Abstract
Human JAK2 tyrosine kinase mediates signaling through numerous cytokine receptors. The JAK2 JH2 domain functions as a negative regulator and is presumed to be a catalytically inactive pseudokinase, but the mechanism(s) for its inhibition of JAK2 remains unknown. Mutations in JH2 lead to increased JAK2 activity contributing to myeloproliferative neoplasms (MPNs). Here, we show that JH2 is a dual-specificity protein kinase that phosphorylates two negative regulatory sites in JAK2, Ser523 and Tyr570. Inactivation of JH2 catalytic activity increased JAK2 basal activity and downstream signaling. Importantly, different MPN mutations were found to abrogate JH2 activity in cells, and in MPN (V617F) patient cells, phosphorylation of Tyr570 was reduced, suggesting that loss of JH2 activity contributes to the pathogenesis of MPNs. These results identify the catalytic activity of JH2 as a previously unrecognized mechanism to control basal activity and signaling of JAK2.
Introduction
JAK2 belongs to the Janus family of cytoplasmic tyrosine kinases (JAK1-3, TYK2) and functions as a critical mediator of signaling for hematopoietic cytokines and hormones including erythropoietin (Epo), thrombopoietin (Tpo), interferon-γ (IFN-γ), several interleukins, growth hormone, prolactin, leptin and granulocyte-macrophage colonystimulating factor1,2. JAK2 serves as a triggering kinase for cytokine receptors, and phosphorylation and activation of downstream signaling proteins and progression of signal transduction are dependent on JAK2 activity. JAK2 associates with the cytoplasmic domains of cytokine or hormone receptors, and ligand-induced receptor rearrangement facilitates JAK2 trans-phosphorylation of activation-loop tyrosines 1007–1008 in JH1 (tyrosine kinase domain), leading to its activation. Subsequent phosphorylation of tyrosine residues in the receptors by JAK2 creates docking sites for SH2-containing signaling proteins such as STATs (Signal Transducer and Activator of Transcription)3, 4.
Phosphorylation plays an important role in regulation of JAK2, both positively and negatively. In the absence of cytokine stimulation, JAK2 is constitutively phosphorylated on Ser5235, but upon activation becomes phosphorylated on as many as 20 tyrosine residues. In addition to Tyr1007-Tyr1008, phosphorylation of Tyr637, Tyr813, Tyr868, Tyr966 and Tyr972 potentiate JAK2 activity, whereas phosphorylation of Ser523, Tyr119, Tyr221, Tyr317, Tyr570 and Tyr913 negatively regulate JAK26–11. Because JAK2 mediates critical physiological functions such as cell proliferation, the kinase activity of JAK2 is tightly regulated via various mechanisms, including trans-acting proteins (tyrosine phosphatases, SOCS proteins) and by JH2 (pseudokinase domain)12–15.
Both biochemical and clinical evidence have demonstrated an important regulatory function for JH2 in JAKs, and at present, 32 different mutations in JH2 of JAK2 have been shown to cause, or are linked to, hematological diseases16. The most frequent somatic mutation, V617F, results in constitutively active JAK2, and is responsible for >95% of polycythemia vera cases and ~50% of essential thrombocythemia and primary myelofibrosis cases17–19. The mechanism(s) by which JH2 negatively regulates the tyrosine kinase activity of JH1 is currently not known, but it is likely to involve an intramolecular interaction between JH2 and JH113. In JAK2 and JAK3, deletion of JH2 increases basal JAK activity, and the JH2 domain has been shown to co-immunoprecipitate with the JH1 domain (in trans)14, 20. At present, crystal structures are available only for JH1 of JAKs21, 22. JH2 is predicted to adopt a canonical protein kinase fold, but to be catalytically inactive due to amino-acid substitutions of key catalytic residues conserved in active protein kinases, particularly, an aspartic acid in the catalytic loop (HRD motif). Therefore, JH2 in JAKs has been classified as a pseudokinase. Pseudokinases make up ~10% of the kinome and have been implicated in the regulation of a variety of cellular functions including tumorigenesis23.
The understanding of JAK kinase function, regulation and structure has been hampered by the difficulty of producing and purifying recombinant, soluble JAKs and their domains. This holds true also for JH2, and production and purification of JH2 has not been previously reported. We have now produced recombinant JAK2 JH2 domain using a baculovirus expression system, which allowed us to address questions regarding the function of the pseudokinase domain of JAK2. We show that, contrary to the generally accepted belief, JH2 is an active protein kinase that phosphorylates two sites in JAK2, Ser523 and Tyr570, which serves to maintain a low basal level of JAK2 activity. Moreover, JAK2 mutations that cause MPNs were found to abrogate JH2 activity. These results identify a previously unrecognized mechanism in regulation of normal and pathogenic JAK2 and cytokine signaling.
Results
Phosphorylation of purified JAK2 JH2 in vitro
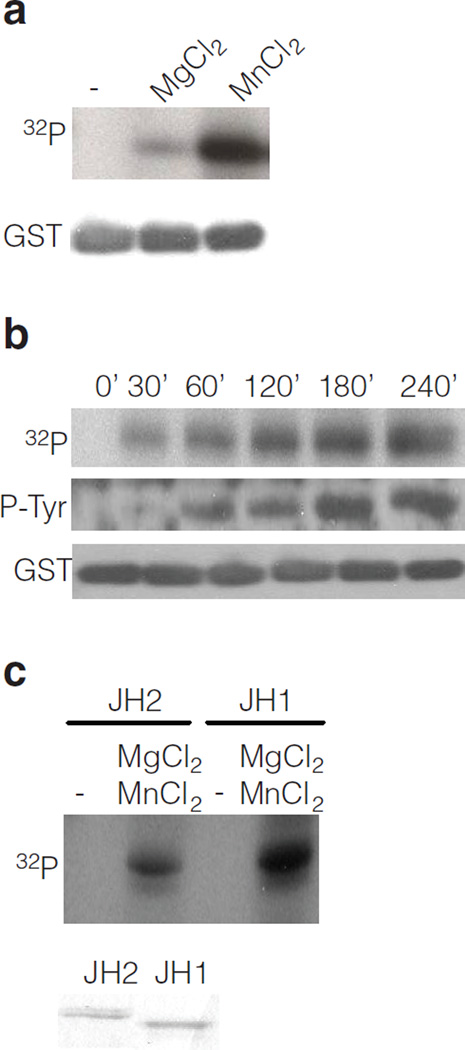
JH2 was expressed as a GST-fusion protein in insect cells (Sf9) (Supplementary Fig. 1, 2). Purified GST-JH2 was used in an in vitro kinase assay, which showed a time-dependent phosphorylation of JH2 with a strong preference for Mn2+ as divalent cation (Fig. 1a and b). A comparison of the autophosphorylation activity of purified JH1 versus JH2 of JAK2 indicates that JH2 has ~ 10% of JH1 activity (Fig. 1c), which could explain why JH2 activity has previously gone unnoticed. To verify the autophosphorylation activity of JH2, a kinase-inactivating mutation, K581A, was introduced in JH2. This lysine (in β-strand 3 of the JH2 N-lobe) serves to coordinate the α-and β-phosphates of ATP in active protein kinases24. GST-JH2 wild-type (WT) and K581A mutant were produced and purified sideby- side from insect cells. In vitro kinase assay (Supplementary Fig. 3a) shows that the kinase-inactive JH2 mutant is devoid of autophosphorylation activity.
Figure 1.
Identification of JAK2 JH2 catalytic activity in vitro. (a) In vitro kinase assay with purified JAK2 GST-JH2 with [32P] γ-ATP in the absence or presence of divalent cations. (b) Time-course kinase assay with purified JAK2 GST-JH2 in the presence of [γ- 32P] ATP or unlabeled ATP. (c) Autoradiography of kinase assay (30 min) using purified JAK2 JH2 and JH1 domain and [γ-32P] ATP, in the absence or presence of cations. Coomassie staining shows the protein levels of JH1 and JH2.
To further confirm that the observed kinase activity was due to JH2 autophosphorylation and not to phosphorylation by a contaminating protein kinase, JH2 was in vitro translated (Supplementary Methods) and analyzed in a kinase assay. Western blotting showed that in vitro translated JH2 of JAK2 is autophosphorylated on tyrosine (Supplementary Fig. 3b). Next, JH2 wild-type (WT) and JH2 K581A were in vitro translated, His-tag purified and subjected to an in vitro kinase assay in the presence of [γ-32 P] ATP. Autophosphorylation was detected in JH2 domain but not in JH2 K581A mutant (Supplementary Fig. 3c). Taken together, these results demonstrate that JH2 possesses autophosphorylation activity.
Purified JAK2 JH2 becomes autophosphorylated on Ser523 and Tyr570 residues
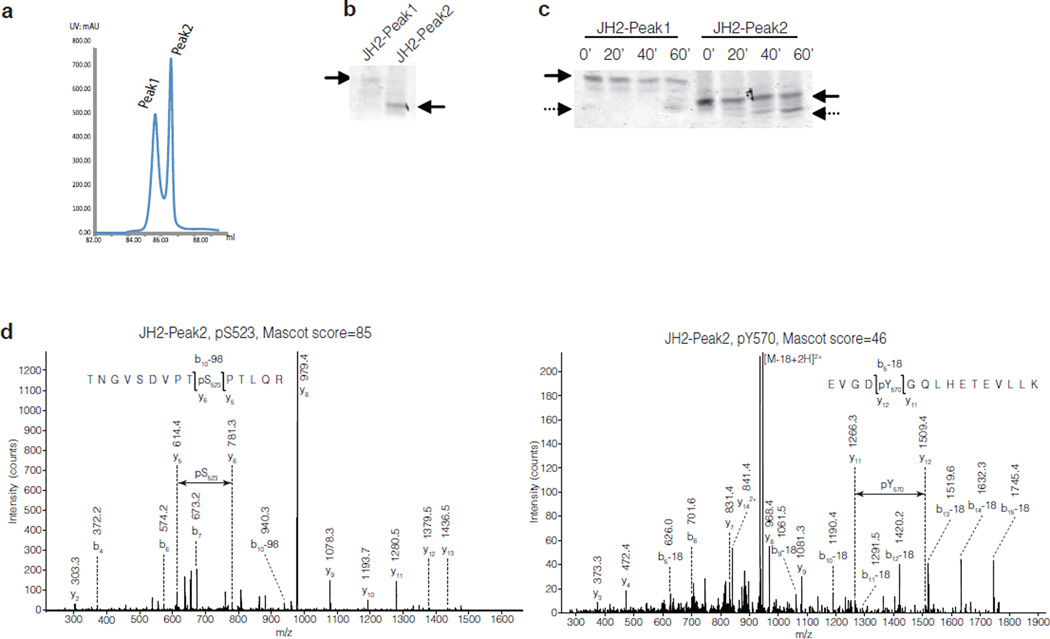
To study the kinase activity of the JAK2 pseudokinase domain in more detail, His-tagged JH2 was expressed in insect cells and purified using Ni-NTA affinity and anion-exchange chromatography. JH2 eluted in two closely spaced peaks on an anion-exchange column (Fig. 2a and Supplementary Fig. 4). In native-gel electrophoresis, JH2 in peak 2 (JH2-Peak2) migrated faster than JH2-Peak1 (Fig. 2b). The chromatography and electrophoresis data are suggestive of a higher phosphorylation state for JH2-Peak2 than for -Peak1. The autophosphorylation activities of the two JH2 samples were analyzed in an in vitro kinase assay. Native-gel electrophoresis showed the appearance of a faster-migrating band for both samples at later time points of the reaction, consistent with an increase in phosphorylation state (Fig. 2c). LC-ESI-MS and MS LTQ-Orbitrap mass spectrometry was used to identify the phosphorylated residues in JH2. The analysis showed that JH2-Peak1 was unphosphorylated at time zero and underwent autophosphorylation on Ser523 during the kinase reaction (data not shown). In contrast, JH2-Peak2 was robustly (stoichiometrically) phosphorylated on Ser523 at time zero, hence explaining the migration difference between the proteins in the two peaks, and became phosphorylated additionally on Tyr570 during the kinase reaction (Fig. 2d).
Figure 2.
Identification of phosphorylated residues in JAK2 JH2. (a) Chromatogram of JAK2 JH2 purification showing the peaks from anion-exchange chromatography. (b) Coomassie staining of a native-gel electrophoresis of JH2-Peak1 and JH2-Peak2 proteins. (c) Coomassie staining of a native-gel electrophoresis of purified JH2-Peak1 and JH2- Peak2 after kinase reaction. (d) MS-MS spectra of the phosphorylated residues in JAK2 JH2-Peak2 4h kinase assay. Left: JH2-Peak2 is stoichiometrically phosphorylated at Ser523. Right: JH2-Peak2 is partially phosphorylated at Tyr570.
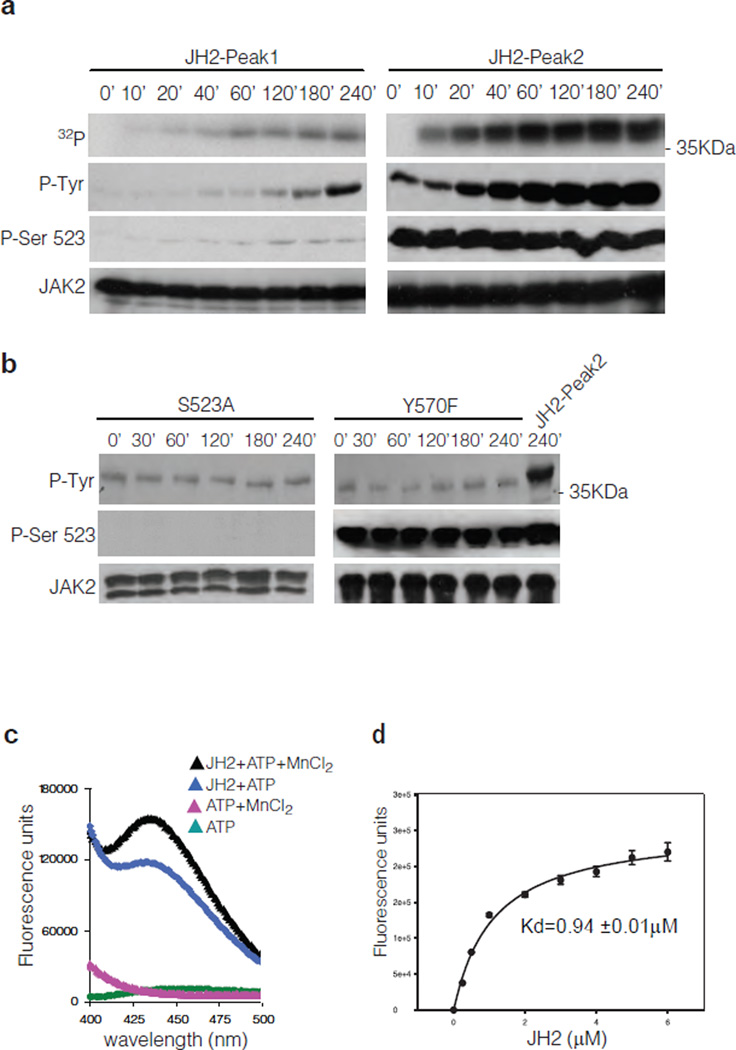
Further analysis of the JH2 autophosphorylation activity in kinase assays demonstrated that JH2-Peak2 has substantially higher tyrosine kinase activity than JH2-Peak1 (Fig. 3a). To investigate the basis for this difference, the phosphorylation state of Ser523 was monitored by Western blotting using an anti-pSer523 specific antibody5. Consistent with the mass spectrometry results, Ser523 phosphorylation increased in JH2-Peak1 during the kinase reaction, whereas JH2-Peak2 was fully phosphorylated already at time zero and the pSer523 level remained constant during the reaction (Fig. 3a). Moreover, GST-JH2 K581A mutant purified from insect cells does not show any phosphorylation on Ser523 (Supplementary Fig. 5) demonstrating that Ser523 is de facto autophosphorylation site of JH2. These results, together with the results in Fig. 2c, suggest that phosphorylation of Ser523 regulates the tyrosine kinase activity of JH2. To address this possibility, Ser523 was mutated to alanine, and, consistent with the hypothesis, S523A did not undergo tyrosine phosphorylation during an in vitro kinase assay (Fig. 3b). Mutation of Tyr570 to phenylalanine did not affect Ser523 phosphorylation, but abolished tyrosine phosphorylation. These phospho-specific antibody data also confirm the mass spectrometry identification of the two autophosphorylated residues in JH2, Ser523 and Tyr570. Importantly, these two residues have previously been identified as negative regulatory sites in JAK25, 7, 9, 10.
Figure 3.
Analysis of JAK2 JH2 autophosphorylation and ATP binding activity. (a) Timecourse kinase assay of purified JH2-Peak1 and JH2-Peak2. (b) Time-course kinase assay of purified JH2 S523A and Y570F mutants compared to JH2-Peak2. (c) Fluorescence measurement of ATP binding assay of JAK2 JH2-Peak2. (d) Kd measurement of mant-ATP binding to JH2-Peak2. Graph, mean ± s.d. of three independent experiments.
If the pseudokinase domain of JAK2 is an active protein kinase, utilizing ATP as a phosphate donor, it should bind ATP with a physiologic Kd value. The binding of ATP to JH2 of JAK2 was evaluated using the fluorescent ATP-analogue mant [2’-(3’)-O-(N-methylanthraniloyl)]-ATP. The fluorescence emission scan showed a peak at ~440 nm only when MnCl2 and JH2 were present along with mant-ATP (Fig. 3c). Mant-ATP bound to JH2 with a Kd of ~1 µM (Fig. 3d), which is ~10% of the reported Kd for JH1 of JAK225. Taken together, the in vitro data demonstrate that the pseudokinase domain of JAK2 is a dual-specificity serine and tyrosine kinase. Autophosphorylation of Ser523 is the primary event in JH2 activation, which enhances subsequent autophosphorylation of Tyr570.
Analysis of JAK2 JH2 in mammalian cells
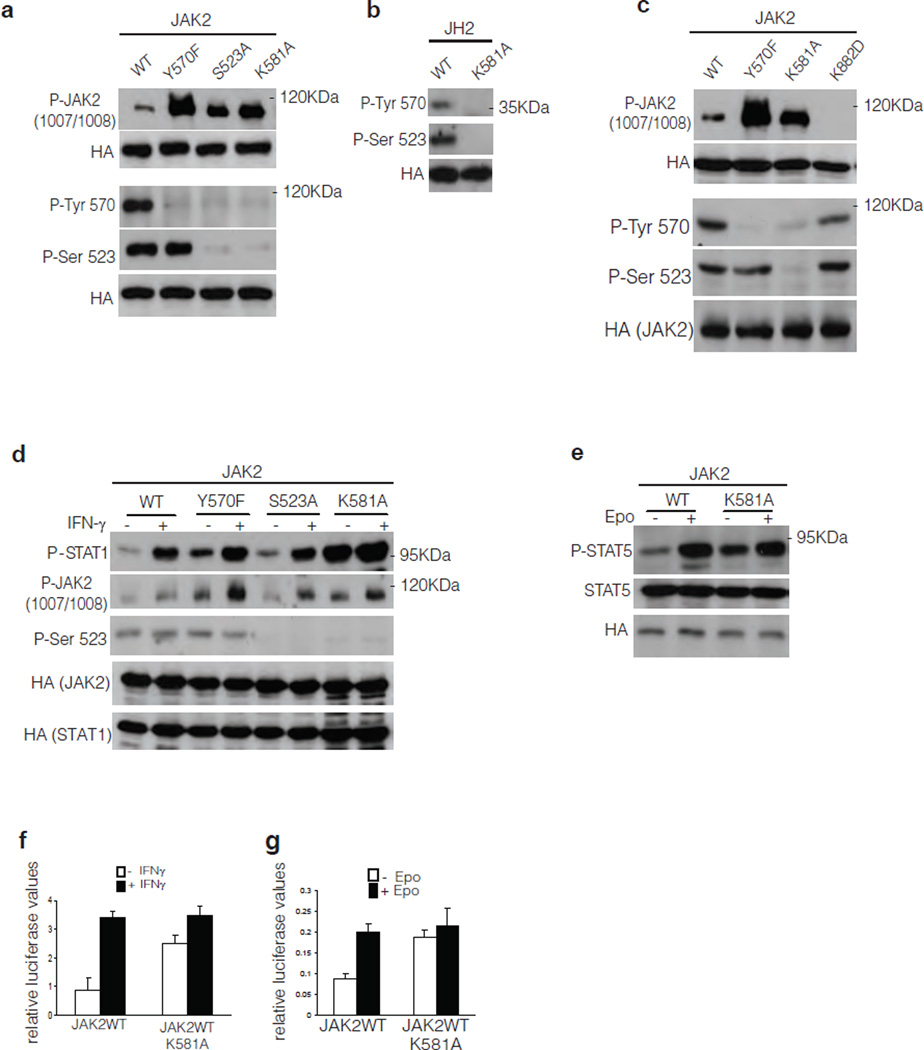
To analyze the function of the catalytic activity of JH2 in a cellular context, the kinase-inactivating point mutation in JH2, K581A, was introduced into JAK2. Various JAK2 constructs were expressed in JAK2-deficient γ2A cells, and JAK2 phosphorylation was analyzed by Western blotting. JAK2 WT was tyrosine phosphorylated at a low level, and consistent with previous studies5, 7, 9, 10, mutation of either Ser523 or Tyr570 increased JAK2 tyrosine phosphorylation of the JH1 activation loop (Tyr1007-Tyr1008), an indicator of JAK2 activation. Similar to the data for S523A and Y570F, JAK2 K581A displayed a higher level of tyrosine phosphorylation than JAK2 WT and, importantly, Ser523 and Tyr570 sites were not phosphorylated in K581A (Fig. 4a). In addition, these results corroborate the in vitro results (Fig. 3b) and show that, in cells, phosphorylation of Tyr570 is dependent on Ser523 phosphorylation of JH2.
Figure 4.
Analysis of JAK2 signaling in mammalian cells. (a, c) Phosphorylation of JAK2WT and mutants thereof in JAK2-deficient γ2A cells. HA-tagged JAK2 proteins were immunoprecipitated with anti-HA antibody and JAK2 phosphorylation is shown by Western blotting. Anti-HA Western blots show protein levels for each independent experiment. (b) Phosphorylation of JAK2 JH2 in γ2A cells. (d, e) Phosphorylation of STAT1 in response to IFN-γ stimulation and phosphorylation of STAT5 in response to Epo stimulation in γ2A cells. (f) Effect of JAK2 K581A mutation on STAT1 transcription activation using IF–γ-dependent GAS luciferase reporter. Graph, mean ± s.d. of three independent experiments, P < 0.05. (g) Effect of JAK2 K581A mutation on STAT5 transcription activation using SPI-Luc2 luciferase reporter. The basal JAK2 WT activity was set to 1 for all experiments, graph, mean ± s.d. of six independent experiments are shown, P < 0.05.
To confirm the role of JH2 catalytic activity in phosphorylation of Ser523 and Tyr570, JH2 alone, WT or the K581A mutant, were expressed in γ2A cells. In JH2 WT, both Ser523 and Tyr570 were phosphorylated, and the K581A mutation abolished their phosphorylation (Fig. 4b). Moreover, in the context of full-length JAK2 bearing a point mutation that abrogates JH1 activity (K882D), phosphorylation of Ser523 occurred to the same extent as in JAK2 WT, and K882D mutation did not markedly affect phosphorylation of Tyr570 (Fig. 4c). Finally, in JAK2 constructs lacking the entire JH1 domain (JAK2del.JH1) phosphorylation of Ser523 and Tyr570 occurred to the same level as in JAK2 WT (Supplementary Fig. 6). Finally, we wanted to verify that the effects of the K581A mutation were due to abrogation of JH2 catalytic activity and minimize the possibility that they were caused by secondary conformational alterations in JH2. To this end, a more conservative mutation, K581R, and, separately, a distinct inactivating mutation, N678A (catalytic loop) were introduced into the full length protein. The K581R mutant showed clear decreases in Ser523 and Tyr570 phosphorylation and increase in JAK2 Y1007-1008 phosphorylation. Similar effects, albeit less pronounced, were observed with N678A mutant (Supplementary Fig. 7). These in-cell data substantiate the conclusion that JH2 is a dual-specificity protein kinase that autophosphorylates Ser523 and Tyr570.
JH2 catalytic activity is required to maintain low basal activity of JAK2
JH2 activity was next investigated in cytokine receptor-mediated signaling by analyzing STAT activation in γ2A cells in response to cytokine stimulation. Compared to JAK2 WT, the JAK2 mutants S523A, Y570F and K581A showed increased basal phosphorylation of STAT1, but the mutations did not influence the IFN-γ-induced STAT1 phosphorylation (Fig. 4d). There was some variation in the level of STAT1 phosphorylation between the experiments (Fig. 4d and Supplementary Fig. 8), but increased basal phosphorylation of STAT1 was a consistent finding that was also observed with K581A in EpoR-induced STAT5 phosphorylation (Fig. 4e). The JH2 activity was investigated in cytokine-induced transcriptional response by using reporter-gene analysis. Introduction of K581A in JAK2 increased the basal STAT1- and STAT5-dependent reporter-gene activation, but did not affect the IFN-γ or Epo induced responses (Fig. 4f and g). Taken together, these results indicate that JH2 catalytic activity is required to maintain a low basal level of JAK2 (JH1) activity.
JAK2 MPN-causing mutations affect the catalytic activity of JH2
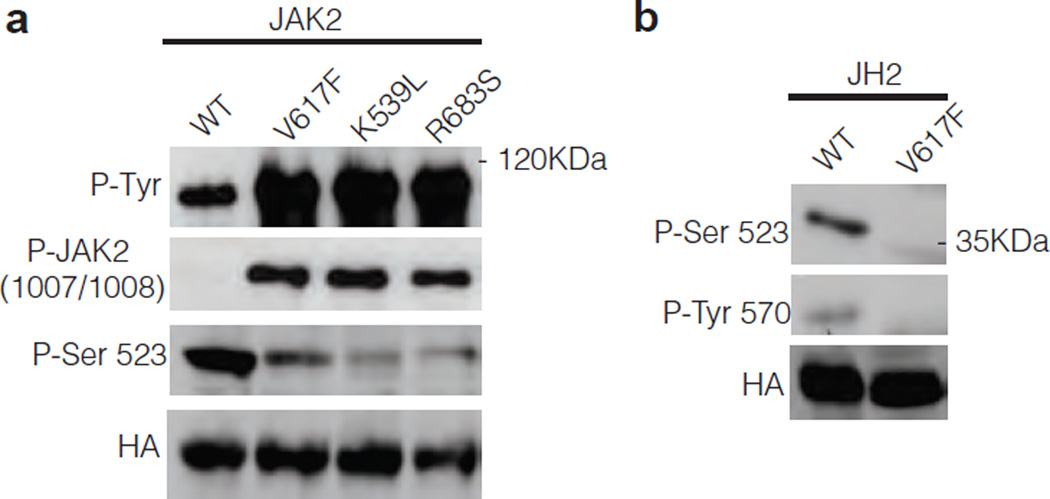
Our results showing that the catalytic activity of JH2 regulates the basal activity of JAK2 raises the question of the possible connection of this activity to human JAK2 mutants and disease pathogenesis. We were interested in understanding whether the catalytic activity of JH2 was involved in the pathogenesis of JAK2 MPN mutants. For this analysis, we chose three distinct MPN-causing JH2 mutants: V617F (exon 14, predominant MPN-causing mutation), K539L (exon 12)26 and R683S (exon 16)27. Consistent with previous studies, these mutants showed high levels of tyrosine phosphorylation and activation of JAK2 in γ2A cells when compared to JAK2 WT (Fig. 5a). Interestingly, all three mutants showed significantly decreased Ser523 phosphorylation. These results suggest that the JH2 mutations that cause MPN reduce or abrogate JH2 catalytic activity. To test this hypothesis directly, JH2 alone and its V617F counterpart were analyzed in γ2A cells. The results show that V617F, like K581A, abrogates Ser523 and Tyr570 phosphorylation (Fig. 5b). Finally, it was of interest to determine whether the altered JH2 function is also observed in clinical samples from MPN patients, and thus could be a causative mechanism for the disease. To this end, platelets from three MPN patients carrying the V617F mutation and from a healthy control were isolated and subjected to Tpo stimulation (Supplementary Methods). As a readout for JH2 activity, the phosphorylation of JAK2 Tyr570 was analyzed. Tpo stimulation readily induced Tyr570 phosphorylation in control cells, while in patient samples Tyr570 phosphorylation was significantly reduced, and the reduction correlated with the V617F allelic burden of the patient cells (Supplementary Fig. 9). Taken together, these results show that MPN-causing mutations disturb the catalytic activity of JH2 and abrogate phosphorylation of negative regulatory residues that lead to increased basal activation of JAK2.
Figure 5.
Phosphorylation of different JAK2 MPN mutants. (a) Phosphorylation of JAK2WT and MPN mutants in JAK2-deficient γ2A cells. (b) Phosphorylation of JAK2 JH2 in γ2A cells.
Discussion
Protein kinases have been classified as pseudokinases if they lack conserved residues thought to be required for phosphoryl transfer, and if catalytic activity has not been detected23, 28. Recent studies have provided important new information and insights into the functions of this protein family. Some of the structurally characterized proteins such as VRK3, is unable to bind ATP and obtains a pseudoactive conformation by filling the ATP binding pocket by hydrophobic residues, and thus is retaining the pseudokinase status29. However, for several other proteins the functional status has been overturned, and proteins including CASK, haspin, WNK1, HER3(ErbB3), and STRADα have been shown to have ATP-binding and (or) catalytic activity that can be achieved through non-canonical mechanisms30–34. Each of these pseudokinases utilizes a distinct mechanism to carry out its cellular functions. For example, WNK1 compensates for the missing ATP-binding lysine in β-strand 3 by employing instead a lysine residue in the nucleotide binding loop32. The calcium calmodulin-activated serine-threonine kinase CASK displays atypical catalytic activity in that Mg2+ inhibits its activity30. HER3 lacks the catalytic base aspartate and the crystal structure reveal that it assumes an atypical conformation for active kinases, particularly in αC helix and activation segment33, 35. However, HER3 was found to retain low levels kinase activity and phosphorylate its intracellular region in vitro, but the physiological role of this activity remains to be determined33. These results argue that each alleged pseudokinase needs to be functionally analyzed and scrutinized for possible catalytic activity. In this study, we have shown that, both in vitro and in cells, the pseudokinase domain of JAK2 is an active dual-specificity protein kinase that phosphorylates two previously identified negative regulatory sites in JAK2, Ser523 and Tyr570. Phosphorylation of these sites is required to maintain low basal activity of JAK2. Our results on the catalytic activity of JH2 provide novel insights into the regulation of JAK activation in signaling by a variety of cytokines such as Epo, Tpo, IFN-γ, growth hormone, prolactin, IL-3, IL-5, and GM-CSF. In unstimulated cells, Ser523 has been shown to be the only constitutively phosphorylated residue in JAK25, and phosphorylation of other sites, including Tyr570, occurs only upon cytokine stimulation and activation of JAK29, 10. The kinases responsible for phosphorylation of Ser523 and Tyr570 have not been identified, but the activity of JH1 was not required for these phosphorylation events.5,7,9,10 We show here that JH2 phosphorylates Ser523 and Tyr570, and that autophosphorylation of Ser523 is the primary event in JH2 activation and it is observed in unstimulated conditions (Fig. 4d). Cytokine-induced receptor dimerization and juxtapositioning of the JAKs lead to other regulatory trans-phosphorylation events, including phosphorylation of Tyr570. Ser523 resides in the linker region between the SH2-like domain of JAK2 and JH2 (Supplementary Fig. 10), and from steric considerations could be phosphorylated in cis. Tyr570, predicted to be in the β2-β3 loop of JH2, is distal to the JH2 active site and is presumed to be phosphorylated in trans by JH2 in another JAK2 molecule. A crystal structure of JH1-JH2 will be required to understand the mechanisms by which JH2 sterically inhibits JH1 and by which JH2-mediated phosphorylation of Ser523 and Tyr570 suppresses JH1 activity. Our results are consistent with a model whereby phosphorylation of Ser523 and Tyr570 strengthens the JH1–JH2 autoinhibitory interaction. The relatively low catalytic activity of JH2 is in accordance with autophosphorylation of regulatory residues as a physiological function for JH2 while the JH1 is mainly responsible for phosphorylation of substrate proteins. The low catalytic activity, together with the critical regulatory role of Ser523 and atypical requirement for Mn2+ for catalysis, have probably hampered the detection of JH2 activity.
The discovery of somatic mutations in JH2 of JAK2 in the majority of Philadelphia chromosome-negative MPNs and other hematological malignancies have focused attention on the functional role of JH2 and turned JAKs into important therapeutic targets. However, the underlying mechanisms for JAK2 hyperactivation in MPNs have remained obscure. Currently, several inhibitors targeting the JAK2 tyrosine kinase domain are in clinical trials for MPNs36. The JAK2 inhibitors show beneficial clinical effects and alleviate symptoms, but they do not substantially reduce the JAK2-mutant tumor load, and the inhibitors do not discriminate between normal and mutated JAK2. We show that MPN-causing JAK2 mutations disturb JH2 catalytic activity and remove the negative regulatory effects of Ser523 and Tyr570 phosphorylation in cell lines and in primary cells from MPN patients. These results identify a molecular pathogenic mechanism in MPNs and suggest that loss of JH2 function is involved in the hyperactive JAK2 MPN phenotype.
In conclusion, these studies have identified an unexpected regulatory mechanism for JH2 in JAK2. JH2 is an active protein kinase that autophosphorylates two negative regulatory residues, which is required to maintain a low-activity level of JAK2 in the absence of cytokine stimulation. The discovery of JH2 catalytic activity and its connection to MPNs may also afford novel approaches for the development of targeted therapies to combat JAK-mediated diseases.
Methods
Protein expression and purification
JH2 from JAK2 was amplified and cloned into pFASTBAC1 vector (Invitrogen) with a Thrombin-cleavable N-terminus GST-tag or a Cterminus 6xHis-tag and expressed as a fusion protein in insect cells (Sf9) cells. For protein expression, cells were infected with 10% (v/v) virus supernatant and grown for 48h and harvested by centrifugation. Cell pellets containing GST-JH2 or JH2-His fusion protein were resuspended in lysis buffer containing 20mM TRIS-HCl, pH 8.5, 500mM NaCl, 15% (v/v) glycerol, 0.5mM TCEP and 20mM imidazole (for JH2-His protein only), supplemented with protease inhibitors cocktail (Roche), lysed using cell-disruptor (Avensis) and clarified by centrifugation for 1h at 45000×g. The supernatant was incubated for 2h with pre-washed GST beads (GE Healthcare) or Ni-NTA beads (Qiagen) with gentle rotation at 4°C. The beads were extensively washed and the fusion protein was eluted with 10mM glutathione (Sigma-Aldrich) for GST-JH2 or 250mM imidazole (Fluka) for JH2-His protein. Fractions containing the fusion protein were pooled and dialyzed for 2h at 4°C in buffer (20mM TRIS-HCl, pH 8.5, 250mM NaCl, 15% (v/v) glycerol and 0.5mM TCEP). For JH2-His fusion protein, after dialysis samples were incubated with 10U ml−1 Thrombin (Enzyme Research Laboratory) overnight. Proteins were loaded onto a MonoQ column (GE Healthcare) equilibrated with 20mM TRIS-HCl, pH 8.5, 25mM NaCl, 15% (v/v) glycerol and 0.5mM TCEP and eluted with a linear gradient 1–200mM NaCl. Fractions containing purified GST-JH2 or JH2 were analyzed by Coomassie staining and pooled and concentrated to 1 mg ml−1 for further use. JAK2 JH1 kinase domain (aa 836–1132) was cloned by PCR amplification into pFASTBAC1 plasmid with an N-terminus GST-tag and purified as previously described21.
Autophosphorylation Reaction
The autophosphorylation reactions were carried out using 1µg µl−1 of JH2, 10mM ATP (Sigma-Aldrich) or 10µCi [γ-32 P] ATP (PerkinElmer), 20 mM MnCl2, 300mM NaCl, 10% (v/v) glycerol, 0.5mM TCEP, and 20mM Tris-HCl (pH 8.0) at room temperature. The reactions were stopped by adding EDTA to a final concentration of 100 mM. The phosphorylation states of JH2 were monitored by autoradiography and native-PAGE (PhastGel System, GE Healthcare) and Western blotting.
Mass Spectrometry
JAK2 gel bands were processed for in-gel digestion as previously reported37. Phosphopeptide enrichment was performed with titanium dioxide microcolumns, with eluates desalted with Poros R3 microcolumns as previously described38. LC-MS MS was conducted on an EASY-nLC system (Thermo Fisher Scientific) coupled to an LTQ - Orbitrap XL mass spectrometer (Thermo Fisher Scientific) as previously reported39 except that chromatography was conducted with a 30 min gradient. Raw data files were submitted for Mascot searches (Matrix Science) using Proteome Discoverer 1.1 software (Thermo Fisher Scientific). Databases containing the human JAK2 protein sequence were searched with the following parameters: ESI-TRAP was selected as the instrument setting, with specified mass tolerances of 10 ppm (precursor) and 0.6 Da (fragment). Serine, threonine and tyrosine phosphorylation, along with methionine oxidation, were set as variable modifications. Cysteine carbamidomethylation was included as a fixed modification and Trypsin -P specified with a maximum of two missed cleavages. Only MS-MS spectra from JAK2 phosphopeptides possessing Mascot ion scores above 20 were manually validated for the sites of phosphorylation.
Mant-ATP binding assay
The fluorescence intensity of mant-ATP (Invitrogen) complex with JH2 was measured using FluoroMax-2 spectrofluorimeter. Mant-ATP (1µM) was added to a buffer solution (20mM TRIS-HCl, pH 8, 200mM NaCl, 10% (v/v) glycerol and 0.5mM TCEP) along with 5mM MnCl2 and 1µM JH2 (from peak 2 fraction). The excitation and emission wavelengths were 280nm and 440nm, respectively, and emission was scanned from 400–500nm. For kd measurements, increased concentration of purified JH2 (0.25µM – 6µM) was added to buffer solution with 5mM MnCl2 and 1µM mant-ATP.
Transfection, Western blotting and Luciferase assay
Human JAK2 wild-type, JAK2 JH2 domain and human EpoR were obtained by PCR amplification and cloned with a C-terminus HA-tag into pCI-neo mammalian expression plasmid (Promega). JAK2 mutations were done using QuickChange Site-Directed Mutagenesis method (Stratagene) and verified by sequencing. STAT1 and STAT5 plasmids were previously described14. JAK2-deficient γ2A cells (fibrosarcoma cells) were transfected with different JAK2 mutants using Fugene (Roche) according to manufacturer’s instructions. After 8h, cells were lysed in lysis buffer (50mM TRIS-HCl, pH 8, 150mM NaCl, 100mM NaF, 10% (v/v) glycerol, 1% (v/v) Triton-X and protease inhibitors cocktail) and protein phosphorylation was analyzed by Immunoprecipitation and Western blotting with anti-phosphotyrosine (4G10) antibody (Millipore), anti-pJAK2 1007–1008 (Cell Signaling Technology), anti-pSer5235, antipTyr5709 and anti-HA (Covance) antibody. STAT1 and STAT5 phosphorylation was analyzed in γ2A cells transfected with different JAK2 constructs together with STAT1 or STAT5 as indicated, and after 8h cells were starved for 12h in serum-free media followed by stimulation with hIFN-γ (100U ml−1, R&D Systems) or hEpo (50U ml−1, Janssen-Cilag). After cell lysis, STAT1 phosphorylation was analysed by Western Blotting with antipSTAT1 antibody or anti-pSTAT5 antibody (Cell Signalling Technology). STAT1 and STAT5 transcriptional activity of JAK2 wild type and K581A mutant was measured in γ2A cells using GAS-luc STAT1 reporter or or SPI-luc2 STAT5 reporter as previously described14. After stimulations, cells were lysed in 1× reporter lysis buffer (Promega). Luminescence was recorded using Luminoscan Ascent 96-well plate luminometer (Thermo Labsystem) and the transfection efficiency was normalized using β-GAL values.
Supplementary Material
Acknowledgments
We thank Dr. Martin Myers (University of Michigan Medical School) for generous provision of reagents, Elina Koskenalho, Paula Kosonen, and Merja Lehtinen for technical assistance, and Biocenter Finland protein production platform (Dr. Vesa Hytönen, Ulla Kiiskinen) for technological support. This study was supported by the Medical Research Council of Academy of Finland (O.S.), the Sigrid Juselius Foundation (O.S.), the Finnish Cancer Foundation (O.S.), EU Research Training Network ReceptEur (O.S.), Competitive Research Funding and Centre of Laboratory Medicine of the Tampere University Hospital (O.S.), and the Tampere Tuberculosis Foundation (O.S.), NIH core grant CA016087 (S.R.H.), Danish Research Agency and the Danish National Research Foundation (Centre for Epigenetics) (C.Y. and O.N.J.).
Footnotes
Author contributions
D.U. performed the experiments and wrote the paper. O.S. and S.R.H. designed the experiments and wrote the paper. J.W. performed the in vitro experiments with recombinant proteins. T.P. and Y.N. performed the mutagenesis experiments in mammalian cells. C.Y., O.J., T.A.N., and C.F.X. performed the experiments for MS analysis. R.S. performed the experiments for clinical sample analysis.
References
- 1.Gadina M, et al. Signaling by type I and II cytokine receptors: ten years after. Curr. Opin. Immunol. 2001;13:363–373. doi: 10.1016/s0952-7915(00)00228-4. [DOI] [PubMed] [Google Scholar]
- 2.Imada K, Leonard WJ. The Jak-STAT pathway. Mol. Immunol. 2000;37:1–11. doi: 10.1016/s0161-5890(00)00018-3. [DOI] [PubMed] [Google Scholar]
- 3.Silvennoinen O, et al. Structure of the murine Jak2 protein-tyrosine kinase and its role in interleukin 3 signal transduction. Proc. Natl. Acad. Sci. U. S. A. 1993;90:8429–8433. doi: 10.1073/pnas.90.18.8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamaoka K, et al. The Janus kinases (Jaks) Genome Biol. 2004;5:253. doi: 10.1186/gb-2004-5-12-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishida-Takahashi R, et al. Phosphorylation of Jak2 on SeR523) inhibits Jak2-dependent leptin receptor signaling. Mol. Cell. Biol. 2006;26:4063–4073. doi: 10.1128/MCB.01589-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsuda T, Feng J, Witthuhn BA, Sekine Y, Ihle JN. Determination of the transphosphorylation sites of Jak2 kinase. Biochem. Biophys. Res. Commun. 2004;325:586–594. doi: 10.1016/j.bbrc.2004.10.071. [DOI] [PubMed] [Google Scholar]
- 7.Mazurkiewicz-Munoz AM, et al. Phosphorylation of JAK2 at serine 523: a negative regulator of JAK2 that is stimulated by growth hormone and epidermal growth factor. Mol. Cell. Biol. 2006;26:4052–4062. doi: 10.1128/MCB.01591-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robertson SA, et al. Regulation of Jak2 function by phosphorylation of Tyr317 and Tyr637 during cytokine signaling. Mol. Cell. Biol. 2009;29:3367–3378. doi: 10.1128/MCB.00278-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feener EP, Rosario F, Dunn SL, Stancheva Z, Myers MG., Jr Tyrosine phosphorylation of Jak2 in the JH2 domain inhibits cytokine signaling. Mol. Cell. Biol. 2004;24:4968–4978. doi: 10.1128/MCB.24.11.4968-4978.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Argetsinger LS, et al. Autophosphorylation of JAK2 on tyrosines 221 and 570 regulates its activity. Mol. Cell. Biol. 2004;24:4955–4967. doi: 10.1128/MCB.24.11.4955-4967.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Argetsinger LS, et al. Tyrosines 868, 966, and 972 in the kinase domain of JAK2 are autophosphorylated and required for maximal JAK2 kinase activity. Mol. Endocrinol. 2010;24:1062–1076. doi: 10.1210/me.2009-0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shuai K, Liu B. Regulation of JAK-STAT signalling in the immune system. Nat. Rev. Immunol. 2003;3:900–911. doi: 10.1038/nri1226. [DOI] [PubMed] [Google Scholar]
- 13.Saharinen P, Takaluoma K, Silvennoinen O. Regulation of the Jak2 tyrosine kinase by its pseudokinase domain. Mol. Cell. Biol. 2000;20:3387–3395. doi: 10.1128/mcb.20.10.3387-3395.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saharinen P, Silvennoinen O. The pseudokinase domain is required for suppression of basal activity of Jak2 and Jak3 tyrosine kinases and for cytokine-inducible activation of signal transduction. J. Biol. Chem. 2002;277:47954–47963. doi: 10.1074/jbc.M205156200. [DOI] [PubMed] [Google Scholar]
- 15.Saharinen P, Vihinen M, Silvennoinen O. Autoinhibition of Jak2 tyrosine kinase is dependent on specific regions in its pseudokinase domain. Mol. Biol. Cell. 2003;14:1448–1459. doi: 10.1091/mbc.E02-06-0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haan C, Behrmann I, Haan S. Perspectives for the use of structural information and chemical genetics to develop inhibitors of Janus kinases. J. Cell. Mol. Med. 2010;14:504–527. doi: 10.1111/j.1582-4934.2010.01018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baxter EJ, et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365:1054–1061. doi: 10.1016/S0140-6736(05)71142-9. [DOI] [PubMed] [Google Scholar]
- 18.James C, et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434:1144–1148. doi: 10.1038/nature03546. [DOI] [PubMed] [Google Scholar]
- 19.Kralovics R, et al. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N. Engl. J. Med. 2005;352:1779–1790. doi: 10.1056/NEJMoa051113. [DOI] [PubMed] [Google Scholar]
- 20.Chen M, et al. Complex effects of naturally occurring mutations in the JAK3 pseudokinase domain: evidence for interactions between the kinase and pseudokinase domains. Mol. Cell. Biol. 2000;20:947–956. doi: 10.1128/mcb.20.3.947-956.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lucet IS, et al. The structural basis of Janus kinase 2 inhibition by a potent and specific pan-Janus kinase inhibitor. Blood. 2006;107:176–183. doi: 10.1182/blood-2005-06-2413. [DOI] [PubMed] [Google Scholar]
- 22.Boggon TJ, Li Y, Manley PW, Eck MJ. Crystal structure of the Jak3 kinase domain in complex with a staurosporine analog. Blood. 2005;106:996–1002. doi: 10.1182/blood-2005-02-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boudeau J, Miranda-Saavedra D, Barton GJ, Alessi DR. Emerging roles of pseudokinases. Trends Cell Biol. 2006;16:443–452. doi: 10.1016/j.tcb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 24.Hubbard SR, Till JH. Protein tyrosine kinase structure and function. Annu. Rev. Biochem. 2000;69:373–398. doi: 10.1146/annurev.biochem.69.1.373. [DOI] [PubMed] [Google Scholar]
- 25.Hall T, et al. Expression, purification, characterization and crystallization of non- and phosphorylated states of JAK2 and JAK3 kinase domain. Protein Expr. Purif. 2010;69:54–63. doi: 10.1016/j.pep.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 26.Scott LM, et al. JAK2 exon 12 mutations in polycythemia vera and idiopathic erythrocytosis. N. Engl. J. Med. 2007;356:459–468. doi: 10.1056/NEJMoa065202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bercovich D, et al. Mutations of JAK2 in acute lymphoblastic leukaemias associated with Down's syndrome. Lancet. 2008;372:1484–1492. doi: 10.1016/S0140-6736(08)61341-0. [DOI] [PubMed] [Google Scholar]
- 28.Zeqiraj E, van Aalten DM. Pseudokinases-remnants of evolution or key allosteric regulators? Curr. Opin. Struct. Biol. 2010;20:772–781. doi: 10.1016/j.sbi.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scheeff ED, Eswaran J, Bunkoczi G, Knapp S, Manning G. Structure of the pseudokinase VRK3 reveals a degraded catalytic site, a highly conserved kinase fold, and a putative regulatory binding site. Structure. 2009;17:128–138. doi: 10.1016/j.str.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mukherjee K, et al. CASK Functions as a Mg2+-independent neurexin kinase. Cell. 2008;133:328–339. doi: 10.1016/j.cell.2008.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eswaran J, et al. Structure and functional characterization of the atypical human kinase haspin. Proc. Natl. Acad. Sci. U. S. A. 2009;106:20198–20203. doi: 10.1073/pnas.0901989106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Min X, Lee BH, Cobb MH, Goldsmith EJ. Crystal structure of the kinase domain of WNK1, a kinase that causes a hereditary form of hypertension. Structure. 2004;12:1303–1311. doi: 10.1016/j.str.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 33.Shi F, Telesco SE, Liu Y, Radhakrishnan R, Lemmon MA. ErbB3/HER3 intracellular domain is competent to bind ATP and catalyze autophosphorylation. Proc. Natl. Acad. Sci. U. S. A. 2010;107:7692–7697. doi: 10.1073/pnas.1002753107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeqiraj E, Filippi BM, Deak M, Alessi DR, van Aalten DM. Structure of the LKB1-STRAD-MO25 complex reveals an allosteric mechanism of kinase activation. Science. 2009;326:1707–1711. doi: 10.1126/science.1178377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jura N, Shan Y, Cao X, Shaw DE, Kuriyan J. Structural analysis of the catalytically inactive kinase domain of the human EGF receptor 3. Proc. Natl. Acad. Sci. U. S. A. 2009;106:21608–21613. doi: 10.1073/pnas.0912101106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santos FP, Verstovsek S. JAK2 inhibitors: What's the true therapeutic potential? Blood Rev. 2010 doi: 10.1016/j.blre.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods references
- 37.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 38.Thingholm TE, Jorgensen TJ, Jensen ON, Larsen MR. Highly selective enrichment of phosphorylated peptides using titanium dioxide. Nat. Protoc. 2006;1:1929–1935. doi: 10.1038/nprot.2006.185. [DOI] [PubMed] [Google Scholar]
- 39.Ye J, et al. Optimized IMAC-IMAC Protocol for Phosphopeptide Recovery from Complex Biological Samples. J. Proteome Res. 2010;9:3561–3573. doi: 10.1021/pr100075x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.