Abstract
Surgical treatment is suggested for seizure control in medically intractable epilepsy patients. Detailed pre-surgical evaluation and lateralization using Magnetic Resonance Images (MRI) is expected to result in a successful surgical outcome. In this study, an optimized pattern recognition approach is proposed for lateralization of mesial Temporal Lobe Epilepsy (mTLE) patients using asymmetry of imaging indices of hippocampus. T1-weighted and Fluid-Attenuated Inversion Recovery (FLAIR) images of 76 symptomatic mTLE patients are considered. First, hippocampus is segmented using automatic and manual segmentation methods; then, volumetric and intensity features are extracted from the MR images. A nonlinear Support Vector Machine (SVM) with optimized Gaussian Radial Basis Function (GRBF) kernel is used to classify the imaging features. Using leave-one-out cross validation, this method results in a correct lateralization rate of 82%, a probability of detection for the left side of 0.90 (with false alarm probability of 0.04) and a probability of detection for the right side of 0.69 (with zero false alarm probability). The lateralization results are compared to linear SVM, multi-layer perceptron Artificial Neural Network (ANN), and volumetry and FLAIR asymmetry analysis. This lateralization method is suggested for pre-surgical evaluation using MRI before surgical treatment in mTLE patients. It achieves a more correct lateralization rate and fewer false positives.
Keywords: Magnetic Resonance Imaging (MRI), Hippocampus, Epilepsy, Lateralization, Support Vector Machine (SVM)
I. Introduction
Epilepsy affects about three million Americans and sixty-six million people around the world. A group of disorders in which patients suffer from recurrent epileptic seizures arising in one or both temporal lobes of the brain is called Temporal Lobe Epilepsy (TLE). Surgical resection of the epileptogenic temporal lobe is the last resort to achieve long-term seizure freedom for drug-resistant TLE patients. According to the International League Against Epilepsy, TLE is categorized in two main groups: mesial TLE (mTLE) and lateral TLE. The most common form of epilepsy is the mesial TLE while hippocampal sclerosis considered as its pathophysiological substance [1].
Segmentation of right and left hippocampi from T1-weighted images is necessary for volumetric analysis aimed at lateralization of TLE. Different energy-minimizing models guided by internal-shape forces and external-image forces like discrete contour models, classic snakes, and deformable-contour models can be applied for automatic segmentation of brain structures [2-5]. Atlas registration as well as minimization of energy function with intensity and prior terms for hippocampus segmentation is presented in [6]. Moreover, automated and manual methods for hippocampal volumetry in TLE patients are compared by Akhondi et al. in [7].
After hippocampus segmentation, the shape and volume of hippocampus can be estimated and used for lateralization. Lateralization of epileptic patients is an essential step before performing the surgery which is defined as localization of function attributed to either the left or right side of brain by the American Heritage Medical Dictionary. For this purpose, standard analysis of MRI is done as a part of pre-surgical lateralization. T1-weighted and Fluid Attenuated Inversion Recovery (FLAIR) MR images are used for pre-surgical lateralization. Volumetric and FLAIR intensity features of the hippocampus extracted from MR images are useful to find abnormality and candidacy of pre-surgery evaluation in temporal lobe epilepsy [8]. According to the literature, the reduction of hippocampal volume in T1-weighted images and the side of mTLE are correlated to each other. In the other side, in association with a focal epileptogenicity, the hippocampal FLAIR signal intensity in mTLE increased in FLAIR images [9]. Some studies have evaluated several visual and architectural features of hippocampus. Jafari-Khouzani et al. proposed texture features for lateralization [10]. In their study, mean and standard deviation of FLAIR signal intensities, wavelet transform-derived energy and texture analysis of the hippocampal body were shown to provide a highly sensitive method for lateralization. In another study, the MR volumetric features of hippocampus are discussed for TLE patients [11].
Pattern recognition algorithms were applied for data classification in many areas of healthcare such as planning, diagnosis, and treatment. Structural Risk Minimization (SRM) criterion in statistical learning theory finds a balance between suitable empirical performance and proper generalization ability. Support Vector Machine (SVM) is considered a popular application of SRM. The SVM algorithm and its modified alternations, which are considered as supervised learning models for machine learning and pattern recognition, have been used for classification of a variety of medical data. The SVM algorithm and its modified alternations are employed for TLE applications using both linear and nonlinear kernels. For nonlinear classification, the kernel trick has been used to map the data into a high dimensional feature space. In [12] the volume of brain structures is estimated using multi-atlas segmentation and SVM classification. In [13], a fusion method based on SVM and on least square optimization is proposed for classification of some TLE features extracted from electroencephalography (EEG). Also, SVM is applied to determine the motor state from EEG features in Wistar rats with TLE in [14].
In this study, the hippocampus is first segmented using both automatic and manual methods. Then, the volumetric and intensity features are extracted from T1-weighted FLAIR images. Finally, to analyze the extracted features, a model is developed using SVM with optimized Gaussian Radial Basis Function (GRBF) kernel. The lateralization results of this model are compared with its linear counterpart and with the previously introduced lateralization methods based on volumetry and FLAIR asymmetry analysis using hippocampal T1 volumetric and FLAIR intensity [15]. Since Artificial Neural Networks (ANNs) have been successfully used in many areas of medicine, the results of SVM classification is compared to a feed-forward Multi-Layer Persectron Neural Network (MLPNN) using a three-layer structure and back-propagation learning algorithm.
The rest of the paper is organized as follows. In Section II, the material and methods including subjects, image acquisition, hippocampus segmentation, feature selection, and SVM classification are explained. In Section III, results of lateralization using the proposed method are compared against those from previously suggested methods and MLPNN. Finally, the paper is concluded in Section IV.
II. Material and Methods
A. Subjects
We use images of mTLE patients operated by a single surgeon between June 1993 and June 2009 at Henry Ford Hospital, Detroit, MI. Seventy-six symptomatic patients (31 males) achieved an Engel class I outcome are used in this study.
B. Image Acquisition
Preoperative FLAIR and T1-weighted images are obtained using a 1.5T or a 3T GE Excite HD MRI system (GE Healthcare, Waukesha, WI) with a standard eight-channel phased-array RF coil and receiver system. Spoiled gradient-echo (SPGR) sequence with an adequate spatial resolution appropriate for hippocampal volumetry was used. Figure 1 shows coronal and sagittal views of T1 images of a TLE patient.
Figure 1.
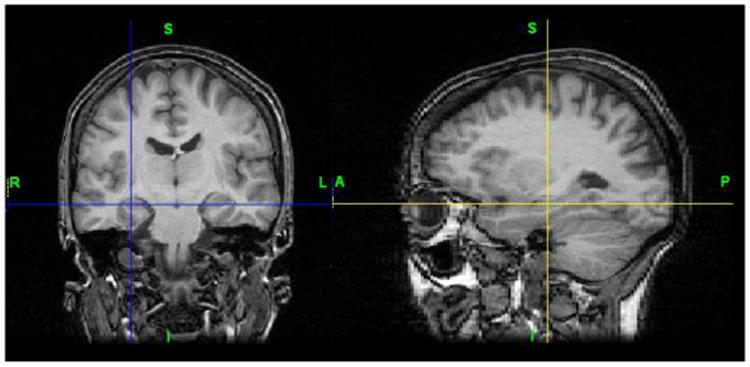
T1 brain MRI with epileptogenic hippocampus.
C. Hippocampus Segmentation
Automatic and manual segmentation methods are used to extract hippocampus in this step. An automatic segmentation algorithm proposed by Akhondi et al. in 2011 [7] is used to extract hippocampus. Left and right hippocampi are segmented with Local information-based multiple atlas method (LocalInfo). In addition of automatic segmentation, manual segmentation of the hippocampus was also performed (to be used as benchmark). For this purpose, hippocampi were outlined in the coronal plane and then fine-tuning steps were done in the sagittal view. Figure 2 shows the results of manual hippocampus segmentation in coronal and sagittal views.
Figure 2.
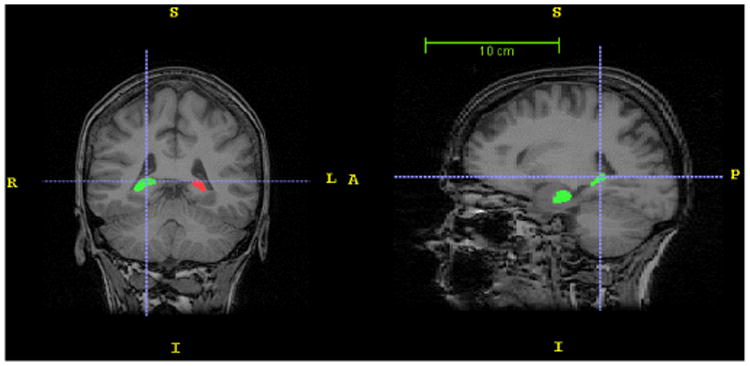
Results of manual hippocampus segmentation.
D. Feature Extraction
After segmentation, images are skulls stripped by MRIcro software [16]. T1-weighted images that are segmented and skulls stripped are co-registrated to FLAIR images by FSL [17] software in order to locate the hippocampus locations in FLAIR images. Then for each patient in both manual and automatic procedures, volumes (from T1-weighted images), mean, and standard deviation of FLAIR intensities (from FLAIR images) of hippocampi are extracted. For all the feature sets, the differences of right and left features are obtained and normalized by the mean value of the right and left sides. Finally, a database of normalized volumes, mean, and standard deviation of each side of hippocampi is created to have an organized collection of the extracted features for all seventy-six patients. Figure 3 shows the right and left hippocampal volumes extracted from T1-weighted images. The lateralization using SVM with nonlinear kernel is done in the next step.
Figure 3.

Right and left hippocampi extracted in T1-weighted images for volumetric analysis.
E. Classification
A nonlinear SVM with the GRBF kernel is used for classification of the extracted features. To improve the results, the Gaussian kernel parameters are optimized by maximizing a classical class separability criterion as the trace of the scatter ratio. Then, a quasi-Newton algorithm is used by exploiting a recently proposed criterion of decomposition of the objective [18]. The GRBF kernel K(x,y) is defined as,
| (1) |
where x and y are two feature vectors, and is the squared Euclidean distance between these two vectors. The Objective function J(σ) is defined as in [15]. The quasi-Newton optimization method defines the sequence of σ(n) in the objective function as follows,
| (2) |
| (3) |
| (4) |
In summary, for the purpose of kernel optimization in a SVM classifier, the developed differentiable version of the classical class separability criterion is used in isotropic Gaussian kernel.
The results of proposed method are compared with a multi-layer perceptron neural network (MLPNN). The network structure is designed in three layers with 6, 18, and 1 neurons in the first, second, and third layers, respectively. The Levenberg-Marquardt optimization for the training phase was used. Different optimization schemes are applied for the training phase and the best optimization are reported. For each data using leave-one-out cross validation, we evaluated the performance of different methods by comparing their lateralization results. The results of the proposed method are evaluated with the correct TLE side. SVM with Gaussian kernel optimization, SVM with linear kernel, MLPNN, hippocampal volumetry asymmetry analysis, and FLAIR asymmetry analysis are evaluated by the correct lateralization percentage (CL%) as well as by the probability of detection and false alarm for the left and right sides ( , , , and , respectively).
III. Results
The mean and standard error of the volumetric and FLAIR features for the right and left hippocampi of 76 mTLE patients by automatic and manual methods are listed in TABLE I. The correct lateralization and probability of detection for the proposed method is shown in TABLE II. The correct lateralization percentage with automatic and manual segmentation methods are 80% and 82%, respectively.
Table I.
The mean and standard error (se) of the hippocampal volumetric and FLAIR features for the right and left hippocampi of 76 mTLE patients in automatic and manual methods.
| Volume (mm3) (mean±se) | Mean intensity (mean±se) | StD intensity (mean±se) | ||||
|---|---|---|---|---|---|---|
| Right | Left | Right | Left | Right | Left | |
| Automatic Method | 2528 ± 62 | 2503 ± 57 | 258 ± 12 | 261 ± 12 | 44 ± 2 | 44 ± 2 |
| Manual Method | 2482 ± 79 | 2203± 71 | 273 ± 11 | 278 ± 11 | 36 ± 1 | 36 ± 1 |
Table II.
Comparison of the lateralization results of the proposed method using automatic and manual segmentation of hippocampus.
| Automatic Segmentation | Manual Segmentation | |
|---|---|---|
| CL % | 80% | 82% |
| PrLD | 0.84 | 0.90 |
| PrRD | 0.76 | 0.70 |
TABLE III shows the results of lateralization by SVM with Gaussian kernel optimization compared against different methods using manual segmentation. By leave-one-out cross validation, 62 out of 76 TLE patients match the clinical diagnosis (p-value<0.05). The correct lateralization percentage for volumetry and FLAIR asymmetry analysis, feed-forward MLPNN, linear SVM, and proposed method are, 76%, 70%, 75%, 78%, and 82%, respectively. The proposed method obtained a correct lateralization rate of 82%, a probability of detection for the left side of 0.90 (with false alarm probability of 0.04) and a probability of detection for the right side of 0.69 (with zero false alarm probability).
Table III.
Correct lateralization percentage (CL%) as well as probability of detection and false alarm for the left ( , and ) and right side ( , and ) are compared to the previously reported methods for manual segmentation.
| Method: | M 1 | M 2 | M 3 | M 4 | M 5 |
|---|---|---|---|---|---|
| CL % | 76% | 70% | 75% | 78% | 82% |
| PrLD | 0.84 | 0.63 | 0.81 | 0.86 | 0.90 |
| PrRD | 0.67 | 0.79 | 0.67 | 0.69 | 0.70 |
| PrLFA | 0.09 | 0.07 | 0.05 | 0.07 | 0.04 |
| PrRFA | 0.03 | 0.03 | 0.00 | 0.04 | 0.00 |
M 1= volumetry asymmetry analysis
M 2= FLAIR asymmetry analysis
M 3= feed forward MLPNN
M 4= SVM with linear kernel
M 5= SVM with Gaussian kernel optimization
IV. Conclusion
Support Vector Machine (SVM) has become one of the most applicable algorithms in medical applications for supervised-based pattern recognition because of its generalization power. In this paper, we proposed a nonlinear SVM with optimized Gaussian Radial Basis Function kernel for lateralization in mesial Temporal Lobe Epilepsy (mTLE). First, hippocampus is segmented in T1-weighted images by automatic and manual methods. Then, skull stripping is done in these images. T1-weighted images after segmentation are co-registered to FLAIR images by FSL software to extract the hippocampus in FLAIR images. According to previous work of our team, the volumetric feature of T1-weighted images as well as the FLAIR mean signal intensity and its standard deviation are effective in lateralization of mTLE. So, these features are extracted using segmented MR images and all features are normalized according to right and left sides. Finally, SVM classifier with Gaussian kernel optimization is used for lateralization. The results of lateralization are compared with previous models reported in literature. In addition, a feed-forward Multi-Layer Perceptron Neural Network (MLPNN) was designed to compare with proposed method. In comparison with MLPNN, volumetric asymmetry method, and FLAIR asymmetry method, our proposed lateralization method achieved more correct lateralization percentage and fewer false positives.
Acknowledgments
We would like to thank Prof. Dario Pompili, Rutgers University, for revision and comments, and Dr. Kourosh Jafari-Khozani, Harvard Medical School, for helping with image segmentation and skull stripping in this study. We also thank Dr. Kost V. Elliserich, Henry Ford Hospital, for referring epileptic cases to us for this study.
This research is supported in part by the NIH grant R01-EB013227.
Contributor Information
Mohammad-Parsa Hosseini, Email: parsah@rad.hfh.edu, parsa.hosseini@wayne.edu, the Medical Image Analysis Lab., Radiology Department, Henry Ford Health System, Detroit, MI, USA and the Department of Electrical and Computer Engineering, Wayne State University, Detroit, MI, USA (phone: +13133200462; fax: +13138744494.
Mohammad R. Nazem-Zadeh, Email: mohamadn@rad.hfh.edu, The Medical Image Analysis Lab., Radiology Department, Henry Ford Health System, Detroit, MI, USA.
Fariborz Mahmoudi, Email: fariborzm@rad.hfh.edu, The Medical Image Analysis Lab., Radiology Department, Henry Ford Health System, Detroit, MI, USA.
Hao Ying, Email: hao.ying@wayne.edu, The Department of Electrical and Computer Engineering, Wayne State University, Detroit, MI, USA.
Hamid Soltanian-Zadeh, Email: hszadeh@ut.ac.ir, hamids@rad.hfh.edu, The Control and Intelligent Processing Center of Excellence, School of Electrical and Computer Engineering, University of Tehran. Tehran, Iran and with the Medical Image Analysis Lab., Radiology Department, Henry Ford Health System, Detroit, MI, USA.
References
- 1.Engel J. Mesial temporal lobe epilepsy: what have we learned? The Neuroscientist. 2001;7(4):340–352. doi: 10.1177/107385840100700410. [DOI] [PubMed] [Google Scholar]
- 2.Shen L, Saykin AJ, Kim S, Firpi HA, West JD, Risacher SL, Mcdonald BC, Mchugh TL, Wishart HA, Flashman LA. Comparison of manual and automated determination of hippocampal volumes in MCI and early AD. Brain imaging and behavior. 2010;4(1):86–95. doi: 10.1007/s11682-010-9088-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hosseini MP, Soltanian-zadeh H, Akhlaghpoor S. Three cuts method for identification of COPD. Acta Medica Iranica. 2013;51(11):771–778. [PubMed] [Google Scholar]
- 4.Hosseini MP, Soltanian-zadeh H, Akhlaghpoor S. Detection and severity scoring of chronic obstructive pulmonary disease using volumetric analysis of lung CT images. Iranian Journal of Radiology. 2012;9(1):22. doi: 10.5812/iranjradiol.6759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hosseini MP, Soltanian-zadeh H, Akhlaghpoor S. A new schem for evaluation of air-trapping in CT images. IEEE. 2010 [Google Scholar]
- 6.Van der Lijn F, den Heijer T, Breteler M, Niessen WJ. Hippocampus segmentation in MR images using atlas registration, voxel classification, and graph cuts. Neuroimage. 2008;43(4):708–720. doi: 10.1016/j.neuroimage.2008.07.058. [DOI] [PubMed] [Google Scholar]
- 7.Akhondi-Asl A, Jafari-Khouzani K, Elisevich K, Soltanian-Zadeh H. Hippocampal volumetry for lateralization of temporal lobe epilepsy: automated versus manual methods. Neuroimage. 2011;54:S218–S226. doi: 10.1016/j.neuroimage.2010.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jafari-Khouzani K, Soltanian-Zadeh H, Elisevich K. Hippocampus volume and texture analysis for temporal lobe epilepsy. :394–397. [Google Scholar]
- 9.Davatzikos C, Barzi A, Lawrie T, Hoon AH, Melhem ER. Correlation of corpus callosal morphometry with cognitive and motor function in periventricular leukomalacia. Neuropediatrics. 2003;34(5):247–252. doi: 10.1055/s-2003-43259. [DOI] [PubMed] [Google Scholar]
- 10.Jafari-Khouzani K, Elisevich K, Patel S, Smith B, Soltanian-Zadeh H. FLAIR signal and texture analysis for lateralizing mesial temporal lobe epilepsy. Neuroimage. 2010;49(2):1559–1571. doi: 10.1016/j.neuroimage.2009.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernasconi N, Bernasconi A, Caramanos Z, Antel S, Andermann F, Arnold D. Mesial temporal damage in temporal lobe epilepsy: a volumetric MRI study of the hippocampus, amygdala and parahippocampal region. Brain. 2003;126(2):462–469. doi: 10.1093/brain/awg034. [DOI] [PubMed] [Google Scholar]
- 12.Keihaninejad S, Heckemann R, Gousias IS, Aljabar P, Hajnal JV, Rueckert D, Hammers A. Automatic volumetry can reveal visually undetected disease features on brain MR images in temporal lobe epilepsy. :105–108. [Google Scholar]
- 13.Rezaie M, Soltanian-Zadeh H, Siadat M, Elisevich K. Soft computing approaches to computer aided decision making for temporal lobe epilepsy. :42–45. [Google Scholar]
- 14.Wang YL, Liang SF, Shaw FZ, Su AW, Chen YL, Wu SY. Use of accelerometers to detect motor states in a seizure of rats with temporal lobe epilepsy. :372–375. [Google Scholar]
- 15.Nazem-Zadeh MR, Chapman CH, Lawrence TS, Tsien CI, Cao Y. Uncertainty in assessment of radiation-induced diffusion index changes in individual patients. Physics in medicine and biology. 2013;58(12):4277. doi: 10.1088/0031-9155/58/12/4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rorden C. MRIcro. University of South Carolina. 2005 [Google Scholar]
- 17.http://fsl.fmrib.ox.ac.uk/
- 18.Wang J, Lu H, Plataniotis KN, Lu J. Gaussian kernel optimization for pattern classification. Pattern recognition. 2009;42(7):1237–1247. [Google Scholar]


