Abstract
Background
Genome-wide association studies focusing on European-ancestry populations have identified ALL risk loci on: IKZF1, ARID5B, and CEBPE. To capture the impacts of these genes on ALL risk in the California Hispanic population, we comprehensively assessed variation within the genes and further assessed the joint effects between the genetic variation and surrogates for early life infections (presence of older siblings, daycare attendance, and ear infections).
Methods
Genotypic data for 323 Hispanic ALL cases and 454 controls from the California Childhood Leukemia Study (CCLS) were generated using Illumina OmniExpress v1 platform. Logistic regression assuming a log-additive model estimated odds ratios (OR) associated for each SNP, adjusted for age, sex, and the first five principal components. In addition, we examined potential interactions between six ALL risk alleles and surrogates for early life infections using logistic regression models that included an interaction term.
Results
Significant associations between genotypes at IKZF1, ARID5B, and CEBPE and ALL risk were identified; rs7780012, OR=0.50, 95% confidence interval (CI): 0.35-0.71 (p=0.004); rs7089424, OR=2.12, 95% CI: 1.70-2.65 (p=1.16 x 10-9); rs4982731, OR=1.69, 95% CI: 1.37-2.08 (p=2.35 x 10-6), respectively. Evidence for multiplicative interactions between genetic variants and surrogates for early life infections with ALL risk was not observed.
Conclusions
Consistent with findings in non-Hispanic White population, our study showed that variants within IKZF1, ARID5B, and CEBPE were associated with increased ALL risk, and the effects for ARID5B and CEBPE were most prominent in the high-hyperdiploid ALL subtype in the California Hispanic population.
Impact
Results implicate the ARID5B, CEBPE and IKZF1 genes in the pathogenesis of childhood ALL.
Keywords: cancer, genetic association, early life infections, childhood leukemia, gene-environment interaction
INTRODUCTION
Leukemia is the most common malignancy under the age of 15, accounting for 31% of all cancer cases (1). Several risk factors for acute lymphoblastic leukemia (ALL) have been established, including sex, age, race, prenatal exposure to x-rays, therapeutic radiation, and specific genetic syndromes (2). Direct evidence for inherited genetic susceptibility is demonstrated by the high risk of ALL associated with Bloom's syndrome, neurofibromatosis, ataxia telangiectasia and constitutional trisomy 21(3). These predisposing disorders only account for <5% of all diagnosed cases. Most genetic association studies of ALL have focused on candidate genes, primarily those implicated in the metabolism of carcinogens, folate metabolism, immune function, and cell-cycle regulation (4, 5). Recent genome-wide association studies (GWAS) have identified common genetic variation near IKZF1 (7p12.2), ARID5B (10q21.2), and CEBPE (14q11.2) that influences ALL risk in non-Hispanic White populations (6-9). However, only one study has explored these loci in Hispanic populations using a genome-wide approach (9). Given the observation of high childhood ALL incidence in California Hispanics, understanding the influence of variants within these genes with comprehensive adjustment for genetic ancestry is important for characterizing the relationship between candidate loci and ALL, and furthering our understanding of disease pathogenesis.
Epidemiologic studies have provided indirect evidence for an infectious etiology of ALL, though no direct evidence has been shown (10). Greaves hypothesizes that the absence of an early immune challenge and priming during early childhood combined with ‘delayed’ exposures to infection might subsequently result in adverse immune responses to common infectious agents, thereby increasing the risk of childhood ALL (10). This suggests that exposure to infections early in life may provide a protective effect of childhood ALL compared to children who lack such exposure. Associations of ALL have been observed with proxy measures of exposure to infections including daycare attendance (11-14), birth order (15, 16), and child's history of infections (17, 18). The most compelling evidence regarding the relationship between early life infection and development of childhood ALL is from studies of daycare attendance, which is considered a surrogate for exposure to multiple microbial agents (19). The results from a recent meta-analysis of published studies conducted by Urayama et al. showed a significantly reduced ALL risk for children who attend daycare facilities among non-Hispanic Whites (13).
Ethnic differences in the risk of ALL are well-recognized as the incidence of ALL is nearly 20% higher among Hispanics than non-Hispanic Whites in California (20). This higher risk is possibly due to an increased prevalence of ALL risk alleles in populations with Native American ancestry, as well as ethnic differences in exposure to environmental risk factors (10, 21). In the current study, we examine associations between ALL risk in Hispanic children and genetic variation in candidate B-cell development genes previously linked to ALL risk in GWAS of non-Hispanic White populations (IKZF1, ARID5B, and CEBPE). We further investigate potential interactions between these genetic variants and surrogates for early life exposure to infections, including presence of older siblings, daycare attendance, and ear infections during infancy, in California Hispanics.
MATERIALS AND METHODS
Study populations
The California Childhood Leukemia Study (CCLS) is a population-based case-control study which began in 1995. Incident cases of newly diagnosed childhood leukemia (age 0–14 years) were rapidly ascertained from major clinical centers in the study area, usually within 72 hours of diagnosis. Cases were initially identified from four hospitals (later expanded to nine) in the San Francisco Bay Area and Central Valley. For each case, one or two healthy controls matched on child's age, sex, Hispanic status (a child is considered Hispanic if either parent is Hispanic) and maternal race (White, Black, Asian/Pacific Islander, Native American, and Other/Mixed) were randomly selected from the state birth registry maintained by the Center for Health Statistics of the California Department of Public Health (CDPH). A detailed description of control selection in the CCLS has been previously reported (22). A total of 86% of case subjects determined eligible consented to participate and 86% of controls subjects participated among those contacted and considered eligible (23).
Cases and controls were eligible to enter the study if they were under 15 years of age, resided in the study area at the time of diagnosis, had at least one parent who speaks either English or Spanish, and had no prior history of malignancy. The current analysis included 777 Hispanics (323 cases and 454 controls) in the CCLS who consented to participate and were interviewed between 1995 and 2008, and for whom archived newborn blood (ANB) spots were available. Immunophenotype was determined for ALL using flow cytometry profiles (CD10 and CD19 for B-cell lineage) (24). When fluorescence in situ hybridization (FISH) assays conducted at University of California Berkeley identified extra copies of chromosomes 21 and X, assignment of high hyperdiploid status (51-67 chromosomes) was made (24).
This study was reviewed and approved by the institutional review committees at the University of California Berkeley, the CDPH, and the participating hospitals. Written informed consent was obtained from all parent respondents.
Genotyping and Quality Control
Samples were genotyped at the Genetic Epidemiology and Genomics Laboratory, School of Public Health, University of California, Berkeley, using the Illumina OmniExpress v1 platform which contains 730,525 markers. Quality control filtering removed SNPs that were not on autosomal chromosomes, were missing in >2% of samples, had minor allele frequency (MAF) of <2% or showed significant deviation from Hardy-Weinberg equilibrium in controls (P < 1 × 10−5). The resulting data set of 634,037 SNPs was then subjected to additional quality control filtering in all samples. We excluded samples for which < 98% of loci were successfully genotyped, samples with discordant sex profiles (birth certificate vs. genetically determined sex) and samples displaying cryptic relatedness (based on identity-by-descent calculations with pi-hat cutoff of 0.15). Ten pairs of duplicate samples were included to assess assay reproducibility, with average concordance >99.99%. In the current study, we included all SNPs that passed quality control and were located within ±10 kb of selected candidate genes.119 SNPs were included in these analyses.
Proxy measures of early childhood exposure to infections
Information on daycare attendance, birth order and history of infections was collected through in-person interview with the biological mothers. Detailed information on collection early childhood exposure to infections was presented previously (11). Briefly, the child's birth order was determined based on a detailed pregnancy history obtained for the biological mother. Information on the child's social contacts outside the home was obtained through a history of daycare attendance before the date of diagnosis for cases and reference date for controls, or before age six, whichever occurred first. To examine the influence of daycare attendance during a specific time window of exposure, daycare attendance was censored at six months and one year of age. For each daycare the child attended, information on age attended, duration of time attended, hours per week, and numbers of other children were obtained (11, 25). These data was used to calculated “total child-hours of exposure” for each child. Child-hours at each daycare facility was calculated as follows: (number of months attending the day care) x (mean hours per week at this day care) x (number of other children at this day care) x (4.35 weeks per month) (11, 25). In this study, we dichotomized daycare attendance variable into ever/never due to the distribution of child-hour attendance is binomial for the Hispanic population included in the study.
Respondents were also asked for a history of common infectious illnesses the child had during the first year of life, including severe diarrhea/vomiting, ear infection, persistent cough, mouth and eye infection, influenza, and unspecified “other infection.
Statistical Analysis
Single SNP analyses were conducted assuming a log-additive model (0, 1, or 2 copies of the minor allele), using unconditional logistic regression in PLINK v1.07 (26). To adjust for potential population stratification in study samples, a principal component analysis approach was implemented in EIGENSTRAT (27). The odds ratio (OR) and 95% confidence intervals (CI) for each SNP were calculated to estimate the risk of ALL associated with each additional copy of the minor allele, adjusted for: age, sex, and the first five genetic principal components. Significance criteria based on the Benjamini and Hochberg (BH) procedure for controlling the false discovery rate (FDR) were determined by using PLINK v1.07 with a type I error rate of 5% (12). Significant regions were plotted using the online tool SNAP (28). Logistic regression adjusting for age, sex, and income was used to estimate the OR and 95% CI associated with proxies for early life infections (presence of older siblings, daycare attendance, and ear infections) and ALL risk.
Gene expression analysis
Association of genotypes and mRNA expression in IKZF1, ARID5B, and CEBPE was examined using mRNA data from lymphoblastoid cell lines derived from 45 MEX (Mexican ancestry in Los Angeles, California) HapMap individuals available from the database of the Gene Expression Variation (GENEVAR) project (29). Spearman's rank correlation coefficient was used to estimate the strength of relationship between genotypes and intensity of gene expression (30). Alternatively, non-parametric permutation P-values were also provided to further evaluate the significance of nominal P-values as implemented in GENEVAR (29).
Gene-environment interaction analysis
To determine which SNPs in the three candidate regions contribute independently to disease susceptibility for subsequent geneXenvironment interaction analysis, we performed conditional haplotype analysis on all significant SNPs within the three gene regions as implemented in PLINK v1.07. To evaluate whether the signals from these three candidate genes were independent from each other, the association between childhood ALL and each significant locus was tested using logistic regression, conditioning on all other SNPs in the region. To test for heterogeneity (interaction), we focused on the association between daycare attendance by critical development periods (age 6 months and one year of age), presence of older siblings, and ear infections in infancy and ALL. The joint effects of proxies for early life infections and the six SNPs selected from conditional haplotype analysis were evaluated using logistic regression, while adjusting for age, sex, income and the first five genetic principal components (PCs). The three infection variables were chosen for evaluation in the joint effect analysis based on previous CCLS publications (14). A dominant genetic model was assumed given small sample size. A P-value of 0.2 or less for interaction was considered statistically significant given the available sample size, i.e. <350 observations (31). The study has 80% power to detect an interaction odds ratio of 2.0, assuming a minor allele frequency of 30 %, exposure prevalence of 20%, an estimated OR of 1.3 for the genotype and the exposure, and a two-sided significance level of 0.05.
RESULTS
Quality control filtering yielded 777 Hispanic individuals (323 ALL cases and 454 controls) and 119 SNPs were relevant to this analysis. Study characteristics for the Hispanic participants are described in Table 1. The distributions of child's sex, age, and race/ethnicity were similar between cases and controls. Cases generally had lower annual household income compared to controls. Since Hispanics are a recently admixed group (32), a proportion (34%-37%) of our Hispanic population reported “ Mixed or Other” race and 49% -51% of them reported “White and Caucasian” race. In our data, the frequency of B-cell precursor (BCP) ALL is 91.9% among Hispanics (N=297) and the frequency of BCP high-hyperdiploid ALL (>50 chromosomes) for Hispanics is 30% (N=97).
Table 1.
Characteristics of Hispanic case-control study subjects, CCLS, 1995-2008
| Cases, n (%) | Controls, n (%) | |
|---|---|---|
| Study subjects | 323 (41.6) | 454 (58.4) |
| Sex | ||
| Male | 173 (53.6) | 240 (52.9) |
| Female | 150 (46.4) | 214 (47.1) |
| Age | ||
| Mean age, y(SE) | 5.3(3.4) | 5.3 (3.4) |
| Income | ||
| <$15,000 | 79 (24.5) | 74 (16.3) |
| $15,000-$29,999 | 88 (27.2) | 106 (23.35) |
| $30,000-$44,999 | 64 (19.8) | 87 (19.2) |
| $45,000-$59,999 | 41 (12.7) | 63 (13.9) |
| $60,000-$74,999 | 17 (5.3) | 42 (9.3) |
| ≥$75,000 | 34 (10.5) | 82 (18.1) |
| Race | ||
| White/Caucasian | 161 (49.8) | 235 (51.8) |
| African American | 14 (4.3) | 15 (3.3) |
| Native American | 0 (0) | 4 (0.9) |
| Asian or Pacific Islander | 26 (8.1) | 40 (8.8) |
| Mixed or others | 120 (37.2) | 156 (34.4) |
| Cytogenetics (case-only) | ||
| B-cell precursor (BCP) ALL | 297 (91.9) | - |
| BCP high-hyperdiploid ALL (>50 chromosome) | 97 (30.0) | - |
| Daycare attendance (Mean and SE) | ||
| Thousand child hours before 6 mo of age (SE) | 0.24 (1.15) | 0.18 (0.85) |
| Thousand child hours before 1 yr of age (SE) | 0.94 (3.83) | 0.65 (2.46) |
Single SNP analyses
A summary of IKZF1 gene information and childhood ALL association results for SNPs with P< 0.05 (based on correction for FDR using BH procedure) is provided in Table 2. Ten of thirty-four SNPs examined in the IKZF1 gene showed evidence of associations among the Hispanic population (PFDR <0.05). The most significant single SNP result is rs7780012 (OR=0.50, 95% CI 0.35-0.71, PFDR = 0.004), which maps to intron 2 of the IKZF1 gene (Supplementary Fig. S1). The association remained significant when analyses were restricted to BCP ALL (OR=0.52, 95% CI 0.36-0.74) or BCP high-hyperdiploid ALL (OR=0.56, 95% CI 0.40-0.77) (Table 5).
Table 2.
Odds ratio (95%CI) and p-values† for significant* IKZF1 SNPs associated with childhood ALL risk in the Hispanic population, CCLS, 1995-2008
| Chr. | SNP | Minor Allele (frequency) | Base-pair location | ORa | 95% CI | Pvalues | Pvaluesb |
|---|---|---|---|---|---|---|---|
| 7 | rs7780012 | A (0.12) | 50438720 | 0.50 | 0.35, 0.71 | 0.00013 | 0.004426 |
| 7 | rs11980379 | G (0.28) | 50469981 | 1.47 | 1.17, 1.85 | 0.00093 | 0.01054 |
| 7 | rs4132601 | C (0.28) | 50470604 | 1.47 | 1.17, 1.85 | 0.00093 | 0.01054 |
| 7 | rs716719 | A (0.46) | 50325717 | 1.38 | 1.12, 1.69 | 0.002102 | 0.01787 |
| 7 | rs6952409 | A (0.23) | 50462935 | 1.41 | 1.11, 1.78 | 0.004368 | 0.0297 |
| 7 | rs6964823 | A (0.31) | 50460096 | 0.73 | 0.59, 0.91 | 0.00585 | 0.03315 |
| 7 | rs7781977 | G (0.49) | 50346134 | 0.75 | 0.61, 0.93 | 0.007108 | 0.03452 |
| 7 | rs9886239 | A (0.48) | 50336551 | 0.76 | 0.62, 0.93 | 0.008676 | 0.03531 |
| 7 | rs4917017 | G (0.49) | 50335232 | 0.76 | 0.62, 0.94 | 0.009347 | 0.03531 |
| 7 | rs12719019 | A (0.28) | 50476139 | 0.75 | 0.60, 0.95 | 0.01467 | 0.04988 |
Odds ratio (OR) and 95% confidence interval (CI) calculated using log additive models adjusting for age, sex and top 5 genetic principal components (PCs).
P-values based on correlation for False Discovery Rate (FDR) using Benjamini and Hochberg (BH) procedure.
Adjusted and unadjusted for multiple comparisons.
Ten of thirty-four SNPs had significant p-values adjusted for FDR.
Table 5.
Odds ratio (95% CI) for association with ALL by subsets of significant* SNPs in ARID5B, CEBPE and IKZF1 and immunological subtypes of B-cell precursor (BCP) ALL and BCP high-hyperdiploid ALL in the Hispanic population, CCLS, 1995-2008
| Gene | ALL (n=323) | B-cell precursor (BCP) ALL (n=297) | BCP high-hyperdiploid ALL (n=97) |
|---|---|---|---|
| ORa (95% CI) | ORa (95% CI) | ORa (95% CI) | |
| IKZF1 | |||
| rs7780012 | 0.50 (0.35, 0.71) | 0.52 (0.36,0.74) | 0.56 (0.17,0.40) |
| rs11980379 | 1.47 (1.17, 1.85) | 1.47 (1.17,1.86) | 1.50 (0.18,1.06) |
| rs4132601 | 1.47 (1.17, 1.85) | 1.47 (1.17,1.86) | 1.50 (0.18,1.06) |
| rs716719 | 1.38 (1.12, 1.69) | 1.37 (1.11,1.69) | 1.72(1.25,2.38) |
| rs6952409 | 1.41 (1.11, 1.78) | 1.40 (1.10,1.78) | 1.42 (0.99,2.02) |
| rs694823 | 0.73 (0.59, 0.91) | 0.73 (0.58-0.91) | 0.67 (0.47,0.95) |
| ARID5B | |||
| rs7089424 | 2.12 (1.70, 2.65) | 2.30(1.83,2.94) | 3.05 (2.13,4.36) |
| rs7090445 | 2.08 (1.67, 2.60) | 2.24 (1.78,2.81) | 3.07 (2.14,4.39) |
| rs4506592 | 0.48 (0.38, 0.60) | 0.45 (0.36,0.56) | 0.35 (0.25,0.50) |
| rs7073837 | 0.55 (0.45, 0.69) | 0.51 (0.41,0.64) | 0.42 (0.30,0.60) |
| rs10821938 | 0.56 (0.45, 0.69) | 0.53 (0.42,0.66) | 0.42 (0.30-0.60) |
| rs10994981 | 0.65 (0.52, 0.80) | 0.61 (0.49,0.76) | 0.49 (0.35-0.70) |
| rs6479778 | 1.52 (1.21,1.90) | 1.55 (1.23,1.95) | 1.77 (1.26-2.46) |
| rs2893881 | 1.47(1.18,1.84) | 1.51 (1.20,0.89) | 1.71 (1.20-2.42) |
| CEBPE | |||
| rs4982731 | 1.69 (1.37,2.08) | 1.77 (1.43,2.20) | 2.47 (1.76,3.48) |
| rs10143875 | 1.67 (1.36,2.07) | 1.75 (1.41,2.18) | 2.45(1.75,3.45) |
| rs6572981 | 0.53 (0.39,0.70) | 0.53 (0.39,0.71) | 0.40 (0.24,0.67) |
| rs2144827 | 0.50 (0.36,0.68) | 0.49 (0.35,0.68) | 0.32 (0.17,0.60) |
| rs17794251 | 1.53 (1.24,1.88) | 1.54 (1.24,1.89) | 2.00 (1.44,2.78) |
Odds ratio (OR) and 95% confidence interval (CI) calculated using log additive models adjusting for age, sex and top 5 genetic principal components (PCs).
SNPs had significant p-values adjusted for FDR with ALL risk and immunological subtypes of BCP ALL and BCP high-hyperdiploid ALL.
A summary of ARID5B gene information and childhood ALL association results for SNPs with P < 0.05 (based on correction for FDR using BH procedure) is provided in Table 3. Eleven of fifty-seven SNPs examined in ARID5B showed evidence of association among the Hispanic population (PFDR <0.05). The most significant single SNP association, which maps to intron 3 of the gene ARID5B, is rs7089424 (OR= 2.12, 95% CI 1.70-2.65, PFDR = 1.16 × 10−9) (Supplementary Fig. S2). The association signal remained significant when analyses were restricted to BCP ALL (OR=2.30, 95% CI 1.83-2.94) or BCP high-hyperdiploid ALL (OR=3.05, 95% CI 2.13-4.36) (Table 5). Two additional SNPs in ARID5B (rs7090445 and rs4506592) were associated with ALL at a significance level of 10−8 after FDR adjustment and are in linkage disequilibrium (LD) (r2 =0.77). Interestingly, the positive associations between ALL subtypes and ARID5B SNPs were stronger and remained significant when analyses were restricted to ALL cases with BCP high-hyperdiploid ALL (rs7090445, OR= 3.07, 95% CI 2.14-4.39; rs4506592, OR=0.35, 95% CI 0.25-0.50) (Table 5).
Table 3.
Odds ratio (95% CI) and p-values† for significant* ARID5B SNPs associated with childhood ALL risk in the Hispanic population, CCLS, 1995-2008
| Chr. | SNP | Minor Allele (frequency) | Base-pair location | ORa | 95% CI | Pvalues | Pvaluesb |
|---|---|---|---|---|---|---|---|
| 10 | rs7089424 | C (0.49) | 63752159 | 2.12 | 1.70, 2.65 | 2.04E-11 | 1.16E-09 |
| 10 | rs7090445 | G (0.49) | 63721176 | 2.08 | 1.67, 2.60 | 6.24E-11 | 1.57E-09 |
| 10 | rs4506592 | G (0.49) | 63727187 | 0.48 | 0.38, 0.60 | 8.24E-11 | 1.57E-09 |
| 10 | rs7073837 | C (0.48) | 63699895 | 0.55 | 0.45, 0.69 | 6.84E-08 | 9.75E-07 |
| 10 | rs10821938 | C (0.45) | 63724773 | 0.56 | 0.45, 0.69 | 1.24E-07 | 1.42E-06 |
| 10 | rs10994981 | A (0.41) | 63708007 | 0.65 | 0.52, 0.80 | 7.94E-05 | 0.000754 |
| 10 | rs6479778 | A (0.29) | 63689077 | 1.52 | 1.21, 1.90 | 0.000326 | 0.002651 |
| 10 | rs2893881 | G (0.30) | 63688672 | 1.47 | 1.18, 1.84 | 0.000759 | 0.005406 |
| 10 | rs4948491 | G (0.46) | 63696889 | 1.43 | 1.16, 1.76 | 0.000941 | 0.005514 |
| 10 | rs4948488 | G (0.40) | 63685154 | 1.44 | 1.16, 1.79 | 0.000967 | 0.005514 |
| 10 | rs12249208 | A (0.03) | 63730012 | 0.36 | 0.18, 0.72 | 0.003687 | 0.01911 |
Odds ratio (OR) and 95% confidence interval (CI) calculated using log additive models adjusting for age, sex and top 5 genetic principal components (PCs).
P-values based on correlation for False Discovery Rate (FDR) using Benjamini and Hochberg (BH) procedure.
Adjusted and unadjusted for multiple comparisons.
Eleven of fifty-seven SNPs had significant p-values adjusted for FDR.
A summary of CEBPE gene information and childhood ALL association results for SNPs with P < 0.05 based on correction for FDR using the BH procedure is provided in Table 4. Eleven of twenty-eight SNPs showed evidence of association among Hispanic population (PFDR <0.05). The most significant single SNP association, which maps to 3’ region of CEBPE, is rs4982731 (OR= 1.69, 95% CI 1.37-2.08, PFDR = 2.35 × 10−5) (Supplementary Fig. S3). SNP rs10143875 in CEBPE was also associated with ALL risk (OR= 1.67, 95% CI 1.36-2.07, PFDR = 2.35× 10−5) and is in complete LD with rs4982731 (r2=1). When restricted to BCP high-hyperdiploid ALL, results for variants rs4982731 and rs10143875 were more strongly associated with this ALL subtype (OR= 2.47, 95% CI 1.76-3.48; OR=2.45, 95% CI 1.75-3.45) (Table 5).
Table 4.
Odds ratio (95% CI) and p-values† for significant* CEBPE SNPs associated with childhood ALL risk in the Hispanic population, CCLS, 1995-2008
| Chr. | SNP | Minor Allele (frequency) | Base-pair location | ORa | 95% CI | Pvalues | Pvaluesb |
|---|---|---|---|---|---|---|---|
| 14 | rs4982731 | G (0.41) | 23585333 | 1.69 | 1.37, 2.08 | 1.15E-06 | 2.35E-05 |
| 14 | rs10143875 | A (0.41) | 23584265 | 1.67 | 1.36, 2.07 | 1.68E-06 | 2.35E-05 |
| 14 | rs6572981 | A (0.18) | 23599252 | 0.53 | 0.39, 0.70 | 1.64E-05 | 0.000123 |
| 14 | rs2144827 | A (0.14) | 23587231 | 0.50 | 0.36, 0.68 | 1.76E-05 | 0.000123 |
| 14 | rs17794251 | A (0.41) | 23593442 | 1.53 | 1.24, 1.88 | 6.50E-05 | 0.000364 |
| 14 | rs7155790 | C (0.27) | 23589586 | 0.62 | 0.49, 0.79 | 9.06E-05 | 0.000423 |
| 14 | rs2236135 | G (0.11) | 23595721 | 0.56 | 0.39, 0.80 | 0.001201 | 0.004785 |
| 14 | rs2239629 | C (0.11) | 23598128 | 0.57 | 0.40, 0.81 | 0.001545 | 0.004785 |
| 14 | rs17198995 | G (0.05) | 23595423 | 0.44 | 0.26, 0.73 | 0.001674 | 0.004785 |
| 14 | rs2180395 | A (0.10) | 23596621 | 0.56 | 0.39, 0.80 | 0.001709 | 0.004785 |
| 14 | rs2073305 | A (0.09) | 23601073 | 0.59 | 0.41, 0.85 | 0.004994 | 0.01271 |
Odds ratio (OR) and 95% confidence interval (CI) calculated using log additive models adjusting for age, sex and top 5 genetic principal components (PCs).
P-values based on correlation for False Discovery Rate (FDR) using Benjamini and Hochberg (BH) procedure.
Adjusted and unadjusted for multiple comparisons.
Eleven of twenty-eight SNPs had significant p-values adjusted for FDR.
Expression quantitative trait loci (eQTLs) analysis
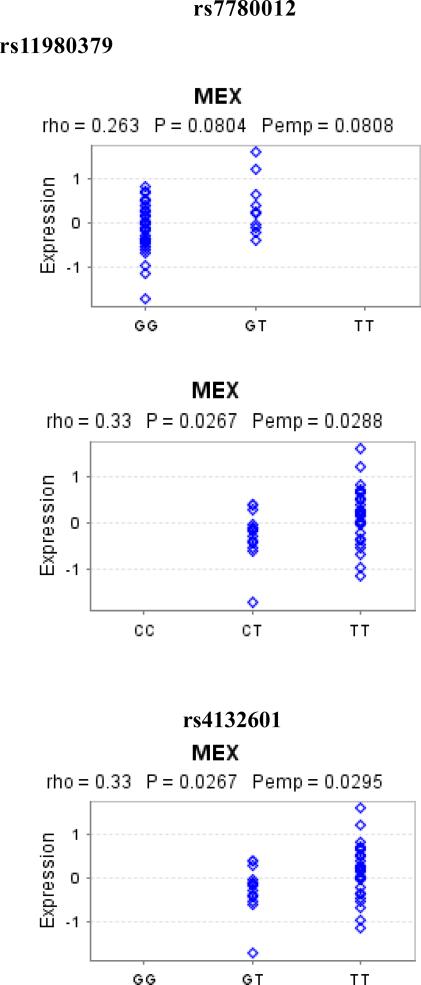
To explore whether the observed SNP associations might influence gene expression, we investigated the correlation between the most significant SNP within three candidate genes and publicly available mRNA expression data (Figure 1). Associations between both rs11980379 and rs4132601 risk genotypes and reduced IKZF1 expression were observed (P=0.028 and P=0.029 respectively; Figure 1), with lower expression being associated with the risk alleles. Associated variants within ARID5B and CEBPE did not appear to influence gene expression (data not shown).
Figure 1.
The correlation between rs7780012, rs11980379, and rs4132601 and IKZF1 expression using publicly available dataset*
The correlation between normalized IKZF1 gene expression and alleles of rs7780012, rs11980379 and rs4132601 were examined using Spearman rank correlation.
* Publicly available Sentrix Human-6 Expression BeadChips (Illumina) data on 45 lymphoblastoid cell lines derived from healthy Mexican ancestry in Los Angeles, California as part of the HapMap 3 project as of June, 2009 (GENEVAR project). Pemp=Empirical p-value derived from 10,000 permutations
Gene-environment interaction
To minimize the numbers of statistical tests, six independent SNPs from the three genes were selected using conditional haplotype analysis for further gene-environment interaction analysis, including three SNPs from IKZF1, one SNP from ARID5B, and two SNPs from CEBPE. Tests of association for the six SNPs stratified by daycare attendance (ever/never) were performed using a multiplicative model of interaction to estimate the joint effects between daycare attendance for different time periods and the six genetic variants (Supplementary Table S1 and Supplementary Table S2). Daycare attendance censored at age of 6 months and rs4982731in CEBPE showed suggestive evidence for interaction on a multiplicative scale (P interaction=0.07; Supplementary Table S1); however, after controlling for multiple comparisons, results were not significant. We did not find any evidence of interaction between the other SNPs and daycare attendance censored at age of one year after FDR correction (P interaction >0.20). Similarly, presence of older siblings and ear infections in infancy showed no evidence of multiplicative interactions with the genetic variants in the risk of childhood ALL (data not shown).
DISCUSSION
Our study is among the first to comprehensively assess genetic variation within the previously identified genes (IKZF1, ARID5B, and CEBPE) and further examine the joint effects between the genetic variation and proxies for early life infections. There is no effect modification of genetic associations by proxies for early life infections in the Hispanic population. In addition, our study also examines the functional relevance on gene expression levels in the Hispanic population. Among these genetic variants tested, rs11980379 and rs4132601 risk alleles within IKZF1 were correlated with reduced IKZF1 mRNA expression. Our results align well with previous observations of frequent IKZF1 somatic deletion in leukemic cells, indicating that a reduction in IKZF1 levels is pro-leukemic (6).
To date, most research on childhood ALL has focused on non-Hispanic White populations. GWAS have established that inherited genetic variants are associated with childhood ALL, including IKZF1 (encoding the early lymphoid transcription factor IKAROS), ARID5B (encoding the AT-rich interactive domain 5B transcription factor), and CEBPE (encoding the transcription factor CCAAT/enhancer-binding protein, epsilon) (6-9, 33). Subsequently, follow-up studies have shown consistent genetic associations with the risk of childhood ALL in different populations, including a large German replication study (34), a Thai population (35), a Polish population (36), a French-Canadian cohort (37), and African American children from St. Jude's Children's Hospital (38).
In our previous publication, we replicated previously identified GWAS SNPs from non-Hispanic White population in the CCLS Hispanic population (39). Our current study take a more comprehensive approach and use GWAS data to look at these three gene regions. The results are consistent with these previous published studies, indicating that causal SNPs are likely to be within or nearby these B-cell development-related genes. In particular, family members of the Ikaros gene are crucial for regulation of cell-fate decisions during hematopoiesis (40, 41). In our California Hispanic population, we were able to confirm the association between IKZF1 and childhood ALL risk. IKZF1, which encodes Ikaros, is the founding member of a family of zinc finger transcription factors required for the development of all lymphoid lineages (40). IKZF1 alterations are present in more than 70% of BCR-ABL1 lymphoid leukemias and have been associated with poor prognoses in BCR-ABL ALL (42, 43). Evidence from homozygous mutant mice have shown that deletion of IKZF1 leads to a rapid development of leukemia (44). Interestingly, the observation of the correlation between the rs4132601 genotype and IKZF1 mRNA expression level in Epstein-Barr virus transformed lymphocytes in our study is consistent with previous findings, and with the hypothesis that the variant may influence ALL risk by impacting on early B-cell differentiation (6, 45).
We observed the association at 10q21.2 encoding the AT-rich interactive domain 5B (ARID5B) with childhood ALL in the Hispanic population. Given the biological heterogeneity of ALL, risk variants are likely to have differential effects on ALL risk depending on cell lineage and phenotype (46). Subtype analysis of B-cell precursor ALL provides strong evidence that variants at 10q21.2-ARID5B are highly associated with the risk of developing high hyperdiploid childhood ALL in the Hispanic population, consistent with prior findings (6, 47). One of the possible biological explanations is that constitutional variants of ARID5B are associated with trisomy 10 in high hyperdiploid ALL. Although ARID5B has not been studied extensively, it is highly conserved and plays a key role in embryonic development. In addition, ARID5B homozygous (Arid5b−/−) mice show immune abnormalities including a reduction in the B-cell progenitor (48). In the present study, we also note the association between SNPs in CEPBE and childhood ALL risk. A few studies suggest a role for CEBPE (CCAAT/enhancer-binding protein, epsilon) in the development of childhood ALL. CEBPE is a suppressor of myeloid leukemogenesis (49, 50). CEBPE, along with other CEBP family members, is targeted by recurrent IGH (immunoglobulin heavy) translocation in B-cell precursor ALL, supporting a role in susceptibility to ALL (51). Even though the variants in ARID5B and CEBPE are not associated with their gene expression in our study, it is possible that trans-eQTL influences the gene expression from genes in further distance. More functional studies will be needed to examine the effects of trans-eQTL.
While the risk of developing ALL may be determined by complex interactions between genetic and environmental factors (10), epidemiologic studies have thus far provided only indirect evidence that ALL has an infectious etiology, and no specific agent has been implicated (17). In the current analysis, we investigated the joint effect of the genetic variants and surrogates of exposure to early life infections on ALL risk. To minimize the number of statistical tests, six SNPs within the three genes (IKZF1, ARID5B and CEBPE) were selected using conditional haplotype analysis. As in previous CCLS studies, we used daycare attendance, presence of older siblings, and ear infections in infancy as proxies for early life infectious exposures in our Hispanic population (14). No evidence of interaction was observed between these proxies for early life exposure to common infections with six genetic variants based on a multiplicative scale. Compared with the non-Hispanic White population in the CCLS, Hispanic children have fewer hours of daycare attendance, have more children living in the same household, have lower family income and lower parental education (data not shown). All of these factors might contribute to the different patterns of childhood exposures to infection, as well as response to infections. A more refined measure of early life infections among Hispanics may be a better proxy, such as the total number of people living in the household at the time of child's birth and/or parental or other child's social contacts. However, such measures are not available in the study.
In an admixed population such as Hispanics, population stratification is a potential problem. However, the effect is likely to be minimal in the CCLS due to the careful matching of race and ethnicity obtained from the subjects (52). Further, we used a principal components analysis (PCA) to successfully reduce the effects of potential population stratification (genomic control factor λ = 1.02) (Supplementary Fig. S4) (27). Another major strength of the present study is the detailed assessment of early life infections exposures such as daycare attendance. We calculated a composite variable “child-hours” to measure the duration of each individual attending daycare facility and number of other children at the facility (11). However, the distribution of child-hour attendance is binomial for the Hispanic population included in the study; therefore, we used a dichotomous daycare attendance variable (ever/never) in the study.
One limitation of this study for assessing gene-environment interaction is the sample size. Even though the CCLS is one of the largest case-control studies in the United States with relevant biospecimens and environmental data, gene-environment interactions with moderate to small effect sizes may not be detected. Another potential limitation of gene-environment analyses in our study is the influence of uncontrolled or residual confounding on risk estimates. The consideration of other surrogate indicators such as the total number of children/adults living in the household or definitive serologic evidence may be more suitable to address the gene-environment interaction in the Hispanic population. Furthermore, while performing gene-environment analyses, we focused on only six SNPs selected from conditional haplotype analysis; however, it is possible that other SNPs within the genes or the combination of these SNPs might have a joint effect with daycare attendance on ALL risk.
Our study showed that variants within IKZF1, ARID5B, and CEBPE were associated with increased ALL risk, and the effects for ARID5B and CEBPE were most prominent in the high-hyperdiploid ALL subtype in the California Hispanic population. Even though we did not observe significant gene-environment interactions in the study, identification of interactions between genetic variants and environmental risk factors may require much larger datasets. Replication studies with larger sample sizes in other Hispanic populations will be desirable to extend our results. Additional functional studies and re-sequencing of the relevant genetic regions are needed to better understand the role of these genes in early hematopoiesis.
Supplementary Material
Acknowledgements
Participating hospitals and clinical collaborators included University of California Davis Medical Center (Dr Jonathan Ducore), University of California San Francisco (Dr Mignon Loh and Dr Katherine Matthay), Children's Hospital of Central California (Dr Vonda Crouse), Lucile Packard Children's Hospital (Dr Gary Dahl), Children's Hospital Oakland (Dr James Feusner), Kaiser Permanente Sacramento (Dr Vincent Kiley), Kaiser Permanente Santa Clara (Dr Carolyn Russo and Dr Alan Wong), Kaiser Permanente San Francisco (Dr Kenneth Leung), Children's Hospital of Los Angeles (Dr Cecilia Fu) and Kaiser Permanente Oakland (Dr Stacy Month). We also acknowledge our collaborators at the Northern California Cancer Center and the entire California Childhood Leukemia Study staff for their effort and dedication.
Grant support
This study was supported by grants from the National Institute of Environmental Health Sciences (PS42 ES04705 and R01 ES09137), the National Cancer Institute (R25CA112355), and Children with Cancer, United Kingdom.
REFERENCES
- 1.Kaatsch P. Epidemiology of childhood cancer. Cancer Treat Rev. 2010;36(4):277–85. doi: 10.1016/j.ctrv.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 2.Pui CH, Relling MV, Downing JR. Acute lymphoblastic leukemia. N Engl J Med. 2004;350(15):1535–48. doi: 10.1056/NEJMra023001. [DOI] [PubMed] [Google Scholar]
- 3.Eden T. Aetiology of childhood leukaemia. Cancer Treat Rev. 2010;36(4):286–97. doi: 10.1016/j.ctrv.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 4.Urayama KY, Chokkalingam AP, Manabe A, Mizutani S. Current evidence for an inherited genetic basis of childhood acute lymphoblastic leukemia. Int J Hematol. 2013;97(1):3–19. doi: 10.1007/s12185-012-1220-9. [DOI] [PubMed] [Google Scholar]
- 5.Vijayakrishnan J, Houlston RS. Candidate gene association studies and risk of childhood acute lymphoblastic leukemia: a systematic review and meta-analysis. Haematologica. 2010;95(8):1405–14. doi: 10.3324/haematol.2010.022095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papaemmanuil E, Hosking FJ, Vijayakrishnan J, Price A, Olver B, Sheridan E, et al. Loci on 7p12.2, 10q21.2 and 14q11.2 are associated with risk of childhood acute lymphoblastic leukemia. Nat Genet. 2009;41(9):1006–10. doi: 10.1038/ng.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orsi L, Rudant J, Bonaventure A, Goujon-Bellec S, Corda E, Evans TJ, et al. Genetic polymorphisms and childhood acute lymphoblastic leukemia: GWAS of the ESCALE study (SFCE). Leukemia. 2012;26(12):2561–4. doi: 10.1038/leu.2012.148. [DOI] [PubMed] [Google Scholar]
- 8.Trevino LR, Yang W, French D, Hunger SP, Carroll WL, Devidas M, et al. Germline genomic variants associated with childhood acute lymphoblastic leukemia. Nat Genet. 2009;41(9):1001–5. doi: 10.1038/ng.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu H, Yang W, Perez-Andreu V, Devidas M, Fan Y, Cheng C, et al. Novel Susceptibility Variants at 10p12.31-12.2 for Childhood Acute Lymphoblastic Leukemia in Ethnically Diverse Populations. J Natl Cancer Inst. 2013 doi: 10.1093/jnci/djt042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greaves M. Infection, immune responses and the aetiology of childhood leukaemia. Nat Rev Cancer. 2006;6(3):193–203. doi: 10.1038/nrc1816. [DOI] [PubMed] [Google Scholar]
- 11.Ma X, Buffler PA, Wiemels JL, Selvin S, Metayer C, Loh M, et al. Ethnic difference in daycare attendance, early infections, and risk of childhood acute lymphoblastic leukemia. Cancer Epidemiol Biomarkers Prev. 2005;14(8):1928–34. doi: 10.1158/1055-9965.EPI-05-0115. [DOI] [PubMed] [Google Scholar]
- 12.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9(7):811–8. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 13.Urayama KY, Buffler PA, Gallagher ER, Ayoob JM, Ma X. A meta-analysis of the association between day-care attendance and childhood acute lymphoblastic leukaemia. Int J Epidemiol. 2010;39(3):718–32. doi: 10.1093/ije/dyp378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Urayama KY, Ma X, Selvin S, Metayer C, Chokkalingam AP, Wiemels JL, et al. Early life exposure to infections and risk of childhood acute lymphoblastic leukemia. Int J Cancer. 2011;128(7):1632–43. doi: 10.1002/ijc.25752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Westergaard T, Andersen PK, Pedersen JB, Olsen JH, Frisch M, Sorensen HT, et al. Birth characteristics, sibling patterns, and acute leukemia risk in childhood: a population-based cohort study. J Natl Cancer Inst. 1997;89(13):939–47. doi: 10.1093/jnci/89.13.939. [DOI] [PubMed] [Google Scholar]
- 16.Dockerty JD, Draper G, Vincent T, Rowan SD, Bunch KJ. Case-control study of parental age, parity and socioeconomic level in relation to childhood cancers. Int J Epidemiol. 2001;30(6):1428–37. doi: 10.1093/ije/30.6.1428. [DOI] [PubMed] [Google Scholar]
- 17.Roman E, Simpson J, Ansell P, Kinsey S, Mitchell CD, McKinney PA, et al. Childhood acute lymphoblastic leukemia and infections in the first year of life: a report from the United Kingdom Childhood Cancer Study. Am J Epidemiol. 2007;165(5):496–504. doi: 10.1093/aje/kwk039. [DOI] [PubMed] [Google Scholar]
- 18.Rudant J, Orsi L, Menegaux F, Petit A, Baruchel A, Bertrand Y, et al. Childhood acute leukemia, early common infections, and allergy: The ESCALE Study. Am J Epidemiol. 2010;172(9):1015–27. doi: 10.1093/aje/kwq233. [DOI] [PubMed] [Google Scholar]
- 19.O'Connor SM, Boneva RS. Infectious etiologies of childhood leukemia: plausibility and challenges to proof. Environ Health Perspect. 2007;115(1):146–50. doi: 10.1289/ehp.9024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Campleman SL WW. Childhood cancer in California 1988 to 1999 Volume I: birth to age 14 Sacramento. Vol. 2004. California Department of Health Services, Cancer Surveillance Section; CA: pp. 16–17. [Google Scholar]
- 21.Walsh KM, Chokkalingam AP, Hsu LI, Metayer C, de Smith AJ, Jacobs DI, et al. Associations between genome-wide Native American ancestry, known risk alleles and B-cell ALL risk in Hispanic children. Leukemia. 2013 doi: 10.1038/leu.2013.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma X, Buffler PA, Layefsky M, Does MB, Reynolds P. Control selection strategies in case-control studies of childhood diseases. Am J Epidemiol. 2004;159(10):915–21. doi: 10.1093/aje/kwh136. [DOI] [PubMed] [Google Scholar]
- 23.Bartley K, Metayer C, Selvin S, Ducore J, Buffler P. Diagnostic X-rays and risk of childhood leukaemia. Int J Epidemiol. 2010;39(6):1628–37. doi: 10.1093/ije/dyq162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aldrich MC, Zhang L, Wiemels JL, Ma X, Loh ML, Metayer C, et al. Cytogenetics of Hispanic and White children with acute lymphoblastic leukemia in California. Cancer Epidemiol Biomarkers Prev. 2006;15(3):578–81. doi: 10.1158/1055-9965.EPI-05-0833. [DOI] [PubMed] [Google Scholar]
- 25.Ma X, Buffler PA, Selvin S, Matthay KK, Wiencke JK, Wiemels JL, et al. Daycare attendance and risk of childhood acute lymphoblastic leukaemia. Br J Cancer. 2002;86(9):1419–24. doi: 10.1038/sj.bjc.6600274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 28.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O'Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24(24):2938–9. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang TP, Beazley C, Montgomery SB, Dimas AS, Gutierrez-Arcelus M, Stranger BE, et al. Genevar: a database and Java application for the analysis and visualization of SNP-gene associations in eQTL studies. Bioinformatics. 2010;26(19):2474–6. doi: 10.1093/bioinformatics/btq452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stranger BE, Nica AC, Forrest MS, Dimas A, Bird CP, Beazley C, et al. Population genomics of human gene expression. Nat Genet. 2007;39(10):1217–24. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jewell N. Statistics for Epidemiology. Chapman&Hall/CRC; Boca Raton, Florida: 2004. [Google Scholar]
- 32.Baye TM, Wilke RA. Mapping genes that predict treatment outcome in admixed populations. Pharmacogenomics J. 2010;10(6):465–77. doi: 10.1038/tpj.2010.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han S, Lee KM, Park SK, Lee JE, Ahn HS, Shin HY, et al. Genome-wide association study of childhood acute lymphoblastic leukemia in Korea. Leuk Res. 2010;34(10):1271–4. doi: 10.1016/j.leukres.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 34.Prasad RB, Hosking FJ, Vijayakrishnan J, Papaemmanuil E, Koehler R, Greaves M, et al. Verification of the susceptibility loci on 7p12.2, 10q21.2, and 14q11.2 in precursor B-cell acute lymphoblastic leukemia of childhood. Blood. 2010;115(9):1765–7. doi: 10.1182/blood-2009-09-241513. [DOI] [PubMed] [Google Scholar]
- 35.Vijayakrishnan J, Sherborne AL, Sawangpanich R, Hongeng S, Houlston RS, Pakakasama S. Variation at 7p12.2 and 10q21.2 influences childhood acute lymphoblastic leukemia risk in the Thai population and may contribute to racial differences in leukemia incidence. Leuk Lymphoma. 2010;51(10):1870–4. doi: 10.3109/10428194.2010.511356. [DOI] [PubMed] [Google Scholar]
- 36.Pastorczak A, Gorniak P, Sherborne A, Hosking F, Trelinska J, Lejman M, et al. Role of 657del5 NBN mutation and 7p12.2 (IKZF1), 9p21 (CDKN2A), 10q21.2 (ARID5B) and 14q11.2 (CEBPE) variation and risk of childhood ALL in the Polish population. Leuk Res. 2011;35(11):1534–6. doi: 10.1016/j.leukres.2011.07.034. [DOI] [PubMed] [Google Scholar]
- 37.Healy J, Richer C, Bourgey M, Kritikou EA, Sinnett D. Replication analysis confirms the association of ARID5B with childhood B-cell acute lymphoblastic leukemia. Haematologica. 2010;95(9):1608–11. doi: 10.3324/haematol.2010.022459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang W, Trevino LR, Yang JJ, Scheet P, Pui CH, Evans WE, et al. ARID5B SNP rs10821936 is associated with risk of childhood acute lymphoblastic leukemia in blacks and contributes to racial differences in leukemia incidence. Leukemia. 2010;24(4):894–6. doi: 10.1038/leu.2009.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chokkalingam AP, Hsu LI, Metayer C, Hansen HM, Month SR, Barcellos LF, et al. Genetic variants in ARID5B and CEBPE are childhood ALL susceptibility loci in Hispanics. Cancer Causes Control. 2013;24(10):1789–95. doi: 10.1007/s10552-013-0256-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.John LB, Ward AC. The Ikaros gene family: transcriptional regulators of hematopoiesis and immunity. Mol Immunol. 2011;48(9-10):1272–8. doi: 10.1016/j.molimm.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 41.Schmitt C, Tonnelle C, Dalloul A, Chabannon C, Debre P, Rebollo A. Aiolos and Ikaros: regulators of lymphocyte development, homeostasis and lymphoproliferation. Apoptosis. 2002;7(3):277–84. doi: 10.1023/a:1015372322419. [DOI] [PubMed] [Google Scholar]
- 42.Kuiper RP, Waanders E, van der Velden VH, van Reijmersdal SV, Venkatachalam R, Scheijen B, et al. IKZF1 deletions predict relapse in uniformly treated pediatric precursor B-ALL. Leukemia. 2010;24(7):1258–64. doi: 10.1038/leu.2010.87. [DOI] [PubMed] [Google Scholar]
- 43.Mullighan CG, Su X, Zhang J, Radtke I, Phillips LA, Miller CB, et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med. 2009;360(5):470–80. doi: 10.1056/NEJMoa0808253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Virely C, Moulin S, Cobaleda C, Lasgi C, Alberdi A, Soulier J, et al. Haploinsufficiency of the IKZF1 (IKAROS) tumor suppressor gene cooperates with BCR-ABL in a transgenic model of acute lymphoblastic leukemia. Leukemia. 2010;24(6):1200–4. doi: 10.1038/leu.2010.63. [DOI] [PubMed] [Google Scholar]
- 45.Greaves MF, Wiemels J. Origins of chromosome translocations in childhood leukaemia. Nat Rev Cancer. 2003;3(9):639–49. doi: 10.1038/nrc1164. [DOI] [PubMed] [Google Scholar]
- 46.Walsh KM, de Smith AJ, Chokkalingam AP, Metayer C, Dahl GV, Hsu LI, et al. Novel childhood ALL susceptibility locus BMI1-PIP4K2A is specifically associated with the hyperdiploid subtype. Blood. 2013;121(23):4808–9. doi: 10.1182/blood-2013-04-495390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paulsson K, Forestier E, Lilljebjorn H, Heldrup J, Behrendtz M, Young BD, et al. Genetic landscape of high hyperdiploid childhood acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 2010;107(50):21719–24. doi: 10.1073/pnas.1006981107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sherborne AL, Houlston RS. What are genome-wide association studies telling us about B-cell tumor development? Oncotarget. 2010;1(5):367–72. doi: 10.18632/oncotarget.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Akagi T, Thoennissen NH, George A, Crooks G, Song JH, Okamoto R, et al. In vivo deficiency of both C/EBPbeta and C/EBPepsilon results in highly defective myeloid differentiation and lack of cytokine response. PLoS One. 2010;5(11):e15419. doi: 10.1371/journal.pone.0015419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bedi R, Du J, Sharma AK, Gomes I, Ackerman SJ. Human C/EBP-epsilon activator and repressor isoforms differentially reprogram myeloid lineage commitment and differentiation. Blood. 2009;113(2):317–27. doi: 10.1182/blood-2008-02-139741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Akasaka T, Balasas T, Russell LJ, Sugimoto KJ, Majid A, Walewska R, et al. Five members of the CEBP transcription factor family are targeted by recurrent IGH translocations in B-cell precursor acute lymphoblastic leukemia (BCP-ALL). Blood. 2007;109(8):3451–61. doi: 10.1182/blood-2006-08-041012. [DOI] [PubMed] [Google Scholar]
- 52.Chokkalingam AP AM, Bartley K, Hsu LI, Metayer C, et al. Matching on Race and Ethnicity in Case-Control Studies as a Means of Control for Population Stratification. Epidemiol. 2011;1:101. doi: 10.4172/2161-1165.1000101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.