Abstract
Objective: A high level of RGS17 expression is observed in diverse human cancers and correlates with tumor progression. Herein, we aim to investigate its expression and function in breast cancer.
Methods: The expression of RGS17 was detected by immunohistochemical analysis and western blot analysis. The level of miR-32 expression was investigated by qRT-PCR. Western blot analysis was used to determine the relationship between RGS17 and miR-32. A series of loss or gain of function assays was performed to measure the effects of RGS17 or miR-32 on tumor migration, invasion, and proliferation.
Results: Compared to that in normal breast specimen, the expression of RGS17 had a significantly higher expression level in breast cancer tissues and cell lines. Although the potential relationship of RGS17 expression with clinicopathological features was not observed, there was a significant correlation of RGS17 expression with p63 expression. In cells, inhibition of RGS17 expression impaired cell migration, invasion, and proliferation. Further, RGS17 was identified as a direct and functional target of miR-32. Overexpression of miR-32 in cells could decrease the expression of RGS17 and inhibit cell migration, invasion, and proliferation. In contrast, ectopic expression of RGS17 could attenuate phenotypes caused by miR-32 overexpression.
Conclusion: The expression of RGS17 was upregulated in breast cancer, which could enhance cell migration, invasion, and proliferation. Moreover, the RGS17 was identified as a target of miR-32. Our results suggest that RGS17 might play an important role in breast cancer progression and could be a potential target for human breast cancer treatment.
Keywords: breast cancer, RGS17, p63, has-miR-32
Introduction
Breast cancer is by far the most common female malignancy diagnosed and the most frequent cause of cancer death among women worldwide 1. Despite recent advances in diagnosis and experimental oncology, the prognosis of breast cancer is still unfavorable due to its metastatic nature. Previous studies have emphasized that breast cancer is a highly heterogeneous group of diseases that differ in their prognosis and response to treatment 1, 2. The development of breast cancer is thought to occur through a multi-step process and a large number of molecules are shown to be involved in the tumorgenesis and progression 1. The clarification of its molecular mechanisms will promote early diagnosis and allow an effective scheme to target specific pathway.
G protein-coupled receptors (GPCR) constitute a large protein family of receptors that sense molecules outside cells and activate signal transduction pathways and, ultimately, cellular responses 3. Consequently, GPCRs are linked to many diseases including cancers. Accumulating research indicates that GPCRs are expressed in cancerous tissues and associated with proliferation, survival from apoptotic signals, invasion, and metastasis 4-6. Therefore, GPCRs serve as direct and indirect targets for approximately 50% of currently marketed drugs to cancers 7. G proteins consist of a guanine nucleotide-binding Ga and Gβγ dimer, which provide signal coupling to GPCRs and act as signal transduction proteins through a cycle of guanine nucleotide exchange and hydrolysis 8. Thus, G proteins mediate a wide variety of signals and their activity is finely tuned by some modulators. One of those modulators is the regulator of G-protein signaling (RGS) protein family. RGS proteins have been identified as negative modulators in G protein-dependent signaling through accelerating GTP hydrolysis up to 2000 times that of inherent GTPase activity 4. Thus, RGS proteins could critically regulate the magnitude and duration of G protein signaling 9.
RGS proteins comprise over 20 different proteins and are divided into 8 subfamilies (RZ/A, R4/B, R7/C, R12/D, RA/E, RGEF/F, RGRK/G and RSNX/H) based on the homology of RGS domains and the construction of proteins 4. Considering the prevalence of GPCRs for targeted therapeutics and the role of RGS proteins in G protein signaling, some RGS proteins are determined as deregulated genes in a variety of cancers including breast cancer, ovarian cancer, and prostate cancer 10-12. For example, two members of the R4/B subfamily (RGS2, RGS5) are upregulated in breast cancer 4, 13, 14, and one member of the RZ/A subfamily (RGS19) is also upregulated in ovarian cancer 15. RGS17, the most recently discovered member of the RZ/A subfamily, has been frequently reported as an overexpressed gene in human lung adenocarcinomas, prostate cancer, and hepatocellular carcinoma 4, 8. Furthermore, increased RGS17 expression has been identified to positively associate with tumor cell proliferation through the cyclic AMP-PKA-CREB pathway in human lung and prostate cancer. Hence, RGS17 is regarded as an emerging therapeutic target for lung and prostate cancers 16. These data strongly suggest a potentially important role of RGS17 in oncogenesis. Instead, RGS17 is downregulated at significantly low level in ovarian tumor compared to normal ovary and chemotherapy exposure triggers the loss of RGS17 expression, which leads to the development of chemotherapeutic resistance possibly through amplification of endogenous AKT signals 16, 17. However, it is little known about the mechanisms for the discrepant role of RGS17 in different tumors, and whether RGS17 has a role in breast cancer progression remains to be understood.
In the present study, we focused on the expression and function of RGS17 in breast cancer. We found that RGS17 was upregulated in breast cancer tissues than that in normal breast tissues. Increased RGS17 expression was significantly correlated with p63 expression. In vitro functional assays demonstrated that inhibition of RGS17 expression reduced breast cancer cell migration, invasion, and proliferation. Finally, we identified miR-32 as a regulator that could inhibit the expression of RGS17 in breast cancer cell lines.
Materials and Methods
Tissue specimens
Human breast tissues were retrieved from Department of Pathology, Huashan Hospital, Shanghai, China. The patients had not previously received chemotherapy or radiation therapy. All cases were diagnosed by two experienced pathologists without discrepancy. The study was approved by the institutional review board at Huashan Hospital, and signed informed consent was obtained from all patients and the bereaved.
Cell culture
Breast cancer cell lines (MCF-7, MDA-MB-231, SK-BR-3, and ZR-75-30) and HEK 293T cells were preserved in our laboratory and maintained at 37℃ in a humidified 5% CO2 incubator in high glucose, complete DMEM (Hyclone, UT, USA) supplemented with 10% FBS. All transfections were carried out according to the manufacturer's instructions.
Protein extraction and western blot
Tissues and cells were lysed using a RIPA lysis buffer (Beyotime, Hangzhou, China) supplemented with inhibitors of proteases. The whole lysates were stored at -20℃ until the western blot analysis. For western blot analysis, proteins were loaded on to a 10% SDS-PAGE and transferred to nitrocellulose membrane (Bio-Rad, CA, USA). After blocking in 5% nonfat milk, the membranes were incubated overnight at 4℃ with the following primary antibodies: rabbit RGS17 polyclonal antibody (1:1000; Proteintech, IL, USA), mouse monoclonal GAPDH antibody (1:1000; Biotech Well, Shanghai, China). After that, membranes were washed and incubated with HRP-labeled secondary antibody in TBST for 1h. Immunoreactivity was detected with enhanced chemoluminescent autoradiography (ECL kit, Pierce, IL, USA).
Immunohistochemical (IHC) analysis
IHC staining was done according to standard procedures. The paraffin-embedded tissues were first stained with hematoxylin and eosin (HE) for histological examination. Subsequently, pfraffin sections in 0.1M citric acid buffer (pH6) were boiled in a microwave at 100℃, and then pre-incubated in blocking solution (normal goat sera, Biotech Well, Shanghai, China) at 37℃ for 30min. The slides were then incubated at 4℃ overnight with rabbit anti-RGS17 mAb (1: 400; Proteintech, IL, USA). Five high-power fields (×200) were selected at random. Specimens were scored according to the intensity of the dye color and the number of positive cells. The intensity of the dye color was graded as 0 (no color), 1 (light yellow), 2 (light brown), or 3(brown); The number of positive cells were graded as 0 (<5%), 1 (5-25%), 2 (26-50%), 3 (51-75%), or 4 (>75%). The two grades were added together and specimens were assigned to one of 4 levels: 0-1 score (-), 2 scores (+), 3-4 scores (++), more than 5 scores (+++).
Knockdown of RGS17
The knockdown of RGS17 was achieved using two short hairpin RNA (shRNA) fragments strategy. The RGS17 shRNA construct with target sequences of RGS17 (GGCTTGCTTGTGAA GACTTAA and GCTGGCTCTTCTTCTGAATCT, respectively) and non-targeting shRNA construct were generated using pLVX-shRNA1 system (Takara Co., Dalian, China). Cells were selected in medium containing puromycin (1ng/ml) and a stable cell pool was obtained for further study. The RGS17 expression was verified by western blot analysis. Specific miRNA (has-miR-32) to RGS17 was purchased from GeneChem Co., Ltd (Shanghai, China).
RNA extraction and Quantitative real-time PCR (qRT-PCR)
Total RAN was extracted using the Trizol reagent (Invitrogen, CA, USA) and dissolved in DEPC water according to the manufacturer's instructions. Reverse transcription reaction was carried out starting from 100ng of total RNA using the looped primers. Real-time PCR was performed using the standard TaqMan MicroRNA assays protocol on ABI7500 real-time PCR detection system. U6 small nuclear RNA was used as an internal control. The threshold cycle (Ct) was defined as the fractional cycle number at which the fluorescence passed the fixed threshold. Each sample was measured in triplicate, and the relative amount of miR-32 to U6 was calculated using the equation 2-△Ct, where △Ct=(CTmiR-32-CTU6).
Overexpression of RGS17
For construction of RGS17 plasmid, the RGS17 cDNA was generated by RT-PCR (Roche, Swiss) from breast cancer tissue sample using RGS17 forward primer (5'-CCCAA GCTTATGCGAAAAAGGCAGCAGTC-3') and reverse primer (5'-GGGGAATTCGAAGATTCAGAAGAAGAGCC-3'). Then full-length RGS17 cDNA fragment was cloned into the pcDNA3.1 vector (GeneChem Co., Ltd, Shanghai, China).
Wound-healing assay
For wound-healing assay, wounds were made with sterile pipette tips. Exfoliated cells were washed off using phosphate-buffered saline (PBS), and culture was continued in fresh medium without FBS. Wound closure was monitored by microscopy per 24 hours. Migration activity was calculated as the mean distance between the edges of three points. Healing rate = (mean original distance-mean distance at a time point)/mean original distance×100. Each test group was assayed in triplicate.
CCK-8 proliferation assay
Cells were digested, resuspended, and seeded on 96-well plates at an initial density of 4×103/well. At each monitored time point, cells of each well were stained with CCK-8 (Dojindo, Japan) for 30min at 37℃. Spectrometric absorbance was measured using a microplate reader (Bio Tek Instruments, Winooski, VT, USA) at 450nm. All experiments were carried out in triplicate.
Boyden chamber assay
Migration assay was carried out using Boyden chambers (8-μm; Corning, NY, USA) containing polycarbonate membrane. For invasion assay, 60μl matrigel (BD Biosciences, NJ, USA) was used to mimic basement membrane. Briefly, upper chamber was added with 100μl of 1×106 cells in serum-free medium and lower chamber was added with 600μl of appropriate medium with 10% FBS. Cells were incubated for 24h. Migratory cells on the under-surface of the random regions were fixed and stained with crystal violet for 30s at room temperature. Photographs of 5 random regions were taken and the number of cells was counted to calculate the average number of migrated cells per plate.
Statistical analysis
All statistical analysis was performed with SPSS 17.0 software package (SPSS, Chicago, IL, USA). Data were presented as mean ± standard deviation. Differences between groups were analyzed using the Student's t-test or Chi-square test. Relationship between RGS17 expression and clinical characteristics in breast cancers specimens were explored by Kruskal-Wallis tests and Spearman tests. *p<0.05 was considered to be statistically significant.
Results
RGS17 is highly expressed in breast cancer
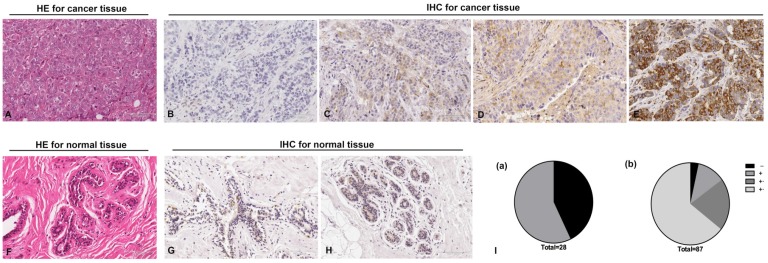
To investigate the role of RGS17 in breast cancer, we examined RGS17 expression in 87 human breast cancer tissues and 28 normal breast tissues. Histological examination was first carried out on breast cancer tissues and normal tissues, respectively (Fig. 1A and 1F). Then all tissues were analyzed by IHC using RGS17 antibody and graded based on intensity of the dye color and the number of positive cells (Fig. 1B, 1C, 1D, 1E, 1G and 1H). We found that RGS17 protein was mainly localized in the cytoplasm, and the positive rate in breast cancer tissues (96.55%, 84/87) is significantly higher than that in normal breast tissues (57.14%, 16/28, p<0.001). Among 28 normal breast tissues, only 16 samples presented to be RGS17-weak positive (57.14%), whereas the other 12 samples were all RGS17-negative. In contrast, 3 out of 87 cases of breast cancer tissues were negative, 10 were weakly positive, 18 were moderately positive and 56 were strongly positive (Table 1 and Fig. 1I). These data strong support that RGS17 expression is significantly increased in breast cancer tissues.
Fig 1.
Immunohistochemistry staining for RGS17 protein with RGS17 antibody in the cancerous and normal tissues. (A) Validation of breast cancer tissues by Hematoxylin-Eosin (HE) stain. (B, C, D, E) Representative results of negative staining (-, B), weakly positive staining (+, C), moderate staining (++, D) and strong staining (+++, E) of RGS17 protein in breast cancer tissues are shown respectively. (F) HE staining of normal tissues. (G, H) Representative results of the negative staining (G) and weakly positive staining (H) of RGS17 protein in normal tissues. (I) Distributions of RGS17 staining grades (-, +, ++ and +++) in both normal tissues (I (a)) and breast cancer tissues (I (b)). Bar=100um.
Table 1.
Distributions of RGS17 staining grades (-, +, ++ and +++) in both normal breast tissues and breast cancer tissues.
| Total | RGS17 | Positive rate | P-value | ||||
|---|---|---|---|---|---|---|---|
| - | + | ++ | +++ | ||||
| Normal breast tissue | 28 | 12 | 16 | 0 | 0 | 57.14% | 0.000*** |
| Breast cancer tissue | 87 | 3 | 10 | 18 | 56 | 96.55% | |
***p<0.001
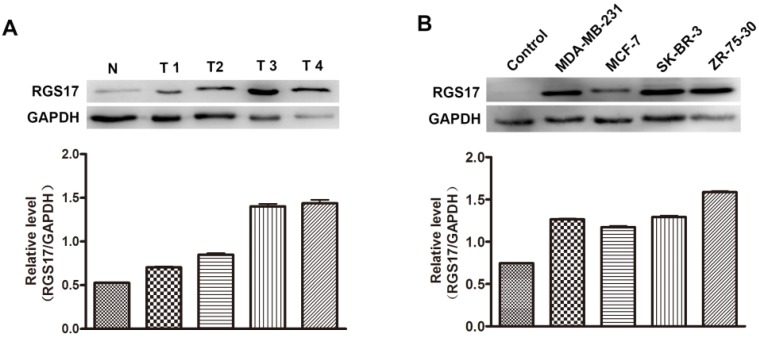
Additionally, we randomly selected 4 breast cancer tissues and one normal breast tissue to investigate the expression level of RGS17 by western blot analysis. As illustrated in Fig.2A, the protein level of RGS17 was higher in the breast cancer tissues than that in the normal breast tissues, which is consistent with results from staining analysis. The results indicate that RGS17 was frequently overexpressed in breast cancer, and might play an important role in breast cancer progression.
Fig 2.
Expression level of RGS17 in breast cancer tissues and cell lines. (A) Western blot analysis of RGS17 protein from 4 random human primary breast cancer (T) and normal breast tissues (N). Expression levels were normalized for GAPDH. (B) Expression of RGS17 in breast cancer cell lines was detected by western blot analysis. Expression levels were normalized for GAPDH.
Expression of RGS17 is correlated with p63
To determine whether the expression level of RGS17 was associated with progression of breast cancer, the expressions of RGS17 were determined in 4 human breast cancer cell lines (MDA-MB-231, MCF-7, SK-BR-3, and ZR-75-30). Results from western blot analysis showed that the expression of RGS17 was significantly higher in cell lines than that in one random normal breast tissue (Fig. 2B), which implies a role of RGS17 in the tumor progression. We then carried out Kruskal-Wallis and Spearman correlation analysis to determine the relationship between RGS17 expression and clinicopathological features (Table 2 and Table 3). Statistical analysis revealed that RGS17 expression was not significantly correlated with the TNM stage, tumor size, lymph node metastasis, age, ERα status, PR status, HER2 status, or Ki67 status. Interestingly, we found that the RGS17 expression was positively correlated with p63 expression (p=0.014, r=0.262).
Table 2.
Association between RGS17 expression and disease parameters in 87 cases of breast cancers.
| Total | RGS17 | P-value | |||||
|---|---|---|---|---|---|---|---|
| - | + | ++ | +++ | ||||
| n=3 | n=10 | n=18 | n=56 | ||||
| TNM stage | |||||||
| I and Ⅱ | 80 | 2 | 9 | 17 | 52 | 0.497 | |
| Ⅲ and Ⅳ | 7 | 1 | 1 | 1 | 4 | ||
| Tumor size | |||||||
| ≤2cm | 66 | 3 | 8 | 13 | 42 | 0.578 | |
| 2cm<T≤5cm | 19 | 0 | 2 | 5 | 12 | ||
| >5cm | 2 | 0 | 0 | 0 | 2 | ||
| Lymph node metastasis | |||||||
| Negative | 53 | 2 | 6 | 10 | 35 | 0.755 | |
| Positive | 34 | 1 | 4 | 8 | 21 | ||
| Age | |||||||
| <50 | 27 | 2 | 2 | 5 | 18 | 0.859 | |
| ≥50 | 60 | 1 | 8 | 13 | 38 | ||
TNM stage is a classification of malignant tumors used by the International Federation of Gynecology and Obstetrics (FIGO). T describes the size of primary tumor, N describes nearby lymph nodes that are involved, and M describes distant metastasis.
Table 3.
Association between RGS17 expression and clinicopathological parameters in 87 cases of breast cancers.
| Total | RGS17 | P-value | r | ||||
|---|---|---|---|---|---|---|---|
| - | + | ++ | +++ | ||||
| n=3 | n=10 | n=18 | n=56 | ||||
| ER | |||||||
| Negative | 26 | 2 | 2 | 5 | 17 | 1.000 | -0.001 |
| Positive | 61 | 1 | 8 | 13 | 39 | ||
| PR | |||||||
| Negative | 41 | 2 | 3 | 8 | 28 | 0.478 | -0.077 |
| Positive | 46 | 1 | 7 | 10 | 28 | ||
| HER2 | |||||||
| Negative | 1 | 0 | 1 | 0 | 0 | 0.097 | 0.179 |
| Positive | 86 | 3 | 9 | 18 | 56 | ||
| p63 | |||||||
| Negative | 77 | 3 | 10 | 18 | 46 | 0.014* | 0.262 |
| Positive | 10 | 0 | 0 | 0 | 10 | ||
| Ki67 | |||||||
| ≤30% | 32 | 3 | 2 | 5 | 22 | 0.708 | -0.041 |
| >30% | 55 | 0 | 8 | 13 | 34 | ||
*p<0.05. ER, estrogen receptor; PR, progesterone receptor
Downregulation of RGS17 is associated with cell migration, invasion, and proliferation
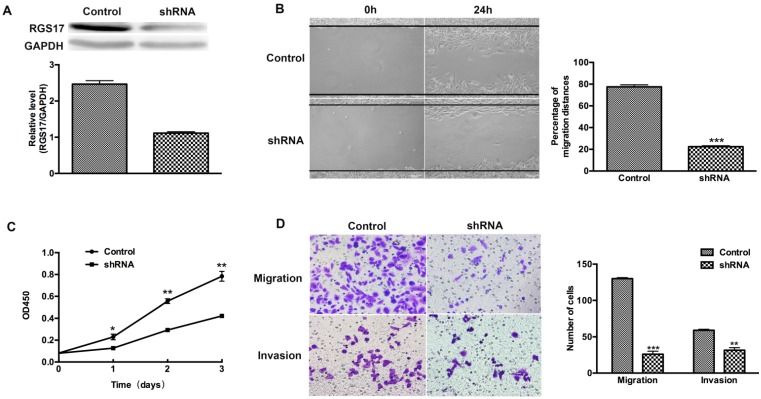
To determine whether RGS17 is a positive regulator of cell migration, invasion and proliferation, we used shRNA to inhibit the expression of RGS17 in ZR-75-30 cells that have the highest basal level of RGS17 among tested breast cancer cell lines (Fig. 2B). We knocked down endogenous RGS17 continuously by specific shRNA in ZR-75-30 cells and scramble sequence shRNA as the control (Fig. 3A). In wound-healing assay, we observed that knockdown of RGS17 in ZR-75-30 cells markedly inhibited the migration (Fig. 3B). To further confirm the effect of RGS17 on migration and invasion of ZR-75-30 cells, we carried out a migration assay and an invasion assay using Boyden chamber. Our results showed the significant inhibition of migration and invasion in ZR-75-30 cells by knockdown of RGS17. These results indicate that RGS17 has a promotional role in cell migration and invasion (Fig. 3D). To examine the proliferation potential of these cells, we carried out a CCK assay. As shown in Fig. 3C, knockdown of RGS17 inhibited cell growth. Taken together, these data indicate that RGS17 expression level was positively correlated with the aggressive phenotype of breast cancer cells.
Fig 3.
RGS17 promotes migration, invasion, and proliferation of breast cancer cells (ZR-75-30). (A) Western blot analysis of RGS17 in cells stably transfected with specific shRNA or scramble sequence shRNA (Control). (B) Photographs at 0h and 24h after wounding were made in control and RGS17 knockdown cells as results of a wound-healing assay. The mean ± SD of percentage of migration distances obtained from three parallel experiments. ***p<0.001. (C) Detection of cell proliferation by CCK assay in the control and RGS17 knockdown cells. Data represent the mean ± SD from triplicate samples and were representative of three independent experiments. *p<0.05, **p<0.01. (D) Effects of RGS17 on the cell migration and invasion by Boyden chamber assay. The mean ± SD shows the number of migrated and invaded cells obtained from three parallel experiments. **p<0.01, ***p<0.001.
RGS17 is a target gene of miR-32
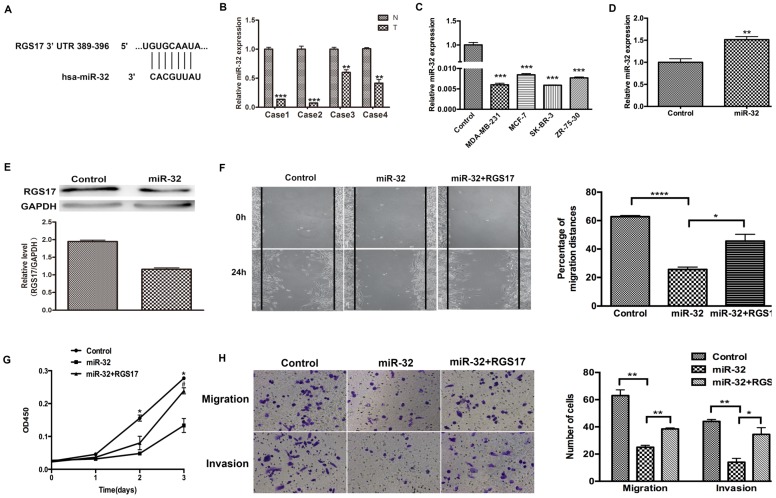
To investigate the potential microRNAs that target RGS17 mRNA, we used several online bioinformatics softwares including TargetScanHuman, PicTar, miRBase and CancerNum to predict the miRNA binding sites in the 3'UTR of RGS17 mRNA. Several candidates, such as miR-32 and miR-182, were evaluated in the inhibition of RGS17 expression, which suggested that the binding site of miR-32 had a higher probability of preferential conservation. In fact, the miR-32 binding site (389-396) was found in RGS17 3'UTR (Fig. 4A). To determine the expression levels of miR-32 in breast cancer tissues and cell lines, qRT-PCR was performed and the expression of miR-32 was normalized to U6 small nuclear RNA. The miR-32 expression was clearly downregulated in breast cancer specimens than that in the corresponding adjacent non-cancerous specimens (Fig. 4B). As in Fig. 4C, the results showed that the expression level of miR-32 were significantly lower in 4 breast cancer cell lines.
Fig 4.
RGS17 is a target gene of miR-32. (A) The predicted binding site of miR-32 in 3'UTR of RGS17. (B) Detection of miR-32 expression in breast cancer tissues (T) and corresponding non-cancerous tissues (N) by qRT-PCR. miR-32 expression level was calculated by 2-ΔCt methods and normalized to U6 small nuclear RNA. **p<0.01, ***p<0.001. (C) Expression of miR-32 in cell lines was detected by qRT-PCR. ***p<0.001. (D) Detection of miR-32 expression in breast cancer cell line (ZR-73-30). **p<0.01. (E) Expression of RGS17 in cells transfected with miR-32 was detected by western blot. (F) Cell migration was analyzed by wound-healing assay in cells transfected with indicated constructs. Data represent the mean ± SD of experiments performed in triplicate. *p<0.05, ****p<0.0001. (G) CCK-8 assay was performed to detect cell proliferation. *p<0.05 (Control vs. miR-32), ♯p<0.05 (miR-32 vs. miR-32+RGS17). (H) Boyden chamber assay was photographed and migrated/invaded cell numbers of three parallel experiments were counted to calculate average number of cells that transmigrated. *p<0.05, **p<0.01.
To further confirm that RGS17 is the direct target of miR-32 in breast cancer, we first transfected GV268-miR-32 plasmid into ZR-75-30 cells and found that miR-32 significantly reduced RGS17 protein level in these cells (Fig. 4D and 4E). Cell migration, invasion, and proliferation were significantly impaired in ZR-75-30 cells transfected with GV268-miR-32 plasmid. When RGS17 plasmid and GV268-miR-32 plasmid were co-transfected, RGS17 could attenuate phenotypes caused by miR-32 overexpression (Fig. 4F, 4G and 4H).
Discussion
In this study, we investigated the expression level of RGS17 in breast cancer tissues and cell lines. Also, the relationship between the expression of RGS17 and several clinicopathological features were evaluated. Although the potential relationships of RGS17 expression with clinicopathological features were not observed, there is a significant correlation of RGS17 expression with p63 expression. In cells, inhibition of RGS17 expression impaired cell migration, invasion, and proliferation. Further, RGS17 was identified as a direct and functional target of miR-32. Overexpression of miR-32 in cells could decrease the expression of RGS17 and inhibit cell migration, invasion, and proliferation, which could be attenuated by ectopic expression of RGS17. To the best of our knowledge, this is the first study to analyze the clinical significance and biological function of RGS17 in breast cancer.
Previous reports have shown that upregulation of RGS17 is a frequent event in cancers, which suggests that RGS17 may play an important role in tumorigenesis and tumor progression 16, 18. In this study, we also found that RGS17 was frequently overexpressed in breast cancer, and 96.55% (84/87) of the breast cancer had about 2-fold increase of RGS17 expression compared with the normal breast tissues. Compared with one random normal tissue, breast cancer cell lines had a higher expression level of RGS17. These results suggest that a higher expression of RGS17 might tend to have more advanced clinical stage. Unexpectedly, we found RGS17 expression had no significant association with TNM stage, tumor size, lymph node metastasis, and so on. It is possible that many cases are diagnosed at the early stage of tumor progression, which results in the deviation of clinicopathological features with RGS17 expression. In fact, inhibition of RGS17 expression could decrease cell migration, invasion, and proliferation. Therefore, a detailed evaluation of RGS17 expression with clinicopathological features requires a larger sample size.
The gene expression can be regulated at different levels, such as transcription, mRNA stability, and protein stability. How the expression of RGS17 is regulated remains to be understood. P63, a member of p53 family of transcription factors, is highly expressed in stratified epithelia such as cervix, skin, prostate, and breast 19. Two major protein isoforms that either contain or lack the N-terminal transactivation domain (TAp63 and △Np63, respectively) are reported to have distinctly different role in tumor progression. Previous studied have suggested that p63 act as a suppressor of tumorgenesis and metastasis by interacting with mutant p53 20. Instead, recent studies suggested that △Np63 promotes stem cell activity in mammary gland development and basal-like breast cancer 21. However, it is difficult to determine which isoform is associated with RGS17 expression in that antibodies against p63 could react with both isoforms. Since △Np63 contributes to the progression of cancers, it is possible that △Np63 might be a regulatory factor that can promote the expression of RGS17.
It's known that microRNA (miRNA) is a class of small, endogenous, noncoding RNAs. They could negatively regulate their target genes by triggering either translation repression or mRNA degradation. Emerging studies have shown that miRNAs are aberrantly expressed or mutated in cancer, suggesting that they may play an essential regulatory role as a novel class of oncogenes or tumor suppressor genes 22, 23. With regard to breast cancer, several important miRNAs have been confirmed in recent research, including miR-21/484/191 22-24. To understand the regulation of RGS17 by miRNAs, we evaluated the efficiency of candidates predicted by online tools in the inhibition of RGS17 expression, which suggests that miR-32 had a preferential performance. In fact, current studies have shown that miR-32 plays as a tumor suppressor in various cancers, and decreased miR-32 expression was significantly associated with aggressive clinicopathological features and poor survival 25, 26. In this study, miR-32 tends to have a low expression level in breast cancers and cell lines. Overexpression of miR-32 in cells could decrease the expression of RGS17 and inhibit cell migration, invasion, and proliferation. Furthermore, ectopic expression of RGS17 could attenuate phenotypes caused by miR-32 overexpression. Therefore, the decreased expression of miR-32 should be a considerable force in the deregulation of RGS17 expression.
RGS proteins can promote tumor progression by several mechanisms. Previous studies show that various RGS proteins, including RGS2, RGS4, and RGS13, can change their localization to nuclear after synthesis in the cytoplasm 27. In addition, cAMP-PKA, a downstream effector of Gas, could promote nuclear translocation of RGS13 28. Increasing evidence suggests that the functions of RGS proteins require the interactions with receptors or intracellular signaling proteins 4. Upregulation of RGS2 could inhibit androgen receptor (AR) and MAP kinase activity in androgen-independent prostate cancer cell lines, suggesting RGS2 may act as a role in regulation of androgen-independent signaling 29. In this study, we did not observe the change of RGS17 localization in tissue staining. Previous studies show that RGS17 can activate PKA signaling and promote CREB phosphorylation in the maintenance of tumor cell proliferation 16. Therefore, RGS17 as a regulator might activate the downstream signaling of GPCR to promote tumor progression.
In summary, RGS17 as a target of miR-32 were frequently overexpressed in breast cancer and has a positive role in the progression of breast cancer, which will help further investigation of RGS17 as a potential treatment target for breast cancer.
Acknowledgments
This work was supported by the National Natural Science Fund (31270862, 81172897 and 21202021) and National Science Foundation for Fostering Talents in Basic Research of China (J1210041).
References
- 1.Stewart BWW. World cancer report 2014. World Health Organization; 2014. [Google Scholar]
- 2.Zhou W, Shi G, Zhang Q, Wu Q, Li B, Zhang Z. MicroRNA-20b promotes cell growth of breast cancer cells partly via targeting phosphatase and tensin homologue (PTEN) Cell & bioscience. 2014;4:62. doi: 10.1186/2045-3701-4-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moreno JL, Holloway T, Gonzalez-Maeso J. G protein-coupled receptor heterocomplexes in neuropsychiatric disorders. Progress in molecular biology and translational science. 2013;117:187–205. doi: 10.1016/B978-0-12-386931-9.00008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hurst JH, Hooks SB. Regulator of G-protein signaling (RGS) proteins in cancer biology. Biochemical pharmacology. 2009;78:1289–97. doi: 10.1016/j.bcp.2009.06.028. [DOI] [PubMed] [Google Scholar]
- 5.Dorsam RT, Gutkind JS. G-protein-coupled receptors and cancer. Nature reviews Cancer. 2007;7:79–94. doi: 10.1038/nrc2069. [DOI] [PubMed] [Google Scholar]
- 6.Li S, Huang S, Peng SB. Overexpression of G protein-coupled receptors in cancer cells: involvement in tumor progression. International journal of oncology. 2005;27:1329–39. [PubMed] [Google Scholar]
- 7.Monroy CA, Mackie DI, Roman DL. A high throughput screen for RGS proteins using steady state monitoring of free phosphate formation. PloS one. 2013;8:e62247. doi: 10.1371/journal.pone.0062247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sokolov E, Iannitti DA, Schrum LW, McKillop IH. Altered expression and function of regulator of G-protein signaling-17 (RGS17) in hepatocellular carcinoma. Cellular signalling. 2011;23:1603–10. doi: 10.1016/j.cellsig.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ross EM. Coordinating speed and amplitude in G-protein signaling. Current biology: CB. 2008;18:R777–R83. doi: 10.1016/j.cub.2008.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsang S, Woo AY, Zhu W, Xiao RP. Deregulation of RGS2 in cardiovascular diseases. Frontiers in bioscience. 2010;2:547–57. doi: 10.2741/s84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boss CN, Grunebach F, Brauer K, Hantschel M, Mirakaj V, Weinschenk T. et al. Identification and characterization of T-cell epitopes deduced from RGS5, a novel broadly expressed tumor antigen. Clinical cancer research: an official journal of the American Association for Cancer Research. 2007;13:3347–55. doi: 10.1158/1078-0432.CCR-06-2156. [DOI] [PubMed] [Google Scholar]
- 12.Wiechec E, Overgaard J, Hansen LL. A fragile site within the HPC1 region at 1q25.3 affecting RGS16, RGSL1, and RGSL2 in human breast carcinomas. Genes, chromosomes & cancer. 2008;47:766–80. doi: 10.1002/gcc.20578. [DOI] [PubMed] [Google Scholar]
- 13.Smalley MJ, Iravani M, Leao M, Grigoriadis A, Kendrick H, Dexter T. et al. Regulator of G-protein signalling 2 mRNA is differentially expressed in mammary epithelial subpopulations and over-expressed in the majority of breast cancers. Breast cancer research: BCR. 2007;9:R85. doi: 10.1186/bcr1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boss CN, Grunebach F, Brauer K, Hantschel M, Mirakaj V, Weinschenk T. et al. Identification and characterization of T-cell epitopes deduced from RGS5, a novel broadly expressed tumor antigen. Clinical cancer research: an official journal of the American Association for Cancer Research. 2007;13:3347–55. doi: 10.1158/1078-0432.CCR-06-2156. [DOI] [PubMed] [Google Scholar]
- 15.Hurst JH, Mendpara N, Hooks SB. Regulator of G-protein signalling expression and function in ovarian cancer cell lines. Cellular & molecular biology letters. 2009;14:153–74. doi: 10.2478/s11658-008-0040-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.James MA, Lu Y, Liu Y, Vikis HG, You M. RGS17, an overexpressed gene in human lung and prostate cancer, induces tumor cell proliferation through the cyclic AMP-PKA-CREB pathway. Cancer research. 2009;69:2108–16. doi: 10.1158/0008-5472.CAN-08-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hooks SB, Callihan P, Altman MK, Hurst JH, Ali MW, Murph MM. Regulators of G-Protein signaling RGS10 and RGS17 regulate chemoresistance in ovarian cancer cells. Molecular cancer. 2010;9:289. doi: 10.1186/1476-4598-9-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bodle CR, Mackie DI, Roman DL. RGS17: an emerging therapeutic target for lung and prostate cancers. Future medicinal chemistry. 2013;5:995–1007. doi: 10.4155/fmc.13.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crum CP, McKeon FD. p63 in epithelial survival, germ cell surveillance, and neoplasia. Annual review of pathology. 2010;5:349–71. doi: 10.1146/annurev-pathol-121808-102117. [DOI] [PubMed] [Google Scholar]
- 20.Melino G. p63 is a suppressor of tumorigenesis and metastasis interacting with mutant p53. Cell death and differentiation. 2011;18:1487–99. doi: 10.1038/cdd.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chakrabarti R, Wei Y, Hwang J, Hang X, Andres Blanco M, Choudhury A. et al. DeltaNp63 promotes stem cell activity in mammary gland development and basal-like breast cancer by enhancing Fzd7 expression and Wnt signalling. Nature cell biology. 2014;16:1004–15. doi: 10.1038/ncb3040. 1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S. et al. MicroRNA gene expression deregulation in human breast cancer. Cancer research. 2005;65:7065–70. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 23.Mosakhani N, Raty R, Tyybakinoja A, Karjalainen-Lindsberg ML, Elonen E, Knuutila S. MicroRNA profiling in chemoresistant and chemosensitive acute myeloid leukemia. Cytogenetic and genome research. 2013;141:272–6. doi: 10.1159/000351219. [DOI] [PubMed] [Google Scholar]
- 24.Hu Z, Dong J, Wang LE, Ma H, Liu J, Zhao Y. et al. Serum microRNA profiling and breast cancer risk: the use of miR-484/191 as endogenous controls. Carcinogenesis. 2012;33:828–34. doi: 10.1093/carcin/bgs030. [DOI] [PubMed] [Google Scholar]
- 25.Zhang J, Kuai X, Song M, Chen X, Yu Z, Zhang H. et al. microRNA-32 inhibits the proliferation and invasion of the SGC-7901 gastric cancer cell line. Oncology letters. 2014;7:270–4. doi: 10.3892/ol.2013.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang D, Ni Z, Xu X, Xiao J. MiR-32 functions as a tumor suppressor and directly targets EZH2 in human oral squamous cell carcinoma. Medical science monitor: international medical journal of experimental and clinical research. 2014;20:2527–35. doi: 10.12659/MSM.892636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burchett SA. In through the out door: nuclear localization of the regulators of G protein signaling. Journal of neurochemistry. 2003;87:551–9. doi: 10.1046/j.1471-4159.2003.02047.x. [DOI] [PubMed] [Google Scholar]
- 28.Xie Z, Geiger TR, Johnson EN, Nyborg JK, Druey KM. RGS13 acts as a nuclear repressor of CREB. Molecular cell. 2008;31:660–70. doi: 10.1016/j.molcel.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao X, Qin J, Xie Y, Khan O, Dowd F, Scofield M. et al. Regulator of G-protein signaling 2 (RGS2) inhibits androgen-independent activation of androgen receptor in prostate cancer cells. Oncogene. 2006;25:3719–34. doi: 10.1038/sj.onc.1209408. [DOI] [PubMed] [Google Scholar]