Abstract
Aiming at generating a comprehensive genomic database on Elaeis spp., our group is leading several R&D initiatives with Elaeis guineensis (African oil palm) and Elaeis oleifera (American oil palm), including the whole-genome sequencing of the last. Genome size estimates currently available for this genus are controversial, as they indicate that American oil palm genome is about half the size of the African oil palm genome and that the genome of the interspecific hybrid is bigger than both the parental species genomes. We estimated the genome size of three E. guineensis genotypes, five E. oleifera genotypes, and two interspecific hybrids genotypes. On average, the genome size of E. guineensis is 4.32 ± 0.173 pg, while that of E. oleifera is 4.43 ± 0.018 pg. This indicates that both genomes are similar in size, even though E. oleifera is in fact bigger. As expected, the hybrid genome size is around the average of the two genomes, 4.40 ± 0.016 pg. Additionally, we demonstrate that both species present around 38% of GC content. As our results contradict the currently available data on Elaeis spp. genome sizes, we propose that the actual genome size of the Elaeis species is around 4 pg and that American oil palm possesses a larger genome than African oil palm.
Keywords: African oil palm, American oil palm, FCM—flow cytometry, GC content, DNA sequencing
Introduction
The Elaeis genus is composed of two species. The African oil palm (Elaeis guineensis) is native from Africa and can be found in spontaneous populations or in cultivated fields in all tropical regions of Africa, Southeast Asia, and South and Central America. The American oil palm, also known as Caiaué (Elaeis oleifera) is endemic to the humid tropical zone of Latin America. It occurs in spontaneous populations from the south of Mexico to Amazon areas in Brazil and Colombia. The oil and fatty acids produced by these species are versatile and are currently being used in the cosmetic industry as well as in the biofuel industries.1,2
The global market for oil and fatty acids grows each year, and only African oil palm, soybean, and canola answer for nearly 60% of the demand.3 Among these species, African oil palm is considered to be the most profitable, since comparative studies have demonstrated that while soybean produces on average 0.46 tons·ha−1·yr−1, African oil palm can produce up to 6 tons·ha−1·yr−1.4 Not surprisingly, current data indicate that soybeans occupy nearly 42% of the area planted with oil-producing species, while African oil palm occupy only 4.2%, even though both species produce approximately the same amount of oil, ie, 33 million tons per year, accounting for 33% of vegetable oil and 45% of edible oil worldwide.5 This advantage over soybean and other oil-producing species is due to the enhanced photosynthetic capacity of oil palm and its continuous production of fruits.6 Currently, the larger, worldwide producers of oil palm are Indonesia, Malaysia, Thailand, Colombia, and Nigeria. Indonesia and Malaysia together correspond to nearly 87% of the world’s production (Wahid et al 2004).6 Globally, Brazil currently occupies the 15th position with the production estimated to be approximately 154 thousand tons per year. This scenario, however, is likely to change as there are thousands of hectares available for oil palm plantation in Brazil. The Brazilian oil palm cultivation is concentrated in the state of Pará, in the northern region of Brazil. This region holds 80% of the Brazilian oil palm fields. The vast majority of the Brazilian oil palm production is destined to the food industry, even though the biofuel market is increasing rapidly and demanding more crude oil each year (Chia et al 2009).4
The expansion of the oil palm plantations in Brazil and other parts of the continent, driven basically by a need to meet the increasing oil demand, has been, however, hampered by the occurrence of an abnormality known as bud rot. The etiology of this abnormality is still unknown, what somewhat limits its control. One of the strategies used to stop the advancement of bud rot in Brazil is the use of interspecific hybrids between African oil palm and American oil palm, as American oil palm has been shown to be resistant/tolerant to this abnormality. Since the species hybridizes well with African oil palm, producing fertile offspring, hybrid varieties can be easily developed and released as an alternative to traditional African oil palm cultivars. Currently, the hybrid is planted in Brazil only in the areas of bud rot occurrence, since it presents some problems that limit its wide adoption.7,8 Some examples are: (i) lower oil production when compared to the ‘Tenera’ oil palm hybrid; (ii) occurrence of abnormalities in the male inflorescences; and (iii) lower amount of pollen, therefore requiring the implementation of assisted pollination as a management practice.2 Besides these limitations, American oil palm represents an important source of genetic variability for oil palm breeding programs.7
Due to the increasing demand for biodiesel in Brazil9 and given the fact that oil palm can produce more oil than soybean (which currently supply more than 85% of the crude oil used for biodiesel production in Brazil), the Brazilian National Agricultural Research Corporation (Embrapa) has decided to focus its breeding efforts in the development of interspecific hybrids. The current breeding strategy relies on the generation and evaluation of F1 interspecific hybrids, followed by the selection of the best to be used as parents in seeds production fields, or in backcrossing schemes with African oil palm.4 Clonal propagation of the best genotypes is also being considered as a strategic action. In support to that and aiming at generating a comprehensive genomic database on the Elaeis spp., our group is also leading several initiatives with African oil palm and American oil palm genetics and genomics. One of these initiatives is the whole-genome sequencing of an American oil palm genotype native to Brazil. Genome size and content information are therefore paramount, as it may allow us to better define the sequencing strategy needed to generate the E. oleifera genome draft, eg, the sequence depth needed to reach a predefined coverage.
However, some inconsistencies in the studies related to Elaeis spp. genome size previously published make it difficult to draw final conclusions about their actual genome sizes. Genome size estimates currently available for the Elaeis spp. indicate that American oil palm genome is about half the size of the African oil palm genome and that the interspecific hybrid genome is larger than both the parental species’ genomes.10,11 This is intriguing since the close relationship between E. guineensis and E. oleifera and especially the fact that both species hybridize well suggest that their genomes would be similar in size. Moreover, the genome size of hybrid is expected to be about the average of the parents’ genome. Moreover, Singh et al5 sequenced the genome of E. guineensis to be approximately 1.8 Gb and the total length of the assembly was estimated to be 1.5 Gbp [very close to the data previously generated by flow cytometry (FCM) in the species]. While reporting a draft sequence for a genotype of E. oleifera distinct from the one we are interested in, the same authors did not detected any significant difference in size while comparing both species. This further supports our hypothesis of inconsistencies of the FCM data earlier reported for the species of the Elaeis genus.
Since we needed high-quality data on that respect to support our sequencing project (currently underway), we decided to reassess the genome sizes of the two Elaeis species and its hybrid. The objective of this work was then to reestimate, through the use of FCM, the genome size of E. guineensis, E. oleifera, and F1 hybrids, as well as to perform an initial evaluation of the genome content (ie, nucleotide content) so that we could draw an adequate strategy to fully fulfill our goal of sequencing a native genotype of E. oleifera.
Materials and Methods
Biological material
The experimental work was carried out in the Plant Genetics Lab of Embrapa Dairy Cattle, Juiz de Fora, Minas Gerais (Brazil). The sample sources were young leaves from plants of 10 genotypes previously grown in vitro: three E. guineensis (Eg2301, Eg1210, and Eg0920), five E. oleifera (Eo0507, Eo0726, Eo0312, Eo0213, and Eo0610), and two interspecific hybrids (H1619 and H413). The genotypes were previously collected in the germplasm collection maintained by Embrapa Western Amazon—Manaus, Amazonas (Brazil).
Estimation of the genome size of E. guineensis, E. oleifera, and their hybrids through FCM
Young leaves of Glycine max cv. ‘Polanka’ (soybean) and Solanum lycopersicum L. ‘Stupické polní rané’ (tomato) were used as internal and external standards. Young leaf samples were crushed in Petri dishes with 800 μL of cold buffer LB01 for nuclear suspension. The solution was aspirated through two layers of bandage and then filtered through a 42-μm mesh, and then 25 μL of propidium iodide and 25 μL of RNAse were added. For each sample, at least 10,000 nuclei were analyzed, both in linear and log scales.12,13 The analyses were performed in FacScalibur cytometer (Becton Dickinson), and histograms were generated by Cell Quest software and analyzed using the software WinMDI version 2.8 (http://facs.scripps.edu/software.html). To increase the reliability of the results, only histograms with variation coefficients below 3% were used. Plant nuclear DNA quantification was estimated in picograms (pg), by comparing G1 peaks from samples to G1 peaks of standard, both internal and external. Tomato 2C is equivalent to 1.96 pg of DNA and soybean 2C to 2.25 pg of DNA.14,15 Calibration was performed by comparing the patterns as follows: G. max × S. lycopersicum (primary pattern) and S. lycopersicum × G. max (primary pattern). The values obtained in the calibration for S. lycopersicum and G. max were 1.96 pg and 2.41 pg, respectively. These values are in accordance to the patterns described by Dolezel et al12,13 and Praça-Fontes et al.16
Nucleotides content
Sequences from four oil palm genotypes (defined based on the regions from where they were collected: BR174, Coari, Manicoré—three E. oleifera, and ‘Tenera’—E. guineensis) were sequenced using Illumina HiSeq2000 platform and cleaned for quality control (Formighieri et al, unpublished data). Nucleotide content was then evaluated using FASTX-toolkit (http://hannonlab.cshl. edu/fastx_toolkit).
Experimental design and data analysis
The experiment was carried out in a completely randomized design with 10 treatments [three E. guineensis genotypes (Eg2301, Eg1210, and Eg0920); five E. oleifera genotypes (Eo0507, Eo0726, Eo0312, Eo0213, and E00610); and two hybrid (E. guineensis × E. oleifera) genotypes (H1619 and H413) with four replicates]. Each experimental repetition consisted of one grounded leaf. The treatment means were compared on the basis of the genome size estimated based on different standard/evaluation combinations (tomato and soybean, internal and external standard, linear and logarithmic scale) by an analysis of variance (ANOVA) with P ≤ 0.05. Means obtained for genome size of the species and hybrids were further differentiated based on Tukey’s Honest Significant Difference (HSD) post hoc test (P ≤ 0.05). To gain some insights of whether there is some significant difference in using internal or external standards while measuring the genome size based either on a linear or on a logarithmic scale, we also compared the means obtained for each genotype in each of these situations against each order with a t-test (P ≤ 0.05) (eg, genome size estimated for Eg2301 based on tomato external standard with linear scale versus genome size estimated for Eg2301 based on tomato external standard with logarithmic scale).
Results
FCM pattern choice
In FCM studies it is imperative to select the best reference pattern in order to obtain accurate estimates of the sample genome size. As a number of references genotypes are currently available (eg, different maize, soybean, tomato genotypes),16 here we have opted to test soybean and tomato as our FCM standards prior to the quantification of the E. guineensis and E. oleifera (and their interspecific hybrid) genome sizes. As can be noted in Figure 1, soybean genome is larger than the tomato genome, and most importantly, closer in size to both Elaeis species. This figure shows the well-defined G1 peaks from nuclei isolated from tomato (yellow), soybean (blue), and samples (red), ie, E. guineensis, E. oleifera, or hybrid sample, combined in a single image. In principle, the use of a reference standard, whose genome is closer in size to the sample genome, should give more accurate estimates.12,13 In fact, we found differences in results. When we estimated E. guineensis and E. oleifera genome sizes, the average results were higher when we used soybean as a reference pattern when compared to using tomato as our reference choice (Table 1). This difference was expected to occur and may potentially indicate that because soybean genome is closer in size to the oil palm genome, it should be preferably used as a reference standard.
Figure 1.
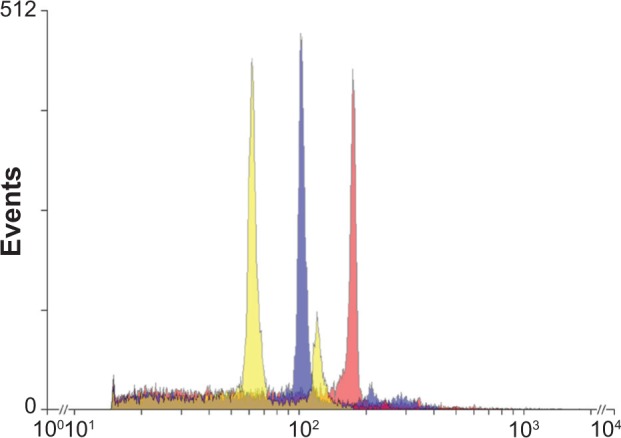
Schematic representation of the quantification of Elaeis spp. DNA content through FCM. Fluorescence intensity histograms were first generated for each species separately, and then the peaks were differentially colored (using yellow for tomato, blue for soybean, and red for the Elaeis spp. sample) and then superimposed. Transparency of the peaks was regulated to 50%. On the X axis the relative fluorescence (channel number) is shown, while on the Y axis the number of nuclei is shown.
Table 1.
Results of Tukey tests of the nuclear DNA quantification (pg) for species Elaeis guineensis (Eg), E. oleifera (Eo) and the interspecific hybrid (H) demonstrating the non-existence of specific patterns of grouping.a
| GENOTYPES | DNA CONTENT 2C (MEANS ± STANDARD DEVIATION) | |||||||
|---|---|---|---|---|---|---|---|---|
| SEbLIN | SEbLOG | SIcLIN | SlcLOG | TEdLIN | TEdLOG | TIeLIN | TIe LOG | |
| H1619 | 4.019 ± 0.259a | 3.955 ± 0.259a | 4.390 ± 0.013a,b,c | 4.438 ± 0.086d,e | 3.793 ± 0.029a | 3.876 ± 0.104a | 3.916 ± 0.051b | 4.006 ± 0.038a,b,c |
| H413 | 3.883 ± 0.164a | 4.199±0.335a | 4.414 ± 0.008a,b | 4.547 ± 0.072c,d | 3.890 ±0.079a | 4.007 ± 0.084a | 3.915 ± 0.058b | 4.051 ± 0.080a,b,c |
| Eg2301 | 4.073 ± 0.079a | 4.008 ± 0.265a | 4.261 ± 0.029b,c,d | 4.363 ± 0.049e | 3.889 ±0.046a | 3.923 ± 0.184a | 3.898 ± 0.039b | 3.900 ± 0.176b,c |
| Eg1210 | 3.965 ± 0.208a | 4.135 ± 0.187a | 4.156 ± 0.022a | 4.353 ± 0.029e | 3.842 ± 0.154a | 4.007 ± 0.175a | 3.876 ± 0.053b | 3.863 ± 0.089c |
| Eg0920 | 3.908 ± 0.197a | 4.058 ± 0.304a | 4.194 ± 0.069c,d | 4.388 ± 0.051e | 3.951 ± 0.175a | 3.952 ± 0.111a | 3.875 ± 0.088b | 3.939 ± 0.057a,b,c |
| Eo0507 | 4.032 ± 0.218a | 4.333 ± 0.316a | 4.544 ±0.029a | 4.686 ±0.059a,b | 3.839±0.111a | 4.223 ±0.274a | 3.983 ±0.058a,b | 4.124 ± 0.025a |
| Eo0726 | 4.022 ± 0.403a | 4.329 ± 0.278a | 4.533 ± 0.063a | 4.758 ± 0.035a | 3.886 ± 0.139a | 4.191 ± 0.115a | 4.060 ± 0.043a | 4.099 ± 0.053a,b |
| Eo0312 | 3.848 ± 0.105a | 4.196 ± 0.288a | 4.361 ±0.244a,b,c,d | 4.575 ± 0.054b,c | 3.781 ± 0.149a | 4.030 ± 0.311a | 3.975 ± 0.036a,b | 4.090 ± 0.117a,b |
| Eo213 | 4.077 ± 0.352a | 4.411 ± 0.464a | 4.561 ± 0.058a | 4.670 ± 0.044a,b,c | 3.743 ± 0.269a | 4.293 ± 0.219a | 4.086 ± 0.038a | 4.080 ± 0.031a,b |
| Eo610 | 4.213 ± 0.274a | 4.203 ± 0.163a | 4.530 ± 0.062a | 4.714 ± 0.041a | 3.757 ± 0.014a | 4.064 ± 0.150a | 3.974 ± 0.065a,b | 4.107 ± 0.067a |
Notes:
Averages at the same column followed by different letters are considered significantly different according to Tukey test (p < 0.05). Standards: Glycine max cv. “Polanka” 2C = 2.50 pg; Solanum lycopersicum L. “Stupické polní rané” 2C = 1.96 pg.
Abbreviations
SE, Soybean External
SI, Soybean Internal
TE, Tomato External
TI, Tomato Internal; Scale: Lin, Linear; Log, Logarithmic.
We have also tested whether there were differences in using nuclei isolated from the reference species as internal or external standards. Many current data have indicated that internal patterns outperform external patterns for a number of reasons.17,18 By doing so, we found that soybean and tomato behave differentially when used as internal or external reference patterns. Significant differences between genome size estimates based on soybean and tomato were systematically found (ie, differences occurred in all evaluated genotypes) when these species were used as internal patterns (Table 2). On the other hand, when these same species were used as external patterns, only three significant differences between genome size estimates based on soybean and tomato were found (one E. guineensis—Eg2301, and one E. oleifera—Eo0610) (Table 2). This might be potentially related to the fact that external patterns seem to be less precise than internal pattern. This can be easily noted by the higher standard deviations of the average genome sizes produced by external patterns (Table 1). This clearly indicates that an internal reference sample is more sensitive than an external reference sample, regardless of the species used as standard. The use of internal reference patterns appears to be then the best strategy to accurately estimate the genome size of the Elaeis species and hybrids, as its precision is more elevated.
Table 2.
Results of pair-wise t-tests demonstrating that the estimates of the Elaeis spp. genotypes genome size obtained through FCM depends upon the type reference pattern used (ie, tomato vs soybean; internal vs external standard and logarithm vs linear reading scale).a
| COMPARISON | GENOTYPES | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Hh1619 | H413 | EGi2301 | EG1210 | EG0920 | EOj0507 | EO0726 | EO0312 | EO0213 | EO0610 | |
| TIb Logf × SIc Log | 0.000659* | 0.000098* | 0.010284* | 0.000747* | 0.000025* | 0.000058* | 0.000003* | 0.001358* | 0.000002* | 0.000021* |
| TEd Log × SEe Log | 0.601495 | 0.339884 | 0.619082 | 0.357502 | 0.549129 | 0.618983 | 0. 412731 | 0.463939 | 0.666777 | 0. 25 6 415 |
| TI Ling × SI Lin | 0.000172* | 0.000340* | 0.000011* | 0.000602* | 0.001494* | 0.000031* | 0.000082* | 0.048873* | 0.000027* | 0.000017* |
| TE Lin × SE Lin | 0.179866 | 0.942835 | 0.010976* | 0.383935 | 0.757525 | 0.18 4 519 | 0.562531 | 0.491574 | 0.185901 | 0.044824* |
| TI Log × TE Log | 0.082531 | 0.484956 | 0.858306 | 0.208082 | 0.850087 | 0.520570 | 0.214994 | 0.737210 | 0.146199 | 0.626609 |
| SI Log × SE Log | 0.027796* | 0.128113 | 0.073304 | 0.101710 | 0.116738 | 0.110345 | 0.052298 | 0.076037 | 0.347001 | 0.006235* |
| TI Lin × TE Lin | 0.009818* | 0.635097 | 0.789899 | 0.696997 | 0.476712 | 0.076298 | 0.082865 | 0.076737 | 0.082748 | 0.005698* |
| SI Lin × SE Lin | 0.064433 | 0.0074188* | 0.012378* | 0.162868 | 0.056907 | 0.017326 * | 0.082649 | 0.017440* | 0.068929 | 0.101045 |
| TI Log × TI Lin | 0.033100* | 0.036858* | 0.985485 | 0.799902 | 0.270413 | 0.010607* | 0.292730 | 0.1416 2 0 | 0.806982 | 0.029244* |
| TE Log × TE Lin | 0.213535 | 0.088742 | 0.741616 | 0.207705 | 0.993592 | 0.060984 | 0.015528* | 0.216437 | 0.020492* | 0.025854* |
| SI Log × SI Lin | 0.345195 | 0.033804* | 0.016748* | 0.000061* | 0.003385* | 0.010435* | 0.003157* | 0.17 76 3 8 | 0.025836* | 0.003796* |
| SE Log × SE Lin | 0.737943 | 0.160012 | 0.670204 | 0.269967 | 0.444839 | 0.173605 | 0.261382 | 0.089561 | 0.297511 | 0.952695 |
Note:
Asterisks indicate means significantly different based on a t-test (p < 0.05).
Abbreviations:
TI, Tomato Internal
SI, Soybean Internal
TE, Tomato External
SE, Soybean External
Log, Logarithmic scale
Lin, linear scale.
H, hybrid
Eg, Elaeis guineensis
Eo, Elaeis oleifera.
FCM is now universally adopted for DNA size quantification, and generally histograms are conventionally plotted using linear scale, instead of logarithmic scales (log amplifiers are designed to expand the smaller signals). However, we have also looked for possible differences that may arise depending on the scale in which readings were recorded, ie, using linear or logarithmic scale. It can be noted based on Tables 1 and 2 that no significant difference among genome sizes estimated using linear or logarithmic scales were detected when soybean was used as an external reference. In all other cases, the results seem to be genotype dependent (Table 2), as a discernible pattern seems to be absent. In general, however, there is a slight tendency to the logarithmic scale to return larger genomes sizes (Table 1) and to better differentiate the genotypes according to the species to which it belongs (more details on that in the next section).
Genome size of Elaeis species and hybrids
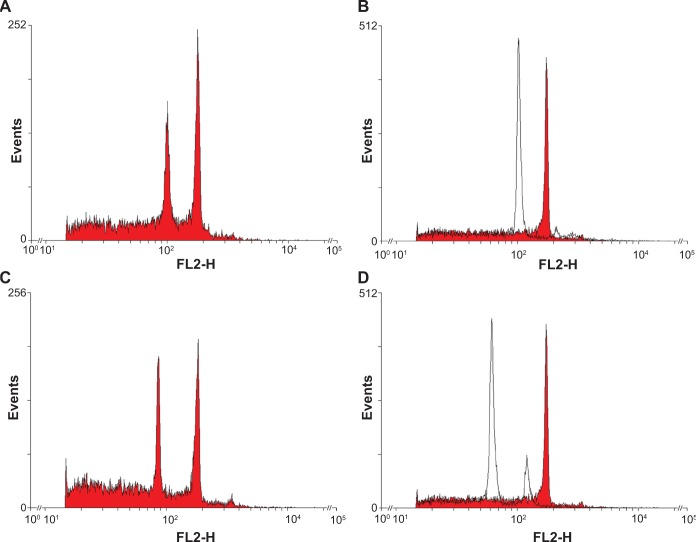
Using different combinations of reference patterns, we have estimated the genome size of three E. guineensis genotypes, five E. oleifera genotypes, and two interspecific hybrids. In general, our results indicate that the genomes of both species are very close in size (around 4 pg, on average) (Table 1) and that the genome of the hybrid is of intermediate size (Table 1). Given the elevated standard deviation produced by external patterns (Table 1), regardless of the species used as reference and of the reading scale, no significant differences were found among the estimated genome sizes for the three groups (E. guineensis, E. oleifera, and interspecific hybrids) when using such patterns. On the other hand, and in accordance to the indication that internal patterns outperform external patterns, several significant differences were found among genome sizes for the three groups, for both linear and log scales, when we considered the internal pattern (Table 1). Table 1 summarizes the results of a series of ANOVAs/Tukey’s tests for genome size averages, considering the different reference patterns. In general, it can be noted that in the cases where significant differences among the three groups were found, the E. oleifera genotypes presented a slightly larger genome, while the E. guineensis genotypes and interspecific hybrids presented smaller genomes (Table 1). This fact is more easily noted with the logarithmic scale that seemed to allow a better differentiation of the genome sizes according to the group to which each genotype belongs. Another interesting thing to note is that the sharp peaks displayed in all FCM histograms (Fig. 2) indicate good data quality, and as consequence, high confidence of the results reported here. This figure, in contrast to Figure 1, illustrates the actual results obtained when soybean and tomato were used as internal and external references for sample evaluation.
Figure 2.
Examples of fluorescence intensity histograms actually used for evaluation of samples genome size. (A) Soybean (internal) + sample, (B) soybean (external, empty) + sample (red), (C) tomato (internal) + sample, and (D) Tomato (external, empty) + sample (red). On the X axis the relative fluorescence (channel number) is shown, while on the Y axis the number of nuclei is shown.
Concerning now specifically the genome size of E. guineensis, E. oleifera, and its interspecific hybrids, assuming that the combination soybean internal is most suited for the quantification of the DNA content in Elaeis species (as indicated by our initial results), on average the genome size of E. guineensis (‘Tenera’) is 4.32 ± 0.173 pg, while that of E. oleifera is 4.43 ± 0.18 pg. As expected, and contrary to what is currently published,11 the hybrid genome presents an intermediate size considering the two parental genomes, with an average of 4.40 ± 0.016 pg. This result contradicts the currently available data in literature (see Discussion section for more details).
Genome content of Elaeis species and hybrids
Aiming at a more refined understanding of the structure of the Elaeis spp. genomes, and especially to help planning our sequencing strategy and to select the best genotype to be sequenced, we partially sequenced the genome of three American oil palm and one African oil palm genotypes, using Illumina’s HiSeq2000 (one lane per genotype). The American oil palm genotypes were chosen based on their importance to the oil palm breeding program at Embrapa and on their geographical distribution. The African oil palm genotype was chosen because it represents the most planted oil palm material in Brazil: the ‘Tenera’ intraspecific hybrid (E. guineensis ‘Dura’ × E. guineensis ‘Pisifera’). The first analysis we carried out related to nucleotide composition. Figure 3 shows the average nucleotide composition for all positions on reads from one of the lanes. The occurrence of an unexpected variation [considering the Chargaff Second Parity Rule (CSPR)] in the beginning of the sequence can be easily noted in this figure. Such variations also smoothly occur at the end of the sequence. Because of this, in Table 3 the nucleotide composition and GC content are presented considering different read segments: one that spans the whole sequenced fragment (1–100 bases), one that excludes the initial and final parts of the sequence (10–80), and finally one that focuses only on the first part of the sequence. In general, besides some small differences, considering the range 10–80, the GC content for both species is around 38% (the ‘Tenera’ was the only sample to present a slightly lower GC content).
Figure 3.
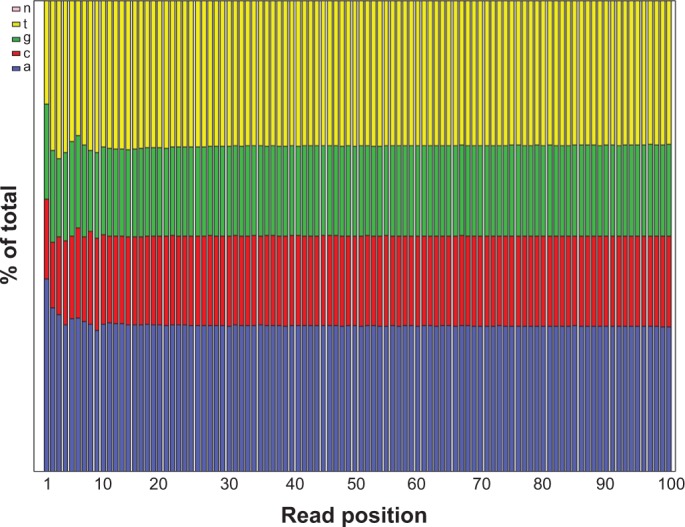
Nucleotide composition in the sequenced reads from Coari sample. Each read position presents the average for all reads nucleotide composition.
Table 3.
General estimates of nucleotide and GC content for Elaeis spp. based on nucleotide sequences obtained from plants of four distinct “groups”.
| “GROUP” | POSITION | AVERAGES | ||||
|---|---|---|---|---|---|---|
| A% | C% | G% | T% | GC% | ||
| BR1741 | 01 to 100 | 38.83% | 19.22% | 19.23% | 30.71% | 38.46% |
| 10 to 80 | 30.89% | 19.15% | 19.14% | 30.83% | 38.29% | |
| 01 to 10 | 32.96% | 17.65% | 19.43% | 29.96% | 37.08% | |
| 01 to 100 | 30.73% | 19.30% | 19.28% | 30.68% | 38.58% | |
| Manicoré1 | 10 to 80 | 30.78% | 19.24% | 19.18% | 30.79% | 38.43% |
| 01 to 10 | 32.16% | 18.37% | 19.12% | 30.35% | 37.50% | |
| 01 to 100 | 30.92% | 19.12% | 19.12% | 30.83% | 38.25% | |
| Coari1 | 10 to 80 | 30.96% | 19.06% | 19.02% | 30.95% | 38.08% |
| 01 to 10 | 32.93% | 17.85% | 18.81% | 30.41% | 36.66% | |
| 01 to 100 | 31.00% | 19.02% | 19.00% | 30.98% | 38.01% | |
| Tenera1 | 10 to 80 | 31.04% | 18.97% | 18.91% | 31.08% | 37.88% |
| 01 to 10 | 32.21% | 18.17% | 19.01% | 30.61% | 37.18% | |
Notes:
BR174, Manicoré and Coari are geographic regions of the Amazon basin, whereas Tenera refers to a type of intra-specific hybrid between Elaeis guineensis Dura × Elaeis guineensis Pisifera.
Discussion
We have successfully obtained high-confidence estimates for genome sizes of E. guineensis, E. oleifera, and its interspecific hybrid. Our estimates are strikingly different from the ones currently available.11,12,20 However, as we have employed a meticulous strategy, first determining the most suited reference pattern to be used in FCM studies of the Elaeis spp. and then using this reference standard to accurately estimate the genome sizes, we believe that our estimates are, to the extent of our knowledge, the most accurately available. We have also designed, based on our genome size estimates, an adequate sequencing strategy to fully sequence the genome of Brazilian E. oleifera genotype. If we have not revisited the genome size of the Elaeis spp. theme, we would probably be focusing our efforts on an inefficient assembly process. Below we discuss the reasons why we are confident that the estimates presented here are accurate and how these new estimates impacted our initial plan to obtain a draft genome sequence for a native E. oleifera genotype.
The choice of reference pattern influences the FCM estimates of Elaeis spp. genome size
As previously mentioned, it is imperative in FCM studies, to select the best reference pattern to be used. Currently, a number of reference genotypes are available16 and one has to select the one more suited to its needs. Estimates have indicated that more than 6,000 plant species have their 2C values determined.20,21 The majority of these estimates (84.5%) were obtained by FCM, while the remaining was obtained using Feulgen methods. Recently Praça-Fontes et al,16 using image cytometry, reassessed in a cascade-like manner the 2C value of eight plants, from Arabidopsis thaliana through Raphanus sativus, S. lycopersicum, G. max, Zea mays, Pisum sativum, Vicia faba, to Allium cepa, which are widely used as standards in DNA quantifications. The authors not only proposed a mean 2C value for each of the eight species but also, based on statistical comparisons, indicated G. max cv. ‘Polanka’ as the most adequate primary standard. However, as highlighted by the authors, besides possessing numerous interesting features, researchers should carefully consider if soybean is indeed the best reference pattern, as it is widely known that 2C DNA content of the reference standard should be as close to that of the sample as possible.22 Here we have opted to test both soybean and tomato as our FCM standards prior to defining the E. guineensis and E. oleifera (and its interspecific hybrids) genome sizes. In order to obtained unbiased 2C estimates for Elaeis species, we also considered using both species as external or internal patterns. Current data have indicated that internal patterns outperform external patterns.17,18 A first consideration is related to the difference in precision of internal or external standards. In our data, the results for external standard are clearly less precise than those for internal standard (Table 1), such as the standard deviation for external pattern is around two to five times bigger in average to the standard deviation for the internal pattern. This corroborates current data, which indicates that internal patterns outperform external patterns. In fact, significant differences in genome size estimates were found between estimates obtained based on soybean and on tomato only when internal patterns were used (Table 2). In striking contrast, only few differences were found between genotypes when external standard was adopted, indicating that internal standard is more sensitive and should be used to estimate the genome size of the Elaeis species and hybrids.
Once we had undoubtedly determined that internal patterns were more suited for 2C value estimation, we moved forward to determine if soybean was indeed the most indicated species to be used as reference pattern as indicated by Praça-Fontes et al.16 For FCM, it has been indicated that standards must have a genome size about 0.4 to 2.5 times the size of the unknown specimen,23 including in this range both soybean, ‘Polanka’ with 2C = 2.50 pg and AT = 63.6%12,13,24 and tomato, S. lycopersicum L. ‘Stupické polní rané’, 2C = 1.96 pg.12,13 However, we actually did not find many significant differences in precision (measured by the standard deviations) when comparing soybean and tomato (Table 1). Figure 1 shows that soybean peak is closer to the sample peak than tomato peak. According to Dolezel et al,12,13 more distant standard peaks may increase the error in estimating 2C values due to linearity problems with FCM. The reported linearity problems can be due to the fact that, the greater the distance between reference and sample is, the bigger can be the extrapolation error. This might not be the only reason, and we are assuming that researchers must be aware that some kind of bias will be included in its estimates. In fact, the occurrence of such bias when using different reference patterns can be easily noted by the differences found among 2C values estimated for Elaeis spp. When using soybean and tomato, estimates obtained based on tomato were fairly underestimated when compared to the estimates obtained based on soybean (Table 1). To minimize such linearity problem, the reference sample must be chosen in a way that its genome is the closest as possible to the investigated sample genome, without overlapping. Then, due to its peak closeness (Fig. 1), more accurate estimates are expected to be obtained when soybean is used as reference standard instead of tomato. Besides the soybean genome size, soybean has been indeed reported as the most stable and appropriate standard for FCM analysis when compared to other species such as R. sativus L. ‘Saxa’, S. lycopersicum L. ‘Stupické polní rané’, Z. mays L. ‘CE-777’, P. sativum L. ‘Ctirad’, V. faba L. ‘Inovec’, and A. cepa L. ‘Alice’ ‘Inovec’ e A. thaliana (L.) Heynh ‘Columbia’.16 It is also interesting to note that this same genotype (‘Polanka’) was recently used to obtain the genome size of macaw palm (Acrocomia aculeate),25 another Arecaceae species and another promising plant for biodiesel production.
So far, we had determined that soybean was the most stable and appropriate standard for FCM analysis of Elaeis spp., and that it should be used as internal standard to minimize precision issues. Our next step was then to perform a comparison between the results obtained using soybean as internal standard in linear or logarithmic scales. With regards to this, we noted that there is a tendency to log scale to return estimates a bit larger than the linear scale. On average, the standard deviation that resulted from using soybean as an internal standard and measuring the results in log or linear scales is similar. However, in one case, the standard deviation produced by the linear scale, that averaged 0.060, was unexpectedly elevated (Eo0312—0.244), while the maximum value for log standard deviation was 0.088, for Eg0920 (log standard deviations averaged 0.052). We considered, however, this to be a minor issue.
Taken together, these results indicated that for the correct estimation of the genome size of Elaeis species, soybean should be used as internal standard and readings recorded preferentially based on linear scales, since this kind of procedure is widely adopted in the research community.
New genome size estimates contradict currently published data
To our knowledge, only few10,11,19 studies were conducted to date using FCM to determine the genome size of E. guineensis, E. oleifera, and hybrids produced from crosses between those species. Genome size estimates currently available for the Elaeis spp. are, however, controversial, as they indicate that American oil palm genome is about half the size of the African oil palm genome and that the interspecific hybrid genome is far bigger than both the parental species genome.10,11 We believe that a number of reasons may explain the inconsistencies detected, including the standard patterns used and intrinsic factors of the species used for determining the genome size through FCM.
The first study that aimed at determining the genome size of E. guineensis was conducted by Rival et al.10 These authors, by using hybrid petunia as reference standard, reported a 2C value of 3.76 ± 0.09 pg for plants grown in vitro. The first study that aimed at determining the genome size of E. guineensis cv. ‘Dura’, ‘Pisifera’, and ‘Tenera’ was, however, conducted by Srisawat et al.19 These authors reported for E. guineensis cv. ‘Dura’, ‘Pisifera’, and ‘Tenera’ 2C values of 3.46 ± 0.02; 3.24 ± 0.01, and 3.76 ± 0.02 pg, respectively. Latter, Madon et al,11 using a similar procedure, ie, FCM with soybean as external standard, estimated 2C values of 4.10 ± 0.02, 3.64 ± 0.28, and 3.83 ± 0.31 pg for ‘Dura’, ‘Pisifera’, and ‘Tenera’, respectively. As can be noted, 2C values obtained by Rival et al,10 Srisawat et al,19 and Madon et al11 are similar, even though the results presented by Madon et al11 are slightly larger. Madon et al11 was also the first to estimate the 2C value of E. oleifera and interspecific hybrids genomes. They estimated 2C values of 2.08 ± 0.04 for E. oleifera (Suriname) and 4.16 ± 0.32 for the interspecific hybrid. Srisawat et al19 also tested tomato as reference sample. In this case, these authors reported for E. guineensis a 2C value of 4.25 ± 0.09 pg. However, due to the fact that this value was considerably larger than those obtained when using soybean (3.77 ± 0.09 pg)19 and hybrid petunia (3.76 ± 0.09 pg)10 as references, Srisawat et al19 argued that it was not reliable. The same problem was detected when Srisawat et al19 used maize as reference standard, ie, the 2C value was considerably inflated (4.72 ± 0.23 pg).
The estimates obtained by Rival et al,10 Srisawat et al,19 and Madon et al11 have been widely adopted by the oil palm research community. The community, however, focuses its research efforts in E. guineensis cv. ‘Dura’, ‘Pisifera’, and ‘Tenera’. For this species, the genome size estimates seem to be quite good. However, when it comes to American oil palm (E. oleifera), FCM results seem to be controversial. Genome size estimates currently available indicate, as previously mentioned, that American oil palm genome is about half the size of the African oil palm genome and that the interspecific hybrid genome is far larger than both the parental species’ genome.10,11 This is intriguing since the close relationship between E. guineensis and E. oleifera and especially the fact that both species hybridize well suggest that their genomes would be similar in size. Moreover, the genome size of the hybrid is expected to be about the average of the parents’ genome. This fact must have remained unnoticed for quite a long time due to the fact that the oil palm research community, until recently did not pay much attention to American oil palm. Only recently E. oleifera has received more attention (especially in Brazil) due to spreading of bud rot.7,8 American oil palm genotypes are apparently the best tolerance/resistance source to this abnormality currently available.
Since we needed high-quality data on the genome size of E. oleifera, to support our sequencing experiment (currently underway) and because the inconsistencies in the studies related to Elaeis spp. genome size previously published make difficult to draw final conclusions about their actual genome sizes, we decided to reestimate E. guineensis, E. oleifera, and F1 hybrids genome sizes through FCM. By using soybean as internal standard and recording data based on linear scales, we determined that, on average, the genome size of E. guineensis is 4.32 ± 0.173 pg, while that of E. oleifera is 4.43 ± 0.18 pg. Comparing our data with those published by Rival et al,10 Srisawat et al,19 and Madon et al,11 it becomes clear that the use of more adequate reference sample resulted in different results. All previously cited papers used external patterns, which we have shown to be less precise than internal standards. Another interesting thing to notice is that considering only E. guineensis, our estimates of genome size are clearly the larger ones. This of course can be due to methodological issues, here addressed, or alternatively due to variations of genome size between plants from different origins or variations due to tissue and stage of development. It has been shown that, the organ type, plant’s nature (in vitro versus seed-germinated plants), and stage of development can largely influence FCM results.12,13 However, the most intriguing and controversial comparison arises when we compare our results for E. oleifera and F1 hybrids and those obtained by Madon et al.11 We estimated the American oil palm genome size to be 4.57 pg while Madon et al11 estimated it to be 2.08 pg, ie, our estimate of E. oleifera genome size is 105% larger than the one obtained by Madon et al.11 It is known, based on diversity studies26 that African oil palm and American oil palm are close relatives and that members of the Arecaceae family possess large genomes. Accordingly, Abreu et al25 determined the macaw palm (A. aculeate) mean 2C value and base composition corresponded to 2C = 5.81 pg and AT = 58.3%. Thus, the 2C value of 2.08 pg presented by Madon et al11 seems to be unrealistic. Perhaps the most plausible explanation of this discrepancy is that Madon et al11 determined the 2C value of a spontaneous haploid individual, as it has been shown that such kind of individuals may naturally occur in African oil palm populations.27 One cannot also rule out some sort of poor isolation of nuclei, which subsequently resulted in the lower genome size estimated for E. oleifera. The results of the recently published African oil palm genome and of the draft sequence of a distinct E. oleifera genotype5 further support this conclusion, as no consistent difference in genome size was detected between the species. Another fact that further supports our estimates are the estimates of genome size for the F1 interspecific hybrids. As expected given the parental species genome sizes, we report that the hybrid genome size is around the average of the two genomes, ie, 4.40 ± 0.016 pg. Madon et al,11 on the other hand, reported that the F1 hybrids genome corresponded to 4.16 pg, even though the African oil palm genome was around 4 pg and the genome of the American oil palm was around 2 pg. Our results are in accordance to what is expected based on the widely known meiotic mechanisms. This behavior of the hybrids in relation to their parents (2C values after crossing) was described and confirmed in previous works with Alstroemeria L.28 and Cirsum.29 Then we propose that the values presented here should be adopted henceforth by the oil palm research community.
Drawing a suitable strategy to sequence the whole genome of native E. oleifera genotype based on the new genome size estimates and GC content information
The recognition that the E. oleifera genome is in fact two times larger than it was previously thought, and in fact even larger than the E. guineensis genome, and had a profound impact in the sequencing strategy employed in the E. oleifera wholegenome sequencing effort that is being currently carried out by Embrapa. Below, we briefly discuss the implications of the new estimates and the sequence strategy ultimately adopted by our research group.
Aiming at generating basic genomic information that would support a well-designed sequence experiment, besides looking at the genome size we checked nucleotide composition. In that respect, we emphasize that data were evaluated for nucleotides composition only after quality evaluation and disposal of low-quality reads. Although CSPR classically demands long reads (100 kb), we applied it here for short reads considering that with a large number of small reads, the parities A-T and G-C are expected. Figure 3 shows the disparities with CSPR, since for over 70 million reads a uniform behavior in the proportions was expected. The final positions usually present lower quality, even after quality control, explaining the conservative upper cut in position 80 in Table 3. However, the quality of initial positions is usually high and the probable cause for initial variation is a bias in sequencing. In fact, Aird et al30 described a bias in Illumina Polymerase Chain Reaction (PCR) amplification for sequencing, and such bias can satisfactorily explain the initial variation. The range between positions 10 and 80 is consistent with CSPR and allows us to estimate values close to real one, despite bias and quality issues. The GC content is around 38% for both E. guineensis and E. oleifera, presenting a small variation among the groups. Even though we cannot detect a significant difference, a trend to a smaller value in ‘Tenera’ (37.88) and a greater value in Manicoré (38.43) seems to exist. These data are in accordance with the results recently published regarding the oil palm genome in Nature. Singh et al5 determined that guanine–cytosine content of the E. guineensis genome is 37% but that genes were conspicuous for having a much higher guanine–cytosine content (50%). This further supports our results, and our approach of evaluating the GC content of different genotypes based on short reads.
Other analyses, like genotypes assembly comparisons, are being performed to define the best genotype to generate the assembly and the best strategy. We can, however, based on the results presented here, foresee that we will need to double our sequencing and assembly efforts, considering that the sequence strategy was initially thought based on the results of Madon et al.11 We also anticipate that we will need to take special care with transposable elements (analyses with present data are in progress), since bigger plant genomes tend to present high level of repetitive regions. Accordingly, Singh et al5 predicted 158,946 gene candidates covering 92 Mb of exonic gene space for the African oil palm genome (5% of the 1.8-Gb genome sequence). Known retroelements and other transposons accounted for almost 70% of the predicted candidates. This means that greater genome coverage must be achieved, with an increase in importance of mate-pairs libraries with large insert sizes.
In summary, this work sheds new light on genome size of the Elaeis species and its hybrids and helped to correctly determine the sequence strategy we are employing in our E. oleifera sequencing effort.
Acknowledgments
We are indebted to the support staff of Embrapa Agroenergy and Embrapa Dairy Cattle for their assistance during the experiments. We also wish to thank the anonymous reviewers for the detailed revision of the manuscript and useful comments.
Footnotes
Author Contributions
Conceived and designed the experiments: JC, APL, AAA, ALSA, JDN, MTSJ. Analyzed the data: JC, APL, AAA, EFF, GC, JKAM, MTSJ. Wrote the first draft of the manuscript: JC, APL, AAA, EFF, MTSJ. Contributed to the writing of the manuscript: JC, APL, AAA, EFF, ALSA, JDN, GC, JKAM, MTSJ. Agree with manuscript results and conclusions: JC, APL, AAA, EFF, ALSA, JDN, GC, JKAM, MTSJ. Jointly developed the structure and arguments for the paper: JC, APL, AAA, EFF, GC, MTSJ. Made critical revisions and approved final version: JC, APL, AAA, MTSJ. All authors reviewed and approved of the final manuscript.
ACADEMIC EDITOR: Gustavo Caetano-Anollés, Editor in Chief
FUNDING: We acknowledge the financial support provided by the Brazilian Ministry of Science, Technology and Innovation, through a Financiadora de Estudos e Projetos (FINEP) grant Dinamização do banco ativo de germoplasma de dendê (Elaeis guineesis) e apoio ao melhoramento genético (PRODENDE). This work was also supported by the Brazilian National Research Council, CNPq, with a research fellowship to M.T.S. Junior.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
This paper was subject to independent, expert peer review by a minimum of two blind peer reviewers. All editorial decisions were made by the independent academic editor. All authors have provided signed confirmation of their compliance with ethical and legal obligations including (but not limited to) use of any copyrighted material, compliance with ICMJE authorship and competing interests disclosure guidelines and, where applicable, compliance with legal and ethical guidelines on human and animal research participants.
Internet Resources
FASTX-Toolkit, http://hannonlab.cshl.edu/fastx_toolkit (July 30, 2012).
WinMDI version 2.8 software, http://facs.scripps.edu/software.html (April 18, 2012).
REFERENCES
- 1.Corley RHV. The genus Elaeis. In: Corley RHV, Hardon JJ, Wood BJ, editors. Oil Palm Research. 2nd ed. New York: Elsevier Scientific Publishing Company Inc; 1982. pp. 03–05. [Google Scholar]
- 2.Cunha RNV, Lopes R, Barcelos E. Domesticação e Melhoramento de caiaué. In: Borém A, Lopes MTG, Clement CR, editors. Domesticação e Melhoramento: Espécies Amazônicas. Viçosa: Editora da Universidade de Viçosa; 2009. pp. 275–296. [Google Scholar]
- 3.Basiron Y. Palm oil production through sustainable plantations. Eur J Lipid Sci Tech. 2007;109(4):289–295. [Google Scholar]
- 4.Chia GS, Lopes R, Cunha RNV, Rocha RNC, Lopes MTG. Repetibilidade da produção de cachos de hibridos interespecificos entre American oil palm e dendezeiro. Acta Amazon. 2009;39(2):249–254. [Google Scholar]
- 5.Singh R, Ong-Abdullah M, Low ET, et al. Oil palm genome sequence reveals divergence of interfertile species in old and new worlds. Nature. 2013;500:335–339. doi: 10.1038/nature12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wahid MB, Abdullah SNA, Henson IE. Oil palm—achievements and potential; 4th International Crop Science Congress; Brisbane. 2004. pp. 01–13. [Google Scholar]
- 7.Barcelos E, Nunes CDM, Cunha RNV. Melhoramento genético e produção de sementes comerciais de dendezeiro. In: Viégas IJ, Muller AA, editors. A cultura do dendezeiro na Amazônia brasileira. Belém, PA: Embrapa Amazonia Oriental/Embrapa Amazonia Ocidental; 2000. pp. 145–171. [Google Scholar]
- 8.Veiga AS, Furlan J, Junior, Kaltner FJ. Situação atual e perspectivas futures da dendeicultura nas principais regiões produtoras: a experiência do Brasil. In: Muller AA, Furlan J Junior, editors. Agronegócio do dendê: uma alternativa social, econômica e ambiental para o desenvolvimento sustentável da Amazônia. Belém, PA: Embrapa Amazonia Oriental; 2001. pp. 41–66. [Google Scholar]
- 9.Osaki M, Batalha MO. Produção de biodíesel e óleo vegetal no Brasil: realidade e desafio. Organizações Rurais & Agroindustriais. 1997;13(2):227–242. [Google Scholar]
- 10.Rival A, Beule T, Barre P, Hamon S, Duval Y, Noirot M. Comparative flow cytometric estimation of nuclear DNA content in oil palm (Elaeis guineensisJacq) tissue cultures and seed-derived plants. Plant Cell Rep. 1997;16:884–887. doi: 10.1007/s002990050339. [DOI] [PubMed] [Google Scholar]
- 11.Madon M, Phoon LQ, Clyde MM, Mohd Din A. Application of flow cytometry for estimation of nuclear DNA content in Elaeis. J Oil Palm Res. 2008;20:447–452. [Google Scholar]
- 12.Dolezel J, Greilhuber J, Suda J. Estimation of nuclear DNA content in plants using flow cytometry. Nat Protoc. 2007;2(9):2233–2244. doi: 10.1038/nprot.2007.310. [DOI] [PubMed] [Google Scholar]
- 13.Dolezel J, Greilhuber J, Suda J. Flow cytometry with plants: an overview. In: Dolezel J, Greilhuber J, Suda J, editors. Flow Cytometry with Plant Cells: Analysis of Genes, Chromosomes and Genomes. Weinheim: Wiley-VCH Verlag GmbH & Co. KGa; 2007. pp. 41–66. [Google Scholar]
- 14.Dolezel J, Sgorbatti S, Lucretti S. Comparison of three DNA fluorochromes for flow cytometry estimation of nuclear DNA content in plants. Physiol Plant. 1992;85:625–631. [Google Scholar]
- 15.Greilhuber J, Temsch EM, Loureiro JCM. Nuclear DNA content measurement. In: Dolezel J, Greilhuber J, Suda J, editors. Flow Cytometry with Plant Cells—Analysis of Genes, Chromosomes and Genomes. Weinheim: Wiley-VCH Verlag GmbH & Co. KGaA; 2007. pp. 67–101. [Google Scholar]
- 16.Praça-Fontes MM, Carvalho CR, Clarindo WR. C-value reassessment of plant standards: an image cytometry approach. Plant Cell Rep. 2011;30(12):2303–2312. doi: 10.1007/s00299-011-1135-6. [DOI] [PubMed] [Google Scholar]
- 17.Leitch IJ, Bennet MD. Genome size and its uses: the impact of flow cytometry. In: Dolezel J, Greilhuber J, Suda J, editors. Flow Cytometry with Plant Cells. Weinheim: Wiley-VCH Verlag GmbH & Co. KGa; 2007. pp. 153–176. [Google Scholar]
- 18.Loureiro J, Suda J, Dolezel J, Santos C. Flower: a plant DNA cytometry database. In: Dolezel J, Greilhuber J, Suda J, editors. Flow Cytometry with Plant Cells. Weinheim: Wiley-VCH Verlag GmbH & Co. KGa; 2007. pp. 423–438. [Google Scholar]
- 19.Srisawat T, Kanchanapooom K, Pattanapanyasat K, Srikul S, Chuthammathat W. Flow cytometry analysis of oil palm: a preliminary analysis for cultivars and genomic DNA alteration. Songklanakarian J Sci Technol. 2005;27(suppl 3):645–652. [Google Scholar]
- 20.Bennett MD, Bhandol P, Leitch IJ. Nuclear DNA amounts in angiosperms and their modern uses-807 new estimates. Ann Bot. 2000;86:859–909. [Google Scholar]
- 21.Zonneveld BJM, Leitch IJ, Bennett MD. First nuclear DNA amounts in more than 300 angiosperms. Ann Bot. 2005;96:229–244. doi: 10.1093/aob/mci170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dolezel J, Bartos J. Plant DNA flow cytometry and estimation of nuclear genome size. Ann Bot. 2005;95:99–110. doi: 10.1093/aob/mci005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Voglmayr H. DNA flow cytometry in non-vascular plants. In: Dolezel J, Greilhuber J, Suda J, editors. Flow Cytometry with Plant Cells: Analysis of Genes, Chromosomes and Genomes. Weinheim: Wiley-VCH Verlag GmbH & Co. KGa; 2007. pp. 267–286. [Google Scholar]
- 24.Barow M, Meister A. Lack of correlation between AT frequency and genome size in higher plants and the effect of nonrandomness of base sequences on dye binding. Cytometry Part A. 2002;47:1–7. doi: 10.1002/cyto.10030. [DOI] [PubMed] [Google Scholar]
- 25.Abreu IS, Carvalho CR, Carvalho GMA, Motoike SY. First karyotype, DNA C-value and AT/GC base composition of macaw palm (Acrocomia aculeata, Arecaceae)—a promising plant for biodiesel production. Aust J Bot. 2011;59:149–155. [Google Scholar]
- 26.Moretzsohn MC, Ferreira MA, Amaral ZPS, Coelho PJA, Grattapaglia D, Ferreira ME. Genetic diversity of Brazilian oil palm (Elaeis oleiferaHBK) germplasm collected in the Amazon Forest. Euphytica. 2002;124(1):35–45. [Google Scholar]
- 27.Dunwell JM, Wilkinson MJ, Nelson S, et al. Production of haploids and doubled haploids in oil palm. BMC Plant Biol. 2010;10(218):01–25. doi: 10.1186/1471-2229-10-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buittendijk J, Boon EJ, Romanna MS. Nuclear DNA content in twelve species of Alstroemeria L. and some of their hybrids. Ann Bot. 1997;79:343–353. [Google Scholar]
- 29.Bures P, Wang YF, Horivá L, Suda J. Genome size variation in Central European species of Cirsum (Compositae) and their natural hybrids. Ann Bot. 2004;94:353–363. doi: 10.1093/aob/mch151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aird D, Ross MG, Chen WS, et al. Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biol. 2011;12:R18. doi: 10.1186/gb-2011-12-2-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]