Highlights
-
•
IFN- leads to the proteasomal degradation of HMGCR.
-
•
IFN- and 25-HC can transcriptionally and post-translationally alter levels of HMGCR.
-
•
The reduction of HMGCR through the action of IFN- requires the de novo synthesis of 25-HC by CH25H.
Abbreviations: HMGCR, 3-hydroxy-3-methylglutaryl-CoA reductase; 25-HC, 25-hydroxycholesterol; IFN, interferon; SREBP, sterol regulatory element-binding protein; SCAP, SREBP cleavage activating protein; ER, endoplasmic reticulum; ERAD, ER-associated protein degradation; FBS, fetal bovine serum; LPDS, lipoprotein depleted serum; MEV, mevalonate; LS, lanosterol; CHO, cholesterol; CH25H, cholesterol 25-hydroxylase; TLR, Toll-like receptor; BMDMs, bone marrow derived macrophages; DCs, dendritic cells; SBGN, systems biology graphical notation; PRRs, Pattern recognition receptors; IFNAR1, IFN- receptor
Keywords: CH25H, Cholesterol biosynthesis, 25-Hydroxycholesterol, Macrophages, Immunity, Infection
Abstract
Interferons (IFNs) play a central role in immunity and emerging evidence suggests that IFN-signalling coordinately regulates sterol biosynthesis in macrophages, via Sterol Regulatory Element-Binding Protein (SREBP) dependent and independent pathways. However, the precise mechanisms and kinetic steps by which IFN controls sterol biosynthesis are as yet not fully understood. Here, we elucidate the molecular circuitry governing how IFN controls the first regulated step in the mevalonate-sterol pathway, 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR), through the synthesis of 25-Hydroxycholesterol (25-HC) from cholesterol by the IFN-inducible Cholesterol-25-Hydroxylase (CH25H). We show for the first 30-min of IFN stimulation of macrophages the rate of de novo synthesis of the Ch25h transcript is markedly increased but by 120-min becomes transcriptionally curtailed, coincident with induction of the Activating Transcription Factor 3 (ATF3) repressor. We demonstrate ATF3 induction by Toll-like receptors is strictly dependent on IFN-signalling. While the SREBP-pathway dependent rates of de novo transcription of Hmgcr are relatively unchanged in the first 90-min of IFN treatment, we find HMGCR enzyme levels undergo a rapid proteasomal-mediated degradation, defining a previously unappreciated SREBP-independent mechanism for IFN-action. These events precede a sustained marked reduction in Hmgcr RNA levels involving SREBP-dependent mechanisms. We demonstrate that HMGCR proteasomal-degradation by IFN strictly requires the synthesis of endogenous 25-HC and functionally couples HMGCR to CH25H to coordinately suppress sterol biosynthesis. In conclusion, we quantitatively delineate proteomic and transcriptional levels of IFN-mediated control of HMGCR, the primary enzymatic step of the mevalonate-sterol biosynthesis pathway, providing a foundational framework for mathematically modelling the therapeutic outcome of immune-metabolic pathways.
1. Introduction
3-Hydroxy-3-methylglutaryl-CoA reductase (HMGCR) operates as an important regulated step enzyme in the mevalonate-sterol biosynthesis pathway, which is responsible for the production of cholesterol and isoprenoid products [1,2]. HMGCR is also an important therapeutic target for a class of inhibitory drugs, known as statins, which are prescribed to reduce cholesterol levels in the serum. Statin treatment reduces cholesterol but leads to a compensatory increase of HMGCR via the SREBP pathway (schematically shown in Fig. 6) and which also increases the level of low-density lipoprotein (LDL) receptors, leading to reduction in plasma cholesterol levels [1]. Given the importance of HMGCR in the biosynthesis of sterols, HMGCR is subject to stringent and multiple regulatory feedback mechanisms.
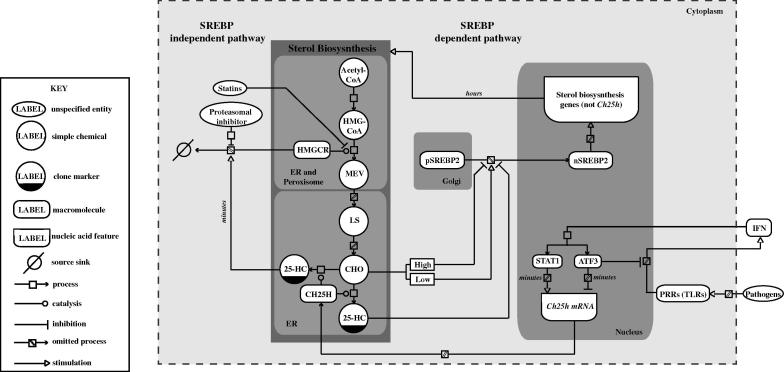
Fig. 6.
Schematic representation of the dual role of 25-HC in the sterol biosynthesis pathway and the IFN--regulated innate immune response using SBGN system (inset shows key).
Regulation of HMGCR at the transcriptional level is exquisitely sensitive to feedback control mediated by the Golgi-nuclear shuttling of the master transcriptional regulator of the sterol pathway, SREBP2 [3–5]. Under conditions of high intra-cellular sterol levels, excess oxysterols and cholesterol bind to INSIG and SCAP proteins, respectively. This blocks transport of mature SREBP2 from the ER to the golgi, where cleavage of SREBP2 and release of the active transcription factor would normally take place [6–9]. Under sterol-limiting conditions, sterol binding to INSIG and SREBP cleavage activating protein (SCAP) does not occur and SCAP becomes free to escort SREBP2 from the ER to the golgi, where SREBP2 undergoes proteolytic cleavage [10–14] to generate an N-terminal domain of SREBP2 which is released into the nucleus to activate multiple sterol biosynthesis related genes, including Hmgcr [15].
Regulation of HMGCR at the post-transcriptional and post-translational level operates independently of the SREBP regulatory pathway and represents an important mechanism for regulating HMGCR in response to changing sterol levels. Post-translational regulation is exerted by oxysterol-dependent degradation, via an INSIG dependent ER-associated protein degradation (ERAD) system, involving an ubiquitin-proteasome degradation pathway [16,17]. Under sterol depleted conditions, the half-life of HMGCR is approximately 12 h. However, in the presence of excess sterols, degradation of HMGCR by the ERAD mechanism results in a half-life of about 40 min [18]. This highly sensitive and responsive proteomic mechanism contrasts with the relatively slow transcriptional regulation mediated by the SREBP pathway which takes several hours to exert an effect on HMGCR protein levels [19].
It is well established that the oxysterol, 25-HC, which is synthesised from cholesterol by the enzyme CH25H, has a potent ability to block cholesterol biosynthesis through INSIG induced ubiquitination and proteasomal degradation of HMGCR [4,20,21] and also by blocking ER translocation of SREBP2 [22]. However, 25-HC has been shown not to play a major role in cholesterol homeostasis but instead has emerged to have key immune modulatory functions [1,23]. We have been the first to report that HMGCR levels and sterol biosynthesis are suppressed by interferon (IFN) and in infected macrophages the reduction of HMGCR is strictly dependent on IFN-induced signalling [24]. Moreover, in this connection 25-HC has also been shown to have a range of effects on immune related host defence systems, including suppressing the differentiation of human monocytes [25], decreasing immunoglobulin A production [26], promoting immune cell guidance [23], moderating inflammasome activity [27] and amplifying inflammatory signalling via AP-1 [28]. Park and Scott reported that the production of CH25H is stimulated by TLR activation in macrophages and DCs [29]. In later studies, we and other laboratories demonstrated that the IFN-mediated production of 25-HC has broad anti-viral and immune modulatory activities [27,28,30,31]. It has also been reported by many groups including our own studies that RNA-interference (RNAi) mediated knockdown of HMGCR protein levels and pharmaceutical inhibition of HMGCR activity can suppress viral infectivity and growth [24,32–34]. In addition, it is worth noting that HMGCR inhibitors (statins) also have immune modulatory effects [35]. Collectively, these findings point to an emerging role for HMGCR as a control point in the regulation of immunomodulatory and host defence mechanisms. However, the precise role of 25-HC in the immune system has not been fully defined. Moreover, roles for SREBP-independent mechanisms for anti-viral and anti-inflammatory effects have yet to be defined.
In this study, we examine further the regulation of HMGCR following early innate immune stimulation by IFN-. We report for the first time systematic quantitative analyses of the IFN regulatory circuit in macrophages for CH25H, its repressor ATF3, and how they couple to HMGCR, revealing a previously unappreciated role for the IFN-mediated degradation of HMGCR enzyme levels that is shown to be critically dependent on the de novo synthesis of 25-HC. Overall, this study contributes to the dissection of the mechanisms underlying the regulation of HMGCR as an emerging effector target during innate-immune-metabolic signalling and provides a foundation for future modelling of pathway feedback control.
2. Experimental
2.1. Reagents and media
25-HC and mevastatin were purchased from Sigma (Sigma-Aldrich, UK). MG132 (Z-Leu-Leu-Leu-al) (CAS number: 133407-82-6) was purchased from Cambridge Bioscience. Murine recombinant IFN- was purchased from Perbio Science or Life Technology (PMC4033). Medium A: DMEM/F12 with GlutaMAX (Gibco, Life technologies) supplemented with 10% (v/v) fetal bovine serum (FBS) (Gibco, Life technologies), 10% (v/v) L929 containing colony-stimulating factor 1 (Csf1) and Penicillin/streptomycin (PS) (Gibco, Life technologies); Medium B: DMEM/F12 with GlutaMAX supplemented with 3% (v/v) lipoprotein depleted serum (LPDS) (Sigma), 10% (v/v) L929 containing Csf1 and PS; Medium C: DMEM/F12 with GlutaMAX supplemented with 3% (v/v) LPDS, 0.01 μM mevastatin, 10% (v/v) L929 containing Csf1 and PS. The following antibodies were used: monoclonal mouse anti-HMGCR (C-1, Santa Cruz Biotechnology), rabbit anti--actin (4967, Cell Signaling) and rabbit anti--tubulin (ab6046, Abcam).
2.2. Primary bone marrow derived macrophage (BMDM) culture generation
Wild-type BMDMs were derived from the femur and tibia isolated from C57BL/6 mice as previously described [24,30]. Ch25h−/− BMDMs were derived from the femur and tibia isolated from B6.129S6-/J mice (The Jackson Laboratory, UK) and grown in Medium A. Ifnb−/− mice were from the Institute of Animal Breeding and Genetics (Veterinary University of Vienna). All procedures were carried out under project and personal licences approved by the Secretary of State for the Home Office, under the United Kingdoms 1986 Animals (Scientific Procedures) Act and the Local Ethical Review Committee at Edinburgh University. All cultures are routinely tested to ensure they are free of mycoplasma and endotoxin contamination.
2.3. Experimental treatment conditions
BMDMs were grown in 24-well plates with Medium A. For the dose titration of mevastatin, cells were treated overnight with Medium B containing various concentrations of mevastatin. For the dose titration of 25-HC, cells were treated as the figure legend described. The cells were then harvested for western blot analysis. For time course experiments, Prior to the experiment, IFN- stock was diluted as described [24]. Culture medium was aspirated from 24-well plates and 1 mL of Medium C containing IFN- (5 ng/mL) was added to wells at 1.5-h, 4-h and 9-h time points, respectively. Meanwhile, a parallel experiment, where the cells were treated with 25-HC (2.5 μM), was performed at the same time. The cells were harvested for western blot analysis. For the measurement of HMGCR protein levels with proteasome inhibitor MG132, BMDMs were pre-treated with Medium B containing MG132 (20 μM) for an hour; and then treated with Medium C plus MG132 (20 μM) containing IFN- (5 ng/mL) or 25-HC (2.5 μM), respectively. The cells were incubated at 37 °C for another 6 h and HMGCR levels were determined by western blot analysis.
2.4. qRT-PCR analysis
QIAGEN Rneasy Plus Mini Kit (QIAGEN, Germany) was used to purify total RNA. The qScript One-Step Fast qRT-PCR kit, Low ROX (Quanta BioSciences, USA), Taqman probe/primer Hmgcr, Atf3, Ch25h, Irf1 and Gapdh (Life technologies, UK) were used for the qRT-PCR measurement. qRT-PCR was undertaken according to manufactures instructions.
2.5. Western blot analysis
After treatments, cells were lysed directly in wells by adding 100 μL of RIPA lysis buffer (9806, Cell Signaling) supplemented with protease cocktail (Complete Protease Inhibitor Cocktail Tablets, Roche) and 1 mM Phenylmethanesulfonyl fluoride (Sigma). Plates were incubated on ice for 20 min. Protein concentrations of whole-cell lysate fractions were then determined using the BCA Protein Assay Reagent kit (Thermo Scientific) according to manufacturers instructions. Prior to SDS–PAGE, whole-cell lysates were mixed with 2X Laemmli sample buffer (Sigma) and incubated at 50 °C for 10 min. Thereafter, an equal amount of protein was subjected to 8% SDS-PAGE, after which proteins were transferred to Hybond ECL Nitrocellulose membranes (Amersham). The membranes were blocked with 5% skimmed milk (Sigma), probed with specific primary antibodies overnight at 4 °C, washed with PBST (Phosphate Buffered Saline with Tween 20), incubated with HRP-conjugated secondary antibodies (Cell Signaling) at room temperature for 1 h. The membranes were then re-washed with PBST and bands were visualised by Hyperfilm ECL (Amersham) or Odyssey Fc Dual-Mode Imaging System (Li-COR Biosciences) with ClarityTM Western ECL Substrate (BIO-RAD). Image Studio Lite (Li-COR Biosciences) was used to analyse the bands.
2.6. BMDM de novo RNA labelling, isolation and microarray analysis
2.6.1. Time course analysis of RNA synthesis and half-life in Control-treated or IFN--treated BMDMs
To enable a simultaneous analysis of RNA synthesis, abundance and decay, BMDMs were control treated or treated with IFN- at a concentration of 5 ng/mL. These treatments were then followed by 16 successive cycles of 4-thiouridine addition to the BMDM cultures and transcriptional termination at 30 min intervals until the end of an 8-h time course. 4-Thiouridine (Sigma) labelling in these experiments was undertaken as described by Dölken et al. [36].
Total RNA was then isolated using an RNeasy Midi kit (Qiagen) according to manufacturers instructions, quantitated using a Nanodrop (Thermo Scientific) and integrity was confirmed using an Agilent Bioanalyser (Agilent UK). Newly transcribed RNA (ntRNA) was then isolated and again quantitated using a Nanodrop.
2.6.2. RNA labelling for time-course microarray analysis
Processing of 100 ng of total RNA samples (8× 1 h for control treated and 8× 1 h for IFN- treated = 16 in total) and ntRNA samples (16× 30 min for control treated and 16× 30 min for IFN- treated = 32 in total) for hybridisation to Affymetrix Mouse Gene 1.0 ST arrays was undertaken according to manufacturers instructions (Affymetrix). Hybridisation, washing, staining and scanning of the arrays were also undertaken following standard Affymetrix protocols.
2.6.3. Data analysis for microarray time-course
After scanning and data capture, open-source R based software “Bioconductor” was used to implement all quality control and statistical analyses.
The RMA method was then used for normalisation, background correction, and probe set summation [37]. To reduce the dataset for further analysis and enrich for reliably detected genes, values for median and 2× the Median Absolute Deviation (2×MAD) for negative control probesets were calculated for each array. Intensity values for each probe on each total RNA array were then compared with their array specific 2×MAD value and were retained if the intensity was equal to or greater than this value. After filtering and removal of all designated Affymetrix control probesets, 12,472 probes were retained for statistical analyses.
Since conventional clustering algorithms do not typically assign significance to differential expression of two conditions over time (on an individual gene basis) the MaSigPro algorithm was used [38]. Using this method, 2086 (ntRNA) or 4482 (total RNA) probes with notable differential profiles of expression across the 8 h of IFN- stimulation were identified.
2.6.4. Microarray analysis of total, pre-existing and newly transcribed RNA in resting BMDMs
Using the protocol of Dölken et al. [36]for de novo RNA labelling and isolation, we used Affymetrix Gene STv1.0 microarrays to analyse total RNA abundance in resting BMDMs at t = 0 h and total, newly transcribed (labelled) and pre-existing (unlabelled) RNA at t = 1 h. This was completed in triplicate. To enrich for reliably detected genes, values for median and 2× the Median Absolute Deviation (2×MAD) for negative control probe-sets were calculated for each array. Intensity values for each probe on each total RNA array were then compared with their array specific 2×MAD value and were retained if the intensity was equal to or greater than this value. After filtering and removal of all designated Affymetrix control probe-sets, 5826 probes were classified as detected on all arrays. Mean values for the total RNA, newly transcribed RNA and pre-existing RNA abundance measurements were then calculated from triplicate measurements and this data was used for the calculation of a median half-life in the resting BMDMs as described below.
2.6.5. Calculation of median transcript half-life in resting BMDMs
If the mean total RNA transcript abundance at the start of a 1 h time interval is T0, the mean total RNA transcript abundance at the end of the interval T1, the mean newly-transcribed RNA transcript abundance N1 and the mean pre-existing RNA transcript abundance P1, we should observe that N1/T1 + P1/T1 = 1. In practice, however, we observed some systematic disagreement with this equality. To maximise agreement, therefore, we introduced two normalisation constants, and , and rewrote the above equation as N1/T1 + βP1/T1 = 1, as proposed in [39]. We then optimised and by linear regression and used the constants obtained in the calculation of half-lives for the time course. To estimate a half-life value for every transcript detected by the microarrays in this experiment, we calculated the values in two ways: (i) using newly transcribed RNA transcript measurements (left equation) and (ii) using the pre-existing RNA transcript measurements (right equation). A mean of the 2 values obtained from these equations was then calculated.
In these calculations, we assumed a delay of 5 min before the initiation of 4-thiouridine incorporation into newly synthesised RNA. The resulting labelling period is, therefore, 55 min. Using the above protocol, a median half-life value for the 5826 RNAs detected in these experiments was estimated. This was subsequently used to assist in the normalisation of half-life data from our time-course analysis of control treated or IFN- treated BMDMs (described in next section).
2.6.6. Estimation of transcript half-life in control or IFN- BMDMs
From our time course experiments we obtained data from 1 h intervals in which total and newly transcribed RNA had been analysed (150–210, 270–330, 330–390 and 390–450 min). This data was used to estimate half-life values for Hmgcr transcript in the control treated or IFN- treated BMDM data. Since we had chosen not to hybridise pre-existing RNA samples to microarrays in our 8-h time course experiment, we used the formulae above to optimise the value of such that the median half-life across all transcripts in the cintrol and IFN- courses was the same as the value calculated for the resting BMDMs. The output of these calculations provided multiple, discrete half-life estimates for each transcript across the 8-h control or IFN- time course.
2.7. Software and statistical analysis
Microarray data analysis was performed by R [40]. The EC50 and IC50 values were calculated by Matlab [41] and Graphpad (GraphPad Software, Inc., La Jolla, CA, USA). Unpaired Student’s t tests were performed by Graphpad. The half-life of Hmgcr mRNA was calculated using Matlab. Adobe illustrator and Graphpad were used to assemble figures.
3. Results
3.1. Molecular circuitry for the rapid induction and inhibition kinetics of de novo RNA synthesis of Ch25h and Atf3 by IFN in primary macrophages.
In resting cells, latent transcription factors (named STATs) associated with the IFN-receptor reside in the cytoplasm in an inactive state and upon IFN binding its receptor, leads within seconds, to phosphorylated active forms that translocates, within minutes, to the nucleus, binding target promoters to transcriptionally activate genes. Our previous studies have shown that IFN activation of BMDMs results in the increased biosynthesis and secretion of 25-HC as a result of a rapid STAT1 recruitment to the promoter of the Ch25h gene within the first hour of treatment [30]. The Ch25h gene encodes an enzyme which is responsible for catalysing the production of 25-HC from cholesterol. The timing and duration of 25-HC synthesis have important implications for its immune functions which are, in part, mediated through the metabolic and immune modulatory re-programming of BMDMs upon infection [42,23]. At the transcriptional level, ATF3 is a well known transcriptional repressor for TLR driven pro-inflammatory cytokines [43,44] including the induction of Ch25h [45]. ATF3 has also been reported to be induced by TLR-IFN signalling [46–48] and more recently has been found to repress the IFN- promoter, establishing a negative feedback loop [49]. A schematic summary is shown in Fig. 6. While previous studies have clearly indicated a rapid up-regulation of Ch25h expression in response to IFN, the precise rate kinetics of IFN- elicited Ch25h mRNA and Atf3 de novo synthesis have not been determined. Furthermore, evidence for a direct functional requirement of IFN signalling in TLR induction of ATF3 in BMDMs has not been shown. Hence, our initial experiments sought to quantitatively analyse de novo synthesis of Ch25h mRNA and its negative regulator Atf3 in IFN- stimulated BMDMs and in BMDMs genetically ablated for the TLR-induction of IFN-.
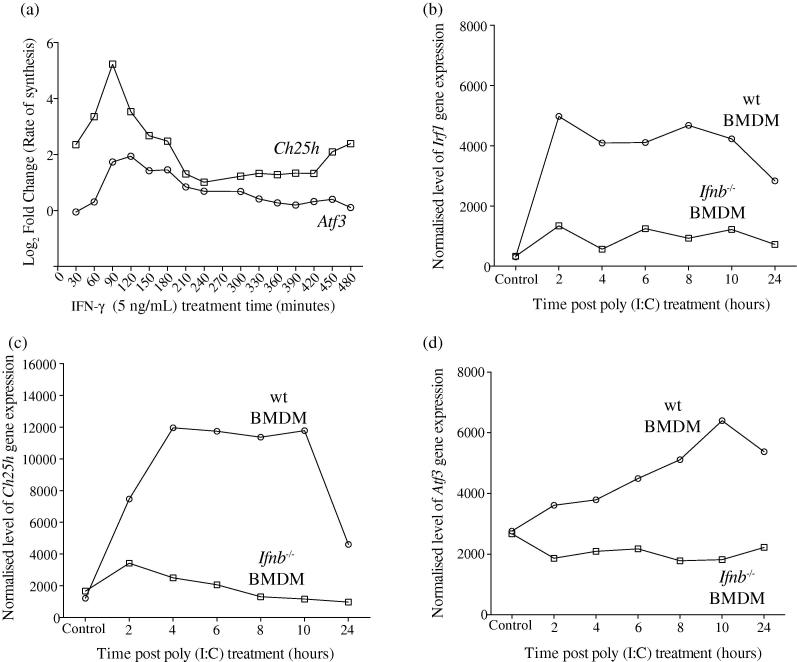
For these experiments, we performed an 8-h high-resolution, systematic microarray analysis of changes in RNA synthesis, abundance and decay in BMDM treated with 5 ng/mL IFN-. Fig. 1a shows how the de novo synthesis of Ch25h and Atf3 RNA altered during this temporal analysis. Ch25h RNA synthesis rate was up-regulated more than 4 fold in treated cells (relative to the control) during the first 30 min after the addition of the cytokine. Synthesis of Ch25h RNA then peaked 60–90 min after treatment and was subsequently down regulated, returning to basal transcriptional levels by 210–240 min (Fig. 1a). The kinetics of the observed transcriptional activation and repression provide an explanation for the observed plateau by 8 h for the amount of intracellular and secreted 25-HC as previously reported [30]. Notably, de novo synthesis of Atf3 RNA, the product of which is a repressor of Ch25h transcription, increased 60 min after the addition of IFN- (Fig. 1a). Atf3 RNA synthesis then peaked at 120 min, a time coincident with a marked reduction in Ch25h RNA synthesis. These experiments show a rapidly initiated, but apparently tightly controlled, transient up-regulation of Ch25h RNA synthesis during the first 4 h of IFN- treatment in BMDMs.
Fig. 1.
Synthesis rates of de novo transcription of Atf3 and Ch25h following IFN- treatment and RNA abundance of Irf1, Ch25h and Atf3 upon Poly (I:C) treatment. (a) Measurement of de novo transcribed RNA of Atf3 and Ch25h upon IFN- treatment in wild-type BMDMs (relative to control treated) in Medium A over an 8-h period. Each point represents transcript synthesis in control treated vs IFN- during a 30-min period. Normalised log fold change values were calculated by subtracting the control treated from the IFN- treated signal values. Atf3: p = ns; Ch25h: p=1.91E−12. (b) Irf1 abundance in wild-type and Ifnb−/− BMDMs (relative to control treated t = 0) in Medium A following 10 μg/mL Poly (I:C) treatment. (c) Ch25h abundance in wild-type and Ifnb−/− BMDMs (relative to control treated t = 0) in Medium A following 10 μg/mL Poly (I:C) treatment. (d) Atf3 abundance in wild-type and Ifnb−/− BMDMs (relative to control treated t = 0) in Medium A following 10 μg/mL Poly (I:C) treatment.
Next, we examined whether the induction of Atf3 by TLR-stimulation depends on the presence of an intact IFN--response in BMDMs. For these experiments, wild-type or Ifnb−/− BMDMs were treated with the TLR-3 agonist, Poly (I:C) (10 μg/mL), and the abundance of Irf1 (as a marker for STAT1 activation), Ch25h, and Atf3 RNA was analysed at 2, 4, 6, 8 ,10 and 24 h after treatment. Fig. 1b shows how overall RNA abundance of the interferon-regulated gene Irf1 varied in our time-course. In wild-type BMDMs, poly(I:C) treatment resulted in a rapid increase in Irf1 RNA abundance during the first 2 h of the time-course. Notably, between 2 and 8 h the abundance of this transcript remained elevated and subsequently decreased from 10 h until 24 h. In Ifnb−/− BMDMs, in the absence of IFN- production, the overall effect of the Poly(I:C) stimulus was negated. Alterations in Ch25h transcript expression elicited by Poly(I:C) followed a similar pattern to those of Irf1 (Fig. 1c). This was expected as Stat1 regulates the transcription of both genes in the context of IFN- activation. After an initial rapid increase of Ch25h RNA abundance, its RNA levels remained elevated between 4 and 10 h and subsequently decreased by 24 h. In these experiments, increases in Ch25h RNA abundance were not completely negated in the knockout BMDMs. This is likely due to the activation of IFNAR1 by other type I interferons [24]. Most notably, Atf3-induced levels were ablated in Poly(I:C)-treated Ifnb−/− BMDMs (Fig. 1d). This demonstrates Atf3 has a strict dependency on the IFN- response for the transcriptional activation of its promoter upon TLR stimulation. The molecular “wiring” for this inflammatory regulatory circuit is shown in Fig. 6, which details the stringent, coordinate control underpinning the regulation of 25-HC synthesis in IFN-activated BMDMs.
3.2. IFN-inducible-25-HC mediates a dose-dependent reduction of HMGCR protein levels in BMDMs
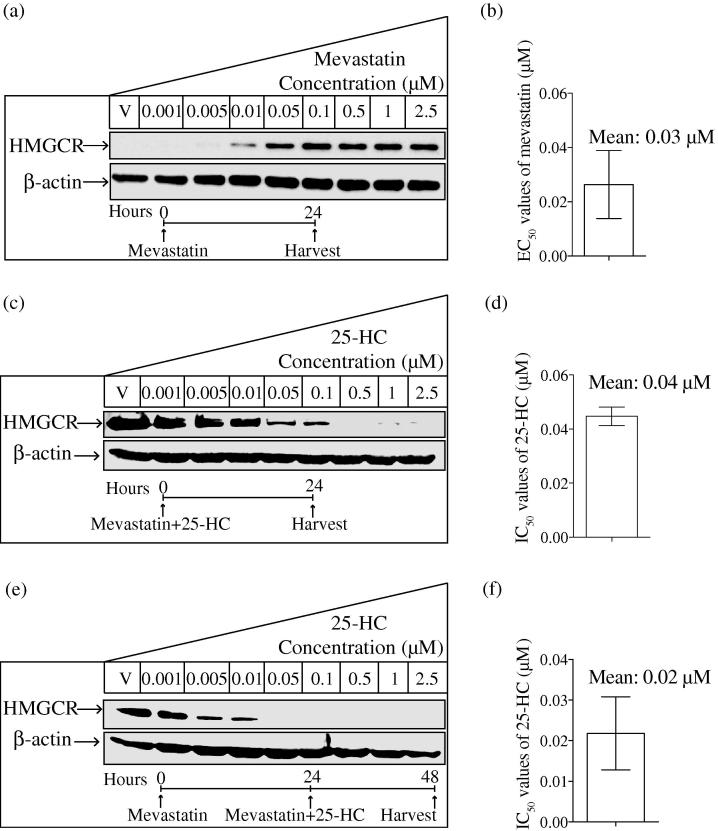
We next sought to quantitatively determine proteomic parameters for HMGCR inhibition that delineate the molecular circuitry for the IFN-inducible 25-HC regulation of the sterol pathway. Whilst the mechanism by which 25-HC regulates the SREBP pathway of the sterol pathway has been extensively characterised (summarised in Fig. 6), it is also known that 25-HC can post-translationally regulate HMGCR protein abundance in a SREBP independent manner [16,17]. We reasoned, therefore, that IFN- treatment might modulate the levels of Hmgcr mRNA and HMGCR protein through SREBP-dependent and independent mechanisms, respectively. To test this hypothesis, we quantified how increasing doses of 25-HC alter HMGCR protein abundance in BMDMs. Under normal serum conditions, endogenous HMGCR protein levels are weakly detected when analysed by western blotting. HMGCR protein levels can, however, be markedly increased upon cholesterol depletion through compensatory homeostatic activity of the SREBP-activated pathway. We first quantified the HMGCR protein level with various concentrations of mevastatin under lipid-deficient condition (Fig. 2a). The EC50 value for mevastatin-induced increase of HMGCR levels in our experimental conditions was estimated to be 0.03 μM (Fig. 2b). This estimate closely agrees with published IC50 data for mevastatin inhibition of HMGCR enzymatic activity (ranging from 0.01 to 0.11 μM) [50–53]. On the basis of these results, we selected a low concentration of mevastatin (0.01 μM), to allow for an increased dynamic range for measuring levels of HMGCR. Note, under these conditions, the SREBP pathway should be mildly activated. The effects of varying 25-HC dose on HMGCR protein abundance in BMDMs were quantitated following concurrent (Fig. 2c) or a 24-h pre-treatment (Fig. 2e) of the cells with 0.01 μM mevastatin. IC50 values for 25-HC-induced reduction of HMGCR under these experimental conditions were 0.04 μM (Fig. 2d) and 0.02 μM (Fig. 2f). Taken together, these data show that there is a dose-dependent decrease in levels of HMGCR protein upon exposure of BMDMs to 25-HC.
Fig. 2.
HMGCR protein levels following mevastatin and 25-HC treatments. (a) Western blot analysis of the HMGCR protein level in mevastatin-treated wild-type BMDMs. Cells were treated overnight with various concentrations of mevastatin in Medium B. (b) Intensity values of HMGCR to -actin were calculated, based on which the EC50 value of mevastatin was then generated. Data are mean ± SEM (n = 3). (c) Western blot analysis of the endogenous HMGCR protein level in 25-HC-treated wild-type BMDMs. Cells were treated with various concentrations of 25-HC for 24 h in Medium C. (d) Intensity values of HMGCR to β-actin were calculated, based on which the IC50 value of 25-HC was then generated. Data are mean ± SEM (n = 4). (e) Western blot analysis of the endogenous HMGCR protein level in 25-HC-treated wild-type BMDMs. Cells were pre-treated with Medium C overnight and then treated with various concentrations of 25-HC in Medium C for another 24 h. (f) Intensity values of HMGCR to -actin were calculated, based on which the IC50 value of 25-HC was then generated. Data are mean ± SEM (n = 4).
3.3. IFN- exerts an immediate early SREBP-independent proteomic layer of regulation of HMGCR followed by a delayed early SREBP-dependent transcriptional mechanism
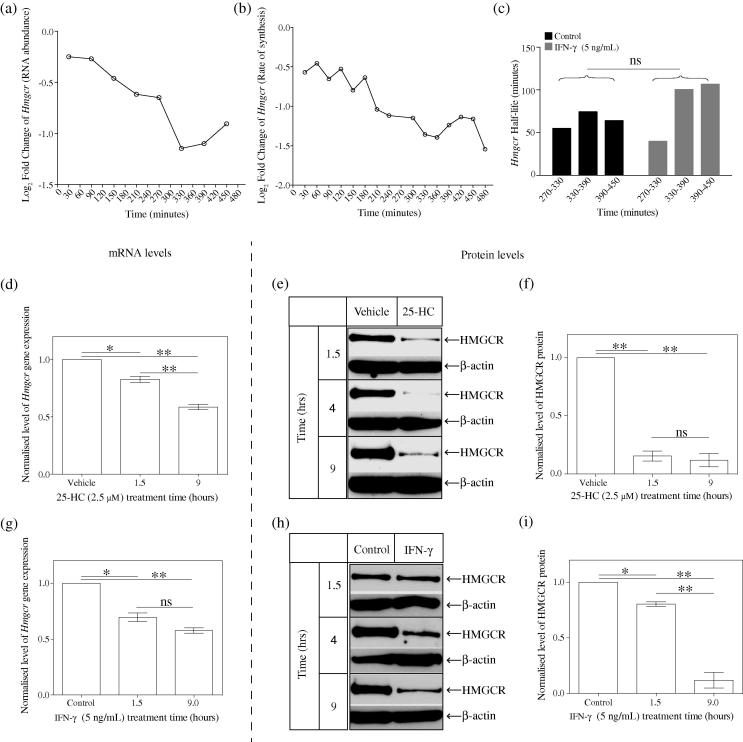
In our next experiments, we aimed to determine and compare the extent of IFN induced reduction in HMGCR RNA levels (dependent on the activity of the SREBP pathway) and protein levels (which we assume involves a combination of SREBP-dependent and SREBP-independent pathways). Time course experiments measuring the reduction of total RNA of Hmgcr after 8 h of IFN treatment revealed RNA abundance for this transcript decreased by approximately 2 fold (Fig. 3a). In separate experiments, measuring the de novo synthesis of RNA over an 8-h period, an approximately 2-fold reduction in the rate of RNA synthesis was also observed (Fig. 3b). Marked changes in Hmgcr abundance and rates of synthesis occur after approximately 200 min. To determine if this is due to rate of transcription or stability of RNA transcript changes we next computed the rates of RNA turnover of Hmgcr RNA from 270 to 450 min in 60-min intervals. Fig. 3c shows there is no statistical difference in the turnover rate. This indicates that the approximately 50% reduction in RNA abundance by 8–9 h is due to changes in the rates of transcription alone and the determined turnover rates (approximately 73 min) suggest at least an hour response time to effect a change in abundance.
Fig. 3.
Time-course showing the Hmgcr mRNA and HMGCR protein levels following IFN- treatment. (a) Temporal alterations in abundance of Hmgcr transcript in IFN- treated BMDMs (relative to control treated) in Medium A over an 8-h period. Each point represents overall abundance of transcript at the indicated time. Relative log fold change values were calculated by subtracting the control treated from the IFN--treated signal values. p = 2.72E−05. (b) Temporal alterations in de novo synthesis rate of Hmgcr transcript in IFN--treated BMDMs (relative to control treated) in Medium A over an 8-h period. Each point represents transcript synthesis rate in control treated vs IFN- during a 30-min period. Normalised log fold change values were calculated by subtracting the control treated from the IFN--treated signal values. p = 2.93E−11. (c) Half-life values of Hmgcr at indicated period time. Based on the RNA abundance and newly transcribed RNA levels of Hmgcr, the half-lives of Hmgcr for each period were calculated as previously described [39]. (d) Wild-type BMDMs were treated with 2.5 μM of 25-HC in Medium C at multiple time points and Hmgcr mRNA level was determined by qRT-PCR. Data are mean ± SEM (n = 3). (e) Western blot analysis of the HMGCR protein level with 2.5 μM of 25-HC treatment in Medium C at multiple time points. (f) Intensity values of HMGCR to -actin. Data are mean ± SEM (n = 3). (g) Wild-type BMDMs were treated with 5 ng/mL of IFN- in Medium C at multiple time points and Hmgcr mRNA level was determined by qRT-PCR. Data are mean ± SEM (n = 3). (h) Western blot analysis of HMGCR protein level with 5 ng/mL of IFN- treatment in Medium C at multiple time points. (i) Intensity values of HMGCR to -actin. Data are mean ± SEM (n = 3). ∗p < 0.05, ∗∗p < 0.01, determined with an unpaired Student’s t test.
We next investigated the quantitative effects of direct 25-HC administration (2.5 μM) to BMDMs on RNA and protein levels under sterol-depleted conditions as discussed above. Consistent with the data in Fig. 3a and b, a significant reduction in RNA levels was detected at 9 h post-treatment revealing a greater magnitude of RNA reduction (approximately 45%) with only a small decrement at 1.5 h (Fig. 3d). In contrast, levels of HMGCR protein were more than 90% reduced by 1.5 h of 25-HC treatment and maintained a similar reduced level at 9-h post-treatment (Fig. 3e and f). In summary, these experiments show that 25-HC rapidly diminishes macrophage HMGCR protein levels within the first hour of treatment. Note, at the later time point (9 h), reductions in RNA levels will also contribute, in an additive manner, to a sustained reduction of HMGCR abundance.
We next quantified the RNA (Fig. 3g) and protein (Fig. 3h and i) levels of HMGCR with IFN- treatment at multiple time points. These experiments were also performed using sterol-depleted conditions. Fig. 3g–i shows that IFN- treatment results in a 20% decrease for both RNA and protein levels of HMGCR at 1.5-h post-treatment. However, at 9-h post-treatment, the reduction in RNA and protein levels is about 50% and 90%, respectively. These results point towards both transcriptional and post-translational mechanisms contributing to the maintenance of reduced HMGCR levels at late times. These data provide evidence showing that IFN- and 25-HC can transcriptionally and post-translationally alter levels of HMGCR. These differential modes of action are temporally coordinated with rapid early effects on HMGCR mediated at the protein level and its subsequent sustained reductions involving equal contributions at the protein and RNA levels.
3.4. IFN- induces proteasomal degradation of HMGCR
It is well documented that 25-HC can promote the degradation of HMGCR protein through an ubiquitin-proteasome system [16,17]. We hypothesised, therefore, that IFN-, acting through an induction of 25-HC, can also promote the proteasomal degradation of HMGCR protein.
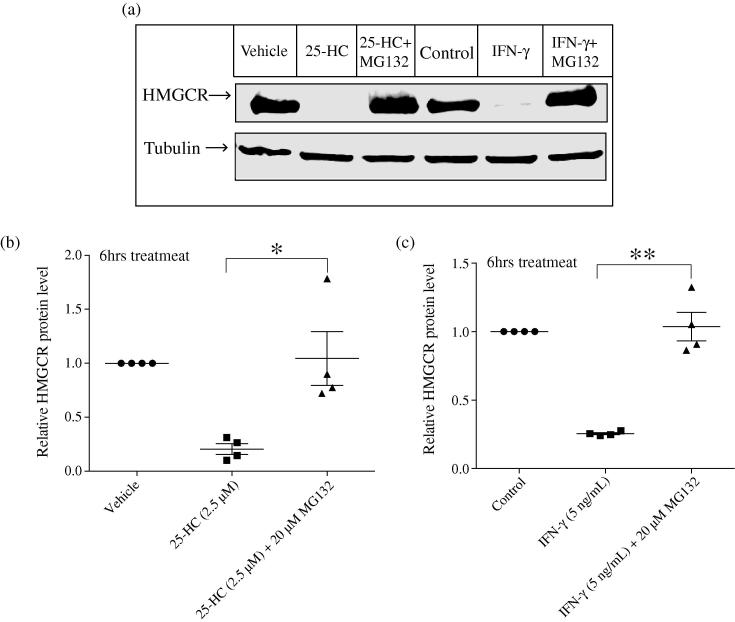
To test whether the reduction of HMGCR protein by IFN- in BMDMs involves proteasomal degradation, we utilised the proteasome specific inhibitor, MG132 [16]. The results shown in Fig. 4a and b demonstrate that MG132 treatment can inhibit the degradation of HMGCR induced by 25-HC as expected. Strikingly, 6 h of IFN- treatment in BMDMs also leads to a significant reduction in HMGCR levels (Fig. 4a and c) that is completely blocked by the proteasome inhibitor, MG132. We conclude from these experiments that 25-HC or IFN- treatment leads to the proteasomal degradation of HMGCR.
Fig. 4.
IFN- induces proteasomal degradation of HMGCR. (a) Wild-type BMDMs were pre-treated with MG132 in Medium C for 1 h and then treated with 25-HC or IFN- in the same culture medium for another 6 h. Western blot was performed to determine HMGCR protein levels. (b) Intensity values of HMGCR to tubulin. As MG132 has an effect on -actin protein levels, tubulin was used as the internal control. Data are mean ± SEM (n = 4). ∗p < 0.05, determined with an unpaired Student’s t test. (c) Intensity values of HMGCR to tubulin. Data are mean ± SEM (n = 4). ∗∗p < 0.01, determined with an unpaired Student’s t test.
3.5. IFN--elicited reduction of HMGCR protein absolutely requires the de novo synthesis of 25-HC
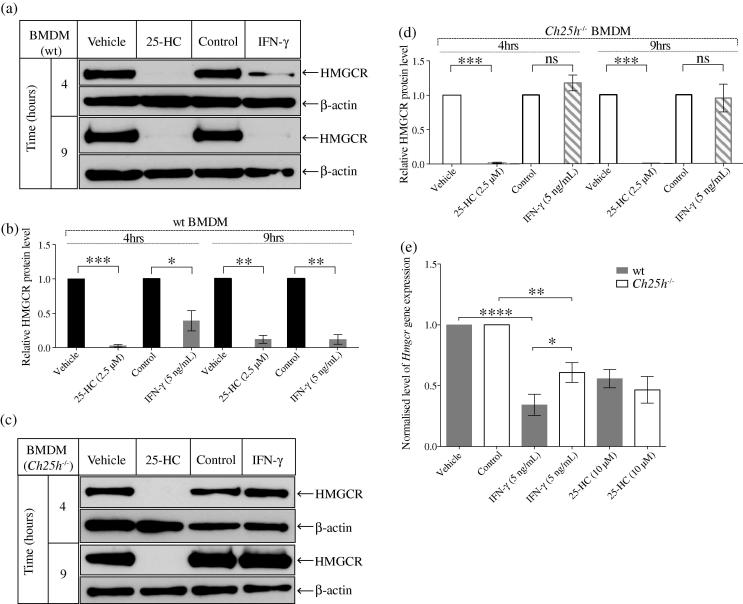
The experiments described above strongly suggest the possibility that IFN- alters levels of HMGCR through de novo synthesis of 25-HC. To unequivocally test this possibility, we utilised BMDMs derived from Ch25h−/− mice. The genetic ablation of Ch25h results in a complete inability of BMDMs from Ch25h−/− mice to produce 25-HC [26]. We anticipated, therefore, that IFN- would be unable to reduce HMGCR abundance in these knockout cells. Accordingly, Ch25h−/− and wild-type BMDMs were treated with, 25-HC (as a positive control) or, IFN- and the levels of HMGCR protein were quantified (Fig. 5a–d). As expected, both 25-HC and IFN- treatment of wild-type BMDMs lead to a greater than 90% decrease in HMGCR protein levels by 9 h post treatment and which temporally reflects both SREBP dependent and independent pathways. However, at earlier times (4 h post-treatment), only a 50% reduction in HMGCR was observed for IFN- treatment in contrast to the nearly 100% reduction observed for 25-HC (Fig. 5a and b). This level of reduction can not be accounted for by the SREBP pathway alone because of the delineated 3–4 h lag period required for optimal induction of CH25H by IFN- and therefore, indicates that the 50% reduction at 4 h of IFN- is mainly due to the SREBP-independent pathway for targeted proteasomal degradation. In Ch25h−/− BMDMs, the reduction in levels of HMGCR by IFN- treatment at both 4 and 9 h were completely abrogated while 25-HC was capable in mediating a decrease of HMGCR protein in these cells at both time points (Fig. 5c and d). These experiments therefore provide direct evidence that the first 9 h of IFN--activation reductions in HMGCR protein abundance require the de novo synthesis of 25-HC by Ch25h.
Fig. 5.
Comparison of Hmgcr mRNA and HMGCR protein levels following 25-HC or IFN- treatment in wild-type and Ch25h−/− BMDMs. (a) Western blot analysis of HMGCR with 2.5 μM of 25-HC or 5 ng/mL of IFN- treatment in Medium C at 4-h and 9-h time points in wild-type BMDMs. (b) Intensity values of HMGCR to -actin. Bars present the mean ± SEM (n = 3). (c) Western blot analysis of HMGCR with 2.5 μM of 25-HC or 5 ng/mL of IFN- treatment in Medium C at 4-h and 9-h time points in Ch25h−/− BMDMs. (d) Intensity values of HMGCR to -actin. Bars present the mean ± SEM (n = 3). (e) HMGCR transcript abundance in wild-type and Ch25h−/− BMDMs with 2.5 μM of 25-HC or 5 ng/mL of IFN- treatment for 24 h in Medium A. The 25-HC-treated groups were normalised by the Vehicle and the IFN--treated groups were normalised by the control treated samples, respectively. Bars present the mean ± SEM (n = 9). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, determined with an unpaired Student’s t test.
As the induction of CH25H is rapidly suppressed we anticipate that the effective time for 25-HC may be limited to the first 12–24 h. Yet we have shown that in IFN activated BMDMs the inhibition of the sterol pathway is maintained for at least a period of 48–72 h [24]. We therefore asked whether the synthesis of 25-HC is also exclusively required for longer-term suppression of HMGCR. Fig. 5e shows the results of experiments measuring Hmgcr RNA levels in wild-type and Ch25h−/− BMDMs treated with IFN- or 25-HC for 24 h. In wild-type BMDMs, an expected 40-50% drop in RNA levels is detected for 25-HC treatment while it is notable for IFN- treatment, Hmgcr levels are further decreased by 60–70%. In Ch25h−/− BMDMs, Hmgcr RNA levels are significantly reduced but statistically higher than the wild-type levels. As a control, in Ch25h−/− BMDMs 25-HC treatment retains a similar level of inhibition of Hmgcr RNA. Therefore, these results demonstrate that while the early protein degradation of HMGCR is completely dependent on the de novo synthesis of 25-HC, the longer-term suppression mediated at the RNA level is only partially (approximately 50%) dependent on this oxysterol.
3.6. A model for the regulatory control of HMGCR by IFN
The above experiments suggest a complex multi-layered regulatory pathway by which IFN- suppresses HMGCR. We have demonstrated that IFN- can transcriptionally and post-translationally regulate HMGCR, mediated through the de novo synthesis of 25-HC. Altogether, our results provide a foundation for further understanding of the complex regulation of the sterol biosynthesis pathway by the immune system through computational modelling. Toward this endeavour, Fig. 6 provides a graphical model for the coordinate regulation of HMGCR by IFN-, constructed using the systems biology graphical notation (SBGN) [54]. Specifically, IFN transcriptionally increases the expression of Ch25h expression via STAT1 , whose translational product catalyses the production of 25-HC. The newly synthesised 25-HC can rapidly promote the proteasomal degradation of HMGCR and retain SREBP2 in the ER, which suppresses the transcription of Hmgcr and with concomitant temporal delay in translation further reduces HMGCR. The induction of Ch25h is curtailed by the repressor ATF3 that also negatively feedback on the IFN response. Therefore, IFN via CH25H regulates both the Hmgcr mRNA and HMGCR protein levels within the first 9 h of treatment through 25-HC mediated SREBP dependent and independent pathways.
4. Discussion
In this study we quantitatively elucidate how IFN- regulates the mevalonate-sterol pathway by targeting HMGCR through both SREBP-dependent and independent pathways. We show that in BMDMs, IFN- elicits a rapid proteasomal degradation of HMGCR. Importantly, the effects of this degradation are later potentiated through transcriptional and translational repression of HMGCR. Further, we demonstrate that IFN- mediates this effect through the de novo induction of CH25H expression and subsequent synthesis of 25-HC. This work provides, therefore, a foundational framework for formal exploitation and advances new mechanistic insights into IFN- mediated macrophage suppression of mevalonate-sterol biosynthesis and the anti-viral response.
The mevalonate-sterol biosynthesis pathway is active in all cells of the body and is responsible for the production of cholesterol, isoprenoids, ubiquinone and downstream metabolites of oxysterols and steroid hormones. These products are involved in a range of core biological processes necessary for cell growth, metabolic and oxidative homeostasis, membrane integrity, vesicular transport and endocrine physiology. In addition, infectious agents, such as microbial, parasitic, and especially viruses, also rely on biosynthetic cellular pathways for their growth [55]. A common feature of many unrelated pathogens is a dependency on cellular lipid metabolism and we, and others, have shown that pharmacological inhibition of the mevalonate-sterol biosynthesis pathway, in particular, can curtail pathogen growth [32,33,24,34]. Increasingly, studies are emerging that the immune system and lipid pathways are tightly coupled. Notably, both upstream and downstream metabolites of this pathway are known to play key roles in immunity and likely explain its long standing association with inflammatory based diseases such as atherosclerosis. Defining how immunity and lipid metabolism are integrated, share resources and cross-regulate one-another during infection is, therefore, of great importance to our understanding of anti-infective immunity and the development of new therapeutic strategies.
Recently, we identified an interferon-induced reduction in sterol biosynthesis as a key component of the very early cellular response to virus-infection in BMDMs [24]. This led to a focus on characterising the close coupling between interferon and mevalonate-sterol metabolism and to the identification of 25-HC biosynthesis as an important component of the interferon-induced anti-viral response in BMDMs [42,24,31]. For over 40 years, 25-HC was recognised to have a potent ability to negatively feedback on the regulation of cholesterol biosynthesis [56]. However, it is only in the last half decade that a physiological role of 25-HC associated with immune-related functions has emerged. These have included the TLR activation of its production in BMDMs and DCs [29], the regulation of immunoglobulin A production [26], as an effector of IFN-response mediating potent cell-autonomous anti-viral immunity in BMDMs [24,31], functional requirement as an immune B-cell guidance cue for adaptive immunity and roles in pro- and anti-inflammatory cytokine production [28,27]. Hence, it is now clear that a key physiological function of 25-HC is as a central effector of immune homeostasis and response to infection.
Our previous studies have shown that 25-HC biosynthesis is regulated by IFN-induced STAT1 binding to the CH25H promoter and, is accordingly dramatically up-regulated in BMDMs in the first hour after infection or IFN activation. Notably, we found that through SREBP-dependent and independent mechanisms, 25-HC can inhibit, in a cell-autonomous manner, a wide range of viruses. The principal pathway for anti-viral activity mediated by 25-HC for a number of viruses consistently maps to the proximal mevalonate arm of the pathway. Importantly, HMGCR represents the first regulatory enzymatic step in this part of the sterol pathway and statins or interference RNA inhibition of HMGCR markedly inhibit viral growth. The role of 25-HC anti-viral pathway in vivo requires further investigation.
How precisely IFN controls the sterol pathway is little understood. Here, in our detailed kinetic and quantitative investigations, to further explore the IFN regulation of sterol biosynthesis, we systematically uncovered evidence that demonstrates a direct coupling between IFN- signalling and the synthesis of 25-HC that mechanistically involves a temporal, multi-layered suppression of the mevalonate arm of the sterol biosynthesis pathway. Within the first 90–200 min of IFN- stimulation of BMDMs, HMGCR is directed by 25-HC for proteasomal degradation and is coordinately sustained for reduced expression through blockade of the restorative SREBP pathway, suppressing both RNA and protein synthesis (Fig. 6). Thus, our findings provide a foundation and framework for future modelling and predictive therapeutic intervention strategies for regulating sterol biosynthesis and its immune effector functions.
Acknowledgements
This work was supported in part by the BBSRC (BB/K019112/1) and BBSRC and EPSRC (SynthSys BB/D019621/1) to P.G. The China Scholarships Council/University of Edinburgh Scholarships supports H.L. We are grateful to Professor David Russell (UT Southwestern, Dallas, Texas, USA) for the supply of the Ch25h−/− mice. We would like to thank Dr. Dick van den Boomen for experimental help and helpful comments, and Marie Craigon and Alan Ross for technical support.
References
- 1.Brown M.S., Goldstein J.L. Multivalent feedback regulation of HMG CoA reductase, a control mechanism coordinating isoprenoid synthesis and cell growth. J Lipid Res. 1980;21(5):505–517. [PubMed] [Google Scholar]
- 2.Goldstein J.L., Brown M.S. Regulation of the mevalonate pathway. Nature. 1990;343(6257):425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 3.Brown M.S., Goldstein J.L. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89(3):331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 4.Goldstein J.L., DeBose-Boyd R.A., Brown M.S. Protein sensors for membrane sterols. Cell. 2006;124(1):35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 5.DeBose-Boyd R.A. Feedback regulation of cholesterol synthesis: sterol-accelerated ubiquitination and degradation of HMG CoA reductase. Cell Res. 2008;18(6):609–621. doi: 10.1038/cr.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang T., Espenshade P.J., Wright M.E., Yabe D., Gong Y. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell. 2002;110(4):489–500. doi: 10.1016/s0092-8674(02)00872-3. [DOI] [PubMed] [Google Scholar]
- 7.Adams C.M., Goldstein J.L., Brown M.S. Cholesterol-induced conformational change in SCAP enhanced by Insig proteins and mimicked by cationic amphiphiles. Proc Natl Acad Sci. 2003;100(19):10647–10652. doi: 10.1073/pnas.1534833100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Radhakrishnan A., Sun L.-P., Kwon H.J., Brown M.S., Goldstein J.L. Direct binding of cholesterol to the purified membrane region of SCAP mechanism for a sterol-sensing domain. Mol Cell. 2004;15(2):259–268. doi: 10.1016/j.molcel.2004.06.019. [DOI] [PubMed] [Google Scholar]
- 9.Sun L.-P., Li L., Goldstein J.L., Brown M.S. Insig required for sterol-mediated inhibition of Scap/SREBP binding to COPII proteins in vitro. J Biol Chem. 2005;280(28):26483–26490. doi: 10.1074/jbc.M504041200. [DOI] [PubMed] [Google Scholar]
- 10.Hua X., Nohturfft A., Goldstein J.L., Brown M.S. Sterol Resistance in CHO cells traced to point mutation in SREBP cleavage-activating protein. Cell. 1996;87(3):415–426. doi: 10.1016/s0092-8674(00)81362-8. [DOI] [PubMed] [Google Scholar]
- 11.Espenshade P.J., Cheng D., Goldstein J.L., Brown M.S. Autocatalytic processing of site-1 protease removes propeptide and permits cleavage of sterol regulatory element-binding proteins. J Biol Chem. 1999;274(32):22795–22804. doi: 10.1074/jbc.274.32.22795. [DOI] [PubMed] [Google Scholar]
- 12.Zelenski N.G., Rawson R.B., Brown M.S., Goldstein J.L. Membrane topology of S2P, a protein required for intramembranous cleavage of sterol regulatory element-binding proteins. J Biol Chem. 1999;274(31):21973–21980. doi: 10.1074/jbc.274.31.21973. [DOI] [PubMed] [Google Scholar]
- 13.Nohturfft A., Yabe D., Goldstein J.L., Brown M.S., Espenshade P.J. Regulated step in cholesterol feedback localized to budding of SCAP from ER membranes. Cell. 2000;102(3):315–323. doi: 10.1016/s0092-8674(00)00037-4. [DOI] [PubMed] [Google Scholar]
- 14.Yang T., Goldstein J.L., Brown M.S. Overexpression of membrane domain of SCAP prevents sterols from inhibiting SCAP. SREBP exit from endoplasmic reticulum. J Biol Chem. 2000;275(38):29881–29886. doi: 10.1074/jbc.M005439200. [DOI] [PubMed] [Google Scholar]
- 15.Edwards P. Regulation of gene expression by SREBP and SCAP. Biochim Biophys Acta. 2000;1529(1–3):103–113. doi: 10.1016/s1388-1981(00)00140-2. [DOI] [PubMed] [Google Scholar]
- 16.Ravid T., Doolman R., Avner R., Harats D., Roitelman J. The ubiquitin-proteasome pathway mediates the regulated degradation of mammalian 3-hydroxy-3-methylglutaryl-coenzyme A reductase. J Biol Chem. 2000;275(46):35840–35847. doi: 10.1074/jbc.M004793200. [DOI] [PubMed] [Google Scholar]
- 17.Song B.-L., DeBose-Boyd R.A. Ubiquitination of 3-hydroxy-3-methylglutaryl-CoA reductase in permeabilized cells mediated by cytosolic E1 and a putative membrane-bound ubiquitin ligase. J Biol Chem. 2004;279(27):28798–28806. doi: 10.1074/jbc.M402442200. [DOI] [PubMed] [Google Scholar]
- 18.Tsai Y.C., Leichner G.S., Pearce M.M.P., Wilson G.L., Wojcikiewicz R.J.H., Roitelman J., Weissman A.M. Differential regulation of HMG-CoA reductase and Insig-1 by enzymes of the ubiquitin-proteasome system. Mol Biol Cell. 2012;23(23):4484–4494. doi: 10.1091/mbc.E12-08-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gil G., Faust J.R., Chin D.J., Goldstein J.L., Brown M.S. Membrane-bound domain of HMG CoA reductase is required for sterol-enhanced degradation of the enzyme. Cell. 1985;41(1):249–258. doi: 10.1016/0092-8674(85)90078-9. [DOI] [PubMed] [Google Scholar]
- 20.Faust J.R., Luskey K.L., Chin D.J., Goldstein J.L., Brown M.S. Regulation of synthesis and degradation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase by low density lipoprotein and 25-hydroxycholesterol in UT-1 cells. Proc Natl Acad Sci. 1982;79(17):5205–5209. doi: 10.1073/pnas.79.17.5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sever N., Yang T., Brown M.S., Goldstein J.L., Debose-Boyd R.A. Accelerated degradation of HMG CoA reductase mediated by binding of Insig-1 to Its sterol-sensing domain. Mol Cell. 2003;11(1):25–33. doi: 10.1016/s1097-2765(02)00822-5. [DOI] [PubMed] [Google Scholar]
- 22.Brown M.S., Goldstein J.L. Cholesterol feedback: from Schoenheimer’s bottle to Scap’s MELADL. J Lipid Res. 2009;50(Supplement):S15–S27. doi: 10.1194/jlr.R800054-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cyster J.G., Dang E.V., Reboldi A., Yi T. 25-Hydroxycholesterols in innate and adaptive immunity. Nat Rev Immunol. 2014;14(11):731–743. doi: 10.1038/nri3755. [DOI] [PubMed] [Google Scholar]
- 24.Blanc M., Hsieh W.Y., Robertson K.A., Watterson S., Shui G., Lacaze P. Host defense against viral infection involves interferon mediated down-regulation of sterol biosynthesis. PLoS Biol. 2011;9(3):e1000598. doi: 10.1371/journal.pbio.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ecker J., Liebisch G., Englmaier M., Grandl M., Robenek H., Schmitz G. Induction of fatty acid synthesis is a key requirement for phagocytic differentiation of human monocytes. 2010;107(17):7817–7822. doi: 10.1073/pnas.0912059107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bauman D.R., Bitmansour A.D., McDonald J.G., Thompson B.M., Liang G., Russell D.W. 25-Hydroxycholesterol secreted by macrophages in response to Toll-like receptor activation suppresses immunoglobulin A production. Proc Natl Acad Sci. 2009;106(39):16764–16769. doi: 10.1073/pnas.0909142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reboldi A., Dang E.V., McDonald J.G., Liang G., Russell D.W., Cyster J.G. Inflammation. 25-Hydroxycholesterol suppresses interleukin-1-driven inflammation downstream of type I interferon. Science. 2014;345(6197):679–684. doi: 10.1126/science.1254790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gold E.S., Diercks A.H., Podolsky I., Podyminogin R.L., Askovich P.S., Treuting P.M. 25-Hydroxycholesterol acts as an amplifier of inflammatory signaling. Proc Natl Acad Sci USA. 2014;111(29):10666–10671. doi: 10.1073/pnas.1404271111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park K., Scott A.L. Cholesterol 25-hydroxylase production by dendritic cells and macrophages is regulated by type I interferons. J Leukoc Biol. 2010;88(6):1081–1087. doi: 10.1189/jlb.0610318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blanc M., Hsieh W.Y., Robertson K.A., Kropp K.A., Forster T., Shui G. The transcription factor STAT-1 couples macrophage synthesis of 25-Hydroxycholesterol to the interferon antiviral response. Immunity. 2013;38(1):106–118. doi: 10.1016/j.immuni.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu S.-Y., Aliyari R., Chikere K., Li G., Marsden M.D., Smith J.K. Interferon-inducible cholesterol-25-hydroxylase broadly inhibits viral entry by production of 25-hydroxycholesterol. Immunity. 2013;38(1):92–105. doi: 10.1016/j.immuni.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.del Real G. Statins inhibit HIV-1 infection by down-regulating rho activity. J Exp Med. 2004;200(4):541–547. doi: 10.1084/jem.20040061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ikeda M., Abe K.-I., Yamada M., Dansako H., Naka K., Kato N. Different anti-HCV profiles of statins and their potential for combination therapy with interferon. Hepatology. 2006;44(1):117–125. doi: 10.1002/hep.21232. [DOI] [PubMed] [Google Scholar]
- 34.Peng J., Zhang D., Ma Y., Wang G., Guo Z., Lu J. Protective effect of fluvastatin on influenza virus infection. Mol Med Rep. 2014;9(6):2221–2226. doi: 10.3892/mmr.2014.2076. [DOI] [PubMed] [Google Scholar]
- 35.Kwak B., Mulhaupt F., Myit S., Mach F. Statins as a newly recognized type of immunomodulator. Nat Med. 2000;6(12):1399–1402. doi: 10.1038/82219. [DOI] [PubMed] [Google Scholar]
- 36.Dolken L., Ruzsics Z., Radle B., Friedel C.C., Zimmer R., Mages J. High-resolution gene expression profiling for simultaneous kinetic – PubMed – NCBI. RNA. 2008;14(9):1959–1972. doi: 10.1261/rna.1136108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Irizarry R.A., Bolstad B.M., Collin F., Cope L.M., Hobbs B., Speed T.P. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31(4):e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Conesa A., Nueda M.J., Ferrer A., Talón M. maSigPro: a method to identify significantly differential expression profiles in time-course microarray experiments. Bioinformatics (Oxford, England) 2006;22(9):1096–1102. doi: 10.1093/bioinformatics/btl056. [DOI] [PubMed] [Google Scholar]
- 39.Friedel C.C., Dölken L., Ruzsics Z., Koszinowski U.H., Zimmer R. Conserved principles of mammalian transcriptional regulation revealed by RNA half-life. Nucleic Acids Res. 2009;37(17):e115–e125. doi: 10.1093/nar/gkp542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2013. R: a language and environment for statistical computing. http://www.R-project.org/ [Google Scholar]
- 41.MATLAB, version 7.10.0, Natick, Massachusetts: The MathWorks Inc.; 2010.
- 42.Shibata N., Carlin A.F., Spann N.J., Saijo K., Morello C.S., McDonald J.G. 25-Hydroxycholesterol activates the integrated stress response to reprogram transcription and translation in macrophages. J Biol Chem. 2013;288(50):35812–35823. doi: 10.1074/jbc.M113.519637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lipopolysaccharide-induced expression of matrix metalloproteinases in human monocytes is suppressed by IFN-gamma via superinduction of ATF-3 and suppression of AP-1. J Imunol (Baltimore, MD: 1950) 2008;181(7):5089–5097. doi: 10.4049/jimmunol.181.7.5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.PIKfyve, a class III lipid kinase, is required for TLR-induced type I IFN production via modulation of ATF3. J Immunol (Baltimore, MD: 1950) 2014;192(7):3383–3389. doi: 10.4049/jimmunol.1302411. [DOI] [PubMed] [Google Scholar]
- 45.Gold E.S., Ramsey S.A., Sartain M.J., Selinummi J., Podolsky I., Rodriguez D.J. ATF3 protects against atherosclerosis by suppressing 25-hydroxycholesterol-induced lipid body formation. J Exp Med. 2012;209(4):807–817. doi: 10.1084/jem.20111202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Drysdale B.E., Howard D.L., Johnson R.J. Identification of a lipopolysaccharide inducible transcription factor in murine macrophages. Mol Immunol. 1996;33(11–12):989–998. doi: 10.1016/s0161-5890(96)00043-0. [DOI] [PubMed] [Google Scholar]
- 47.Taylor M.W., Grosse W.M., Schaley J.E., Sanda C., Wu X., Chien S.-C. Global effect of PEG-IFN-alpha and ribavirin on gene expression in – PubMed – NCBI. J Interferon Cytokine Res. 2004;24(2):107–118. doi: 10.1089/107999004322813354. [DOI] [PubMed] [Google Scholar]
- 48.Rosenberger C.M., Clark A.E., Treuting P.M., Johnson C.D., Aderem A. ATF3 regulates MCMV infection in mice by modulating IFN-gamma expre- PubMed - NCBI. Proc Natl Acad Sci. 2008;105(7):2544–2549. doi: 10.1073/pnas.0712182105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gilchrist M., Thorsson V., Li B., Rust A.G., Korb M., Kennedy K. Systems biology approaches identify ATF3 as a negative regulator of Toll-like receptor 4. Nature. 2006;441(7090):173–178. doi: 10.1038/nature04768. [DOI] [PubMed] [Google Scholar]
- 50.Stokker G.E., Hoffman W.F., Alberts A.W., Cragoe E.J., Deana A.A., Gilfillan J.L. 3-Hydroxy-3-methylglutaryl-coenzyme A reductase inhibitors. I. Structural modification of 5-substituted 3,5-dihydroxypentanoic acids and their lactone derivatives. J Med Chem. 1985;28(3):347–358. doi: 10.1021/jm00381a014. [DOI] [PubMed] [Google Scholar]
- 51.Roth B.D., Blankley C.J., Chucholowski A.W., Ferguson E., Hoefle M.L., Ortwine D.F. Inhibitors of cholesterol biosynthesis. 3. Tetrahydro-4-hydroxy-6-[2-(1H-pyrrol-1-yl)ethyl]-2H-pyran 2-one inhibitors of HMG-CoA reductase. 2. Effects of introducing substituents at positions three and four of the pyrrole nucleus. J Med Chem. 1991;34(1):357–366. doi: 10.1021/jm00105a056. [DOI] [PubMed] [Google Scholar]
- 52.Coppola G.M., Damon R.E., Yu H., Engstrom R.G., Scallen T.J. Design and biological evaluation of a series of thiophene-based 3-hydroxy-3-methylglutaryl coenzyme a reductase inhibitors. Bioorg Med Chem Lett. 1997;7(5):549–554. [Google Scholar]
- 53.Hosoda S., Matsuda D., Tomoda H., Hashimoto M., Aoyama H., Hashimoto Y. Application of a 3,3-diphenylpentane skeleton as a multi-template for creation of HMG-CoA reductase inhibitors. Bioorg Med Chem Lett. 2009;19(15):4228–4231. doi: 10.1016/j.bmcl.2009.05.100. [DOI] [PubMed] [Google Scholar]
- 54.Le Novère N., Hucka M., Mi H., Moodie S., Schreiber F., Sorokin A. The systems biology graphical notation. Nat Biotechnol. 2009;27(8):735–741. doi: 10.1038/nbt.1558. [DOI] [PubMed] [Google Scholar]
- 55.Ghazal P., González Armas J.C., Kurz S., Angulo A. Viruses: hostages to the cell. Virology. 2000;275(2):233–237. doi: 10.1006/viro.2000.0553. [DOI] [PubMed] [Google Scholar]
- 56.Kandutsch A.A., Chen H.W. Inhibition of sterol synthesis in cultured mouse cells by cholesterol derivatives oxygenated in the side chain. J Biol Chem. 1974;249(19):6057–6061. [PubMed] [Google Scholar]