Abstract
The chromatoid body is a granule-like structure of male germ cells, containing many proteins and RNAs, and is important for spermatogenesis. However, the molecular mechanisms for the formation and function of the chromatoid body are still elusive. Here, we report that Ca2+/calmodulin-dependent protein kinase IV (CaMKIV) accumulates in the chromatoid body by immunofluorescence staining, indicating that CaMKIV is a new component of the chromatoid body. Furthermore, we find that CaMKIV can interplay with the other components of the chromatoid body by immunoprecipitation: mouse VASA homologue (MVH), mouse homologue of PIWI, PIWIL1 (MIWI), and kinesin KIF17b. Importantly, interplay between KIF17b and MVH or MIWI can be potentially regulated by CaMKIV. These results imply that CaMKIV plays a role in maintenance the structure of chromatoid body by regulating the associations of proteins in it.
The chromatoid body was first described by Benda in 18911 and, since then, it has attracted the interest of many researchers. Chromatoid bodies can be detected in the cytoplasm of meiotic spermatocytes and are characterized as fibrous-granular structures that are formed between mitochondria clusters2. After meiosis, a mature chromatoid body appears, and the fibrous-granular structure is compacted into a finely filamentous and lobulated granule, bouncing around at the surface of the nucleus of round spermatid2,3. This structure remains in the cytoplasm of the spermatid until the nucleus begins to elongate and finally disappears late in spermiogenesis2,4.
Chromatoid bodies contain many RNA-binding proteins and RNA strands5,6, and on the basis of its structural features and composition, it is considered as a specialized form of germplasm or nuage7. Thus, chromatoid bodies are proposed as RNA-processing centers of male germ cells5. Mouse VASA homologue (MVH), a DEAD-box RNA helicase, localizes in the chromatoid body8,9 and regulates RNA granules10. Another RNA-binding protein, mouse homologue of PIWI, PIWIL1 (MIWI) also localizes in the chromatoid body8 and physically interacts with MVH11. MIWI is an important component of the chromatoid body, because Miwi-null mice lack the normal structural features of a chromatoid body, and the fibrous-granular structure fails to condense into the mature chromatoid body12. Chromatoid bodies also contain many non-RNA-binding proteins, and kinesin KIF17b is one of them. KIF17b has been shown to regulate spermatids transcription activity by shuttling between the nucleus and the cytoplasm13,14. In the cytoplasm of spermatids, KIF17b accumulates in the chromatoid body and associates with MIWI, thus KIF17b may be related to the movement of the chromatoid body12.
Ca2+/calmodulin-dependent protein kinase IV (CaMKIV) is expressed in the testes, brain, thymus and other tissues15. In mouse testes, CaMKIV is expressed in spermatogonia and spermatids but not spermatocytes16, and spermiogenesis is impaired in mice lacking CaMKIV17. Here we report that CaMKIV is localized in the chromatoid body and interacts with the components of the chromatoid body: MVH, MIWI and KIF17b. Both MVH and MIWI bind to the motif of Arg–Lys–Lys–Ser at KIF17b, which is also the substrate recognition motif of CaMKIV18. And more importantly, CaMKIV can regulate the interplay of KIF17b between MVH and MIWI. These results indicate that CaMKIV is a new component of the chromatoid body and plays an important role in maintaining the structure of chromatoid body through regulating interactions of proteins.
Results
CaMKIV is localized in the chromatoid body
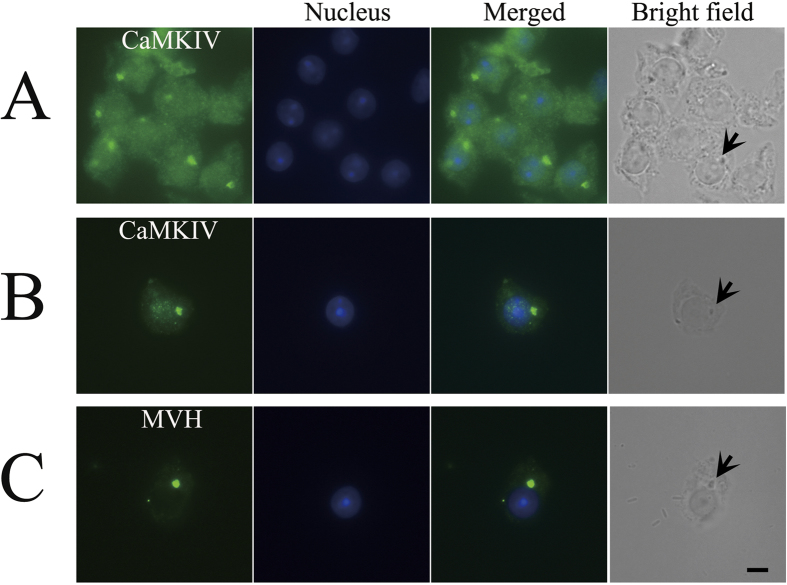
It has been reported that CaMKIV shuttles between the nuclear and cytoplasmic compartments of RAW 264.7 cells19, which indicates that CaMKIV might be localized in the cytoplasmic compartments in germ cells, such as the localization of KIF17b in mouse testes20. Immunofluorescence was carried out on squash preparations to study the localization of CaMKIV in great detail. Interestingly, we found that, in the cytoplasm of spermatids, the signal condensed into one area (Fig. 1A,B) and overlapped with the phase-contrast image of the chromatoid body (Fig. 1A,B). MVH antibody was used as a positive control for chromatoid-body staining (Fig. 1C). These results show that CaMKIV is a new component of the chromatoid body.
Figure 1. Localization of CaMKIV in the chromatoid body.
Testes of adult mice were subjected to squash preparation and then immunofluorescence staining. (A,B) The round spermatids were immunostained with anti-CaMKIV antibody (green). (C) Immunofluorescence staining of MVH (green). The parallel bright-field images demonstrate the location of the chromatoid bodies, which are indicated by arrows. Alexa Fluor 488 anti-rabbit IgG was used as a secondary antibody, and nuclei were stained blue with Hoechst 33342 dye. Scale bar: 5 μm.
CaMKIV interacts with the MVH and MIWI, components of the chromatoid body
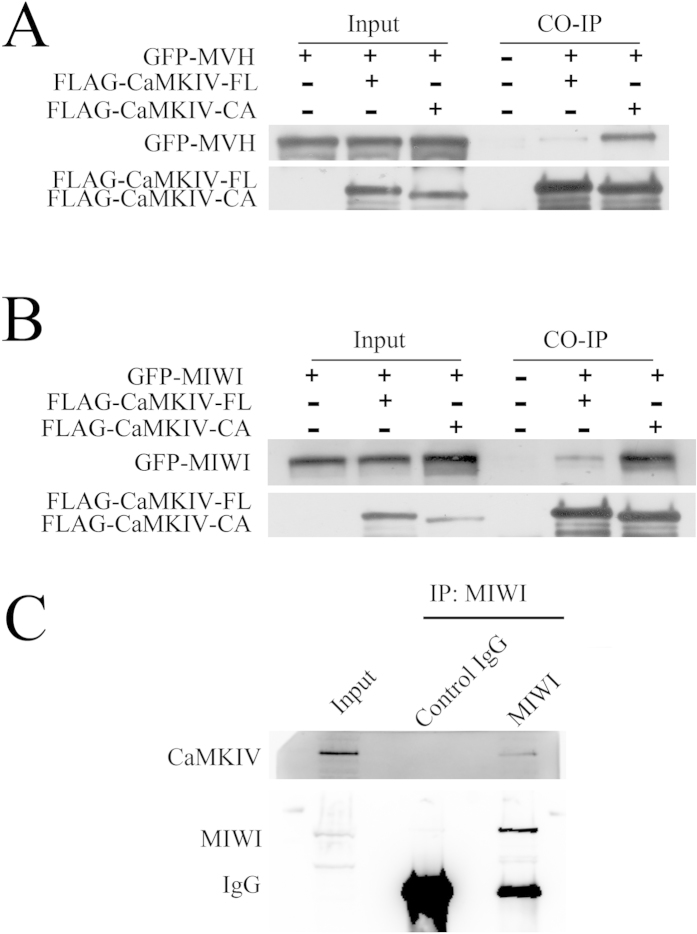
As CaMKIV is a member of the chromatoid body, we guess that CaMKIV may interplay with MVH and MIWI. To confirm this, co-immunoprecipitation experiments were carried out after the co-expression of FLAG-CaMKIV-FL (the full length of CaMKIV) or FLAG-CaMKIV-CA (the active form of CaMKIV) with GFP-MVH or GFP-MIWI in HEK293T cells. FLAG-CaMKIV-FL and FLAG-CaMKIV-CA were immunoprecipitated using FLAG antibody, immunoblotting with GFP antibody revealed that GFP-MVH and GFP-MIWI were co-precipitated with FLAG-CaMKIV-FL, and the association was greatly increased when FLAG-CaMKIV-CA was co-expressed with GFP-MVH or GFP-MIWI (Fig. 2A,B). Mouse testes were immunoprecipitated by MIWI antibody, immunoblotting with CaMKIV antibody showed that CaMKIV associated with MIWI (Fig. 2C). These data show that CaMKIV is a new member of the chromatoid body and associates with MVH and MIWI.
Figure 2. Interaction of CaMKIV with MVH and MIWI.
(A) In HEK293T cells, plasmids of FLAG-CaMKIV-FL (the full length of CaMKIV) or FLAG-CaMKIV-CA (constitutively active form of CaMKIV) were co-transfected with GFP-MVH; (B) plasmids of FLAG-CaMKIV-FL or FLAG-CaMKIV-CA were transfected with GFP-MIWI. Immunoprecipitation from the cell lysates was performed with anti-mouse FLAG antibody, and the samples were immunoblotted by anti-mouse GFP antibody to detect MVH or MIWI, and then by anti-mouse FLAG antibody to detect coimmunoprecipitated CaMKIV. (C) Mouse testes were immunoprecipitated by anti-rabbit MIWI antibody, and the samples were immunoblotted by anti-rabbit MIWI antibody, then by anti-mouse CaMKIV antibody to detect coimmunoprecipitated CaMKIV. Anti-rabbit GFP antibody was used as a negative control.
CaMKIV interplays with another component of the chromatoid body: KIF17b
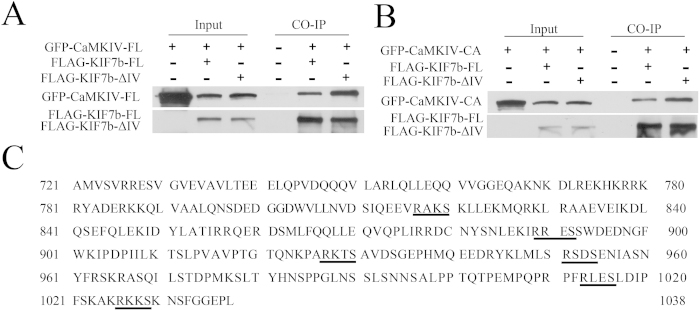
The R-K-K-S sequence is the substrate recognition motif of CaMKII/CaMKIV18, and CaMKII can bind to the Arg-Lys-Lys-Ser (R-K-K-S) sequence of KIF17 in the mouse brain21. Therefore, it is possible that CaMKIV may associate with KIF17b, both of which are components of the chromatoid body. In addition, the Germ Online database was used to analyze the expression of CaMKII in the testes22 and found that CaMKII had a very low expression level in spermatids; therefore, we focused on CaMKIV in this study. As the R-K-K-S sequence is the substrate-recognition motif of CaMKIV, the mutant form of KIF17b (FLAG-KIF17b-ΔIV) with R-K-K-S deletion or the full length of KIF17b (FLAG-KIF17b-FL) was co-transfected with GFP-CaMKIV-FL or GFP-CaMKIV-CA in HEK293T cells. The cell lysate was incubated with the FLAG antibody. Immunoblotting with GFP antibody revealed that FLAG-KIF17b-FL co-precipitated with GFP-CaMKIV-FL and GFP-CaMKIV-CA (Fig. 3A,B). However, the deletion of the R-K-K-S sequence did not reduce the association of KIF17b with CaMKIV (Fig. 3A,B). We analyzed the amino-acid sequence of KIF17b and found that there were multiple R-X-X-S/T motifs at the C-terminal domain (CTD) of KIF17b (Fig. 3C), which were potential binding sites for CaMKIV18; therefore, the deletion of the R-K-K-S sequence did not decrease the interaction of KIF17b with CaMKIV. These data show that KIF17b has multiple binding sites for CaMKIV.
Figure 3. Interaction of CaMKIV with KIF17b.
Plasmids encoding the full length of KIF17b (FLAG-KIF17b-FL) or the mutant form of KIF17b with R-K-K-S deletion (FLAG-KIF17b-ΔIV) were co-expressed with (A) GFP-CaMKIV-FL or (B) GFP-CaMKIV-CA in HEK293T cells. The cell lysates were incubated with anti-mouse FLAG antibody and CaMKIV was immunoblotted by anti-mouse GFP antibody, whereas KIF17b was detected by anti-mouse FLAG antibody, (C) The R-X-X-S/T motifs at the CTD of KIF17b are underlined.
MVH and MIWI associate with the tail of KIF17b
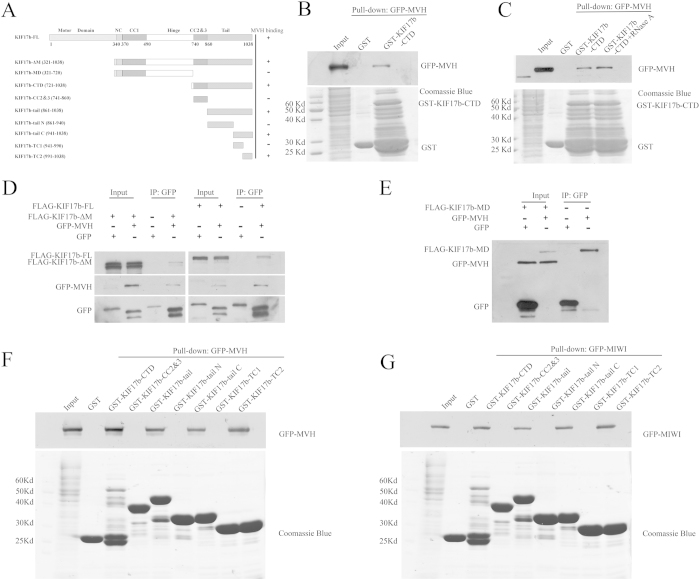
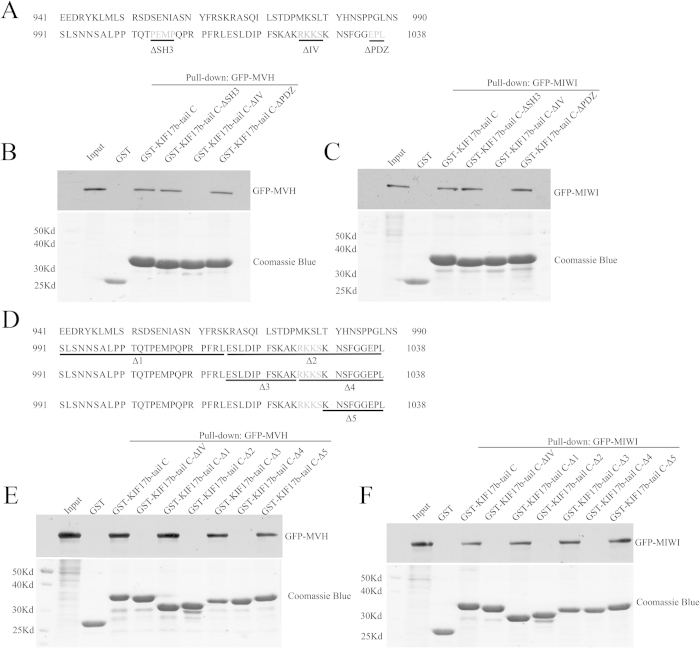
It has been reported that the tail of KIF17 had a cargo binding site and that the cargo could be released by controlling CaMKII21. Therefore, we assumed that the tail of KIF17b had some cargo in which the interactions with KIF17b could be regulated by CaMKIV. To confirm this, a GST pull-down assay was performed to identify the cargoes of KIF17b in testis using the CTD of KIF17b as bait. The truncations of KIF17b are shown in Fig. 4A. A new cargo of KIF17b was identified by 1D LC-MS. The new cargo is MVH, which is also a component of the chromatoid body23. The association was confirmed by the GST pull-down assay (Fig. 4B), and RNase A treatment showed that the association was RNA-independent (Fig. 4C). The interaction of KIF17b with MVH was confirmed by co-immunoprecipitation experiment (Fig. 4D). The motor domain of KIF17b was not necessary for MVH binding at, since the mutant form of KIF17b with deletion of the motor domain (FLAG-KIF17b-ΔM) was still immunoprecipited by GFP-MVH (Fig. 4D), but the middle domain of KIF17b (KIF17b-MD) did not have MVH binding activity (Fig. 4E). These data indicate that MVH mainly associates with the CTD of KIF17b. Then, we mapped the interactions between MVH and KIF17b CTD using constructs that encoded the truncated KIF17b (KIF17b-CC2&3, KIF17b-tail, KIF17b-tail N, KIF17b-tail C, KIF17b-TC1, and KIF17b-TC2) with the GST tag. Purified GST-fusion of KIF17b truncations were used to pull-down 293T cell lysates that were transfected with GFP-MVH and the truncations of KIF17b-tail, KIF17b-tail C, and KIF17b-TC2 associated with MVH (Fig. 4F). As MIWI also interplays with KIF17b14, the GST pull-down assay was performed in order to test the interaction of MIWI with KIF17b by GSTKIF17b-CTD and found that MIWI could interact with the CTD of KIF17b (Fig. 4G). While using the CTD truncations of KIF17b, GST-pull down assay found that KIF17b-tail C and KIF17b-TC2 were associated with MIWI (Fig. 4G), which is the same as MVH. The results of the GST pull-down assay show that the 991–1038 amino acids of KIF17b (KIF17b-TC2) play an important role in the association of MVH, MIWI, and KIF17b. These data indicate that MVH and MIWI are both the cargo of KIF17b and that they both bind to the 991–1038 amino acids of the KIF17b tail.
Figure 4. Interaction of MVH and MIWI with the tail of KIF17b.
(A) Schematic diagram of the full length and truncations of KIF17b; the residue numbers at the domain boundaries are indicated. (B,C) The interaction of KIF17b-CTD with MVH was assayed by the GST pull-down assay. Purified GST-KIF17b was used to absorb GFP-MVH from the lysates of HEK293T cells. The bound materials were then subjected to SDS-PAGE and immunoblotted with anti-GFP antibody or treated with RNase A before SDS-PAGE. The GST-fusion proteins were shown by Coomassie brilliant blue R-250 staining of the gels. (D) Immunoprecipitation of KIF17b by MVH. Expression plasmid of the full length of FLAG-tagged KIF17b, or plasmid of the motor-domain-deleted form of FLAG-tagged KIF17b (FLAG-KIF17b-ΔM) and GFP-tagged MVH were co-transfected to HEK293T cells, and the cell lysates were precipitated by the anti-rabbit GFP antibody, and immunoblotted with anti-mouse GFP antibody or anti-mouse FLAG antibody. (E) Plasmids of the middle domain of KIF17b (FLAG-KIF17b-MD) and GFP-MVH were transfected into HEK293T cells and then the cell lysates were immunoprecipitated by the GFP antibody. (F,G) Mapping of the MVH and MIWI binding sites of KIF17b-CTD was completed using the GST pull-down assay. Purified truncations of GST-KIF17b-CTD were used to precipitate GFP-MVH or GFP-MIWI from the lysates of HEK293T cells. The precipitates were then subjected to SDS-PAGE and immunoblotted with anti-GFP antibody; the gels were then stained by Coomassie brilliant blue R-250.
The sequence of R-K-K-S is the binding site of MVH and MIWI at KIF17b
Previous research has shown that KIF17b-TC2 contained the SH3, CaMKII/IV, and PDZ binding sites21. To identify the exact binding site, GST-fusion of KIF17b-tail C with deletion of the SH3 binding site (GST-KIF17b-tail C-ΔSH3), CaMKIV binding site (GST-KIF17b-tail C-ΔIV), or PDZ binding site (GST-KIF17b-tail C-ΔPZD) (Fig. 5A) were used to pull-down HEK293T cell lysate transfected with GFP-MVH or GFP-MWI. We found that the deletion of the CaMKIV binding site abolished the association of MVH and MIWI between KIF17b, whereas deletion of the SH3 and PDZ binding sites did not affect the association (Fig. 5B,C). Furthermore, truncations of KIF17b-tail C (Fig. 5D) were used to perform the GST pull-down assay, and only the R-K-K-S sequence deletion affected the association of MVH and MIWI between KIF17b (Fig. 5E,F). These data show that MVH and MIWI both bind to the R-K-K-S motif of KIF17b.
Figure 5. The sequence of R-K-K-S is the binding site of MVH and MIWI at KIF17b.
(A) Schematic diagram of the mutations of KIF17b-tail C; the SH3, CaMKII/IV, and PDZ binding sites are underlined and gray. (B,C) Mapping of the MVH and MIWI binding sites of KIF17b-tail C, completed using the GST pull-down assay. (D) Schematic diagram of the mutations of KIF17b-tail C; the mutated sequence is underlined and the CaMKII/IV binding sites are gray. (E,F) The GST pull-down assay was used to certify the binding sites of MVH and MIWI at KIF17b-tail C.
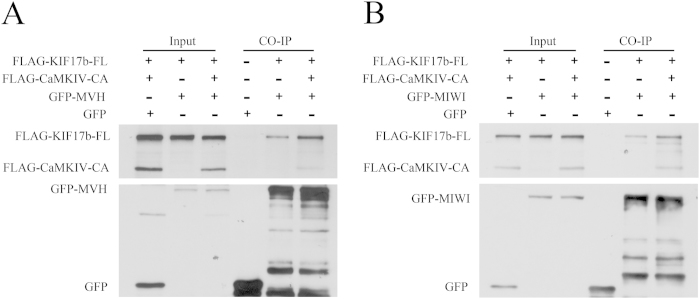
CaMKIV stimulates the association of MVH and MIWI between KIF17b
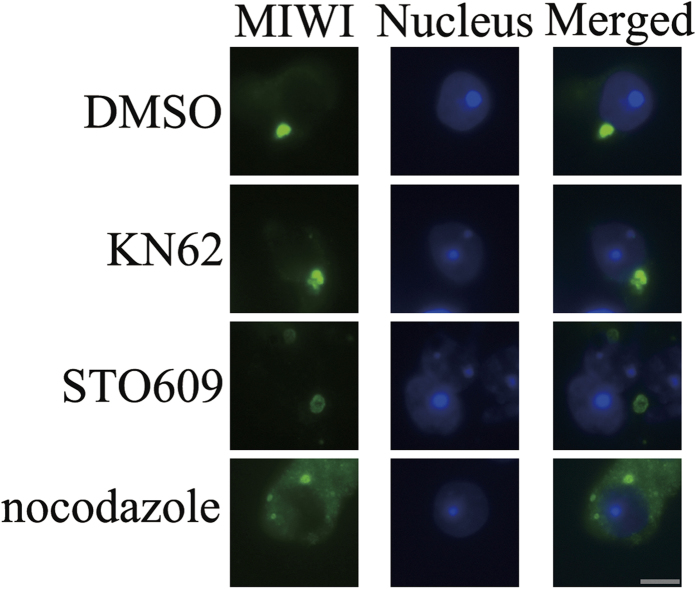
It has been reported that cargo-release from KIF17 was regulated by CaMKII in mouse brain21. Therefore, it is important to find out whether CaMKIV can affect the association of MVH and MIWI between KIF17b. To confirm this, a co-immunoprecipitation experiment was carried out after co-expression of FLAGKIF17b-FL, FLAG-CaMKIV-CA, and GFPMVH or GFP-MIWI in HEK293T cells. The cell lysate was incubated with GFP antibody. Immunoblotting with FLAG antibody revealed that the FLAG-CaMKIV significantly stimulated the association between FLAG-KIF17b-FL and GFP-MVH, as well as FLAG-KIF17b-FL and GFP-MIWI (Fig. 6A,B). To confirm these results, seminiferous tubules were incubated for 48 h with microtubule inhibitor, nocodazole24 and CaMKIV inhibitors, KN62 and STO60919,25,26. Then samples were subjected to immunofluorescence by squash preparation and the chromatoid body was indicated by staining of MIWI. As showed in Fig. 7, chromatoid body was less compacted after incubation with KN62 or STO609, while it disintegrated to form several small spheres after incubation with nocodazole as reported before24. These data indicate that CaMKIV maintains the compact structure of chromatoid body by promoting the associations of MVH and MIWI with KIF17b.
Figure 6. CaMKIV stimulates the association of MVH and MIWI with KIF17b.
HEK293T cells were transfected with the indicated combinations of expression plasmids for FLAG-KIF17b-FL, GFP-CaMKIV-CA, GFP-MVH, and GFP-MIWI. The cell lysates were precipitated by anti-mouse GFP antibody. KIF17b and CaMKIV were immunoblotted by anti-mouse FLAG antibody, whereas MVH and MIWI were immunoblotted by anti-mouse GFP antibody. The total DNA in each group was kept constant using an empty FLAG plasmid.
Figure 7. Inhibitors of CaMKIV affect the structure of chromatoid body.
Seminiferous tubules were treated with with 25 μl/ml DMSO, 60 μM KN62, 10 μM STO609, 20 μg/ml nocodazole for 48 h. DMSO was used as a negative control; CaMKIV inhibitors, KN62 and STO609, were used to treat the samples; microtubule inhibitor, nocodazole, was used as a positive control. After incubation, tubules were performed to immunofluorescence by squash preparation, the chromatoid body was stained by MIWI, Alexa Fluor 488 anti-rabbit IgG was used as a secondary antibody, and nuclei were stained blue with Hoechst 33342 dye. Scale bar: 5 μm.
Discussion
Spermatogenesis is a dynamic and well organized process and is supported by Sertoli cells27. This process is regulated by specialized genetic and epigenetic pathways of gene regulation28. During the late steps of spermatogenesis, transcription of the haploid genome is silenced by the compaction of the haploid genome through histone-to-protamine transition29,30. However, during these steps, the protein synthesis of a large number of specific genes is still ongoing, which are required for the last steps of sperm development31. Thus, mRNA storage and processing are crucial during these steps and, interestingly, many mRNA binding proteins have been identified in male germ cells, regulating the stability and translation of target mRNA strands32,33.
Recent research findings support the hypothesis that the chromatoid body serves as an RNA processing center for male germ cells, based on its structural features and composition5,10,34,35. The chromatoid body contains many RNA-binding proteins, and these proteins form a complicated and dynamic complex through protein interactions12,35,36. However, the mechanism that regulates the formation of the chromatoid body remains unclear. High levels of arginine methylation have been reported in the chromatoid body, and one of the well-known components, MIWI, was arginine-methylated at the N terminus37; the arginine methyl marks could be read by a family of Tudor domain proteins38. The interaction of MIWI with Tudor-domain proteins, mediated by arginine methylation, is crucial for the cytoplasmic granular localization of MIWI and the formation of the chromatoid body in round spermatids39,40,41.
It has been reported that CaMKIV is localized in the nucleus of spermatids16 and that it plays important roles in the histone-to-protamine transition, and spermiogenesis is impaired in mice lacking CaMKIV17. However, Chatila lab finds that CaMKIV-deficient male mice were fertile and did not affect spermatogenesis42. This discrepancy may come from the different gene-targeting strategies. Moreover, just like KIF17b in mouse testes20, CaMKIV has the ability to shuttle between the nucleus and cytoplasm19,43. Therefore, the localization of CaMKIV in mouse testes was studied in great detail. Herein, we report that CaMKIV was localized in the chromatoid body and was a new component of the chromatoid body (Fig. 1). This result reveals that CaMKIV not only plays a role in the nucleus, but also has crucial functions in the cytoplasm of spermatids.
To validate the fact that CaMKIV is a component of the chromatoid body, immunoprecipitation experiments were used to detect whether CaMKIV interacts with MVH and MIWI, which are two well-studied components of the chromatoid body. The experimental results showed that CaMKIV associated with MVH and MIWI; moreover, the constitutively active form of CaMKIV had a stronger interaction with MVH than MIWI (Fig. 2). These results indicate that CaMKIV may function through the active form in the chromatoid body. More interestingly, in mouse brain, CaMKII interacts with KIF17 at the R-K-K-S sequence and regulates the cargo release from KIF1721. CaMKII and CaMKIV have some similar characteristics, such as both of them recognize the motif of R-X-X-S/T, in most cases18. The interaction of CaMKIV with KIF17b was validated by immunoprecipitation experiments and found that the R-K-K-S deletion did not decrease the interaction (Fig. 3A,B). This is possibly due to the fact that there are multiple R-X-X-S/T motifs, that is, the substrate recognition motif of CaMKIV, in the C-terminal domain of KIF17b (Fig. 3C).
GST pull-down experiments were performed to identify the cargoes of KIF17b at the C-terminal domain, and MVH was found as a new cargo (Fig. 4). Through truncations of KIF17b, the MVH binding site was restricted to the 991–1038 amino acids of KIF17b (Fig. 4). In addition, by the mutation assay, the binding site was mapped at the R-K-K-S motif (Fig. 5). It has been reported that MIWI could interact with KIF17b in the chromatoid body12, and GST pull-down experiments showed that MIWI could interact with the tail of KIF17b (Fig. 4). Interestingly, the binding site of MIWI was also mapped at the R-K-K-S motif (Fig. 5). As KIF17 is homodimeric and very similar to KIF17b44, it is possible that, in the chromatoid body, the heterodimer of MVH and MIWI45 interacts with the homodimer of KIF17b to form a heterotetramer. Importantly, the heterotetramer is regulated by CaMKIV, as the results of immunoprecipitation showed that the interaction of KIF17b with MVH and MIWI was enhanced by CaMKIV (Fig. 6); this is a different regulation model compared with CaMKII21. The structure of chromatoid body is disrupted after incubation with inhibitors of CaMKIV (Fig. 7), indicating CaMKIV takes part in the structure maintenance of chromatoid body.
After the analysis of the amino-acid sequence, many proteins in the chromatoid body were found to contain the R-X-X-S/T motif, such as MVH, MIWI, Dcp1a, GW182, and the Tudor-domain proteins. This indicates that CaMKIV may regulate the chromatoid body in a basal and general way, such as the regulation of the interaction of KIF17b with MVH and MIWI. CaMKIV not only plays a crucial role in chromatin compaction in the nucleus of spermatids during the late stages of spermatogenesis, but it also regulates the germ-cell-specific RNA-processing center in the chromatoid body, that is, in the cytoplasm of round spermatids preceding the histone-to-protamine transition and transcriptional silencing.
Methods
Ethics statement
All animal care and experiments of this study were performed in accordance with the guidelines and were approved by the Ethics Committee of International Peace Maternity & Child Health Hospital, School of Medicine, Shanghai Jiaotong University.
Animals
C57BL/6 male mice were originally purchased from Vital River Laboratories in Beijing, China. The mice were kept at temperatures of 22 °C with light cycles of 14 h light and 10 h dark; they were provided food and water ad libitum.
Cell culture and transfection
HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen) that was supplemented with 10% fetal bovine serum (FBS, Invitrogen) and 1% antibiotics (100 U/mL penicillin and 100 μg/mL streptomycin, Invitrogen), and were cultured at 37 °C with 5% CO2. Cells were transfected by the Lipofectamine 2000 reagent (Invitrogen). The transfection procedure was performed according to the manufacturer’s instructions.
Plasmid construction
The full-length mice KIF17b, MVH, MIWI and CaMKIV were amplified by the polymerase chain reaction (PCR) in cells from mouse testes using the primers containing specific restriction sites. To construct the expression vectors, KIF17b was cloned into p3 × FLAG-myc-CMV-24 (Sigma), MIWI into pEGFP-C1 (Clontech), and MVH and CaMKIV into p3xFLAG-myc-CMV-24 and pEGFP-C1, respectively. The following primers were used: KIF17b-FL (aa 1-1038), forward, that is, 5′-TAATGAATTCCATGGCCTCGGAGTCAGTGA-3′, and reverse, 5′-AATAGTCGACATCACAGAGGCTCACCACCG-3′. MVH: forward 5′-GCGCAGATCTAGCTATCATGGGAGATGAAG-3′ and reverse 5′-GCTAGTCGACGCTTTAATCCCATGACTCGT-3′. MIWI: forward, 5′-TATAGAATTCAATGACTGGCCGAGCCCGAG-3′ and reverse 5′-TATAGTCGACGTTAGAGGTAGTAGAGGCGG-3′. Full-length primer CaMKIV (GFP-CaMKIV-FL): forward 5′-ATATGGTACCATGCTCAAAGTCACGGTGCCC-3′ and reverse 5′-ATATTCTAGAGTACTCTGGCTGAATCGCAT-3′. FLAG-CaMKIV-FL: forward 5′-ATATGGTACCGATGCTCAAAGTCACGGTGCC-3′ and reverse 5′-ATATTCTAGAGTACTCTGGCTGAATCGCAT-3′. The active form of CaMKIV (CaMKIV-CA) (Δaa FNARRKLK): forward 5′-CAGAAGAAACTTCAAGAGGCAGCGGTGAAG-3′ and reverse 5′-CTTCACCGCTGCCTCTTGAAGTTTCTTCTG-3′. The truncations of KIF17b were generated by PCR and then subcloned into pGEX-5X-3 (GE Healthcare) and p3 × FLAG-myc-CMV-24 expression vector.
Western blotting
Tissues and cells were lysed in RIPA buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, 1% sodium dodecyl sulfate, 1% sodium deoxycholate, 1 mM EDTA) containing a completely EDTA-free protease inhibitor cocktail (Roche), 1 mM phenylmethylsulfonyl fluoride (PMSF) and phosphatase inhibitors (5 mM sodium orthovanadate). Protein lysates were loaded on SDS-PAGE gels and electroblotted onto nitrocellulose membranes (Amersham Biosciences). The nitrocellulose membranes were blocked for 1 h in 5% nonfat milk in TBST (10 mM Tris, pH 7.5, 200 mM NaCl, and 0.2% Tween 20) followed by incubation with primary antibodies. Mouse anti-GFP antibody (Clontech), rabbit anti-GFP antibody (Clontech), mouse anti-FLAG antibody (Sigma), rabbit anti-FLAG antibody (Sigma) and mouse anti-CaMKIV (Abnova) were used, and the bound antibodies were visualized by Lumi-Phos WB Chemiluminescent Substrate (Thermo Scientific).
GST-pull-down assay
The GST-fusion of KIF17b truncations were expressed in the BL21 strain of Escherichia coli at 28 °C and purified by glutathione-Sepharose 4B beads (GE healthcare). The purified GST-fusion proteins were bound to the Sepharose beads and then incubated with testes lysates or 293 T cell lysates transfected with the indicated plasmids. The GST precipitates were subjected to RNase A (Takara) treatment at 37 °C for 1 min, or directly to SDS–PAGE, and transferred onto nitrocellulose membranes, or the gels were stained with 0.25% Coomassie brilliant blue R-250.
Immunoprecipitation assay
The cells or mice testes were lysed in TNE buffer (10 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA and 1% Nonidet P-40) with protease inhibitor cocktail, 1 mM PMSF and 5 mM sodium orthovanadate. The lysates were centrifuged at 18,000 × g for 10 min at 4 °C, and the supernatants were subjected to preclearing with Protein G Sepharose 4 fast flow (GE Healthcare) for 2 h at 4 °C on a turning wheel. After preclearing, the lysates were immunoprecipitated with the anti-FLAG, anti-GFP or anti-MIWI (Cell Signaling Technology) antibodies overnight at 4 °C on a turning wheel. The immunoprecipitates were separated by SDS-PAGE and transferred onto nitrocellulose membranes.
Immunofluorescence
Squash samples were prepared as previously described46. The testes of adult C57Bl/6 mice were dissected and fixed in freshly prepared 2% formaldehyde in PBS (phosphate-buffered saline) (137 mM NaCl, 2.7 mM KCl, 10.1 mM Na2HPO4, 1.7 mM KH2PO4 pH 7.4) containing 0.05% Triton X-100 (Sigma) and 5 mM sodium orthovanadate for 5–10 min at room temperature. During this time, seminiferous tubules were liberated and dispersed in the fixative. Pieces of tubules were placed on a slide and gently minced with tweezers, and then a coverslip was added and the cells were squashed by exerting pressure on the coverslip. The slides were frozen in liquid nitrogen and the coverslips were removed. The slides were immediately placed in PBS for three subsequent 5 min rinses and blocked 1 h in 5% BSA in PBS. The primary antibody incubation was carried out at 4 °C in 1% BSA solution with CaMKIV antibody (Abcam) and MVH antibody (Abcam). Alexa Fluor 488 donkey anti-rabbit immunoglobulin G (IgG) (Molecular Probes) was used as secondary antibody. The nuclei were stained by Hoechst 33342 (Sigma) and the slides were digitally imaged using a fluorescence microscope (Nikon, T80i, Japan).
Inhibition studies with nocodazole, KN62 and STO609
The drugs were dissolved in dimethyl sulfoxide (DMSO, Sigma). Seminiferous tubules were transferred to 96-well plate containing either DMEM alone supplemented with 25 μl/ml DMSO and 20 μg/ml nocodazole (Sigma), 60 μM KN62 (Sigma) and 10 μM STO609 (Sigma). The tubules were incubated for 48 h at 34 °C in an atmosphere containing 5% CO2 in air. After incubation, tubules were performed to immunofluorescence by squash preparation.
Additional Information
How to cite this article: Wang, G. et al. Ca2+/Calmodulin-Dependent Protein Kinase IV Promotes Interplay of Proteins in Chromatoid Body of Male Germ Cells. Sci. Rep. 5, 12126; doi: 10.1038/srep12126 (2015).
Acknowledgments
This work was supported by National Natural Science Foundation of China Grants 81125005 and 81430027 (to F.S.), the National Basic Research Program of China (2014CB943100).
Footnotes
Author Contributions This study was conceived and designed by G.S.W., H.J.Z., L.W., Y.W., H.F.H., F.S., G.S.W. and H.J.Z. performed the experiments. All authors analyzed the data and discussed the results. G.S.W. wrote the paper, and the other authors commented on the manuscript.
References
- Benda C. Neue mitteilungen über die entwickelung der genitaldrüsen und die metamorphose der samenzellen (Histogenese der Spermatozoen). Verh. Berliner Physiol. Ges. Arch. Anat. Physiol. 1891, 549–522 (1891). [Google Scholar]
- Fawcett D. W., Eddy E. M. & Phillips D. M. Observations on the fine structure and relationships of the chromatoid body in mammalian spermatogenesis. Biol. Reprod. 2, 129–53 (1970). [DOI] [PubMed] [Google Scholar]
- Parvinen M. & Jokelainen P. T. Rapid movements of the chromatoid body in living early spermatids of the rat. Biol. Reprod. 11, 85–92 (1974). [DOI] [PubMed] [Google Scholar]
- Susi F. R. & Clermont Y. Fine structural modifications of the rat chromatoid body during spermiogenesis. Am. J. Anat. 129, 177–91 (1970). [DOI] [PubMed] [Google Scholar]
- Kotaja N. & Sassone-Corsi P. The chromatoid body: a germ-cell-specific RNA-processing centre. Nat. Rev. Mol. Cell Biol. 8, 85–90 (2007). [DOI] [PubMed] [Google Scholar]
- Yokota S. Historical survey on chromatoid body research. Acta Histochem. Cytochem. 41, 65–82 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvinen M. The chromatoid body in spermatogenesis. Int. J. Androl. 28, 189–201 (2005). [DOI] [PubMed] [Google Scholar]
- Kotaja N. et al. The chromatoid body of male germ cells: similarity with processing bodies and presence of Dicer and microRNA pathway components. Proc. Natl. Acad. Sci. USA. 103, 2647–52 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebe M., Onohara Y. & Yokota S. Expression of MAEL in nuage and non-nuage compartments of rat spermatogenic cells and colocalization with DDX4, DDX25 and MIWI. Histochem. Cell Biol. 140, 169–81 (2013). [DOI] [PubMed] [Google Scholar]
- Nagamori I., Cruickshank V. A. & Sassone-Corsi P. Regulation of an RNA granule during spermatogenesis: acetylation of MVH in the chromatoid body of germ cells. J. Cell Sci. 124, 4346–55 (2011). [DOI] [PubMed] [Google Scholar]
- Carmell M. A., Xuan Z., Zhang M. Q. & Hannon G. J. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 16, 2733–42 (2002). [DOI] [PubMed] [Google Scholar]
- Kotaja N., Lin H., Parvinen M. & Sassone-Corsi P. Interplay of PIWI/Argonaute protein MIWI and kinesin KIF17b in chromatoid bodies of male germ cells. J. Cell Sci. 119, 2819–25 (2006). [DOI] [PubMed] [Google Scholar]
- Chuma S. et al. Mouse Tudor Repeat-1 (MTR-1) is a novel component of chromatoid bodies/nuages in male germ cells and forms a complex with snRNPs. Mech. Dev. 120, 979–90 (2003). [DOI] [PubMed] [Google Scholar]
- Kotaja N., Macho B. & Sassone-Corsi P. Microtubule-independent and protein kinase A-mediated function of kinesin KIF17b controls the intracellular transport of activator of CREM in testis (ACT). J. Biol. Chem. 280, 31739–45 (2005). [DOI] [PubMed] [Google Scholar]
- Means A. R. et al. A novel Ca2+/calmodulin-dependent protein kinase and a male germ cell-specific calmodulin-binding protein are derived from the same gene. Mol. Cell Biol. 11, 3960–71 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. Y. & Means A. R. Ca(2+)/calmodulin-dependent protein kinase IV is expressed in spermatids and targeted to chromatin and the nuclear matrix. J. Biol. Chem. 275, 7994–9 (2000). [DOI] [PubMed] [Google Scholar]
- Wu J. Y. et al. Spermiogenesis and exchange of basic nuclear proteins are impaired in male germ cells lacking Camk4. Nat. Genet. 25, 448–52 (2000). [DOI] [PubMed] [Google Scholar]
- White R. R., Kwon Y. G., Taing M., Lawrence D. S. & Edelman A. M. Definition of optimal substrate recognition motifs of Ca2+-calmodulin-dependent protein kinases IV and II reveals shared and distinctive features. J. Biol. Chem. 273, 3166–72 (1998). [DOI] [PubMed] [Google Scholar]
- Zhang X. et al. Calcium/calmodulin-dependent protein kinase (CaMK) IV mediates nucleocytoplasmic shuttling and release of HMGB1 during lipopolysaccharide stimulation of macrophages. J. Immunol. 181, 5015–23 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho B. et al. CREM-dependent transcription in male germ cells controlled by a kinesin. Science. 298, 2388–90 (2002). [DOI] [PubMed] [Google Scholar]
- Guillaud L., Wong R. & Hirokawa N. Disruption of KIF17-Mint1 interaction by CaMKII-dependent phosphorylation: a molecular model of kinesin-cargo release. Nat. Cell Biol. 10, 19–29 (2008). [DOI] [PubMed] [Google Scholar]
- Chalmel F. et al. The conserved transcriptome in human and rodent male gametogenesis. Proc. Natl. Acad. Sci. USA. 104, 8346–51 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara Y. et al. Isolation of a DEAD-family protein gene that encodes a murine homolog of Drosophila vasa and its specific expression in germ cell lineage. Proc. Natl. Acad. Sci. USA. 91, 12258–62 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventela S., Toppari J. & Parvinen M. Intercellular organelle traffic through cytoplasmic bridges in early spermatids of the rat: mechanisms of haploid gene product sharing. Mol. Biol. Cell. 14, 2768–80 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persaud S. J., Liu B., Sampaio H. B., Jones P. M. & Muller D. S. Calcium/calmodulin-dependent kinase IV controls glucose-induced Irs2 expression in mouse beta cells via activation of cAMP response element-binding protein. Diabetologia. 54, 1109–20 (2011). [DOI] [PubMed] [Google Scholar]
- Reece K. M., Mazalouskas M. D. & Wadzinski B. E. The Balpha and Bdelta regulatory subunits of PP2A are necessary for assembly of the CaMKIV.PP2A signaling complex. Biochem. Biophys. Res. Commun. 386, 582–7 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y. et al. Nuclear export factor 3 is involved in regulating the expression of TGF-beta3 in an mRNA export activity-independent manner in mouse Sertoli cells. Biochem. J. 452, 67–78 (2013). [DOI] [PubMed] [Google Scholar]
- Kimmins S. & Sassone-Corsi P. Chromatin remodelling and epigenetic features of germ cells. Nature. 434, 583–9 (2005). [DOI] [PubMed] [Google Scholar]
- Steger K. Transcriptional and translational regulation of gene expression in haploid spermatids. Anat. Embryol. (Berl). 199, 471–87 (1999). [DOI] [PubMed] [Google Scholar]
- Sassone-Corsi P. Unique chromatin remodeling and transcriptional regulation in spermatogenesis. Science. 296, 2176–8 (2002). [DOI] [PubMed] [Google Scholar]
- Steger K. Haploid spermatids exhibit translationally repressed mRNAs. Anat. Embryol. (Berl). 203, 323–34 (2001). [DOI] [PubMed] [Google Scholar]
- Yang J., Medvedev S., Reddi P. P., Schultz R. M. & Hecht N. B. The DNA/RNA-binding protein MSY2 marks specific transcripts for cytoplasmic storage in mouse male germ cells. Proc. Natl. Acad. Sci. USA. 102, 1513–8 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J., Peters A. H., Lee K. & Braun R. E. A double-stranded RNA binding protein required for activation of repressed messages in mammalian germ cells. Nat. Genet. 22, 171–4 (1999). [DOI] [PubMed] [Google Scholar]
- Hussain S. et al. The mouse cytosine-5 RNA methyltransferase NSun2 is a component of the chromatoid body and required for testis differentiation. Mol. Cell Biol. 33, 1561–70 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messina V. et al. The RNA Binding Protein SAM68 Transiently Localizes in the Chromatoid Body of Male Germ Cells and Influences Expression of Select MicroRNAs. PLoS One. 7, e39729 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Saxe J. P., Tanaka T., Chuma S. & Lin H. Mili interacts with tudor domain-containing protein 1 in regulating spermatogenesis. Curr. Biol. 19, 640–4 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin V. V. et al. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev. 23, 1749–62 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida K. M. et al. Functional involvement of Tudor and dPRMT5 in the piRNA processing pathway in Drosophila germlines. EMBO J. 28, 3820–31 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuta Y. et al. TDRD5 is required for retrotransposon silencing, chromatoid body assembly, and spermiogenesis in mice. J. Cell Biol. 192, 781–95 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T. et al. Tudor domain containing 7 (Tdrd7) is essential for dynamic ribonucleoprotein (RNP) remodeling of chromatoid bodies during spermatogenesis. Proc. Natl. Acad. Sci. USA. 108, 10579–84 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasileva A., Tiedau D., Firooznia A., Muller-Reichert T. & Jessberger R. Tdrd6 is required for spermiogenesis, chromatoid body architecture, and regulation of miRNA expression. Curr. Biol. 19, 630–9 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaeser F. et al. CaMKIV/Gr is dispensable for spermatogenesis and CREM-regulated transcription in male germ cells. Am. J. Physiol. Endocrinol. Metab. 281, E931–7 (2001). [DOI] [PubMed] [Google Scholar]
- Kotera I. et al. Importin alpha transports CaMKIV to the nucleus without utilizing importin beta. EMBO J. 24, 942–51 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaulin F. & Kreitzer G. KIF17 stabilizes microtubules and contributes to epithelial morphogenesis by acting at MT plus ends with EB1 and APC. J. Cell Biol. 190, 443–60 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S. et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 131, 839–49 (2004). [DOI] [PubMed] [Google Scholar]
- Page J., Suja J. A., Santos J. L. & Rufas J. S. Squash procedure for protein immunolocalization in meiotic cells. Chromosome Res. 6, 639–42 (1998). [DOI] [PubMed] [Google Scholar]