard deviation
1 65 samples tested positive for Leptospira in the Lepto-MD assay and/or by MAT; 3 samples tested positive by both methods.
Leptospirosis Cases
A total of 65 samples (13.6%) were positive for Leptospira. Leptospira-positive patients were more likely to be male than patients who tested negative (p≤0.001), but the day of disease at sample collection did not differ significantly between these groups. Thirty-five samples (7.3%) tested positive in the Lepto-MD assay and 33 samples (6.9%) were positive by MAT (Table 2). Only three samples tested positive by both the Lepto-MD assay and MAT (Table 3A, kappa statistic 0.02), demonstrating poor agreement between these methods. MAT thresholds of ≥1:100 and ≥1:1600 did not affect concordance between Lepto-MD assay and MAT results (kappa 0.05 and -0.03, respectively; Table 3B and 3C). Information regarding the day of disease at sample collection was available for 282 samples. Samples that tested positive in the Lepto-MD assay were collected significantly earlier than samples positive by MAT [mean day of disease, 9.5 (standard deviation 5.3) vs. 15.0 (standard deviation, 8.8); p ≤ 0.03]. However, poor agreement between the Lepto-MD assay and MAT was consistent regardless of the day of disease at sample collection. Of the samples collected at ≤ 14 days of illness, 14 tested positive in the Lepto-MD assay, 9 were positive by MAT, but none had concordant results between these methods. A single sample collected on day 16 and two samples without recorded day of disease information tested positive by both methods.
Table 3. Comparison of Leptospira results for 478 serum samples tested with the Lepto-MD assay and MAT using titer thresholds for a positive result of ≥1:800 (definition of a confirmed case, ≥1:100, and ≥1:1,600.
| Lepto-MD Assay | ||||
|---|---|---|---|---|
| Positive | Negative | Total | ||
| MAT ≥1:800 | Positive | 3 | 30 | 33 |
| Negative | 32 | 413 | 445 | |
| MAT ≥1:100 | Positive | 5 | 38 | 43 |
| Negative | 30 | 405 | 435 | |
| MAT ≥1:1,600 | Positive | 1 | 23 | 24 |
| Negative | 34 | 420 | 454 | |
| Total | 35 | 443 | 478 | |
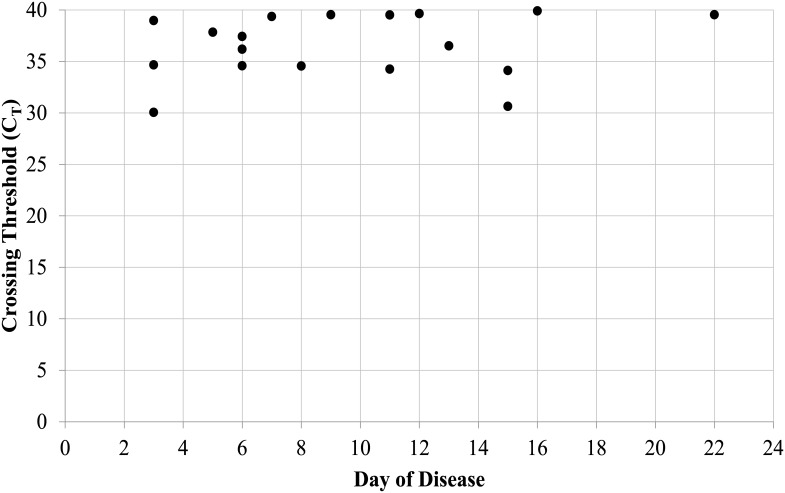
Leptospira was detected in the Lepto-MD assay from samples collected as late as day 22. Rates of detection in the Lepto-MD assay did not differ significantly for samples collected in the acute [8/127 (6.3%)], late acute [6/69 (8.7%)], or convalescent periods [4/86 (4.7%); p≥0.05 for each comparison, Fisher’s exact test]. Cycle threshold (CT) values in the Lepto-MD assay also did not differ based on the day of disease at sample collection (Fig 1), and mean CT values were similar for samples with recorded day of disease information and those without this information [36.5 (standard deviation 3.1) vs 34.4 (standard deviation, 5.1); p ≤ 0.15). In order to confirm Leptospira results from the Lepto-MD assay, 33/35 (94.3%) positive samples were tested using three other molecular tests (see Methods). Two samples had insufficient volume for all comparator tests. 27 of these 33 samples (81.8%) tested positive for Leptospira in at least one other assay, including 25/27 (92.6%) that tested positive for pathogenic species.
Fig 1. Leptospira CT values in the Lepto-MD assay for patients with recorded day of disease information.
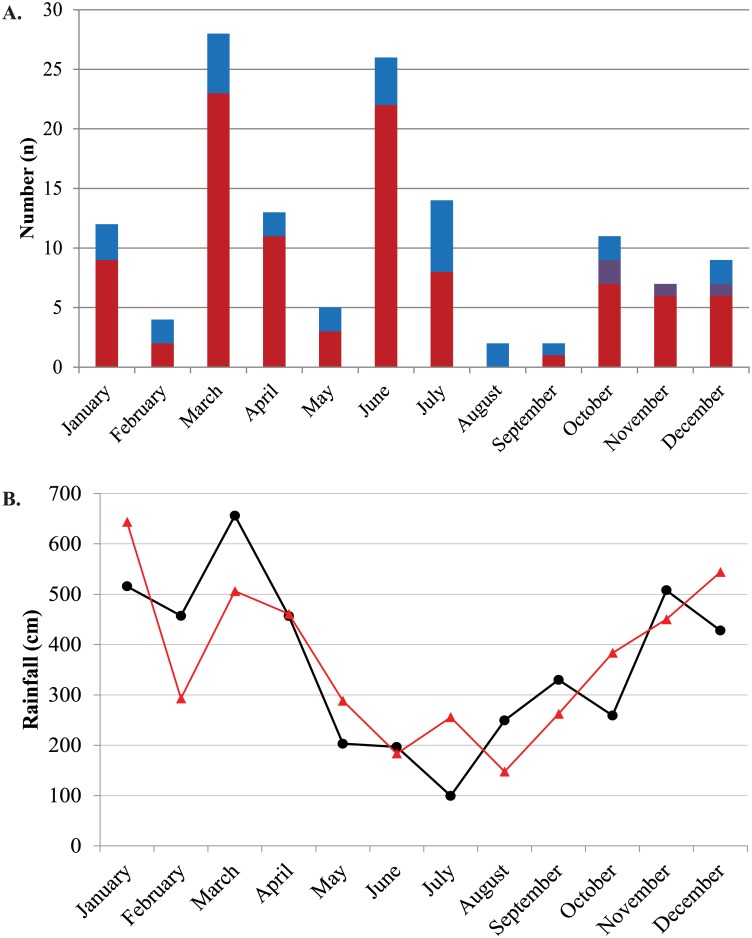
Leptospirosis case detection using the Lepto-MD assay was then added to MAT results for all samples sent to CRNL from Rio de Janeiro during 2008 (Fig 2A). 818 patients had MAT performed, and 102 (12.5%) tested positive. In 77 cases (75.5%), antibody titers were highest to serovars Icterohaemorrhagiae and Copenhageni. From the patients who had samples tested for this study, one additional case was detected by seroconversion when all MAT results were analyzed. The addition of Lepto-MD test results, therefore, increased the number of detected leptospirosis cases by 30.4%, from 102 (12.5%) to 133 (16.3%). The highest number of cases was detected in March, which was also the month with the highest recorded rainfall (Fig 2B). However, there was not a clear seasonality to Leptospira detection using either RT-PCR or MAT despite normal rainfall patterns in Rio de Janeiro during the study period (Fig 2B).
Fig 2. Leptospirosis cases and recorded rainfall in Rio de Janeiro by study month.
(A) Leptospirosis cases diagnosed by RT-PCR (blue), MAT (maroon), or both (purple) are displayed by study month. MAT results are shown for all samples from 2008. (B) Rainfall for 2008 (black circles and line) and the average rainfall for the years 2003–2013 (red triangles and line).
PCR versus RT-PCR
An internally-controlled Leptospira real-time PCR was evaluated using Platinum Taq DNA Polymerase (Life Technologies) and extracted nucleic acids from 6 control Leptospira strains. This PCR uses the same Leptospira primers and probes as the Lepto-MD assay at the same final reaction concentrations. Samples were tested side-by-side with the Lepto-MD assay, which is performed using the SuperScript III Platinum One-Step qRT-PCR kit (Life Technologies). CT values in the PCR were, on average, 5.6 cycles later than CT values in the Lepto-MD RT-PCR assay. Results were similar using the TaqMan Universal PCR Master Mix (also Life Technologies), and the use of three-step versus two-step cycling did not change these findings. To confirm that the improved performance of the Lepto-MD assay was due to the detection of RNA, nucleic acid aliquots from Leptospira controls were treated with RNase A. CT values for treated samples were 10.8 cycles later than untreated controls. RNase A treatment had a larger effect on DENV RNA controls (CT difference 22.7 cycles) and no demonstrable effect on DENV-1 ssDNA or Leptospira plasmid DNA (CT difference 0.1 cycles).
Discussion
This study describes the results of testing 478 serum samples from suspected leptospirosis cases in Rio de Janeiro with the Lepto-MD assay. Thirty-five cases of leptospirosis and 10 dengue cases were identified. It is interesting to note that dengue cases were significantly younger than cases of leptospirosis. This has not been observed during previous studies of concurrent DENV and Leptospira outbreaks [12–14]. The current study involves a relatively small number of cases, however, and this will need to be confirmed in a larger number of patients. No cases of malaria were identified in this population, which is consistent with the epidemiology of malaria in Brazil [15].
Molecular detection of Leptospira in the Lepto-MD assay demonstrated poor agreement with single-specimen MAT. We employed a conservative threshold for calling positive samples by MAT (titer ≥ 1:800), which is consistent with studies by other researchers in Brazil and was selected to maximize specificity [16–18]. Poor agreement between molecular testing and MAT remained consistent when different titer thresholds were evaluated and when results were stratified by the day of disease at sample collection. These findings could result from poor specificity of either the Lepto-MD assay or single-specimen MAT, though this seems unlikely. Of samples that tested positive for Leptospira in the Lepto-MD assay, 27/33 (81.8%) were confirmed with another molecular test. Although six samples were not confirmed by another molecular test, this is consistent with the improved sensitivity of the Lepto-MD assay relative to these comparators [7]. The Lepto-MD assay also demonstrated good specificity during the initial clinical evaluation and when used in patient populations with a low incidence of leptospirosis [6,19]. MAT is regarded as a highly specific test, though few studies report the clinical specificity of MAT performed on a single specimen. In a study by Cumberland, et al., MAT had a specificity of 99% compared to bacterial culture, even in the first, acute phase specimens [20]. This group also used an MAT titer of ≥1:800 to define a positive test. A more recent evaluation of Leptospira diagnostics employed latent class analysis to estimate the specificity of MAT at 98.8% [21]. A titer of ≥1:400 defined as a positive result, but specificity of single-specimen MAT was not evaluated as a separate end-point.
The classic description of leptospirosis is an acute systemic febrile illness with a biphasic course corresponding to an early leptospiremic phase and a later immune phase. The lack of agreement between molecular testing and single-specimen MAT likely resulted from the clearance of leptospiremia coinciding with the appearance of agglutinating antibodies in serum during the immune phase of illness [2,3,22]. This is consistent with the earlier collection of samples that tested positive in the Lepto-MD assay compared to MAT-positive samples. Interestingly, Leptospira detection in the Lepto-MD assay occurred as late as day 22, and samples collected as early as day 1 were positive by MAT. While inaccurate recording of the day of disease at sample collection could affect these numbers, the prolonged detection of Leptospira nucleic acids in serum has been observed by our group as well as others who utilized different molecular assays in varied patient populations [7,23,24]. Taken together, these findings argue that some patients may present acutely, though symptom onset occurred during the immune phase of disease, while others may fail to generate the expected antibody response and develop prolonged bacteremia. Given these two potential disease courses, there is a limit to the clinical sensitivity attainable with molecular diagnostics and a continued role for serologic testing in the workup of acute leptospirosis. Early diagnosis may also be aided by performing molecular testing on non-blood specimen types, such as urine and cerebrospinal fluid, where Leptospira can be detected later in the disease course [2,19].
While testing of acute and convalescent serum is recommended for the diagnosis of leptospirosis by MAT, such specimens can be difficult to obtain in a typical laboratory setting. Of the 818 patients who had serum sent to CRNL for testing in 2008, only 69 (8.4%) had multiple specimens collected. This is similar to findings from the Royal Tropical Institute in The Netherlands, where, in a study detailing testing results from 2001 to 2012, only 18.1% of patients had multiple samples available for serology [5]. However, a majority of patients with confirmed leptospirosis actually had paired specimens (86.9%), illustrating the need for such specimens to improve the sensitivity of MAT. In the absence of paired samples, though, molecular testing can significantly increase leptospirosis case detection rates. If all MAT-negative samples in our population had been tested with the Lepto-MD assay, for example, the predicted incidence of leptospirosis would increase 50%, from 102 cases to 153 cases (12.5% to 18.7%, respectively, p<0.001). It will be useful to evaluate such a combined testing strategy, as it may capture all or almost all of the cases that could be detected using paired samples by MAT [21].
Finally, this study demonstrates the increased sensitivity of Leptospira detection in the Lepto-MD assay, performed as a real-time RT-PCR, compared to an optimized real-time PCR using the same Leptospira primers and probes. The Lepto-MD assay targets the Leptospira 16S rrs gene, and based on our findings using control Leptospira isolates, detection of both RNA and DNA occurs. Further experiments directly comparing PCR and RT-PCR on Leptospira positive clinical specimens with and without RNase A treatment will be required for full characterization. However, the current study lacked sufficient remaining nucleic acids extracted from positive clinical specimens to perform this testing. Of note, nucleic acids from the control Leptospira isolates were extracted using the DNeasy Blood & Tissue Kit (Qiagen). While optimized for DNA extraction, this kit does not specifically remove RNA from the eluted nucleic acids. The increased sensitivity of RT-PCR for the detection of a bacterial 16S target may prove useful in the design of assays for other pathogens that are present at low levels in the blood. Importantly, such assays may not require the use of new or automated extraction protocols.
In conclusion, this study demonstrates a lack of agreement between molecular detection of Leptospira and single-specimen MAT, regardless of the titer cut-off or day of disease at sample collection. Positive results from these methods demonstrate virtually no overlap, and a combined testing strategy for acute leptospirosis, including a sensitive molecular diagnostic and serologic testing, appears necessary to maximize case detection. Furthermore, we have demonstrated improved sensitivity of Leptospira detection using real-time RT-PCR; similar test designs may prove useful for other systemic bacterial infections that are challenging to diagnose.
Acknowledgments
This research was supported by National Institutes of Health (NIH) grant K08AI110528 (JW, salary support) and the Thrasher Research Fund Award 11979. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We would like to thank the staff of the Stanford Clinical Virology Laboratory and the Stanford SPARK program for their support over the course of this project.
Data Availability
All relevant data are within the paper.
Funding Statement
This research was supported by National Institutes of Health grant K08AI110528, www.NIH.gov (JJW, salary support) and the Thrasher Research Fund Award 11979, www.thrasherresearch.org (JJW). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bharti AR, Nally JE, Ricaldi JN, Matthias MA, Diaz MM, Lovett MA, et al. (2003) Leptospirosis: a zoonotic disease of global importance. Lancet Infect Dis 3: 757–771. [DOI] [PubMed] [Google Scholar]
- 2. Levett PN (2001) Leptospirosis. Clin Microbiol Rev 14: 296–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Picardeau M, Bertherat E, Jancloes M, Skouloudis AN, Durski K, Hartskeerl RA (2014) Rapid tests for diagnosis of leptospirosis: Current tools and emerging technologies. Diagn Microbiol Infect Dis 78: 1–8. 10.1016/j.diagmicrobio.2013.09.012 [DOI] [PubMed] [Google Scholar]
- 4.World Health Organization (2011) Report of the Second Meeting of the Leptospirosis Burden Epidemiology Reference Group.
- 5. Goris MG, Leeflang MM, Loden M, Wagenaar JF, Klatser PR, Hartskeerl RA, et al. (2013) Prospective evaluation of three rapid diagnostic tests for diagnosis of human leptospirosis. PLoS Negl Trop Dis 7: e2290 10.1371/journal.pntd.0002290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Waggoner JJ, Abeynayake J, Balassiano I, Lefterova M, Sahoo MK, Liu Y, et al. (2014) A multiplex nucleic acid amplification test for the diagnosis of dengue, malaria, and leptospirosis. J Clin Microbiol 52: 2011–2018. 10.1128/JCM.00341-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Waggoner JJ, Balassiano I, Abeynayake J, Sahoo MK, Mohamed-Hadley A, Liu Y, et al. (2014) Sensitive Real-Time PCR Detection of Pathogenic Leptospira spp. and a Comparison of Nucleic Acid Amplification Methods for the Diagnosis of Leptospirosis. PLoS One 9: e112356 10.1371/journal.pone.0112356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.World Health Organization (2003) Human leptospirosis: guidance for diagnosis, surveillance and control.
- 9. Smythe LD, Smith IL, Smith GA, Dohnt MF, Symonds ML, Barnett LJ, et al. (2002) A quantitative PCR (TaqMan) assay for pathogenic Leptospira spp. BMC Infect Dis 2: 13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stoddard RA, Gee JE, Wilkins PP, McCaustland K, Hoffmaster AR (2009) Detection of pathogenic Leptospira spp. through TaqMan polymerase chain reaction targeting the LipL32 gene. Diagn Microbiol Infect Dis 64: 247–255. 10.1016/j.diagmicrobio.2009.03.014 [DOI] [PubMed] [Google Scholar]
- 11. Thaipadungpanit J, Chierakul W, Wuthiekanun V, Limmathurotsakul D, Amornchai P, Boonslip S, et al. (2011) Diagnostic accuracy of real-time PCR assays targeting 16S rRNA and lipL32 genes for human leptospirosis in Thailand: a case-control study. PLoS One 6: e16236 10.1371/journal.pone.0016236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bruce MG, Sanders EJ, Leake JA, Zaidel O, Bragg SL, Aye T, et al. (2005) Leptospirosis among patients presenting with dengue-like illness in Puerto Rico. Acta Trop 96: 36–46. [DOI] [PubMed] [Google Scholar]
- 13. Karande S, Gandhi D, Kulkarni M, Bharadwaj R, Pol S, Thakare J, et al. (2005) Concurrent outbreak of leptospirosis and dengue in Mumbai, India, 2002. J Trop Pediatr 51: 174–181. [DOI] [PubMed] [Google Scholar]
- 14. LaRocque RC, Breiman RF, Ari MD, Morey RE, Janan FA, Hayes JM, et al. (2005) Leptospirosis during dengue outbreak, Bangladesh. Emerg Infect Dis 11: 766–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. World Health Organization; (2012) World Malaria Report. Geneva: WHO Press. [Google Scholar]
- 16. Gouveia EL, Metcalfe J, de Carvalho AL, Aires TS, Villasboas-Bisneto JC, Queirroz A, et al. (2008) Leptospirosis-associated severe pulmonary hemorrhagic syndrome, Salvador, Brazil. Emerg Infect Dis 14: 505–508. 10.3201/eid1403.071064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Reis RB, Ribeiro GS, Felzemburgh RD, Santana FS, Mohr S, Melendez AX, et al. (2008) Impact of environment and social gradient on Leptospira infection in urban slums. PLoS Negl Trop Dis 2: e228 10.1371/journal.pntd.0000228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Spichler A, Athanazio DA, Vilaca P, Seguro A, Vinetz J, Leake JA (2012) Comparative analysis of severe pediatric and adult leptospirosis in Sao Paulo, Brazil. Am J Trop Med Hyg 86: 306–308. 10.4269/ajtmh.2012.11-0308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Waggoner JJ, Soda EA, Seibert R, Grant PM, Pinsky BA (2015) Molecular detection of Leptospira in two returned travelers: higher bacterial load in cerebrospinal fluid versus serum or plasma. Am J Trop Med Hyg in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cumberland P, Everard CO, Levett PN (1999) Assessment of the efficacy of an IgM-elisa and microscopic agglutination test (MAT) in the diagnosis of acute leptospirosis. Am J Trop Med Hyg 61: 731–734. [DOI] [PubMed] [Google Scholar]
- 21. Limmathurotsakul D, Turner EL, Wuthiekanun V, Thaipadungpanit J, Suputtamongkol Y, Chierakul W, et al. (2012) Fool's gold: Why imperfect reference tests are undermining the evaluation of novel diagnostics: a reevaluation of 5 diagnostic tests for leptospirosis. Clin Infect Dis 55: 322–331. 10.1093/cid/cis403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Galloway RL, Hoffmaster AR (2015) Optimization of LipL32 PCR assay for increased sensitivity in diagnosing leptospirosis. Diagn Microbiol Infect Dis 82: 199–200. 10.1016/j.diagmicrobio.2015.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Agampodi SB, Matthias MA, Moreno AC, Vinetz JM (2012) Utility of quantitative polymerase chain reaction in leptospirosis diagnosis: association of level of leptospiremia and clinical manifestations in Sri Lanka. Clin Infect Dis 54: 1249–1255. 10.1093/cid/cis035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Merien F, Baranton G, Perolat P (1995) Comparison of polymerase chain reaction with microagglutination test and culture for diagnosis of leptospirosis. J Infect Dis 172: 281–285. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.