Abstract
Numerous studies have investigated the association of IL10 -819C>T and -592C>A polymorphisms with non-Hodgkin lymphoma (NHL) susceptibility, and yet reported conflicting results. With this in mind, we performed the current meta-analysis with an aim to verify actual causative variants underlying lymphomagenesis. Pooled odds ratios (ORs) and 95% confidence intervals (CIs) were calculated to evaluate the strength of the associations. Moreover, to explore the biological function of these polymorphisms, we also performed genotype-based mRNA expression analysis using online database derived from 270 subjects within three ethnicities. The final analysis included 11 studies with a total of 5859 NHL cases and 6893 controls for the IL10 -819C>T polymorphism, and 11 studies with 6277 cases and 7350 controls for the IL10 -592C>A polymorphism. No significant association was observed for these two polymorphisms in either the overall analysis or the stratification analyses by ethnicity and source of controls. Nevertheless, stratification analyses demonstrated a significant decreased risk associated with the IL10 -819C>T polymorphism (homozygous: OR=0.81, 95% CI=0.66-0.99, and recessive model: OR=0.80, 95%CI=0.65-0.98) and IL10 -592C>A polymorphism (homozygous: OR=0.80, 95% CI=0.66-0.99, and recessive model: OR=0.80, 95%CI=0.66-0.97) among patients with diffuse large B-cell lymphoma (DLBCL). Despite some limitations, this meta-analysis indicates that polymorphisms in IL10 gene may contribute to DLBCL susceptibility.
Keywords: IL10, NHL, polymorphism, susceptibility, meta-analysis
Introduction
Cancer is a major public health problem and burden worldwide, which cause remarkable cancer-associated death and disability. Cancer-related deaths account for one eighth of deaths overall - more than combined death toll caused by AIDS, tuberculosis and malaria together. Non-Hodgkin lymphoma (NHL) originated from hematological system is one of the most malignant tumors. According to GLOBOCAN 2008 estimates, approximately 355,900 new cases and 191,400 deaths from NHL occurred that year 1, 2. The developed areas such as North America, Australia/New Zealand, and Northern, Western, and Southern Europe were found with the highest incidence rates, while South-Central and Eastern Asia and the Caribbean with the lowest incidence rates 1. NHL is a heterogeneous group of lymphoproliferative malignancies, with more than 60 recognized specific NHL subtypes. Mostly commonly, NHLs are classified into B-cell and T-cell lymphomas based on cell origins. In particular, B-cell lymphomas account for about 80%-85% of NHL cases.
Although the clear etiology of NHL is warranted extensive investigation, there are some well-documented NHL risk factors, such as immune dysfunction and stimulation, infection, high doses of radiation, family history, and occupational exposures to pesticides and chlorinated organic compounds 3. Numerous studies have indicated that altered immunity may also play important roles in the development and prognosis of NHL 4-6. Besides environmental exposures, chromosomal and genetic alterations that affect immune function may modulate the risk of developing NHL 7-10.
Multifunctional cytokines [e.g., tumor necrosis factor-alpha and interleukin 10 (IL10)] that regulate the immunological development and inflammatory responses have been suggested to partake in carcinogenesis 11. IL10 is a well-known anti-inflammatory cytokine whose principal biologic function includes, but not limited to, the suppression of cytokine synthesis in Th1 cells and down-regulation of cell-mediated and cytotoxic inflammatory responses 12, 13. Interestingly, accumulating evidence has shown that IL10 is also involved in the pathogenesis of lymphoid malignancies 14. Increased serum levels of IL10 were found in NHL patients, which were correlated with adverse clinicopathological features and poor outcomes 15.
The IL10 gene is located on chromosome 1q31-1q32 16. It is highly polymorphic, and there are at least 189 reported single nucleotide polymorphisms (SNPs) in the IL10 gene region (http://www.ncbi.nlm.nih.gov/projects/SNP). Among all the indentified SNPs, IL10 -819C>T (rs1800871) and -592C>A (rs1800872) in the promoter region have been widely investigated 17. Previous study have found that the -819C>T and -592C>A polymorphisms were associated with increased serum level of IL10 in NHL patients and may influence NHL susceptibility and clinical outcomes 18. Given potential influence on gene transcription and expression, the promoter polymorphisms have undergone the most extensive scrutiny. Numerous studies have investigated the associations of IL10 -819C>T 18-28 and -592C>A 18-25, 27-29 polymorphisms with NHL susceptibility, but the conclusions of these studies remains conflicting rather than conclusive. The disagreements may be ascribed to relatively small sample size in each study as well as ethnic difference. Hence, we performed the present meta-analysis to provide a more precise estimation of the associations of IL10 -819C>T and -592C>A with NHL susceptibility.
Materials and methods
Identification of relevant studies
In order to track down all relevant studies for the given topic to limit bias, we conducted a comprehensive and systematical literature searched in PubMed and Embase, using the following items: “interleukin-10 or IL-10 or IL10”, “polymorphism or variant or variation” and “non-Hodgkin lymphoma or non-Hodgkin's lymphoma or NHL” (prior to December 18, 2014). Reference lists of original articles and review articles were also checked manually to identify additional pertinent studies. The search was limited to investigations written in English. If more than one article was published with the same patient population, only the latest or the largest study would be used in this study.
Inclusion and exclusion criteria
Studies included had to meet the following criteria: (a) evaluating of the association between IL10 -819C>T and/or -592C>A polymorphisms and cancer risk, (b) using a case-control or cohort design (retrospective or prospective and nested case-control), (c) providing sufficient information to estimate odds ratios (ORs) and their corresponding 95% confidence intervals (CIs), and (d) containing available genotype frequency.
Exclusion criteria were: (a) cases only or healthy subjects only studies, (b) duplicate publication, or (c) the genotype frequency distribution in the controls was not in accordance with Hardy-Weinberg equilibrium (HWE).
Data extraction
Two investigators (Ting Zhang and Shang Xie) assessed all retrieved studies following the inclusion criteria, reached a consensus on all items, and then extracted the following information independently from all eligible publications: the first author's surname, year of published, country of origin, ethnicity, source of control [population based (PB) and hospital based (HB)], genotyping methods, subtype of NHL [diffuse large B-cell lymphoma (DLBCL) and follicular lymphoma (FL)], total number of cases and controls, as well as genotype counts of IL10 -819C>T (CC, CT, and TT) and -592C>A (CC, AC, and AA) polymorphisms in cases and controls.
Correlation analysis between Genotype and gene expression
The genotype data for IL10 polymorphisms (rs1800871C>T and rs1800872C>A) were available online from HapMap (http://hapmap.ncbi.nlm.nih.gov/), while corresponding IL10 mRNA expression levels derived from 270 subjects of three ethnicity (Caucasians, Asians and Africans) were located in SNPexp (http://app3.titan.uio.no/biotools/tool.php?app=snpexp) website as described previously 30-32.
Statistical methods
The strength of association between IL10 gene polymorphisms and NHL risk was assessed by calculating ORs with their corresponding 95% CIs. The pooled ORs and 95% CIs were performed for -819C>T using different genetic models: homozygous (TT vs. CC), heterozygous (CT vs. CC), recessive (TT vs. CT+CC), dominant (TT+CT vs. CC) and allele comparing (T vs. C). Likewise, the same models were applied to IL10 -592C>A: homozygous (AA vs. CC), heterozygous (AC vs. CC), recessive (AA vs. AC+CC), dominant (AA+AC vs. CC) and allele comparing (A vs. C). The Chi square-based Q test was performed to calculate the between-study heterogeneity. If P<0.1, the random-effects model would be adopted; otherwise, the fixed-effects model was used. Stratification analysis was performed by race (Asians, Caucasians, and mixed), source of control (PB or HB) and subtype (DLBCL and FL). The chi-square goodness-of-fit test was evaluated for HWE, with a P-value below 0.05 indicating the departure from HWE. Furthermore, we conducted sensitivity analyses by excluding each investigation individually to evaluate the influence of each individual study on summary ORs. Funnel plot and Egger's test were used to assess publication bias. Differences in mRNA expression levels were determined by Student's t test. Moreover, the linear trend of mRNA expression levels among genotypes (0, 1 and 2 variant alleles) was tested by linear regression models. All statistical data manipulations were conducted by using the STATA software (Version 11.0, Stata, College Station, TX). A P-value less than 0.05 was defined as statistical significant.
Results
Study characteristics
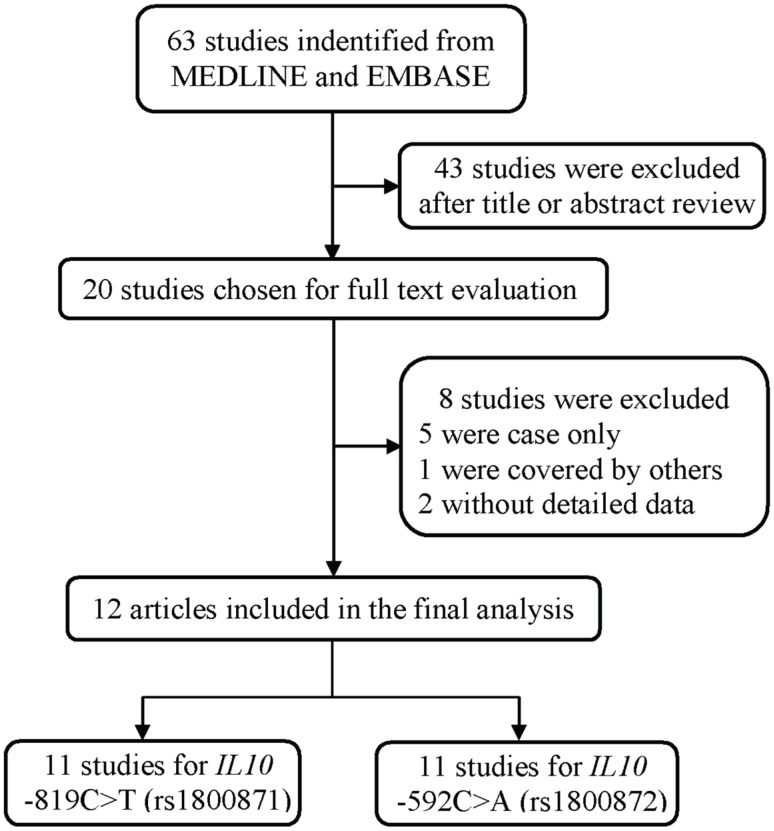
As shown in Figure 1, a total of 63 publications were retrieved from PubMed and Embase database. After the assessment of abstracts and texts, 43 irrelevant publications were excluded, and the remaining 20 publications that met the crude inclusion criteria and were subjected to further evaluation. Among them, eight investigations were excluded, for five 33-37 were case only studies, one 38 was covered by another study 22, and two 39, 40 without detailed data available. Finally, 12 investigations 18-29 met the inclusion criteria and were included in the final meta-analysis (Table 1). Overall, 11 studies with 5859 cases and 6893 controls investigated the IL10 -819C>T polymorphism, and 11 studies with 6277 cases and 7350 controls investigated the IL10 -592C>A polymorphism. Of the remaining studies, six studies provided detailed genotype frequency data for the DLBCL and FL subtype for these two polymorphisms (Supplemental Table S1).
Figure 1.
The flowchart of included studies.
Table 1.
Characteristics of studies included in the current meta-analysis
| Surname | Year | Country | Ethnicity | Source | Genotype method | Case | Control | MAF | HWE | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| -819C>T (rs1800871) polymorphism | CC | CT | TT | All | CC | CT | TT | All | |||||||
| Lech-Maranda | 2004 | France | Caucasian | HB | PCR-RFLP | 107 | 81 | 11 | 199 | 53 | 46 | 13 | 112 | 0.321 | 0.536 |
| Guzowski | 2005 | USA | Caucasian | HB | DHPLC | 9 | 6 | 2 | 17 | 14 | 10 | 1 | 25 | 0.24 | 0.629 |
| Lan | 2006 | USA | Mixed | PB | TaqMan | 274 | 191 | 26 | 491 | 329 | 211 | 34 | 574 | 0.243 | 0.982 |
| Persico | 2006 | Italy | Caucasian | PB | PCR-RFLP | 138 | 100 | 12 | 250 | 53 | 51 | 6 | 110 | 0.286 | 0.159 |
| Wang | 2006 | USA | Mixed | PB | TaqMan | 625 | 427 | 92 | 1144 | 514 | 339 | 81 | 934 | 0.268 | 0.021 |
| Kube | 2007 | Germany | Caucasian | NA | NA | 230 | 167 | 21 | 418 | 111 | 81 | 10 | 202 | 0.25 | 0.325 |
| Lech-Maranda | 2007 | France | Caucasian | HB | PCR-RFLP | 92 | 68 | 15 | 175 | 53 | 46 | 13 | 112 | 0.321 | 0.536 |
| Purdue | 2007 | Australia | Caucasian | PB | TaqMan | 342 | 175 | 41 | 558 | 295 | 170 | 23 | 488 | 0.221 | 0.813 |
| Andrie | 2009 | Greece | Caucasian | HB | ARMS-PCR | 22 | 20 | 6 | 48 | 45 | 35 | 5 | 85 | 0.265 | 0.594 |
| Wong | 2010 | USA | Mixed | HB | TaqMan | 109 | 47 | 2 | 158 | 88 | 57 | 9 | 154 | 0.244 | 0.954 |
| Hosgood | 2013 | Asia | Asian | HB | TaqMan | 1193 | 971 | 237 | 2401 | 2033 | 1700 | 364 | 4097 | 0.296 | 0.749 |
| -592C>A (rs1800872) polymorphism | CC | AC | AA | All | CC | AC | AA | All | |||||||
| Lech-Maranda | 2004 | France | Caucasian | HB | PCR-RFLP | 107 | 81 | 11 | 199 | 53 | 46 | 13 | 112 | 0.321 | 0.536 |
| Guzowski | 2005 | USA | Caucasian | HB | DHPLC | 10 | 5 | 2 | 17 | 13 | 10 | 2 | 25 | 0.280 | 0.968 |
| Lan | 2006 | USA | Mixed | PB | TaqMan | 273 | 174 | 35 | 482 | 331 | 189 | 43 | 563 | 0.244 | 0.032 |
| Persico | 2006 | Italy | Caucasian | PB | PCR-RFLP | 138 | 100 | 12 | 250 | 53 | 51 | 6 | 110 | 0.286 | 0.159 |
| Wang | 2006 | USA | Mixed | PB | TaqMan | 601 | 426 | 93 | 1120 | 515 | 342 | 81 | 938 | 0.269 | 0.027 |
| Kube | 2007 | Germany | Caucasian | NA | NA | 226 | 165 | 21 | 412 | 111 | 81 | 10 | 202 | 0.250 | 0.325 |
| Lech-Maranda | 2007 | France | Caucasian | HB | PCR-RFLP | 92 | 68 | 15 | 175 | 53 | 46 | 13 | 112 | 0.321 | 0.536 |
| Purdue | 2007 | Australia | Caucasian | PB | TaqMan | 343 | 176 | 21 | 540 | 297 | 169 | 23 | 489 | 0.22 | 0.868 |
| Wong | 2010 | USA | Mixed | HB | TaqMan | 107 | 49 | 11 | 167 | 88 | 57 | 9 | 154 | 0.244 | 0.954 |
| Zhang | 2012 | China | Asian | HB | TaqMan | 226 | 228 | 60 | 514 | 269 | 235 | 53 | 557 | 0.306 | 0.872 |
| Hosgood | 2013 | Asia | Asian | HB | TaqMan | 1204 | 961 | 236 | 2401 | 2041 | 1685 | 362 | 4088 | 0.295 | 0.593 |
PB, Population based; HB, Hospital based; PCR-RFLP, polymorphism chain reaction-restriction fragment length polymorphism; DHPLC, denaturing high-performance liquid chromatography; MAF, minor allele frequency; HWE, Hardy-Weinberg equilibrium.
For the IL10 -819C>T polymorphism, sample sizes of cases ranged from 17 to 2401, and those of controls from 25 to 4097. There were seven studies focused on Caucasians, one study on Asians, and three studies on mixed ethnic group. Of these 11 studies, four were PB, six were HB designed and last one did not provide the source of control. As to the IL10 -592C>A polymorphism, sample sizes ranged from 17 to 2401 for the cases, and those of controls from 25 to 4088. There were six studies conducted among Caucasians, two studies among Asians, and three studies among mixed ethnic group. Four studies were PB, and six were HB designed.
Meta-analysis results
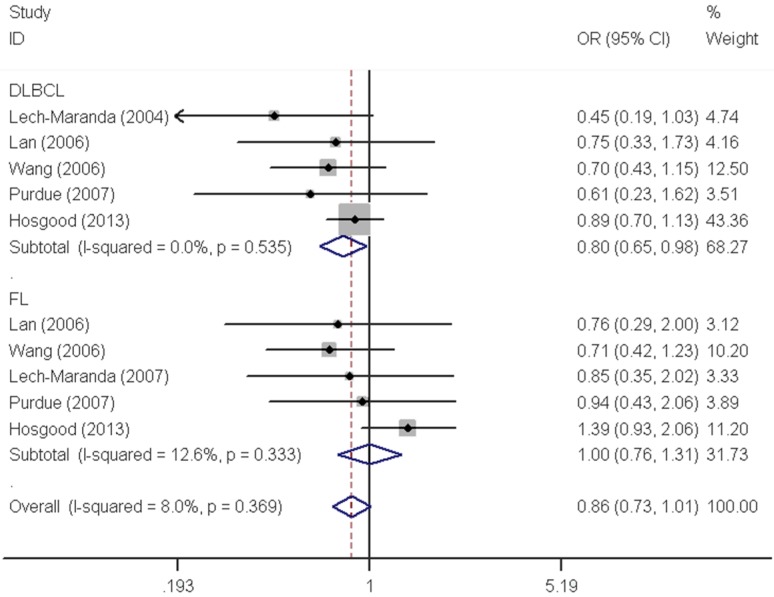
As shown in Table 2, pooled analysis did not yield a significant association between IL10 -819C>T polymorphism and overall NHL risk (homozygous: OR=0.97, 95% CI=0.76-1.22; heterozygous: OR=0.97, 95% CI=0.90-1.04; recessive: OR=1.04, 95% CI=0.92-1.19; dominant: OR=0.98, 95% CI=0.91-1.05 and allele comparing: OR=0.99, 95% CI=0.94-1.05). The subgroup analysis found no significant association by either ethnicity or source of control. However, stratified analysis by tumor subtype demonstrated a significant decreased risk associated with IL10 -819C>T polymorphism for DLBCL (homozygous: OR=0.81, 95% CI=0.66-0.99 and recessive: OR=0.80, 95% CI=0.65-0.98) (Figure 2).
Table 2.
Meta-analysis of the association between IL10 polymorphisms and NHL risk
| Variables | No. of | Sample size Case/control |
Homozygous | Heterozygous | Recessive | Dominant | Allele comparing | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| studies | OR (95% CI) | P het | OR (95% CI) | P het | OR (95% CI) | P het | OR (95% CI) | P het | OR (95% CI) | P het | ||
| -819C>T (rs1800871) | TT vs. CC | CT vs. CC | TT vs. (CT + CC) | (CT + TT) vs. CC | T vs. C | |||||||
| All | 11 | 5859/6893 | 0.97 (0.76-1.22) | 0.094 | 0.97 (0.90-1.04) | 0.821 | 1.04 (0.92-1.19) | 0.108 | 0.98 (0.91-1.05) | 0.558 | 0.99 (0.94-1.05) | 0.184 |
| Ethnicity | ||||||||||||
| Caucasian | 7 | 1665/1134 | 0.99 (0.63-1.55) | 0.118 | 0.90 (0.76-1.06) | 0.964 | 1.07 (0.79-1.46) | 0.132 | 0.92 (0.79-1.07) | 0.800 | 0.96 (0.85-1.09) | 0.365 |
| Asian | 1 | 2401/4097 | 1.10 (0.93-1.33) | / | 0.97 (0.88-1.08) | / | 1.12 (0.95-1.33) | / | 1.00 (0.90-1.10) | / | 1.02 (0.95-1.11) | / |
| Mixed | 3 | 1793/1662 | 0.80 (0.48-1.32) | 0.124 | 1.01 (0.88-1.17) | 0.190 | 0.86 (0.66-1.12) | 0.177 | 0.99 (0.86-1.13) | 0.086 | 0.97 (0.87-1.08) | 0.047 |
| Source of control | ||||||||||||
| PB | 4 | 2443/2106 | 1.02 (0.80-1.29) | 0.392 | 0.99 (0.87-1.12) | 0.430 | 1.02 (0.81-1.29) | 0.306 | 1.00 (0.88-1.12) | 0.603 | 1.00 (0.91-1.10) | 0.689 |
| HB | 6 | 2998/4585 | 0.80 (0.44-1.46) | 0.022 | 0.95 (0.86-1.05) | 0.712 | 1.05 (0.90-1.24) | 0.034 | 0.97 (0.88-1.06) | 0.244 | 0.99 (0.92-1.06) | 0.031 |
| Subtype | ||||||||||||
| DLBCL | 5 | 1997/6205 | 0.81 (0.66-0.99) | 0.510 | 1.02 (0.92-1.14) | 0.396 | 0.80 (0.65-0.98) | 0.535 | 0.99 (0.89-1.09) | 0.369 | 0.95 (0.88-1.03) | 0.374 |
| FL | 5 | 932/6205 | 1.00 (0.76-1.32) | 0.258 | 1.03 (0.88-1.20) | 0.223 | 1.00 (0.76-1.31) | 0.333 | 1.02 (0.89-1.19) | 0.161 | 1.02 (0.90-1.14) | 0.140 |
| -592C>A (rs1800872) | AA vs. CC | AC vs. CC | AA vs. (AC + CC) | (AC + AA) vs. CC | A vs. C | |||||||
| Alla | 11 | 6277/7350 | 1.04 (0.91-1.18) | 0.565 | 0.98 (0.92-1.06) | 0.652 | 1.04 (0.92-1.18) | 0.652 | 0.99 (0.93-1.06) | 0.542 | 1.00 (0.95-1.06) | 0.435 |
| Ethnicity | ||||||||||||
| Caucasian | 6 | 1593/1050 | 0.74 (0.52-1.04) | 0.743 | 0.89 (0.75-1.05) | 0.946 | 0.78 (0.55-1.09) | 0.739 | 0.87 (0.74-1.02) | 0.915 | 0.88 (0.77-1.00) | 0.824 |
| Asian | 2 | 2915/4645 | 1.14 (0.97-1.34) | 0.385 | 0.99 (0.90-1.10) | 0.206 | 1.14 (0.98-1.34) | 0.603 | 1.02 (0.93-1.12) | 0.169 | 1.04 (0.97-1.12) | 0.196 |
| Mixed | 3 | 1769/1655 | 0.99 (0.76-1.27) | 0.999 | 1.04 (0.90-1.20) | 0.235 | 0.97 (0.75-1.24) | 0.936 | 1.03 (0.90-1.18) | 0.329 | 1.01 (0.91-1.13) | 0.565 |
| Source of control | ||||||||||||
| PB | 4 | 2392/2100 | 0.94 (0.74-1.19) | 0.902 | 1.01 (0.89-1.15) | 0.366 | 0.93 (0.74-1.17) | 0.974 | 1.00 (0.89-1.12) | 0.367 | 0.99 (0.90-1.09) | 0.492 |
| HB | 6 | 3473/5048 | 1.08 (0.93-1.26) | 0.209 | 0.97 (0.88-1.06) | 0.508 | 1.09 (0.94-1.27) | 0.286 | 0.99 (0.91-1.08) | 0.336 | 1.01 (0.94-1.08) | 0.187 |
| Subtype | ||||||||||||
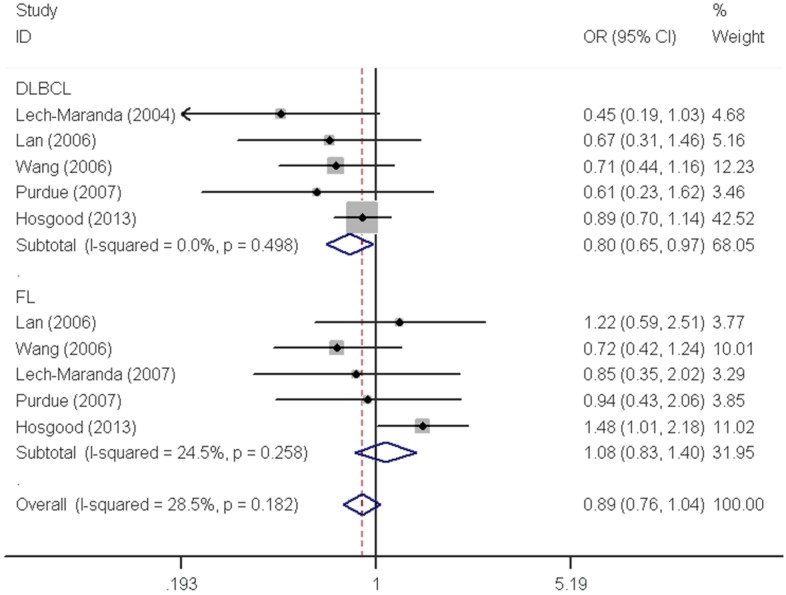
| DLBCL | 5 | 1986/6190 | 0.80 (0.66-0.99) | 0.510 | 1.02 (0.91-1.13) | 0.363 | 0.80 (0.66-0.97) | 0.498 | 0.98 (0.88-1.08) | 0.399 | 0.95 (0.87-1.03) | 0.448 |
| FL | 5 | 934/6190 | 1.08 (0.83-1.42) | 0.164 | 1.02 (0.88-1.19) | 0.375 | 1.08 (0.83-1.40) | 0.258 | 1.03 (0.89-1.19) | 0.199 | 1.03 (0.92-1.16) | 0.098 |
HB, Hospital based; PB, Population based; DLBCL, Diffuse large B-cell lymphomas; FL, Follicular lymphoma.
Figure 2.
Forest plot of NHL risk associated with the IL10 -819C>T polymorphism by subtypes under recessive model. For each study, the estimates of OR and their corresponding 95% CI were plotted with a box and a horizontal line. The symbol filled diamond indicates pooled OR and its corresponding 95% CI.
Similar results were observed for the IL10 -592C>A polymorphism. We did not find any significant association with overall NHL risk (homozygous: OR=1.04, 95% CI=0.91-1.18; heterozygous: OR=0.98, 95% CI=0.92-1.06; recessive: OR=1.04, 95% CI=0.92-1.18; dominant: OR=0.99, 95% CI=0.93-1.06 and allele comparing: OR=1.00, 95% CI=0.95-1.06). Moreover, no any significant association was observed between IL10 -592C>A polymorphism and NHL risk either, when data were stratified by ethnicity and source of control. In contrast, a significant decreased DLBCL risk was found with IL10 -592C>A polymorphism in the stratification analysis by tumor subtype (homozygous: OR=0.80, 95% CI=0.66-0.99 and recessive: OR=0.80, 95% CI=0.66-0.97) (Figure 3).
Figure 3.
Forest plot of NHL risk associated with the IL10 -592C>A polymorphism by subtypes under recessive model. For each study, the estimates of OR and their corresponding 95% CI were plotted with a box and a horizontal line. The symbol filled diamond indicates pooled OR and its corresponding 95% CI.
The correlation between the mRNA expression and genotypes
We explored the correlation between IL10 mRNA expressions levels and the genotypes of IL10 polymorphisms among Caucasians, Africans and Asians as well as the whole group (Table 3). We failed to find any significant difference in the IL10 mRNA expression levels among subjects with three genotypes (wild-type, heterozygote, and homozygote) of either the IL10 -819C>T or -592C>A polymorphism in the Africans, Asians and the whole group. However, among the Caucasians, IL10 mRNA expression level in IL10 -819C>T polymorphism heterozygotes was significantly higher than wild-types (heterozygous: P=0.041), while IL10 mRNA expression levels in heterozygous carriers or combined heterozygous and homozygous carriers of IL10 -592C>A polymorphism were significantly increased, when compared with wild-type control (heterozygous: P=0.030 and dominant model: P=0.023).
Table 3.
IL10 mRNA expression by the genotypes of SNPs, using data from the HapMapa
| Population | -819C>T (rs1800871) | -592C>A (rs1800872) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| genotypes | No. | Mean ± SD | Pb | Ptrendc | genotypes | No. | Mean ± SD | Pb | Ptrendc | |
| Caucasiand | CC | 52 | 7.77 ± 0.49 | 0.106 | CC | 54 | 7.78 ± 0.50 | 0.070 | ||
| CT | 28 | 8.02 ± 0.59 | 0.041 | AC | 33 | 8.04 ± 0.61 | 0.030 | |||
| TT | 1 | 7.55 | 0.668 | AA | 2 | 8.21 ± 0.93 | 0.246 | |||
| Dominant | 29 | 8.00 ± 0.58 | 0.053 | Dominant | 35 | 8.05 ± 0.61 | 0.023 | |||
| Africand | CC | 24 | 7.60 ± 0.53 | 0.369 | CC | 24 | 7.60 ± 0.53 | 0.449 | ||
| CT | 43 | 7.76 ± 0.46 | 0.196 | AC | 48 | 7.75 ± 0.45 | 0.219 | |||
| TT | 17 | 7.64 ± 0.43 | 0.793 | AA | 18 | 7.68 ± 0.44 | 0.617 | |||
| Dominant | 60 | 7.73 ± 0.45 | 0.269 | Dominant | 66 | 7.73 ± 0.45 | 0.252 | |||
| Asiand | CC | 6 | 7.41 ± 0.52 | 0.217 | CC | 6 | 7.41 ± 0.52 | 0.151 | ||
| CT | 33 | 7.57 ± 0.40 | 0.373 | AC | 39 | 7.57 ± 0.40 | 0.375 | |||
| TT | 40 | 7.70 ± 0.45 | 0.153 | AA | 44 | 7.71 ± 0.45 | 0.133 | |||
| Dominant | 73 | 7.64 ± 0.43 | 0.207 | Dominant | 83 | 7.64 ± 0.43 | 0.200 | |||
| Alld | CC | 82 | 7.69 ± 0.51 | 0.403 | CC | 84 | 7.70 ± 0.52 | 0.575 | ||
| CT | 104 | 7.77 ± 0.50 | 0.283 | AC | 120 | 7.77 ± 0.52 | 0.334 | |||
| TT | 58 | 7.68 ± 0.44 | 0.889 | AA | 64 | 7.72 ± 0.46 | 0.843 | |||
| Dominant | 162 | 7.74 ± 0.48 | 0.475 | Dominant | 184 | 7.75 ± 0.50 | 0.433 | |||
a Genotyping data and mRNA expression levels for IL10 by genotypes were obtained from the HapMap phase II release 23 data from EBV-transformed lymphoblastoid cell lines from 270 individuals.
b Two-side Student's t test within the stratum.
c P values for the trend test of IL10 mRNA expression among 3 genotypes for each SNP from a general linear model.
d There were missing data because genotyping data were not available.
Heterogeneity and sensitivity analyses
As shown in Table 2, no heterogeneities were observed among all studies, while evaluating the association between IL10 -819C>T polymorphism and NHL susceptibility (heterozygous: P=0.821, recessive: P=0.108, dominant: P=0.558, and allele comparing: P=0.184), except for the homozygous model with moderate heterogeneity (P=0.094). Likewise, for the IL10 -592C>A polymorphism, no heterogeneity were detected, either (homozygous: P= 0.565, heterozygous: P=0.652, recessive: P=0.652, dominant: P=0.542, and allele comparing: P=0.435).
Publication bias
The shape of the funnel plot seemed symmetrical, indicating that there was no evidence of publication bias. Moreover, the Egger's test further demonstrated the absence of publication bias in this meta-analysis with statistical evidence (for the IL10 -819C>T polymorphism, homozygous: P=0.365; heterozygous: P=0.249; recessive: P=0.413; dominant: P=0.215 and allele comparing P=0.246, and for the IL10 -592C>A polymorphism, homozygous: P=0.099; heterozygous: P=0.225; recessive: P=0.128; dominant: P=0.127 and allele comparing P=0.074).
Discussion
We performed this meta-analysis to comprehensively estimate the association between IL10 gene polymorphisms and NHL risk, including 11 studies with 5859 NHL cases and 6893 controls for the IL10 -819C>T polymorphism, and 11 studies with 6277 cases and 7350 controls for the IL10 -592C>A polymorphism. Overall, we did not observe any significant association between either of two polymorphisms and NHL susceptibility. However, both of these two polymorphisms showed a trend to decrease DLBCL risk.
The IL10 gene is comprised of five exons and four introns. Its protein product is an important anti-inflammatory cytokine that is involved in the regulation of inflammatory responses through directly influencing tumor necrosis factor production 41. IL10 is also a critical immunoregulatory cytokine that regulates many aspects of the immune response. Moreover, it has been implicated in the tumorigenesis of various types of cancers 42, 43. For instance, it might protect malignant cells by inhibiting cytotoxic T lymphocyte-mediated tumour-specific cell lysis 44. Given IL10's important role in carcinogenesis, it is biologically plausible that the genetic variations in its coding gene may modulate the risk of cancers. It has been reported that IL10 gene promoter may influence the IL10 expression, consequentially alter NHL susceptibility and clinical outcomes 18.
There were two recently published meta-analyses focusing on NHL risk and IL10 gene polymorphisms 45, 46. In one study carried out by Wang et al. 45, the association of IL10 -1082A>C polymorphism with general cancer risk was investigated. The stratified analysis by cancer type found that the -1082A>C polymorphism was associated with overall NHL susceptibility, with a total of 2338 NHL cases and 1999 controls from eight studies. However, this study did not investigate the IL10 -819C>T and -592C>A polymorphisms. Moreover, the other study by Cao et al. 46 merely focused on DLBCL, and included only three studies for either of IL10 -819C>T or -592C>A polymorphisms. In the end, no significant association was observed between IL10 -819C>T polymorphism and DLBCL risk under all genetic models, except that a modestly decreased DLBCL risk was observed under recessive model. Furthermore, they found that a significantly decreased DLBCL risk was associated with IL10 -592C>A polymorphism under the recessive and homozygous models. The findings in the present meta-analysis were consistent with the results derived from Cao's meta-analysis. Moreover, we also investigated the associations between IL10 polymorphisms and follicle lymphoma as well as all other types of NHL, though no significant associations were observed. Carcinogenesis is a quite complex multi-step process, and different types of cancer may have different mechanisms. One possible explanation for the discrepancy in the cancer susceptibility between DLBCL and FL is that carcinogenic mechanisms under these two NHL subtypes may be different, and IL10 genetic variants may exert differential effects in different cancers. Additionally, it is possible that the total sample size for the current meta-analysis is not sufficient enough to detect some potential association.
The current study has several merits: first, we included the latest studies, and this is the largest and most comprehensive meta-analysis for the association of IL10 -819C>T and -592C>A polymorphisms with NHL susceptibility; second, we collected data for NHL subtype and performed subgroup analysis by subtype; third, we performed genotype-based mRNA expression analysis to further explore the potential function of these two polymorphisms which are located in the promoter region of IL10 gene. Despite these merits, there remained some limitations in this meta-analysis to be addressed. First, we mainly searched published studies in PubMed and EMBASE, thus, we might have missed some publications, especially the unpublished negative studies. As a result, some degree of bias may occur. Second, due to lacking of original data, our meta-analysis was based on unadjusted estimates. A more precise analysis could have be performed if individual data were available such as age, gender, smoking status, body mass index, environment exposure, and lifestyles, which would allow us to further evaluate adjusted OR and gene-environment interactions. Third, the numbers of studies for these two polymorphisms were relatively small. Moreover, the individual sample sizes for cases in most studies included in the current meta-analysis were relatively small (<500) except for four studies 22, 25, 28, 29, which may lead to a limited statistical power to detect the real association. Finally, meta-analysis is a retrospective study that is subject to methodological limitations.
In conclusion, this meta-analysis indicates that both the IL10 -819C>T and -592C>A polymorphisms may be associated with decreased DLBCL risk. However, well-designed large prospective studies using standardized genotyping methods, enrolling precisely diagnosed NHL patients, especially DLBCL patient, and strictly matched controls are warranted to validate the current findings.
Supplementary Materials
Supplementary Table S1.
Acknowledgments
This study was supported by the grant from Special Financial Grant from the China Postdoctoral Science Foundation (Grant No. 2014T70836), the grant funded by Heilongjiang Education Department of China (1252HQ016) and the grant funded by Harbin Medical University Cancer Hospital (JJ2011-11).
Abbreviations
- NHL
non-Hodgkin lymphoma
- IL10
interleukin 10
- SNP
single nucleotide polymorphisms
- OR
odds ratio
- CI
confidence interval
- HWE
Hardy-Weinberg equilibrium
- DLBCL
diffuse large B-cell lymphoma
- FL
follicular lymphoma
- PB
population based
- HB
hospital based.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Adamson P, Bray F, Costantini AS, Tao MH, Weiderpass E, Roman E. Time trends in the registration of Hodgkin and non-Hodgkin lymphomas in Europe. Eur J Cancer. 2007;43:391–401. doi: 10.1016/j.ejca.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 3.Muller AM, Ihorst G, Mertelsmann R, Engelhardt M. Epidemiology of non-Hodgkin's lymphoma (NHL): trends, geographic distribution, and etiology. Ann Hematol. 2005;84:1–12. doi: 10.1007/s00277-004-0939-7. [DOI] [PubMed] [Google Scholar]
- 4.Grulich AE, Vajdic CM, Cozen W. Altered immunity as a risk factor for non-Hodgkin lymphoma. Cancer Epidemiol Biomarkers Prev. 2007;16:405–8. doi: 10.1158/1055-9965.EPI-06-1070. [DOI] [PubMed] [Google Scholar]
- 5.Silverberg MJ, Chao C, Leyden WA, Xu L, Horberg MA, Klein D. et al. HIV infection, immunodeficiency, viral replication, and the risk of cancer. Cancer Epidemiol Biomarkers Prev. 2011;20:2551–9. doi: 10.1158/1055-9965.EPI-11-0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Leeuwen MT, Grulich AE, Webster AC, McCredie MR, Stewart JH, McDonald SP. et al. Immunosuppression and other risk factors for early and late non-Hodgkin lymphoma after kidney transplantation. Blood. 2009;114:630–7. doi: 10.1182/blood-2009-02-202507. [DOI] [PubMed] [Google Scholar]
- 7.Shen M, Zheng T, Lan Q, Zhang Y, Zahm SH, Wang SS. et al. Polymorphisms in DNA repair genes and risk of non-Hodgkin lymphoma among women in Connecticut. Hum Genet. 2006;119:659–68. doi: 10.1007/s00439-006-0177-2. [DOI] [PubMed] [Google Scholar]
- 8.He J, Liao XY, Zhu JH, Xue WQ, Shen GP, Huang SY. et al. Association of MTHFR C677T and A1298C polymorphisms with non-Hodgkin lymphoma susceptibility: evidence from a meta-analysis. Sci Rep. 2014;4:6159. doi: 10.1038/srep06159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He J, Wang F, Zhu JH, Chen W, Cui Z, Jia WH. No association between MTR rs1805087 A > G polymorphism and non-Hodgkin lymphoma susceptibility: evidence from 11 486 subjects. Leuk Lymphoma. 2015;56:763–7. doi: 10.3109/10428194.2014.935370. [DOI] [PubMed] [Google Scholar]
- 10.He YQ, Zhu JH, Huang SY, Cui Z, He J, Jia WH. The association between the polymorphisms of TNF-alpha and non-Hodgkin lymphoma: a meta-analysis. Tumour Biol. 2014;35:12509–17. doi: 10.1007/s13277-014-2569-6. [DOI] [PubMed] [Google Scholar]
- 11.Smyth MJ, Cretney E, Kershaw MH, Hayakawa Y. Cytokines in cancer immunity and immunotherapy. Immunol Rev. 2004;202:275–93. doi: 10.1111/j.0105-2896.2004.00199.x. [DOI] [PubMed] [Google Scholar]
- 12.Wu MS, Huang SP, Chang YT, Shun CT, Chang MC, Lin MT. et al. Tumor necrosis factor-alpha and interleukin-10 promoter polymorphisms in Epstein-Barr virus-associated gastric carcinoma. J Infect Dis. 2002;185:106–9. doi: 10.1086/324771. [DOI] [PubMed] [Google Scholar]
- 13.Kovacs E. Interleukin-6 leads to interleukin-10 production in several human multiple myeloma cell lines. Does interleukin-10 enhance the proliferation of these cells? Leuk Res. 2010;34:912–6. doi: 10.1016/j.leukres.2009.08.012. [DOI] [PubMed] [Google Scholar]
- 14.Lossos IS, Morgensztern D. Prognostic biomarkers in diffuse large B-cell lymphoma. J Clin Oncol. 2006;24:995–1007. doi: 10.1200/JCO.2005.02.4786. [DOI] [PubMed] [Google Scholar]
- 15.Lech-Maranda E, Bienvenu J, Michallet AS, Houot R, Robak T, Coiffier B. et al. Elevated IL-10 plasma levels correlate with poor prognosis in diffuse large B-cell lymphoma. Eur Cytokine Netw. 2006;17:60–6. [PubMed] [Google Scholar]
- 16.Eskdale J, Kube D, Tesch H, Gallagher G. Mapping of the human IL10 gene and further characterization of the 5' flanking sequence. Immunogenetics. 1997;46:120–8. doi: 10.1007/s002510050250. [DOI] [PubMed] [Google Scholar]
- 17.Turner DM, Williams DM, Sankaran D, Lazarus M, Sinnott PJ, Hutchinson IV. An investigation of polymorphism in the interleukin-10 gene promoter. Eur J Immunogenet. 1997;24:1–8. doi: 10.1111/j.1365-2370.1997.tb00001.x. [DOI] [PubMed] [Google Scholar]
- 18.Lech-Maranda E, Baseggio L, Bienvenu J, Charlot C, Berger F, Rigal D. et al. Interleukin-10 gene promoter polymorphisms influence the clinical outcome of diffuse large B-cell lymphoma. Blood. 2004;103:3529–34. doi: 10.1182/blood-2003-06-1850. [DOI] [PubMed] [Google Scholar]
- 19.Guzowski D, Chandrasekaran A, Gawel C, Palma J, Koenig J, Wang XP. et al. Analysis of single nucleotide polymorphisms in the promoter region of interleukin-10 by denaturing high-performance liquid chromatography. J Biomol Tech. 2005;16:154–66. [PMC free article] [PubMed] [Google Scholar]
- 20.Lan Q, Zheng T, Rothman N, Zhang Y, Wang SS, Shen M. et al. Cytokine polymorphisms in the Th1/Th2 pathway and susceptibility to non-Hodgkin lymphoma. Blood. 2006;107:4101–8. doi: 10.1182/blood-2005-10-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Persico M, Capasso M, Persico E, Masarone M, Renzo A, Spano D. et al. Interleukin-10 - 1082 GG polymorphism influences the occurrence and the clinical characteristics of hepatitis C virus infection. J Hepatol. 2006;45:779–85. doi: 10.1016/j.jhep.2006.07.026. [DOI] [PubMed] [Google Scholar]
- 22.Wang SS, Cerhan JR, Hartge P, Davis S, Cozen W, Severson RK. et al. Common genetic variants in proinflammatory and other immunoregulatory genes and risk for non-Hodgkin lymphoma. Cancer Res. 2006;66:9771–80. doi: 10.1158/0008-5472.CAN-06-0324. [DOI] [PubMed] [Google Scholar]
- 23.Kube D, Hua TD, Kloss M, Kulle B, Brockmoller J, Wojnowski L. et al. The interleukin-10 gene promoter polymorphism -1087AG does not correlate with clinical outcome in non-Hodgkin's lymphoma. Genes Immun. 2007;8:164–7. doi: 10.1038/sj.gene.6364364. [DOI] [PubMed] [Google Scholar]
- 24.Lech-Maranda E, Baseggio L, Charlot C, Rigal D, Berger F, Jamroziak K. et al. Genetic polymorphisms in the proximal IL-10 promoter and susceptibility to non-Hodgkin lymphoma. Leuk Lymphoma. 2007;48:2235–8. doi: 10.1080/10428190701615926. [DOI] [PubMed] [Google Scholar]
- 25.Purdue MP, Lan Q, Kricker A, Grulich AE, Vajdic CM, Turner J. et al. Polymorphisms in immune function genes and risk of non-Hodgkin lymphoma: findings from the New South Wales non-Hodgkin Lymphoma Study. Carcinogenesis. 2007;28:704–12. doi: 10.1093/carcin/bgl200. [DOI] [PubMed] [Google Scholar]
- 26.Andrie E, Michos A, Kalampoki V, Pourtsidis A, Moschovi M, Polychronopoulou S. et al. Genetic variants in immunoregulatory genes and risk for childhood lymphomas. Eur J Haematol. 2009;83:334–42. doi: 10.1111/j.1600-0609.2009.01288.x. [DOI] [PubMed] [Google Scholar]
- 27.Wong HL, Breen EC, Pfeiffer RM, Aissani B, Martinson JJ, Margolick JB. et al. Cytokine signaling pathway polymorphisms and AIDS-related non-Hodgkin lymphoma risk in the multicenter AIDS cohort study. AIDS. 2010;24:1025–33. doi: 10.1097/QAD.0b013e328332d5b1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hosgood HD 3rd, Au WY, Kim HN, Liu J, Hu W, Tse J. et al. IL10 and TNF variants and risk of non-Hodgkin lymphoma among three Asian populations. Int J Hematol. 2013;97:793–9. doi: 10.1007/s12185-013-1345-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, Wang MY, He J, Wang JC, Yang YJ, Jin L. et al. Tumor necrosis factor-alpha induced protein 8 polymorphism and risk of non-Hodgkin's lymphoma in a Chinese population: a case-control study. PLoS One. 2012;7:e37846. doi: 10.1371/journal.pone.0037846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He J, Qiu LX, Wang MY, Hua RX, Zhang RX, Yu HP. et al. Polymorphisms in the XPG gene and risk of gastric cancer in Chinese populations. Hum Genet. 2012;131:1235–44. doi: 10.1007/s00439-012-1152-8. [DOI] [PubMed] [Google Scholar]
- 31.He J, Shi TY, Zhu ML, Wang MY, Li QX, Wei QY. Associations of Lys939Gln and Ala499Val polymorphisms of the XPC gene with cancer susceptibility: a meta-analysis. Int J Cancer. 2013;133:1765–75. doi: 10.1002/ijc.28089. [DOI] [PubMed] [Google Scholar]
- 32.He J, Xi B, Ruiter R, Shi TY, Zhu ML, Wang MY. et al. Association of LEP G2548A and LEPR Q223R polymorphisms with cancer susceptibility: evidence from a meta-analysis. PLoS One. 2013;8:e75135. doi: 10.1371/journal.pone.0075135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee JJ, Kim DH, Lee NY, Sohn SK, Kim JG, Kim HJ. et al. Interleukin-10 gene polymorphism influences the prognosis of T-cell non-Hodgkin lymphomas. Br J Haematol. 2007;137:329–36. doi: 10.1111/j.1365-2141.2007.06570.x. [DOI] [PubMed] [Google Scholar]
- 34.Habermann TM, Wang SS, Maurer MJ, Morton LM, Lynch CF, Ansell SM. et al. Host immune gene polymorphisms in combination with clinical and demographic factors predict late survival in diffuse large B-cell lymphoma patients in the pre-rituximab era. Blood. 2008;112:2694–702. doi: 10.1182/blood-2007-09-111658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park YH, Sohn SK, Kim JG, Lee MH, Song HS, Kim MK. et al. Interaction between BCL2 and interleukin-10 gene polymorphisms alter outcomes of diffuse large B-cell lymphoma following rituximab plus CHOP chemotherapy. Clin Cancer Res. 2009;15:2107–15. doi: 10.1158/1078-0432.CCR-08-1588. [DOI] [PubMed] [Google Scholar]
- 36.Heemann C, Kreuz M, Stoller I, Schoof N, von Bonin F, Ziepert M. et al. Circulating levels of TNF receptor II are prognostic for patients with peripheral T-cell non-Hodgkin lymphoma. Clin Cancer Res. 2012;18:3637–47. doi: 10.1158/1078-0432.CCR-11-3299. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, Chen MB, Zhou XY, Hong XN. Lymphotoxin alpha (LTA) polymorphism is associated with prognosis of non-Hodgkin's lymphoma in a Chinese population. PLoS One. 2013;8:e66411. doi: 10.1371/journal.pone.0066411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Colt JS, Rothman N, Severson RK, Hartge P, Cerhan JR, Chatterjee N. et al. Organochlorine exposure, immune gene variation, and risk of non-Hodgkin lymphoma. Blood. 2009;113:1899–905. doi: 10.1182/blood-2008-04-153858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bogunia-Kubik K, Mazur G, Wrobel T, Kuliczkowski K, Lange A. Interleukin-10 gene polymorphisms influence the clinical course of non-Hodgkin's lymphoma. Tissue Antigens. 2008;71:146–50. doi: 10.1111/j.1399-0039.2007.00984.x. [DOI] [PubMed] [Google Scholar]
- 40.Lim YY, Chin YM, Tai MC, Fani S, Chang KM, Ong TC. et al. Analysis of interleukin-10 promoter single nucleotide polymorphisms and risk of non-Hodgkin lymphoma in a Malaysian population. Leuk Lymphoma. 2015;56:163–8. doi: 10.3109/10428194.2014.907895. [DOI] [PubMed] [Google Scholar]
- 41.Eskdale J, Keijsers V, Huizinga T, Gallagher G. Microsatellite alleles and single nucleotide polymorphisms (SNP) combine to form four major haplotype families at the human interleukin-10 (IL-10) locus. Genes Immun. 1999;1:151–5. doi: 10.1038/sj.gene.6363656. [DOI] [PubMed] [Google Scholar]
- 42.Pisa P, Halapi E, Pisa EK, Gerdin E, Hising C, Bucht A. et al. Selective expression of interleukin 10, interferon gamma, and granulocyte-macrophage colony-stimulating factor in ovarian cancer biopsies. Proc Natl Acad Sci U S A. 1992;89:7708–12. doi: 10.1073/pnas.89.16.7708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kamiya T, Hatanaka H, Abe Y, Kijima H, Yamazaki H, Ohnishi Y. et al. Interleukin-10 expression is closely correlated with the expression of granulocyte-macrophage colony-stimulating factor in non-small cell lung cancer. Anticancer Res. 2003;23:2909–13. [PubMed] [Google Scholar]
- 44.Matsuda M, Salazar F, Petersson M, Masucci G, Hansson J, Pisa P. et al. Interleukin 10 pretreatment protects target cells from tumor- and allo-specific cytotoxic T cells and downregulates HLA class I expression. J Exp Med. 1994;180:2371–6. doi: 10.1084/jem.180.6.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang J, Ding Q, Shi Y, Cao Q, Qin C, Zhu J. et al. The interleukin-10-1082 promoter polymorphism and cancer risk: a meta-analysis. Mutagenesis. 2012;27:305–12. doi: 10.1093/mutage/ger078. [DOI] [PubMed] [Google Scholar]
- 46.Cao HY, Zou P, Zhou H. Genetic association of interleukin-10 promoter polymorphisms and susceptibility to diffuse large B-cell lymphoma: a meta-analysis. Gene. 2013;519:288–94. doi: 10.1016/j.gene.2013.01.066. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1.