Abstract
Microorganisms can form biofilms, which are multicellular communities surrounded by a hydrated extracellular matrix of polymers. Central properties of the biofilm are governed by this extracellular matrix, which provides mechanical stability to the three-dimensional biofilm structure, regulates the ability of the biofilm to adhere to surfaces, and determines the ability of the biofilm to adsorb gasses, solutes, and foreign cells. Despite their critical relevance for understanding and eliminating of biofilms, the materials properties of the extracellular matrix are understudied. Here, we offer the reader a guide to current technologies that can be utilized to specifically assess the permeability and mechanical properties of the biofilm matrix and its interacting components. In particular, we highlight technological advances in instrumentation and interactions between multiple disciplines that have broadened the spectrum of methods available to conduct these studies. We review pioneering work that furthers our understanding of the material properties of biofilms.
Keywords: biofilm matrix, material properties, fluorescence microscopy, microrheology, permeability, mechanical properties
1. Introduction
Biofilms are defined as consortia of single cell microorganisms that are physiologically distinct from their free-swimming counterparts. A distinguishing feature of microbial biofilms is the presence of an adhesive matrix of highly hydrated extracellular polymeric substances (EPSs), comprised of polysaccharides, proteins, lipids, and nucleic acids (for detailed reviews regarding matrix composition we direct the reader to (Flemming and Wingender, 2010, Sutherland, 2001b, Sutherland, 2001a)). The matrix is a primary component of the biofilm, contributing to 50–90% of the total dry biomass (Flemming and Wingender, 2010). Biofilms are typically described as surface attached communities, but microbes can also form suspended aggregates, microbial mats, and flocs with biofilm-like properties, where all arrangements rely on some mixture of EPS for aggregation, structure, and maintenance of the community lifestyle (Flemming and Wingender, 2010, De Beer and Stoodley, 2013).
Over the past decades extensive information has been gained about the genetic traits and physiological processes of biofilm inhabiting cells, and how these properties correlate with the ability to form biofilms. However, important characteristics that are brought about by the extracellular matrix, such as the adherence to physiological or synthetic substrates, adsorption of gasses, solutes, and foreign cells, and resistance to deformation and rupture, are understudied. This is due, in part, to the intrinsic structural heterogeneity and complexity of the matrix, which complicates interpretation of results and identification of underlying mechanisms (Figure 1).
Figure 1.
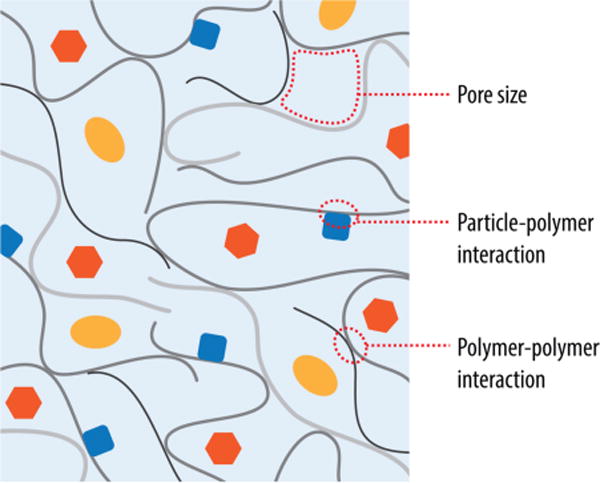
The biofilm matrix is comprised of entangled polymers (polysaccharides, DNA, proteins) that affect the permeability and mechanical properties of the entire biofilm. To understand the biophysical properties of the biofilm several questions need to be addressed. For example, what is the pore size of the matrix? Does a specific substrate interact with the matrix components? Which structural components of the matrix regulate the permeability properties? Is the matrix a static arrangement or do the individual components engage in dynamic rearrangements?
In this review we shall discuss the study of permeability and mechanical properties of biofilms. Molecular gradients of nutrients, dissolved gasses, and signaling molecules that arise within the matrix, as determined by the permeability, lead to genetic and physiological heterogeneity of the inhabiting cells (Stewart and Franklin, 2008). While traditionally studied separately, permeability and mechanical properties in the biofilm matrix are closely linked. For example, the mechanical properties and permeability of the biofilm are both influenced by the pore size, heterogeneity, and internal structure, of the matrix. This overlap between material properties should be considered when determining the appropriate tools required to characterize a specific biofilm system.
To develop greater insight into a structure-function relationship in biofilms, several questions need to be asked (Figure 1). For example, what is the pore size of the matrix? Which structural components of the matrix dominate its permeability properties? What are the dynamics of the matrix, i.e. is it a static arrangement or do the individual components engage in dynamic interactions, leading to structural rearrangements? All of these questions require different methodologies or combinatorial approaches to assess permeability and mechanical properties within a specific biofilm system.
Recent technological advances in instrumentation and the interaction between multiple disciplines have enhanced the availability of tools required for such studies. In this review we shall give an overview of pioneering work and recent findings by investigators that further our understanding of the permeability and mechanics of biofilms. We shall also provide the reader with a guide to current technologies that can be utilized to assess these properties, where these methods are specifically designed to measure properties of the biofilm matrix and its interacting components (Figure 2).
Figure 2.
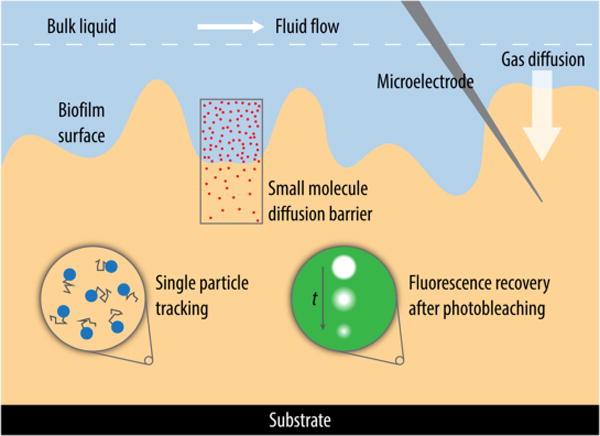
Conceptual scheme of a biofilm with an illustration of tools currently used for studies of biofilm permeability and mechanics. Fluorescence microscopy techniques such as FRAP, FCS, and fluorescence intensity measurements offer methods to visualize the transport of solutes and particles. Microelectrodes can be directly inserted into regions of interest to quantify concentration gradients of gasses and solutes within biofilms. Macroscale rheological measurements provide insight into the mechanical properties of a biofilm in total, where as a microrheological assessment of specific regions within a biofilm from single particle tracking provides information on the local properties of materials.
2. Permeability of Biofilms
2.1 Measuring concentration gradients in biofilms using microsensors
The physiochemical properties of the biofilm matrix influence the exchange of nutrients, dissolved gasses, molecules, and cells between the environment and the biofilm. The transport of water and solutes through a biofilm depends on the structure and composition of the biofilm constituents. Specifically, biofilms reduce both water flow and solute diffusion relative to bulk water, which can result in steep concentration gradients between the biofilm and the surrounding water phase (Jorgensen and Revsbech, 1985, De Beer et al., 1993, De Beer and Stoodley, 1995, Stewart, 2003, Staal et al., 2011). In circumstances where flow is completely absent from the biofilm, transport of solutes is diffusion dominated. Here, molecules move by random walk through the porous matrix or physically interact with matrix components. In other structural arrangements where channels form throughout the biomass, flow is introduced and a combinatory effect of advection within the channels and diffusion within the microcolonies influences mass transport (Yang and Lewandowski, 1995, Stewart, 2012).
One major challenge in measuring the permeability of biofilms, particularly the steep concentration gradients of solutes, is the limited ability to penetrate the biofilm structure without affecting its properties. Microsensor technology has been used to analyze the spatial distributions of solutes in biofilms since the 1960s (Whalen et al., 1969, Bungay et al., 1969), and consists of using a microscale probe at most 10–20 microns in size to measure the concentration of a particular chemical. Probes have been used to measure the concentrations of oxygen, carbon dioxide, sulfide, pH, oxidation-reduction potential, ammonium, nitrate and nitrite in biofilms (Li and Bishop, 2004, Yu and Bishop, 2001, deBeer et al., 1997). These probes are thought to be small enough as to not harm the biofilm significantly upon entry, such that they provide reliable data on chemical concentrations at microscale within the biofilm.
A commonly studied molecule in the context of biofilms is oxygen, due to the large interest in understanding the metabolic activity of biofilm bacteria. Early works on oxygen microsensing described the development of appropriate probes, and provided detailed profiles of oxygen concentration based on the position within a biofilm (Bungay et al., 1969, Whalen et al., 1969). Later studies began using the oxygen profiles to calculate physical constants based on models developed for nutrient transport within the biofilm (Revsbech, 1989, Lewandowski et al., 1993). Oxygen sensing has also been used to understand the transport limitations posed by various types of biofilm macrostructure, and to understand the consequences of void space within these structures (de Beer et al., 1994b, Rasmussen and Lewandowski, 1998). The high level of spatial resolution afforded by microsensors allow for the three-dimensional measurement of oxygen concentration, which provides clues to the structure of biofilms (Yu et al., 2004). By making oxygen the limiting substrate for growth, it has been possible to determine parameters for growth kinetics by fitting growth models to experimental data (Yurt et al., 2003).
Bacterial nitrification is important in several industries and multiple studies have been undertaken to better understand how these biofilms are structured and stratified by metabolic processes (Schramm et al., 1996, Li and Bishop, 2003) as well as how the metabolic profile changes due to the environment (Li and Bishop, 2002). Further investigations of nitrifying bacteria showed spatiotemporal changes of concentrations of measured molecules, to help locate where specific processes occur in a biofilm (Li and Bishop, 2004).
Biofilms often contain multiple species of bacteria, which can be profiled using a wide range of metabolic fingerprints, and the use of multiple sensors allows for a better understanding of how such communities are arranged. Bacterial nitrification is important in several industries and multiple studies have been undertaken to better understand how these biofilms are structured and stratified by metabolic processes (Schramm et al., 1996, Li and Bishop, 2003) as well as how the metabolic profile changes due to the environment (Li and Bishop, 2002). Further investigations of nitrifying bacteria showed spatiotemporal changes of concentrations of measured molecules, to help locate where specific processes occur in a biofilm (Li and Bishop, 2004). Similar work in mixed anaerobic/aerobic biofilms also showed spatially distinct metabolic regions (Yu and Bishop, 2001). By creating an array of microelectrodes on a micro-fabricated device, one group was able to simultaneously measure multiple chemical signatures at once in the same location, which may provide a more detailed and accurate resolution of spatially dependent processes (Lee et al., 2007).
Microsensors enable the online spatial and temporal resolution of chemical concentrations, which can improve understanding of transport within the biofilms and help model biofilm organization. One shortcoming of this technology is its limitation to very small length scales to measure local concentrations of a single parameter (e.g. oxygen, pH, carbon dioxide) surrounding the probe. Nevertheless, microsensor techniques have proven to be a valuable technology when investigating concentration gradients in biofilms.
2.2 Measuring mass transport with imaging-based techniques
Imaging based technologies are widely used to observe and quantify the transport of solutes through biofilms. Perhaps the most common and widely available optical technique is fluorescence-based microscopy. Fluorescence facilitates direct visualization of molecules in real time inside the biofilm. This enables investigators to calculate their effective diffusivity (Deff), determine interactions between the probe and the matrix, and to map voids, channels, and microcolony structures in biofilms. In comparison to microsensor technology, fluorescence imaging can typically cover a larger region of interest, ranging from hundreds of nanometers up to millimeters (Schmolze et al., 2011). In recent years, confocal and two-photon fluorescence microscopy has yielded three-dimensional information of fluorescence distribution through greater depths of biomass with less invasive methods relative to microelectrode probing. Furthermore, fluorescence imaging can be used to visualize the biofilm matrix by directly labeling matrix components (Ma et al., 2007, Barnes et al., 2012, Lawrence et al., 2007, Wrangstadh et al., 1990). This method has provided valuable information on the matrix structure, localization, and composition. Fluorescence imaging techniques provide high spatial resolution for identifying cell micro-clusters, densely packed films, and fluid filled channels and voids. However, imaging is limited to fluorescence labeling of a molecular probe, and the introduction of the label can perhaps perturb the probe’s behavior.
The size of particles used for fluorescence imaging range from small molecules (fluorescent dyes, peptides, or lipids) to larger macromolecules (proteins and glycan chains), up to micro-sized fluorescent beads (Figure 3 a,b) (De Beer et al., 1994a, Stoodley et al., 1994, Stewart, 1998, Wilking et al., 2013). One of the earliest approaches to assess diffusion though the bulk biomass resulted from bathing biofilms with fluorescent tracers. To calculate an internal effective diffusion coefficient (Deff) within a biofilm, several studies have relied on the introduction of fluorescent tracers into the biofilm coupled with time-lapse microscopy to record the change in fluorescence intensity (F) of tracers over time. Fluorescent dyes (fluorescein, Rhodamine B, and BoDipy) and fluorescently labeled dextrans and Ficolls provide a reasonable estimate of diffusion through a biofilm (De Beer et al., 1994a, De Beer et al., 1997, Lawrence et al., 1994, Thurnheer et al., 2003). Due to the variation in matrix polymers and overall biofilm structure, departures from predicted diffusion coefficients may occur. Such deviations may indicate either electrostatic or hydrophobic interactions between the fluorescence probe and the matrix components, or rapid movement of the fluorescent tracer through water filled channels and voids.
Figure 3.
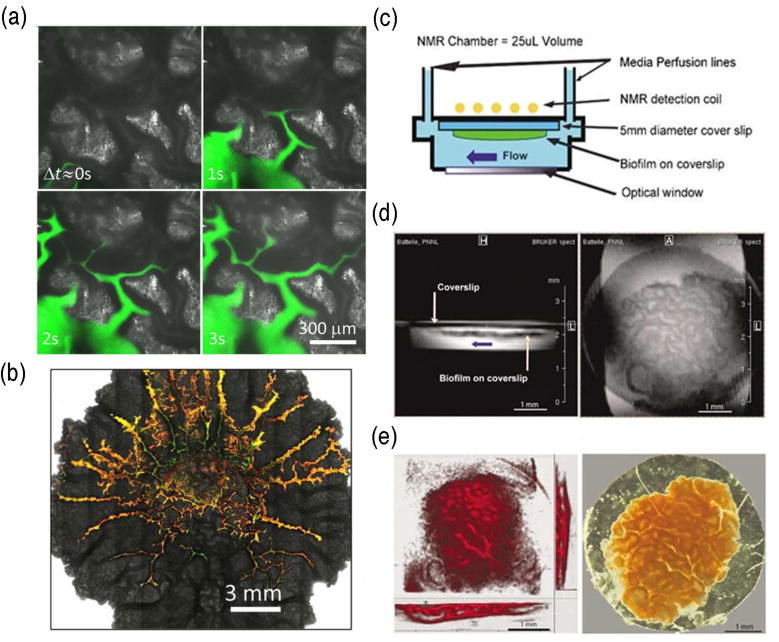
Fluorescence microscopy and nuclear magnetic resonance (NMR) can be used to observe channel development and fluid transport in biofilms. Bacillus subtilis biofilms grown on an agar surface where channels are visualized with fluorescent dye (a,b) Reprinted with permission from (Wilking et al., 2013). Copyright (2012) National Academy of Sciences, U.S.A. An NMR setup for imaging biofilms in situ (c) Horizontal and vertical 2D MRI sections of a Shewanella oneidensis biofilm (d). 3D MRI rendering (left) and transmitted light image (right) of a S. oneidensis biofilm (e). Reprinted by permission from Macmillan Publishers Ltd: The ISME Journal (McLean et al., 2008b), copyright (2008).
One of the pioneering studies in biofilm permeability utilized fluorescence imaging to investigate the behavior of fluorescein plumes microinjected into the biofilm at different locations (De Beer et al., 1994a). When fluorescein was injected into microcolonies, the plume maintained a spherical shape, indicating limited or no fluid flow through these structures even with increasing the liquid velocity within the growth chamber. However, when fluorescein was injected into fluid-filled voids, the plume sphere elongated as the fluid velocity increased, indicating flow within the void (De Beer et al., 1994a). This work highlighted the fact that biofilm structure has significant impact upon the movement of solutes within a biofilm. Fluorescence microscopy has also been utilized to study the transport of antimicrobials through biofilms. An increase in antibiotic tolerance is a common property of biofilm-associated cells (Lewis, 2001, Stewart and Costerton, 2001, Stewart, 2002, Hoiby et al., 2010, Hogan and Kolter, 2002). In some cases, tolerance can increase orders of magnitude above the minimal inhibitory concentration (MIC) of planktonic cells, rendering many of the cells virtually resistant to antibiotic monotherapy (Sandoe et al., 2006, Svensson et al., 1997, Girard et al., 2010). Since the matrix serves as a selective filter of solutes, likely through electrostatic and hydrophobic interactions, the matrix perhaps influences transport of antimicrobial agents. Several groups have experimentally addressed this question by examining transport of fluorescently labeled antibiotics (Rani et al., 2005, Jefferson et al., 2005, Oubekka et al., 2012, Tseng et al., 2013, Billings et al., 2013) or antibiotics that trigger downstream fluorescence upon interaction with the cells (Stone et al., 2002). In addition, antibiotic penetration through biofilms has been intensively studied, even beyond fluorescence based methods (Stewart, 1996, Stewart, 1998, Singh et al., 2010, Pibalpakdee et al., 2012, Walters et al., 2003, Shigeta et al., 1997). The results have demonstrated that transport is dependent upon the composition of the extracellular matrix, the physiochemical properties of the antibiotic, and the overall heterogeneity of the biofilm structure. It is important to recognize that although antimicrobials may rapidly pass through a biofilm by means of channels and pores in the matrix mesh, sorption to the matrix, steric hindrances to its target molecules, and inactivation of the antibiotic may reduce the bioavailability such that an antibiotic cannot reach its target at effective concentrations.
Another property that is worth considering for mass transport within biofilms is the active mobility of cells, where fluorescence microscopy can be used to track cell movement in real time. Conventional paradigm dictates that biofilms are sessile communities of cells and that motility is reserved for active dispersal. However, recent work by Houry et al. demonstrated that some species of motile bacteria infiltrate deep into the biofilm matrix, originating from planktonic subpopulations (Houry et al., 2012). Time-lapse confocal microscopy was used to track single cell movement within the biofilm using the expression of green fluorescent protein (GFP) as a marker. This motile sub-population of cells produce tunnels that collapse after 2–5 seconds or transient pores up to 10 μm in diameter. The behavior was identified in Bacillus thuringiensis and Yersinia entrocolitica. However, this behavior was not observed in the motile species Bacillus subtilis or Pseudomonas aeruginosa. The authors propose that stealth swimming in the biofilm matrix may require a bacterium to overcome the kinetic energy threshold of forces generated by the EPS. This sub-population is perhaps biologically significant since stealth swimmers could potentially facilitate the transport of nutrients and oxygen deep into the biofilm. Of note, the diffusion of FITC-labeled Dextran (250 kDa) into a B. thuringiensis biofilm was enhanced by the presence of the intra-biofilm swimming phenotype. These results suggested that cellular motility within a biofilm should be considered, along with diffusion and advection, when describing mass transfer within biofilms of specific species (Boles and Horswill, 2012).
2.2.1. Laser scanning microscopy techniques
Both fluorescence recovery after photobleaching (FRAP) and fluorescence correlation spectroscopy (FCS) are powerful quantitative tools for measuring local diffusion coefficients for multiple locations within a biofilm beyond initial diffusive rates. These techniques provide a greater spatial resolution to resolve transport properties within a biofilm. Importantly, these methods allow for separation of free diffusion from anomalous diffusion, facilitating the discrimination of solutes that interact with matrix components from those that are nonreactive (Oubekka et al., 2012). Furthermore, these techniques can be applied after the fluorescent probe has reached equilibrium within the biofilm matrix to monitor the reactivity of the solute on extended timescales, which can be on the order of days.
FRAP is a method used to extract transport information of mobile fluorescent molecules in a user defined region of interest (ROI). It is based on the principle of irreversibly quenching fluorophores when exposed to a brief, high intensity pulse of light in the user-defined region. Fluorescence recovery that occurs within the ROI is the result of bleached and non-bleached fluorophores redistributing within the bleached parameter space. Measuring and plotting fluorescence recovery intensity profiles as a function of time can determine transport properties of the fluorescent molecules (Axelrod et al., 1976, Edidin et al., 1976).
In the context of living systems, FRAP has been predominantly used to study mobility of molecules within membranes (Liebman and Entine, 1974, Poo and Cone, 1974) or intracellular space (Cole et al., 1996, White and Stelzer, 1999, Carrero et al., 2003). However, a few studies have managed to use the technique to characterize the diffusion of small molecules and particles within biofilms. One of the first studies by Lawrence and colleagues (Lawrence et al., 1994) developed a FRAP based method to quantify the mobility fluorescently labeled dextrans of different molecular weights through biofilms. With this experimental approach, the authors defined a large, rectangular ROI that measured 800 μm2 of biofilm area. Due to the large area encompassed by the ROI, the authors measured average diffusion coefficients within a heterogenous mix of channels and micro-cell clusters (Lawrence et al., 1994). Although this study demonstrated the utility of FRAP with biofilm studies, other groups reported different diffusion coefficients for the same fluorescent molecules, perhaps due to the macroscale bleaching ROI and the one-dimensional diffusion parameter in the z-direction considered by Lawrence et. al. for fitting of FRAP recovery curves. Other groups have reduced the size of the bleach spot to focus on specific structural regions within the biofilm (e.g. cell clusters) (Bryers and Drummond, 1998, Birmingham et al., 1995). Further, a standard two-dimensional diffusion model, based on the model presented by Axelrod et. al. (Axelrod et al., 1976), serves as a general fitting model. These assume a Gaussian laser beam profile and an infinite homogeneous distribution of fluorescent molecules outside of the ROI (Bryers and Drummond, 1998, Birmingham et al., 1995):
| (1) |
where F(t) is the recorded fluorescence intensity at postbleach time (t); Fo refers to the initial pre-bleach fluorescence intensity, and Rf refers to the mobile fraction of fluorescently labeled fluorophores and is defined as:
| (2) |
In the case of molecular diffusion without reversible binding reactions between the fluorescently labeled molecule and the surrounding environment, f(t) is defined as:
| (3) |
where κ represents the bleach constant, representing the extent of bleaching, and τd is the 2D characteristic diffusion time:
| (4) |
where ω represents the e−2 laser radius and D is the lateral diffusion coefficient. Using appropriate software, the experimental FRAP recovery curves can be fitted to the theoretical model to calculate D. In cases where the fluorescent solute interacts with the biofilm environment, a different mathematical approach must be employed to separate diffusive behavior from bimolecular interactions with the surrounding matrix. Birmingham et al.(Birmingham et al., 1995) derived a series of theoretical FRAP recovery functions that consider differences in the lateral diffusion of the interacting species, along with differences in photobleaching/fluorescent yields between bound versus free labeled molecules.
With increased availability of commercial confocal microscopy systems, a recent method has emerged to improve the accuracy of FRAP measurements within biofilm systems (Waharte et al., 2010). This methodology, first introduced by Waharte et al., includes image acquisition during the bleach phase, high frequency of image capture, and consideration of bacterial movement over the duration of the experiment. The authors are the first to implement a kymogram analysis (xt plot) of a FRAP experiment within the context of bacterial biofilms, which provides the added advantage of eliminating experiments that are unusable due to microbial motion. In contrast to classical FRAP, the authors implement an intensity profile analysis to quantify diffusion of FITC-dextrans within Lactococcus lactis and Stenotrophomonas maltophilia biofilms instead of a single spot ROI. A series of intensity profiles extending from the bleach region are plotted with a Gaussian function (Seiffert and Oppermann, 2005, Waharte et al., 2010):
| (5) |
where K is the bleach constant, x0 is the profile center, and the diffusion coefficient is solved by d2 = 8Dt for 2-dimensional diffusion. The diffusion coefficients calculated from intensity profiles were determined to be on the same order of magnitude as calculated by analytical models (Braga et al., 2004) and fluorescence correlation spectroscopy (Guiot et al., 2002). Other studies have used this method to determine diffusion-reaction behavior of fluorescently labeled vancomycin in Staphylococcus aureus biofilms (Daddi Oubekka et al., 2012, Oubekka et al., 2011).
As an alternative to FRAP, fluorescence correlation spectroscopy (FCS) is a powerful, non-invasive tool to measure diffusion in biofilms based on single molecule fluorescence intensity fluctuations over time. These fluctuations are the result of fluorophore movement through a microspace and are recorded as the molecules pass through the confocal excitation volume. FCS rapidly detects thousands of single molecule diffusion events with low fluorophore concentrations (20–100 nM) in small volumes (20–30 μ l), making the approach ideal for characterizing multiple regions within a single biofilm with high statistical confidence (Zhang et al., 2011). Even though FCS is well suited for confocal microscopy, two-photon excitation (TPE) microscopy has the added advantage of confining excitation to the imaging focal point at femtoliter volumes, resulting in low background, reduction in photobleaching, while limiting cellular photodamage (Briandet et al., 2008). Diffusion times are calculated from raw FCS data by fitting with a normalized autocorrelation function [g(τ)] for the free Brownian motion of molecules, operating under the assumptions that the intensity profiles are approximated using a three dimensional Gaussian distribution and that fluctuations in intensity are only due to diffusion through the excitation volume (Briandet et al., 2008, Guiot et al., 2002):
| (6) |
In this model, N is defined as the number of fluorophores in the excitation volume, ω0 is beam width in the focal plane, z0 is the focal depth, and τ0 is the translational diffusion time. The translational diffusion coefficient is related to τD by:
| (7) |
Diffusion of two separate fluorescent populations or anomalous diffusion, which may occur in complex biological systems, requires alternative models for raw FCS data fitting (Briandet et al., 2008, Guiot et al., 2002).
The first application of FCS with TPE toward characterizing diffusion within bacterial biofilms was implemented by Guiot and colleagues (Guiot et al., 2002). In this study, latex beads and FITC-dextrans ranging in size and charge were introduced into L. lactis and S. maltophilia biofilms and the resulting correlation curves were compared with those acquired in free solution. From strictly a steric perspective (in the absence of charge interactions) the authors conclude that nutrients, antibiotics, or small particles, can penetrate and diffuse within the biofilm matrix with modest hindrance in some cases. However, FCS analysis of charged particles revealed heterogeneous distribution of spatial regions within both L. lactis and S. maltophilia biofilms that reduced or completely inhibited cationic particle diffusion (Guiot et al., 2002). The efficiency of FCS analysis has lead to further investigations of small molecule (Daddi Oubekka et al., 2012, Zhang et al., 2011), bacteriophage(Briandet et al., 2008, Lacroix-Gueu et al., 2005), and nanoparticle (Peulen and Wilkinson, 2011) mobility within biofilms of varying species.
As described above, experimental setups are diverse among many groups, and include static or continuous-flow biofilm systems, an assortment of fluorescence tracers, and different methods of introducing the probes into biofilms. For imaging-based fluorescence experiments, it is important to consider the type of probe required to answer specific questions regarding the material properties of a biofilm. For instance, small molecules and micro-scale fluorescent beads may both provide information regarding the transport through channels and voids, while fluorescent micro-beads can also be utilized for probing local mechanical properties of the biofilm matrix as will be discussed in later sections of this review. Ultimately, careful fluorescence probe design and detailed knowledge of biofilm experimental conditions are required to quantify physical properties, such as probe interactions with matrix polymers and geometric constraints resulting from the matrix mesh size.
2.2.2 Nuclear Magnetic Resonance Imaging
In addition to light microscopy based methods to study transport in biofilms, the phenomenon of nuclear magnetic resonance (NMR) has been employed as a multipurpose, non-invasive technique to study biofilms in situ. Although light microscopy offers an excellent approach to map 3D structures of biofilms and mobility of solutes, it is not possible to directly observe the flow dynamics of water or small molecules without a fluorescent tracer or label. For detailed descriptions on NMR methodology, we refer the reader recent texts by James Keeler and Edme Hardy (Keeler, 2011, Hardy, 2012).
NMR-imaging (e.g. magnetic resonance image; MRI) and NMR spectroscopy methods have been used to map the structure and flow velocity profiles of biofilms without the addition of exogenous fluorescent probes, while providing information on the diffusion of molecules and their interaction with the surrounding biofilm environment. Lewandowski et al (Lewandowski et al., 1993) was the first to report NMR imaging as a technique to measure the flow velocity profiles around a mixed species bacterial biofilm on a polycarbonate substrate under continuous flow. The work addresses the flow properties of the hydrodynamic boundary layer, while challenging the assumption that diffusion is the only property that governs mass transport within biofilms. Specifically, the data revealed that convection and diffusion were not separate properties isolated to the bulk fluid and internal biomass, respectively, but demonstrated that intra-biofilm flow occurs below the biofilm interface, presumably due to intra-biofilm channels and structural heterogeneity (Lewandowski et al., 1993). Pulse-field gradient NMR (PFG-NMR) is the most commonly used method for diffusion analysis of water and small molecules. Using this method, Beuling et al (Beuling et al., 1998) quantified the diffusion of water in natural biofilm systems isolated from industrial settings, while Vogt et al (Vogt et al., 2000) quantified the diffusion of water, glycerol, and unknown chemical constituents in Pseudomonas aeruginosa biofilms. The authors observe a range of diffusion coefficients for these substances at different locations within the biofilm. Manz et al. also employed this method to study the effect of fluid flow at increasing velocities on biofilm structure (Manz et al., 2003). These experiments demonstrate that channels, microcolonies, and extracellular polymeric substances all influence the mobility of dissolved solutes.
With rising interest in utilizing biofilms for applications such as bio-remediation and bio-sensing, recent studies with MRI have focused on probing physical-chemical interactions between the biofilm matrix and metals. Phoenix et al. (Phoenix et al., 2008) mapped diffusion and immobilization of copper in a phototropic biofilm isolated from a hot spring. Their MRI approach revealed a 3-dimensional structural map of the biofilm along with quantitative copper concentration profiles acquired at user defined time intervals (Phoenix et al., 2008). Other reports have described MRI methods to quantify the transport of metal complexes such as Fe-EDTA (Bartacek et al., 2009) and Gd-DTPA (Ramanan et al., 2013). Understanding metal sorption and degradation kinetic in biofilm systems could facilitate the development of efficiently engineered biofilm remediation reactors to vastly improve in situ bioremediation.
While quantification of mass transport helps to understand the physiochemical properties of the biofilm matrix, the consequential physiological response of microbes to mass transport limitations is also of importance. This need for non-invasive studies has prompted the extension of NMR spectroscopy to study metabolic responses in living biofilm systems. Majors et. al. (Majors et al., 2005b)were the first to report that NMR temporally resolved lactate metabolism in S. oneidensis MR1 biofilms grown in a flow cell. Other studies have expanded on this technique where in vivo NMR spectroscopy/imaging was used to quantify the metabolic activity of biofilms in response to environmental perturbations (Figure 3c–e) (McLean et al., 2008a, Cao et al., 2012). The results yielded near real-time kinetic metabolite profiles. Additional studies have demonstrated the functionality of a combined NMR and confocal microscope to temporally and spatially resolve metabolic activity (Majors et al., 2005a, McLean et al., 2008b). Although NMR is considerably less sensitive than optical methods it offers the non-invasive study of living biofilms. NMR does not produce harmful ionizing radiation, measurements can be acquired without structural damage to the system, and NMR has the advantage of using any nucleus with nonzero nuclear spin (i.e. 1H, 13C, 15N, and 13P), which reduces the need for the introduction of tracers.
3. Mechanical Properties of Biofilms
Mechanical properties arise from the internal structural organization of the biofilm. Biofilms consist of bacteria and hydrated macromolecules in water, creating a complex fluid that does not behave as purely viscous or purely elastic. Characterization of their internal structure requires an assessment of both the physical properties of the biofilm on the microscale as well as any structural features that exist within it. Thus, it is important to define specific properties of interest and to determine the appropriate techniques available to measure them.
Rheology is the study of the response of materials to applied forces. Traditionally, rheometry has been performed on bulk materials, and this will be referred to as macroscale or bulk rheology in later sections. More recently, the field of microrheology has arisen, which studies the local properties of materials on the microscale and allows for internally probing the mechanics of a fluid. The techniques used for gathering rheological data will be discussed in later sections, but we begin with a discussion of which properties of materials we are interested in and how they relate to biofilms. These properties are useful in characterizing the ability of a material to either flow or store energy in response to shear stress. For a more thorough discussion of rheology, we guide the reader to the books and reviews cited in this section, all excellent resources.
A Hookean solid is a material that is purely elastic, and can be modeled mechanically as a spring. These materials store energy as they deform under stress and can then relax back to their original shape. Hookean solids exhibit the following stress/strain relationship, where E represents the Young’s modulus of the material, σ is the shear stress, and γ is the shear strain(Morrison, 2001):
| (9) |
A Newtonian liquid acts in a purely viscous manner, meaning that it flows and dissipates energy in response to stress. Such materials can be modeled as dashpots, yielding a different stress/strain relationship (Morrison, 2001):
| (10) |
In this case, η represents the viscosity, and represents shear rate. Most materials, including biofilms and other polymer systems, are neither Hookean nor Newtonian, but are instead viscoelastic. Instead of a lone spring or dashpot, these materials can be modeled most simply as a spring and dashpot in series, though more complex models exist. For these materials, we introduce a complex shear modulus that incorporates elastic storage and viscous loss (Squires and Mason, 2010):
| (11) |
As will be discussed later the storage modulus can be represented via two components: G′(ω), the storage modulus, and G″ (ω), the loss modulus. By determining the values of these moduli, we can understand how a biofilm responds to stresses placed on it, most importantly if it acts in a more viscous or more elastic manner to a given applied stress.
In addition to viscosity, elasticity, and a complex shear modulus, we can also use creep compliance to understand the way in which a material responds to a constant applied stress. Creep compliance is defined as the ratio of strain to stress, where J(t) is the creep compliance, γ(t) is the measured strain of the material, and σ0 is the constant stress (Macosko, 1994):
| (12) |
The higher the creep compliance of a fluid, the more it deforms to a given stress, and thus by evaluating the creep compliance of a biofilm we can better understand how it will react to an applied external force. In a purely Newtonian material, the strain, and therefore compliance will increase linearly with time, whereas for a purely Hookean material there is an instantaneous increase in strain, which then remains constant over time (Morrison, 2001, Macosko, 1994).
Finally, it is also of interest to understand adhesion between biofilms and surfaces, as a key step to biofilm formation is the adhesion of bacteria to a surface. In general, adhesion tells us about the energy of interaction between two materials and may be determined by measuring the force required to separate two surfaces. Historically in the biofilm field, adhesive strength has been defined as follows (Ohashi and Harada, 1994, Chen et al., 1998):
| (13) |
In the above equation, ξ is the adhesive strength in Watts/m2, W is work required to pull a biofilm away from its substrate, A is the total surface area of a test surface, and β is the fraction of that surface covered by biofilm.
3.1. Bulk measurements
The most common tool in rheology is the bulk scale rheometer, which consists of either parallel plates or a cone and plate between which the material of interest is placed (Figure 4a). This setup allows one to apply a known stress to a material and measure the strain or vice versa, from which the complex shear modulus can be calculated. If these tools are used to apply a small strain to a fluid, we can assume that the underlying structure of the material remains unchanged and can assume a linear dependence between stress and strain (Barnes et al., 1989). Therefore, by applying a known, small, oscillatory strain (in a technique known as small angle oscillatory shear), we can measure the linear response of a fluid (Bird et al., 1987):
| (14) |
The stress oscillates with the same frequency (ω) as the strain, but leads by a phase angle (δ) (Rubinstein and Colby, 2003):
| (15) |
These can be substituted into the following relation to determine G′(ω) and G″(ω):
| (16) |
This yields the following for the storage and loss moduli:
| (17) |
Finally:
| (18) |
In a creep test, a constant (rather than oscillatory) stress is applied to a material, and the resultant measurement of strain over time can be measured. Finally, it is also possible to apply a large strain to a fluid, such that the underlying material is physically disrupted, in order to study non-linear rheology. These measurements can provide information about shear-thinning and yield stress phenomena, via the use of step shear rate and large amplitude oscillatory shear tests respectively (Ewoldt et al., 2010, Hyun et al., 2002). While they may not provide insight to the internal structure of biofilms, large strain measurements may be useful in understanding how to externally perturb a biofilm system. The yield stress is of particular interest, as it is a measure of how much force must be applied to an apparently solid material to get it to flow and show liquid-like behavior (Barnes, 1999). Multiple measurement techniques exist for finding a yield stress (Nguyen and Boger, 1992, Barnes, 1999). For example, the y-intercept of a curve fitted to shear stress vs. shear rate data measured in a rheometer is an approximate measure of the yield stress (Nguyen and Boger, 1992, Barnes, 1999). Alternatively, the yield stress can be determined directly by applying a constant stress to a material for some time, and then removing the stress. At stresses below the yield stress, the material returns to a baseline level of zero strain, whereas above the yield stress it will not fully recover from the deformation (Nguyen and Boger, 1992).
Figure 4.
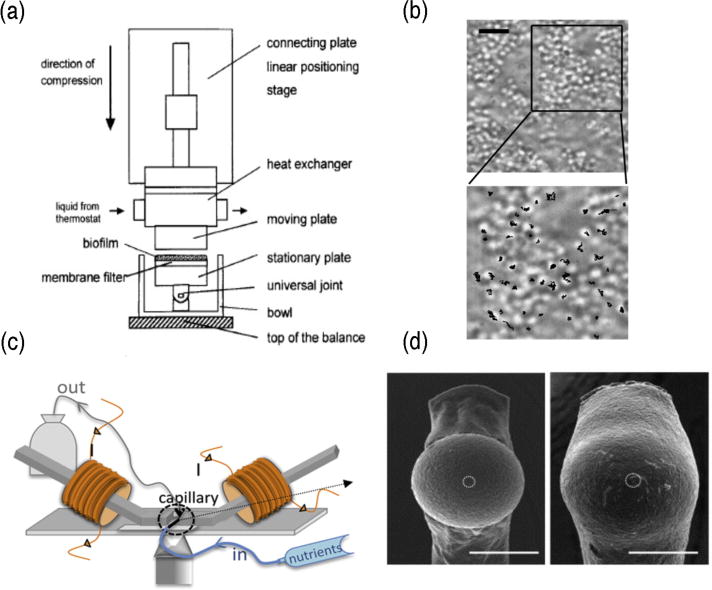
Examples of techniques that can be used to determine biofilm material properties. A rheometer setup in which a natural biofilm sample attached to a membrane can be tested (Korstgens et al., 2001a) (a). Copyright (2001) Institute of Physics Publishing. Staphylococcus aureus biofilm at 8 hours, with the tracks of bacterial motion (b). Scale bar is 5 mm. Reprinted with permission from (Rogers et al., 2008). Copyright (2008) American Chemical Society. A magnetic tweezers setup for monitoring biofilms grown in flow cells (c) Reprinted with permission from (Galy et al., 2012). Copyright (2012) Elsevier. SEM images of biofilm coated beads used for AFM measurements (d). The bead on the left is surrounded by younger biofilm than the bead on the right. Scale bars are 30 μm. Reprinted with permission from (Lau et al., 2009) Copyright (2009) Elsevier.
Several groups have measured the macrorheology of biofilms (Towler et al., 2003, Pavlovsky et al., 2013, Lieleg et al., 2011, Jones et al., 2011, Korstgens et al., 2001a, Shaw et al., 2004, Korstgens et al., 2001b, Houari et al., 2008). These prior studies can be divided into two types: those that scraped biofilms from their original growth locations to place them into a rheometer versus those that grew biofilms directly on a rheometer plate. Scraping biofilms from their original growth location may disrupt their structure, however this approach provides some additional freedom in choosing growth conditions. Several groups have used these methods to fit biofilm viscoelastic behavior to mechanical models that are more complicated than a simple spring and dashpot in series (Towler et al., 2003, Pavlovsky et al., 2013). In addition, macrorheology has been used to assess the effect on different treatments to the properties of biofiolms (Lieleg et al., 2011, Jones et al., 2011, Korstgens et al., 2001a). While the results of multiple groups indicate the biofilm is a shear-thinning fluid, the measured shear moduli range over three to four orders of magnitude, from 10−1 to about 103 Pa. These experiments were performed using different species of bacteria and different growth methods, both of which could help explain the variation in measured moduli. For a comprehensive listing of measured properties and additional discussion of techniques used, we recommend the recent review Bol et al., 2013 (Böl et al., 2013).
Several groups have used less traditional methods for small-scale bulk rheometry to measure biofilm physical properties. One method is to grow biofilms in microfluidic devices and then apply known shear stresses by varying fluid flow (Stoodley et al., 1999, Stoodley et al., 2002, Klapper et al., 2002, Dunsmore et al., 2002). This method allows for the measurement of stress/strain curves and adhesion of specific colonies of bacteria, though still on the bulk scale. Another method is to use a PDMS based microfluidic device through which known stresses can be applied to microscale portions of a biofilm via changes in air pressure applied to a PDMS membrane above it (Hohne et al., 2009). While this technique does provide micrometer scale precision in the x–y plane of a biofilm, the pressure is applied to the top of the material, resulting in the measure of bulk properties.
Macrorheological studies have provided a wide range of insight into biofilm properties, including the discovery of its shear-thinning nature, shear moduli, and the effect of environment on physical properties. However, these are inherently averaged properties. Given the heterogeneous nature of biofilms, techniques that can probe spatial variations within a biofilm are of great use.
3.2. Passive Microrheology Techniques
Microrheology is used to determine the same properties as macrorheology through the use of microscale probes that are generally embedded into the material of interest (Mason and Weitz, 1995, MacKintosh and Schmidt, 1999). This is of particular use for the study of biofilms and other biological materials, as it allows for the probing of the system over small length scales and can be applied without greatly disrupting a system’s natural state. In addition, small sample sizes can be used, which provides great flexibility over macrorheology by allowing the use of young biofilms. The wide range of microrheological tests available allows us to test smaller-scale features of biological systems. To appropriately interpret the results of these tests, it is important to understand the way in which these techniques yield the properties of interest.
3.2.1. Single particle tracking (SPT) and Multiparticle tracking (MPT)
In passive microrheology, beads (single for SPT, and multiple for MPT) are embedded into a material and are not manipulated by any external force. They are therefore assumed to move in response to thermal fluctuations, of energy scale kBT (~10−21 J at room temperature). In a typical experiment, video microscopy will be used to image the beads, and image processing software is then used to track the locations of the particle centers (Crocker and Grier, 1996). These locations can then be converted into individual particle traces, from which a mean-square displacement (MSD) can be extracted using the following definition:
| (19) |
In this equation, r refers to the position of the particle in the x–y plane of an image, t is time, and τ is a lag time. The brackets indicate that this is an ensemble-average value, though it is often practical to use an ensemble and time average. Using more exotic tracking schemes, one can also follow particle trajectories in 3 dimensions. In the discussion that follows, we will assume the MSD is from 2D traces as it is the more common observable. Experimentally, MSDs are determined from trajectories by calculating the change in position for any set of points in a trajectory separated by a given lag time and then calculating the variance of this distribution. Statistical bias may therefore appear for any experiment in which the particles are not embedded in a uniform fluid as trajectory lengths will be dependent on local microenvironment. For a purely Newtonian fluid, it is quite easy to extract a viscosity from the apparent MSD, as given by the following relationship, where r is the position of a particle, D is diffusivity, σsh is the shutter speed of the camera used, and ε is the so-called static error in particle location (Savin and Doyle, 2005):
| (20) |
The static error results from the inability to completely resolve even a completely motionless probe. This error can be corrected for experimentally by measuring the motion of probes embedded in a solid, and subtracting appropriately, as seen in the last term of the equation. Though the above equation is for a Newtonian fluid, the static error correction can be applied to the apparent MSD of beads in any fluid, as it does not depend on the properties of the fluid being measured. The camera shutter speed is introduced in the above equation to account for what is known as dynamic error, which results from the motion of probes while the shutter is open and acquiring light. The longer the shutter is open and the higher the diffusivity of a probe in the fluid of interest, the larger the dynamic error will be. The correction shown above applies only to Newtonian fluids, as the mathematical form of the dynamic error changes with fluid type and is often unknown. The choice of a short enough shutter speed to minimize the effects of static error can be determined by measuring the MSD for several different shutter times and determining when shutter speed no longer significantly affects the measured MSD. For a Newtonian fluid, we can use the Stokes-Einstein relationship to relate D to the particle radius a and fluid viscosity (Rubinstein and Colby, 2003):
| (21) |
A complex fluid does not follow the previous equation, and instead, we must use the generalized Stokes-Einstein relation (GSER), where G*(ω), represents the shear modulus in the Fourier domain, s is equal to i ω, the Laplace frequency, is the shear modulus represented in the Laplace domain, and is the Laplace transform of the MSD, (Mason et al., 1997, Mason, 2000):
| (22) |
This relationship is used to estimate the absolute value |G*(ω)| by using a power law expansion of the MSD to calculate an approximate Laplace transform, and yields the following (Mason, 2000):
| (23) |
From this estimation of the absolute value of the complex shear modulus, we can use the following equations to determine the storage and loss moduli:
| (24) |
| (25) |
| (26) |
Other, more accurate transforms, in which the power law expansion around the MSD includes higher terms have also been published (Zhu et al., 2008).
Finally, the MSD also allows us to calculate the creep compliance of a material without having to deform it externally (Wirtz, 2009):
| (27) |
This equation is the 2-dimensional microrheological equivalent of creep compliance. If we are indeed measuring in the linear regime, where J(τ) of the fluid is the proportionality constant between stress and strain, then all of the MSD curves of a material taken at the same temperature will collapse onto each other when multiplied by probe radius, assuming the probes are large relative to the microstructure of the material. Thus, in order to be sure that all of the above equations relating MSD to physical properties hold true to a set of experimental values, the experiments must repeated for different probe sizes to test the validity of the assumption that the material acts like a continuum. The curves will collapse for probes that are larger than the microstructure of the probed material, so this relationship can also allow us to approximate the mesh size of a gel. The value of the creep compliance also relates to mesh density and crosslinking in a gel, and will likely decrease in value in response to an increase in either factor.
It is important to note that the continuum assumption is not the only assumption that must be verified in order to trust the validity of an MSD to yield physical properties. Another major assumption is that the system is at equilibrium (Squires and Mason, 2010). In a passive particle tracking experiment, where the particle is not being forced, this can be violated via a material that is either internally active or is aging (Squires and Mason, 2010). We must be particularly conscious of both of these assumptions when applying the method to living biofilms, as they can change over time via cell turnover or secretion of new EPS, and they may be internally active if the bacteria are not completely sessile. However, if these events occur over time scales much larger than the probed time scale, which is generally the case in a biofilm where cells are dividing slowly, we can assume a quasi-equilibrium state.
Particle tracking is a versatile technique that can be modified in various ways for use in biofilms. For example, it is possible to use individual bacteria probes for their microenvironment (Rogers et al., 2008). Using the MSDs from tracking bacteria, Rogers et al. found evidence for active motion of flagellated bacteria and were able to determine compliance of the biofilm (Figure 4b). This approach potentially provides a method for separating the effects of bacteria within a biofilm from externally added probes, as the two motions can be compared to one another if measured concurrently.
As described in the section above, macrorheological techniques have provided a wide range of values for measured physical properties of biofilms. In an attempt to reconcile these discrepancies, one group isolated the water soluble and water insoluble polysaccharide fractions from S. mutans biofilm and then separately reconstituted them as gels (Cheong et al., 2009). They found that the shear moduli were orders of magnitude different from each other and suggested that the water-insoluble fraction is likely part of the mechanical scaffold of the biofilm. It should be noted that this system was not applied to biofilms in situ, where the polysaccharide fractions are mixed with other extracellular polymeric substances that may influence the overall mechanical properties.
Recently, SPT has been used to measure the apparent diffusion coefficient of nanoparticles of varying size and surface charge within biofilms of P. aeruginosa, and B. multivorans (Forier et al., 2012). PEGylated particles were found to have apparent diffusion constants similar to those in water, whereas positively and negatively charged particles had lower apparent diffusion constants, attributed to interaction with the biofilm. The diffusion constants similar to those in water may indicate the presence of fluid-filled channels within the biofilm system. The ability to track small particles through biofilms has also been previously used to show differences between cell clusters and voids within the biofilm, and to help measure transport rates through such a system (De Beer et al., 1994a, Stoodley et al., 1997).
3.2.2. Two-Particle Microrheology
The mapping of SPT and MPT data to bulk rheological properties assumes that particles occupy a homogenous, incompressible medium. Biofilms are living systems, and may contain cavities or other internal entrapments, which would violate this assumption. Two-particle microrheology is used to measure the correlation between motion of particles in a sample, which can probe longer length scale interactions (on the order of distance between particles), overcoming the SPT limitation of local interactions (Gardel et al., 2005, Liu et al., 2006). Instead of an MSD, the ensemble average tensor product of displacements between particles is calculated, Laplace transformed, and related to the Laplace transform of the complex shear modulus as detailed in the original paper by Crocker (Crocker et al., 2000). There is no precedent for using two-particle tracking to study biofilms, though the large discrepancies between previously measured bulk properties may be resolved through such a technique. Calculating correlations between particle motions may tease out some of the heterogeneities thought to exist within biofilm systems.
As described above, there is a great advantage to using passive microrheology to study physiological systems as it does not require any external perturbation of the system of interest, save for the addition of the microscale probes. However, there are also limitations to its use, which should be considered when attempting to probe native biological systems. Particle tracking microrheology is performed via video microscopy, which means that any study is temporally limited by the native capture rate of the camera used, providing an upper bound on the frequencies that can be captured to about 1/native frame rate. In addition, errors are introduced by the finite exposure time used to capture particle position, which can alter the calculated MSD and physical properties from their true values (Savin and Doyle, 2007). Camera pixel size and noise limits the ability to resolve particle centers so all MSD values are skewed by a static error, which may be large compared to the MSD at short lag times (Savin and Doyle, 2005). Camera temporal and spatial resolution coupled with the sole use of thermal fluctuations to move the microprobes results in measurable moduli of order 10−5–1 Pa (Waigh, 2005). It is therefore of interest to note other techniques with which to probe biofilms, as bulk measurements indicate that the shear moduli in some species may exceed 1 Pa by several orders of magnitude.
3.3. Active Microrheology Techniques
Active microrheology is defined by the use of external force to move a probe particle through a material, rather than relying solely on fluctuations in thermal energy. Several techniques are commonly used, including atomic force microscopy, optical trapping and magnetic tweezers. These techniques allow us to overcome some of the limitations that may be encountered when attempting to use passive microrheology. In particular, much larger forces can be applied to individual particles using these techniques, which means that the linear rheology of much stiffer materials can be measured (from shear moduli of about 10−3–104 Pa) (Waigh, 2005). However, it is important to note that active forcing of particles through a soft fluid may violate the assumption of equilibrium necessary for linear microrheology, and care should be taken to appropriately calibrate the tools used for such techniques (Squires and Mason, 2010). The ability to deform a material, violating the equilibrium assumption, indicates that these techniques can be used to explore the non-linear microrheology of soft fluids, as has been previously shown (Rich et al., 2011). Another improvement over passive particle tracking is the ability to use laser detection for single particle location, which allows for more precise measurement of location and higher capture frequency (up to 105 Hz), but with fewer measured particles (Waigh, 2005).
Magnetic tweezers are used to apply a known force to a magnetic probe embedded in a material. This force can be calculated as follows, where Χ is the magnetic susceptibility of a particle, V is its volume, represents the magnetic field, x is position, and t is time (Gardel et al., 2005):
| (28) |
This allows for calibration of the field produced at a given distance away from the magnet by observing trajectories of beads in a known fluid and applying Stoke’s Law. By applying a known force to a bead suspended in the biological material of interest and tracking its displacement (xd) over time, it is possible to determine creep using the following relationship (Gardel et al., 2005):
| (29) |
The force applied to a bead depends on the distance of the magnet to the sample as well as on the bead’s size and susceptibility, which allows for a wide experimental dynamic range, dependent upon the choice of probe. Magnetic tweezers have also been used to probe the spatial heterogeneity of creep compliance in biofilms (Figure 4c), and it was found that compliance was higher further away from the surface on which the biofilm was growing (Galy et al., 2012).
Atomic force microscopy (AFM) was developed to measure small forces and has become important in the study of polymers and living systems (Binnig et al., 1986, MacKintosh and Schmidt, 1999). In this technique, a microscale probe is used to scan a surface and its deflections are used to determine topology or interactions between the probe and the surface. The force applied to a surface by AFM depends on the shape of the AFM tip being used, but as before, the technique measures deflection to a known force and can be used to determine rheological data. Multiple AFM and AFM-like techniques have been used to study intact biofilms, most commonly to understand their adhesive and cohesive properties. Non-AFM micromanipulators and microindenters have been used to look at the adhesive strength of biofilms grown in varying physical and chemical conditions (Chen et al., 1998, Chen et al., 2005), as well as to determine storage and loss moduli of biofilm (Cense et al., 2006). In addition, microcantilevers have been used to apply known forces to biofilms to determine how much force is required to pull apart a biofilm, as well as to look at stress/strain curves of biofilms to calculate an elastic modulus (Poppele and Hozalski, 2003, Aggarwal et al., 2010, Aggarwal and Hozalski, 2010). AFM allows for precise force calibration, and a technique has been developed for measuring the force required to disrupt the biofilm in a location-specific manner (Ahimou et al., 2007). A technique has been developed for growing biofilms onto beads attached to microcantilevers (Figure 4d), which allows for AFM to be performed on an intact biofilm (Lau et al., 2009). This technique has been used to measure adhesion and stress/strain relationships of biofilms.
4. Concluding Remarks
The extracellular matrix in biofilms is necessary for microorganisms to establish and maintain a biofilm lifestyle. It is the material properties of the matrix that regulate essential features of the biofilm including adherence, hydration, permeability, and tolerance to mechanical forces. In this review, we have provided the reader with an overview of pioneering work and recent advances toward understanding permeability and mechanics within multiple in vitro biofilm models and biofilms isolated from natural environments. Concomitantly, we have provided a guide to current technologies across multiple disciplines that may serve as useful starting point for researchers seeking to elucidate mechanisms that govern the material properties of biofilms. Microsensors and imaging-based technologies are common tools used dissect concentration gradients and permeability of solutes, gasses, and particles into and within the matrix. Rheology methods across multiple length scales offer an approach to study mechanisms that govern the mechanics of a biofilm.
The studies discussed in this review provide a foundation toward understanding permeability and mechanical properties. Yet, several challenging questions remain. The composition and mechanical properties of the biofilm matrix are sensitive to the environment in which they are formed (Mayer et al., 1999). While many of the studies highlighted in this review were conducted in standard liquid bacterial media, biofilms grown in more physiologically realistic in vitro models or in vivo would provide a more accurate picture of biofilm permeability and mechanical properties. To illustrate this point, we can consider studies of Pseudomonas aeruginosa, an opportunistic pathogen that forms biofilms in the lungs of cystic fibrosis patients, leading to chronic infection. It has recently been observed that the EPS of P. aeruginosa biofilms in the lungs of cystic fibrosis patients incorporates DNA and the structural protein F-actin from dying immune cells (Vu et al., 2009). This likely changes the materials properties of these biofilms, a change that would be missed in studies done on P. aeruginosa biofilms grown in standard laboratory conditions. To address this challenge, researchers have considered growing biofilms in vivo, and then removing them to conduct materials testing ex vivo. In one study, oral biofilms were grown in situ on a dental implant, which was removed to measure biofilm stress relaxation ex situ (Walker et al., 2005). These results were directly compared to in vitro model biofilm systems, yielding insight that could inform the design of improved in vitro models. This study also highlighted subtle differences observed in biofilm material properties that arise due to changes in environmental conditions. While the approach described above represents a step forward, there is still a clear need to study biofilms in native contexts as well as a need for the design of tools to conduct these studies.
A second challenging task that emerges from discussion of measurement techniques is how to integrate structural or permeability data to generate insight regarding biofilm physiology, development, or eradication. A general theoretical framework is needed to connect the different measured properties for a comprehensive understanding of biofilms as a material. While creating such a framework may seem daunting, researchers can take advantage of decades of model development in hydrogels and synthetic polymer gels (Peppas et al., 2000, Peppas, 2004), which may be thought of as ‘synthetic cousins’ of the biofilm matrix. Properties such as gel permeability, dissolution, swelling and self-healing have been mathematically or computationally modeled by several research groups and evaluated for their predictive abilities (for additional resources we direct the reader to (Amsden, 1998, Deen, 1987, Phillips et al., 1989, Phillips, 2000, Zustiak et al., 2010, Balazs, 2007, Wool, 2008). Models of gel permeability and gel dissolution, many of which were created in the context of drug delivery (Hamidi et al., 2008, Bhattarai et al., 2010, Siepmann and Siepmann, 2012), are particularly relevant when considering strategies to dissolve biofilms (i.e. infection eradication) or maintain biofilms (i.e. water purification). Using specific techniques and established theory, researchers could model properties like biofilm permeability and dissolution and compare directly to a well-characterized hydrogel system. Such experiments could facilitate an understanding of similarities and differences between the two materials. Any differences between the well-characterized synthetic gel and a complex biofilm are likely to illuminate interesting physics and biology to investigate further.
In conclusion, to broaden our understanding of the materials properties of biofilms, careful consideration of experimental methodology and sample preparation must be applied. In addition, a general theoretical framework should be constructed to bridge the gap between permeability and mechanics and how this relates to physiological function in the natural environment. Future variations of biofilm experiments may include bottom-up approaches to building biofilm matrices from purified matrix materials or using genetic tools to modulate concentration and structure of matrix components.
Acknowledgments
N.B. is supported by NIH-NIEHS Training Grant in Toxicology 5 T32 ES7020-37. A.B. is supported by Hugh Hampton Young Memorial Fellowship and NIH- NIAID F30 Fellowship 1F30AI110053-01. P.S.D. acknowledges support from Singapore-MIT Alliance for Research and Technology (SMART). This work is funded in part by NIH Grant R01 EB017755.
References
- AGGARWAL S, HOZALSKI RM. Determination of biofilm mechanical properties from tensile tests performed using a micro-cantilever method. Biofouling. 2010;26:479–486. doi: 10.1080/08927011003793080. [DOI] [PubMed] [Google Scholar]
- AGGARWAL S, POPPELE EH, HOZALSKI RM. Development and testing of a novel microcantilever technique for measuring the cohesive strength of intact biofilms. Biotechnology and Bioengineering. 2010;105:924–934. doi: 10.1002/bit.22605. [DOI] [PubMed] [Google Scholar]
- AHIMOU F, SEMMENS MJ, NOVAK PJ, HAUGSTAD G. Biofilm cohesiveness measurement using a novel atomic force microscopy methodology. Appl Environ Microbiol. 2007;73:2897–904. doi: 10.1128/AEM.02388-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AMSDEN B. Solute diffusion within hydrogels. Mechanisms and models. Macromolecules. 1998;31:8382–8395. [Google Scholar]
- AXELROD D, KOPPEL DE, SCHLESSINGER J, ELSON E, WEBB WW. Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophysical Journal. 1976;16:1055–69. doi: 10.1016/S0006-3495(76)85755-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BALAZS AC. Modeling self-healing materials. Materials Today. 2007;10:18–23. [Google Scholar]
- BARNES AM, BALLERING KS, LEIBMAN RS, WELLS CL, DUNNY GM. Enterococcus faecalis produces abundant extracellular structures containing DNA in the absence of cell lysis during early biofilm formation. mBio. 2012;3:e00193–12. doi: 10.1128/mBio.00193-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BARNES HA. The yield stress – a review or ‘pi alpha nu tau alpha rho epsilon iota’ – everything flows? Journal of Non-Newtonian Fluid Mechanics. 1999;81:133–178. [Google Scholar]
- BARNES HA, HUTTON JF, WALTERS K. An Introduction to Rheology. Amsterdam: Elsevier Science Publishers B.V; 1989. [Google Scholar]
- BARTACEK J, VERGELDT FJ, GERKEMA E, JENICEK P, LENS PNL, VAN AS H. Magnetic resonance microscopy of iron transport in methanogenic granules. Journal of Magnetic Resonance. 2009;200:303–312. doi: 10.1016/j.jmr.2009.07.013. [DOI] [PubMed] [Google Scholar]
- BEULING EE, VAN DUSSCHOTEN D, LENS P, VAN DEN HEUVEL JC, VAN AS H, OTTENGRAF SPP. Characterization of the diffusive properties of biofilms using pulsed field gradient-nuclear magnetic resonance. Biotechnology and Bioengineering. 1998;60:283–291. doi: 10.1002/(sici)1097-0290(19981105)60:3<283::aid-bit3>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- BHATTARAI N, GUNN J, ZHANG M. Chitosan-based hydrogels for controlled, localized drug delivery. Advanced drug delivery reviews. 2010;62:83–99. doi: 10.1016/j.addr.2009.07.019. [DOI] [PubMed] [Google Scholar]
- BILLINGS N, MILLAN M, CALDARA M, RUSCONI R, TARASOVA Y, STOCKER R, RIBBECK K. The extracellular matrix Component Psl provides fast-acting antibiotic defense in Pseudomonas aeruginosa biofilms. PLoS pathogens. 2013;9:e1003526. doi: 10.1371/journal.ppat.1003526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BINNIG G, QUATE CF, GERBER C. Atomic Force Microscope. Physical Review Letters. 1986;56:930–933. doi: 10.1103/PhysRevLett.56.930. [DOI] [PubMed] [Google Scholar]
- BIRD RB, ARMSTRONG RC, HASSAGER O. Dynamics of Polymeric Liquids: Volume 1 Fluid Mechanics. New York: John Wiley and Sons; 1987. [Google Scholar]
- BIRMINGHAM JJ, HUGHES NP, TRELOAR R. Diffusion and binding measurements within oral biofilms using fluorescence photobleaching recovery methods. (Series B, Biological sciences).Philosophical transactions of the Royal Society of London. 1995;350:325–43. doi: 10.1098/rstb.1995.0168. [DOI] [PubMed] [Google Scholar]
- BÖL M, EHRET AE, BOLEA ALBERO A, HELLRIEGEL J, KRULL R. Recent advances in mechanical characterisation of biofilm and their significance for material modelling. Critical Reviews in Biotechnology. 2013;33:145–171. doi: 10.3109/07388551.2012.679250. [DOI] [PubMed] [Google Scholar]
- BOLES BR, HORSWILL AR. Swimming cells promote a dynamic environment within biofilms. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:12848–9. doi: 10.1073/pnas.1210297109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRAGA J, DESTERRO JM, CARMO-FONSECA M. Intracellular macromolecular mobility measured by fluorescence recovery after photobleaching with confocal laser scanning microscopes. Molecular biology of the cell. 2004;15:4749–60. doi: 10.1091/mbc.E04-06-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRIANDET R, LACROIX-GUEU P, RENAULT M, LECART S, MEYLHEUC T, BIDNENKO E, STEENKESTE K, BELLON-FONTAINE MN, FONTAINE-AUPART MP. Fluorescence correlation spectroscopy to study diffusion and reaction of bacteriophages inside biofilms. Applied and environmental microbiology. 2008;74:2135–43. doi: 10.1128/AEM.02304-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRYERS JD, DRUMMOND F. Local macromolecule diffusion coefficients in structurally non-uniform bacterial biofilms using fluorescence recovery after photobleaching (FRAP) Biotechnology and Bioengineering. 1998;60:462–73. [PubMed] [Google Scholar]
- BUNGAY HR, WHALEN WJ, SANDERS WM. MICROPROBE TECHNIQUES FOR DETERMINING DIFFUSIVITIES AND RESPIRATION RATES IN MICROBIAL SLIME SYSTEMS. Biotechnology and Bioengineering. 1969;11:765–772. [Google Scholar]
- CAO B, MAJORS PD, AHMED B, RENSLOW RS, SILVIA CP, SHI L, KJELLEBERG S, FREDRICKSON JK, BEYENAL H. Biofilm shows spatially stratified metabolic responses to contaminant exposure. Environmental Microbiology. 2012;14:2901–2910. doi: 10.1111/j.1462-2920.2012.02850.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CARRERO G, MCDONALD D, CRAWFORD E, DE VRIES G, HENDZEL MJ. Using FRAP and mathematical modeling to determine the in vivo kinetics of nuclear proteins. Methods. 2003;29:14–28. doi: 10.1016/s1046-2023(02)00288-8. [DOI] [PubMed] [Google Scholar]
- CENSE AW, PEETERS EAG, GOTTENBOS B, BAAIJENS FPT, NUIJS AM, VAN DONGEN MEH. Mechanical properties and failure of Streptococcus mutans biofilms, studied using a microindentation device. Journal of Microbiological Methods. 2006;67:463–472. doi: 10.1016/j.mimet.2006.04.023. [DOI] [PubMed] [Google Scholar]
- CHEN MJ, ZHANG Z, BOTT TR. Direct measurement of the adhesive strength of biofilms in pipes by micromanipulation. Biotechnology Techniques. 1998;12:875–880. [Google Scholar]
- CHEN MJ, ZHANG Z, BOTT TR. Effects of operating conditions on the adhesive strength of Pseudomonas fluorescens biofilms in tubes. Colloids and Surfaces B: Biointerfaces. 2005;43:61–71. doi: 10.1016/j.colsurfb.2005.04.004. [DOI] [PubMed] [Google Scholar]
- CHEONG FC, DUARTE S, LEE SH, GRIER DG. Holographic microrheology of polysaccharides from Streptococcus mutans biofilms. Rheologica Acta. 2009;48:109–115. [Google Scholar]
- COLE NB, SMITH CL, SCIAKY N, TERASAKI M, EDIDIN M, LIPPINCOTT-SCHWARTZ J. Diffusional mobility of Golgi proteins in membranes of living cells. Science. 1996;273:797–801. doi: 10.1126/science.273.5276.797. [DOI] [PubMed] [Google Scholar]
- CROCKER JC, GRIER DG. Methods of digital video microscopy for colloidal studies. Journal of Colloid and Interface Science. 1996;179:298–310. [Google Scholar]
- CROCKER JC, VALENTINE MT, WEEKS ER, GISLER T, KAPLAN PD, YODH AG, WEITZ DA. Two-point microrheology of inhomogeneous soft materials. Physical Review Letters. 2000;85:888–891. doi: 10.1103/PhysRevLett.85.888. [DOI] [PubMed] [Google Scholar]
- DADDI OUBEKKA S, BRIANDET R, FONTAINE-AUPART MP, STEENKESTE K. Correlative time-resolved fluorescence microscopy to assess antibiotic diffusion-reaction in biofilms. Antimicrobial agents and chemotherapy. 2012;56:3349–58. doi: 10.1128/AAC.00216-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DE BEER D, STOODLEY P. Relation between the structure of an aerobic biofilm and transport phenomena? Water Science and Technology. 1995;32:11–18. [Google Scholar]
- DE BEER D, STOODLEY P. Microbial Biofilms. In: E R, editor. The Prokaryotes: Applied Bacteriology and Biotechnology. 4. Berlin: Springer Reference; 2013. [Google Scholar]
- DE BEER D, STOODLEY P, LEWANDOWSKI Z. Liquid Flow in Heterogeneous Biofilms. Biotechnology and Bioengineering. 1994a;44:636–641. doi: 10.1002/bit.260440510. [DOI] [PubMed] [Google Scholar]
- DE BEER D, STOODLEY P, LEWANDOWSKI Z. Measurement of local diffusion coefficients in biofilms by microinjection and confocal microscopy. Biotechnology and Bioengineering. 1997;53:151–8. doi: 10.1002/(SICI)1097-0290(19970120)53:2<151::AID-BIT4>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- DE BEER D, STOODLEY P, ROE F, LEWANDOWSKI Z. Effects of biofilm structures on oxygen distribution and mass transport. Biotechnology and Bioengineering. 1994b;43:1131–1138. doi: 10.1002/bit.260431118. [DOI] [PubMed] [Google Scholar]
- DE BEER D, VANDENHEUVEL JC, OTTENGRAF SPP. Microelectrode Measurements of the Activity Distribution in Nitrifying Bacterial Aggregates. Applied and Environmental Microbiology. 1993;59:573–579. doi: 10.1128/aem.59.2.573-579.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DEBEER D, SCHRAMM A, SANTEGOEDS CM, KUHL M. A nitrite microsensor for profiling environmental biofilms. Applied and Environmental Microbiology. 1997;63:973–977. doi: 10.1128/aem.63.3.973-977.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DEEN WM. Hindered Transport of Large Molecules in Liquid-Filled Pores. AicheJournal. 1987;33:1409–1425. [Google Scholar]
- DUNSMORE BC, JACOBSEN A, HALL-STOODLEY L, BASS CJ, LAPPIN-SCOTT HM, STOODLEY P. The influence of fluid shear on the structure and material properties of sulphate-reducing bacterial biofilms. Journal of Industrial Microbiology & Biotechnology. 2002;29:347–353. doi: 10.1038/sj.jim.7000302. [DOI] [PubMed] [Google Scholar]
- EDIDIN M, ZAGYANSKY Y, LARDNER TJ. Measurement of membrane protein lateral diffusion in single cells. Science. 1976;191:466–8. doi: 10.1126/science.1246629. [DOI] [PubMed] [Google Scholar]
- EWOLDT RH, WINTER P, MAXEY J, MCKINLEY GH. Large amplitude oscillatory shear of pseudoplastic and elastoviscoplastic materials. Rheologica Acta. 2010;49:191–212. [Google Scholar]
- FLEMMING HC, WINGENDER J. The biofilm matrix. Nature reviews. Microbiology. 2010;8:623–33. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- FORIER K, MESSIAEN AS, RAEMDONCK K, DESCHOUT H, REJMAN J, DE BAETS F, NELIS H, DE SMEDT SC, DEMEESTER J, COENYE T, BRAECKMANS K. Transport of nanoparticles in cystic fibrosis sputum and bacterial biofilms by single-particle tracking microscopy. Nanomedicine (Lond) 2012 doi: 10.2217/nnm.12.129. [DOI] [PubMed] [Google Scholar]
- GALY O, LATOUR-LAMBERT P, ZRELLI K, GHIGO JM, BELOIN C, HENRY N. Mapping of bacterial biofilm local mechanics by magnetic microparticle actuation. Biophysical Journal. 2012;103:1400–8. doi: 10.1016/j.bpj.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GARDEL ML, VALENTINE M, WEITZ DA. Microrheology. In: BREUER K, editor. Microscale Diagnostic Techniques. Springer Verlag; 2005. [Google Scholar]
- GIRARD LP, CERI H, GIBB AP, OLSON M, SEPANDJ F. MIC versus MBEC to determine the antibiotic sensitivity of Staphylococcus aureus in peritoneal dialysis peritonitis. Peritoneal dialysis international: journal of the International Society for Peritoneal Dialysis. 2010;30:652–6. doi: 10.3747/pdi.2010.00010. [DOI] [PubMed] [Google Scholar]
- GUIOT E, GEORGES P, BRUN A, FONTAINE-AUPART MP, BELLON-FONTAINE MN, BRIANDET R. Heterogeneity of diffusion inside microbial biofilms determined by fluorescence correlation spectroscopy under two-photon excitation. Photochemistry and Photobiology. 2002;75:570–578. doi: 10.1562/0031-8655(2002)075<0570:hodimb>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- HAMIDI M, AZADI A, RAFIEI P. Hydrogel nanoparticles in drug delivery. Advanced drug delivery reviews. 2008;60:1638–49. doi: 10.1016/j.addr.2008.08.002. [DOI] [PubMed] [Google Scholar]
- HARDY EH. NMR Methods for the Investigation of Structure and Transport. New York, NY: Springer Publishing; 2012. [Google Scholar]
- HOGAN D, KOLTER R. Why are bacteria refractory to antimicrobials? Current opinion in microbiology. 2002;5:472–7. doi: 10.1016/s1369-5274(02)00357-0. [DOI] [PubMed] [Google Scholar]
- HOHNE DN, YOUNGER JG, SOLOMON MJ. Flexible Microfluidic Device for Mechanical Property Characterization of SoftViscoelastic Solids Such as Bacterial Biofilms. Langmuir. 2009;25:7743–7751. doi: 10.1021/la803413x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOIBY N, BJARNSHOLT T, GIVSKOV M, MOLIN S, CIOFU O. Antibiotic resistance of bacterial biofilms. International journal of antimicrobial agents. 2010;35:322–32. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- HOUARI A, PICARD J, HABAROU H, GALAS L, VAUDRY H, HEIM V, DI MARTINO P. Rheology of biofilms formed at the surface of NF membranes in a drinking water production unit. Biofouling. 2008;24:235–240. doi: 10.1080/08927010802023764. [DOI] [PubMed] [Google Scholar]
- HOURY A, GOHAR M, DESCHAMPS J, TISCHENKO E, AYMERICH S, GRUSS A, BRIANDET R. Bacterial swimmers that infiltrate and take over the biofilm matrix. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:13088–93. doi: 10.1073/pnas.1200791109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HYUN K, KIM SH, AHN KH, LEE SJ. Large amplitude oscillatory shear as a way to classify the complex fluids. Journal of Non-Newtonian Fluid Mechanics. 2002;107:51–65. [Google Scholar]
- JEFFERSON KK, GOLDMANN DA, PIER GB. Use of confocal microscopy to analyze the rate of vancomycin penetration through Staphylococcus aureus biofilms. Antimicrobial agents and chemotherapy. 2005;49:2467–73. doi: 10.1128/AAC.49.6.2467-2473.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JONES WL, SUTTON MP, MCKITTRICK L, STEWART PS. Chemical and antimicrobial treatments change the viscoelastic properties of bacterial biofilms. Biofouling. 2011;27:207–215. doi: 10.1080/08927014.2011.554977. [DOI] [PubMed] [Google Scholar]
- JORGENSEN BB, REVSBECH NP. Diffusive Boundary-Layers and the Oxygen-Uptake of Sediments and Detritus. Limnology and Oceanography. 1985;30:111–122. doi: 10.4319/lo.1990.35.6.1343. [DOI] [PubMed] [Google Scholar]
- KEELER J. Understanding NMR Spectroscopy. Hoboken, NJ: John Wiley and Sons, Inc; 2011. [Google Scholar]
- KLAPPER I, RUPP CJ, CARGO R, PURVEDORJ B, STOODLEY P. Viscoelastic fluid description of bacterial biofilm material properties. Biotechnology and Bioengineering. 2002;80:289–296. doi: 10.1002/bit.10376. [DOI] [PubMed] [Google Scholar]
- KORSTGENS V, FLEMMING HC, WINGENDER J, BORCHARD W. Influence of calcium ions on the mechanical properties of a model biofilm of mucoid Pseudomonas aeruginosa. Water Science and Technology. 2001a;43:49–57. [PubMed] [Google Scholar]
- KORSTGENS V, FLEMMING HC, WINGENDER J, BORCHARD W. Uniaxial compression measurement device for investigation of the mechanical stability ofbiofilms. Journal of Microbiological Methods. 2001b;46:9–17. doi: 10.1016/s0167-7012(01)00248-2. [DOI] [PubMed] [Google Scholar]
- LACROIX-GUEU P, BRIANDET R, LEVEQUE-FORT S, BELLON-FONTAINE MN, FONTAINE-AUPART MP. In situ measurements of viral particles diffusion inside mucoid biofilms. Comptes rendus biologies. 2005;328:1065–72. doi: 10.1016/j.crvi.2005.09.010. [DOI] [PubMed] [Google Scholar]
- LAU PCY, DUTCHER JR, BEVERIDGE TJ, LAM JS. Absolute Quantitation of Bacterial Biofilm Adhesion and Viscoelasticity by Microbead Force Spectroscopy. Biophysical Journal. 2009;96:2935–2948. doi: 10.1016/j.bpj.2008.12.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAWRENCE JR, SWERHONE GD, KUHLICKE U, NEU TR. In situ evidence for microdomains in the polymer matrix of bacterial microcolonies. Canadian journal of microbiology. 2007;53:450–8. doi: 10.1139/W06-146. [DOI] [PubMed] [Google Scholar]
- LAWRENCE JR, WOLFAARDT GM, KORBER DR. Determination of diffusion coefficients in biofilms by confocal laser microscopy. Applied and environmental microbiology. 1994;60:1166–73. doi: 10.1128/aem.60.4.1166-1173.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEE JH, SEO Y, LIM TS, BISHOP PL, PAPAUTSKY I. MEMS needle-type sensor array for in situ measurements of dissolved oxygen and redox potential. Environmental Science & Technology. 2007;41:7857–7863. doi: 10.1021/es070969o. [DOI] [PubMed] [Google Scholar]
- LEWANDOWSKI Z, ALTOBELLI SA, FUKUSHIMA E. Nmr and Microelectrode Studies of Hydrodynamics and Kinetics in Biofilms. Biotechnology Progress. 1993;9:40–45. [Google Scholar]
- LEWIS K. Riddle of biofilm resistance. Antimicrobial agents and chemotherapy. 2001;45:999–1007. doi: 10.1128/AAC.45.4.999-1007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI J, BISHOP PL. In situ identification of azo dye inhibition effects on nitrifying biofilms using microelectrodes. Water Science and Technology. 2002;46:207–214. [PubMed] [Google Scholar]
- LI J, BISHOP PL. Monitoring the influence of toxic compounds on microbial denitrifying biofilm processes. Water Science and Technology. 2003;47:211–216. [PubMed] [Google Scholar]
- LI J, BISHOP PL. Time course observations of nitrifying biofilm development using microelectrodes. Journal of Environmental Engineering and Science. 2004;3 [Online] Available: http://www.icevirtuallibrary.com/content/article/10.1139/s04-027. [Google Scholar]
- LIEBMAN PA, ENTINE G. Lateral diffusion of visual pigment in photorecptor disk membranes. Science. 1974;185:457–9. doi: 10.1126/science.185.4149.457. [DOI] [PubMed] [Google Scholar]
- LIELEG O, CALDARA M, BAUMGARTEL R, RIBBECK K. Mechanical robustness of Pseudomonas aeruginosa biofilms. Soft Matter. 2011;7:3307–3314. doi: 10.1039/c0sm01467b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIU J, GARDEL ML, KROY K, FREY E, HOFFMAN BD, CROCKER JC, BAUSCH AR, WEITZ DA. Microrheology probes length scale dependent rheology. Physical Review Letters. 2006;96 doi: 10.1103/PhysRevLett.96.118104. [DOI] [PubMed] [Google Scholar]
- MA L, LU H, SPRINKLE A, PARSEK MR, WOZNIAK DJ. Pseudomonas aeruginosa Psl is a galactose- and mannose-rich exopolysaccharide. Journal of bacteriology. 2007;189:8353–6. doi: 10.1128/JB.00620-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MACKINTOSH FC, SCHMIDT CF. Microrheology. Current Opinion in Colloid & Interface Science. 1999;4:300–307. [Google Scholar]
- MACOSKO CW. Rheology: Principles, Measurements, and Applications. New York: VCH Publishers, Inc; 1994. [Google Scholar]
- MAJORS PD, MCLEAN JS, FREDRICKSON JK, WIND RA. NMR methods for in-situ biofilm metabolism studies: spatial and temporal resolved measurements. Water Science and Technology. 2005a;52:7–12. doi: 10.1016/j.mimet.2005.04.017. [DOI] [PubMed] [Google Scholar]
- MAJORS PD, MCLEAN JS, PINCHUK GE, FREDRICKSON JK, GORBY YA, MINARD KR, WIND RA. NMR methods for in situ biofilm metabolism studies. Journal of Microbiological Methods. 2005b;62:337–344. doi: 10.1016/j.mimet.2005.04.017. [DOI] [PubMed] [Google Scholar]
- MANZ B, VOLKE F, GOLL D, HORN H. Measuring local flow velocities and biofilm structure in biofilm systems with magnetic resonance imaging (MRI) Biotechnology and Bioengineering. 2003;84:424–432. doi: 10.1002/bit.10782. [DOI] [PubMed] [Google Scholar]
- MASON TG. Estimating the viscoelastic moduli of complex fluids using the generalized Stokes-Einstein equation. Rheologica Acta. 2000;39:371–378. [Google Scholar]
- MASON TG, GANESAN K, VAN ZANTEN JH, WIRTZ D, KUO SC. Particle Tracking Microrheology of Complex Fluids. Physical Review Letters. 1997;79:3282–3285. [Google Scholar]
- MASON TG, WEITZ DA. Optical Measurements of Frequency-Dependent Linear Viscoelastic Moduli of Complex Fluids. Physical Review Letters. 1995;74:1250–1253. doi: 10.1103/PhysRevLett.74.1250. [DOI] [PubMed] [Google Scholar]
- MAYER C, MORITZ R, KIRSCHNER C, BORCHARD W, MAIBAUM R, WINGENDER J, FLEMMING HC. The role of intermolecular interactions: studies on model systems for bacterial biofilms. International journal of biological macromolecules. 1999;26:3–16. doi: 10.1016/s0141-8130(99)00057-4. [DOI] [PubMed] [Google Scholar]
- MCLEAN JS, MAJORS PD, REARDON CL, BILSKIS CL, REED SB, ROMINE MF, FREDRICKSON JK. Investigations of structure and metabolism within Shewanella oneidensis MR-1 biofilms. Journal of Microbiological Methods. 2008a;74:47–56. doi: 10.1016/j.mimet.2008.02.015. [DOI] [PubMed] [Google Scholar]
- MCLEAN JS, ONA ON, MAJORS PD. Correlated biofilm imaging, transport and metabolism measurements via combined nuclear magnetic resonance and confocal microscopy. Isme Journal. 2008b;2:121–131. doi: 10.1038/ismej.2007.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MORRISON FA. Understanding Rheology. New York: Oxford University Press; 2001. [Google Scholar]
- NGUYEN QD, BOGER DV. Measuring the Flow Properties of Yield Stress Fluids. Annual Review of Fluid Mechanics. 1992;24:47–88. [Google Scholar]
- OHASHI A, HARADA H. Adhesion Strength of Biofilm Developed in an Attached-Growth Reactor. Water Science and Technology. 1994;29:281–288. [Google Scholar]
- OUBEKKA SD, BRIANDET R, FONTAINE-AUPART MP, STEENKESTE K. Correlative Time-Resolved Fluorescence Microscopy To Assess Antibiotic Diffusion-Reaction in Biofilms. Antimicrobial agents and chemotherapy. 2012;56:3349–3358. doi: 10.1128/AAC.00216-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OUBEKKA SD, BRIANDET R, WAHARTE F, FONTAINE-AUPART MP, STEENKESTE K. Image-based Fluorescence Recovery After Photobleaching (FRAP) to dissect vancomycin diffusion-reaction processes in Staphylococcus aureus biofilms. Clinical and Biomedical Spectroscopy and Imaging Ii. 2011;8087 [Google Scholar]
- PAVLOVSKY L, YOUNGER JG, SOLOMON MJ. In situ rheology of Staphylococcus epidermidis bacterial biofilms. Soft Matter. 2013 doi: 10.1039/C2SM27005F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PEPPAS NA. Biomaterials Science: An Introduction to Materials in Medicine. New York: Academic Press; 2004. Hydrogels; pp. 100–107. [Google Scholar]
- PEPPAS NA, HUANG Y, TORRES-LUGO M, WARD JH, ZHANG J. Physiochemical Foundations and Structural Design of Hydrogels in Medicine and Biology. Annual Reviews of Biomedical Engineering. 2000;2:9–29. doi: 10.1146/annurev.bioeng.2.1.9. [DOI] [PubMed] [Google Scholar]
- PEULEN TO, WILKINSON KJ. Diffusion of nanoparticles in a biofilm. Environmental science & technology. 2011;45:3367–73. doi: 10.1021/es103450g. [DOI] [PubMed] [Google Scholar]
- PHILLIPS RJ. A hydrodynamic model for hindered diffusion of proteins and micelles in hydrogels. Biophysical Journal. 2000;79:3350–3. doi: 10.1016/S0006-3495(00)76566-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PHILLIPS RJ, DEEN WM, BRADY JF. Hindered Transport of Spherical Macromolecules in Fibrous Membranes and Gels. AicheJournal. 1989;35:1761–1769. [Google Scholar]
- PHOENIX VR, HOLMES WM, RAMANAN B. Magnetic resonance imaging (MRI) of heavy-metal transport and fate in an artificial biofilm. Mineralogical Magazine. 2008;72:483–486. [Google Scholar]
- PIBALPAKDEE P, WONGRATANACHEEWIN S, TAWEECHAISUPAPONG S, NIUMSUP PR. Diffusion and activity of antibiotics against Burkholderia pseudomallei biofilms. International journal of antimicrobial agents. 2012;39:356–9. doi: 10.1016/j.ijantimicag.2011.12.010. [DOI] [PubMed] [Google Scholar]
- POO M, CONE RA. Lateral diffusion of rhodopsin in the photoreceptor membrane. Nature. 1974;247:438–41. doi: 10.1038/247438a0. [DOI] [PubMed] [Google Scholar]
- POPPELE EH, HOZALSKI RM. Micro-cantilever method for measuring the tensile strength of biofilms and microbial flocs. Journal of Microbiological Methods. 2003;55:607–615. doi: 10.1016/s0167-7012(03)00198-2. [DOI] [PubMed] [Google Scholar]
- RAMANAN B, HOLMES WM, SLOAN WT, PHOENIX VR. Magnetic Resonance Imaging of Mass Transport and Structure Inside a Phototrophic Biofilm. Current Microbiology. 2013;66:456–461. doi: 10.1007/s00284-012-0292-3. [DOI] [PubMed] [Google Scholar]
- RANI SA, PITTS B, STEWART PS. Rapid diffusion of fluorescent tracers into Staphylococcus epidermidis biofilms visualized by time lapse microscopy. Antimicrobial agents and chemotherapy. 2005;49:728–32. doi: 10.1128/AAC.49.2.728-732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RASMUSSEN K, LEWANDOWSKI Z. Microelectrode measurements of local mass transport rates in heterogeneous biofilms. Biotechnology and Bioengineering. 1998;59:302–309. doi: 10.1002/(sici)1097-0290(19980805)59:3<302::aid-bit6>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- REVSBECH NP. Diffusion characteristics of microbial communities determined by use of oxygen microsensors. Journal of Microbiological Methods. 1989;9:111–122. [Google Scholar]
- RICH JP, LAMMERDING J, MCKINLEY GH, DOYLE PS. Nonlinear microrheology of an aging, yield stress fluid using magnetic tweezers. Soft Matter. 2011;7:9933–9943. [Google Scholar]
- ROGERS SS, VAN DER WALLE C, WAIGH TA. Microrheology of Bacterial Biofilms In Vitro: Staphylococcus aureus and Pseudomonas aeruginosa. Langmuir. 2008;24:13549–13555. doi: 10.1021/la802442d. [DOI] [PubMed] [Google Scholar]
- RUBINSTEIN M, COLBY RH. Polymer Physics. Oxford: Oxford University Press; 2003. [Google Scholar]
- SANDOE JA, WYSOME J, WEST AP, HERITAGE J, WILCOX MH. Measurement of ampicillin, vancomycin, linezolid and gentamicin activity against enterococcal biofilms. The Journal of antimicrobial chemotherapy. 2006;57:767–70. doi: 10.1093/jac/dkl013. [DOI] [PubMed] [Google Scholar]
- SAVIN T, DOYLE PS. Static and dynamic errors in particle tracking microrheology. Biophysical Journal. 2005;88:623–638. doi: 10.1529/biophysj.104.042457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAVIN T, DOYLE PS. Statistical and sampling issues when using multiple particle tracking. Physical Review E. 2007;76 doi: 10.1103/PhysRevE.76.021501. [DOI] [PubMed] [Google Scholar]
- SCHMOLZE DB, STANDLEY C, FOGARTY KE, FISCHER AH. Advances in microscopy techniques. Archives of pathology & laboratory medicine. 2011;135:255–63. doi: 10.5858/135.2.255. [DOI] [PubMed] [Google Scholar]
- SCHRAMM A, LARSEN LH, REVSBECH NP, RAMSING NB, AMANN R, SCHLEIFER KH. Structure and function of a nitrifying biofilm as determined by in situ hybridization and the use of microelectrodes. Applied and Environmental Microbiology. 1996;62:4641–4647. doi: 10.1128/aem.62.12.4641-4647.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SEIFFERT S, OPPERMANN W. Systematic evaluation of FRAP experiments performed in a confocal laser scanning microscope. Journal of microscopy. 2005;220:20–30. doi: 10.1111/j.1365-2818.2005.01512.x. [DOI] [PubMed] [Google Scholar]
- SHAW T, WINSTON M, RUPP CJ, KLAPPER I, STOODLEY P. Commonality of elastic relaxation times in biofilms. Physical Review Letters. 2004;93 doi: 10.1103/PhysRevLett.93.098102. [DOI] [PubMed] [Google Scholar]
- SHIGETA M, TANAKA G, KOMATSUZAWA H, SUGAI M, SUGINAKA H, USUI T. Permeation of antimicrobial agents through Pseudomonas aeruginosa biofilms: a simple method. Chemotherapy. 1997;43:340–5. doi: 10.1159/000239587. [DOI] [PubMed] [Google Scholar]
- SIEPMANN J, SIEPMANN F. Modeling of diffusion controlled drug delivery. Journal of controlled release: official journal of the Controlled Release Society. 2012;161:351–62. doi: 10.1016/j.jconrel.2011.10.006. [DOI] [PubMed] [Google Scholar]
- SINGH R, RAY P, DAS A, SHARMA M. Penetration of antibiotics through Staphylococcus aureus and Staphylococcus epidermidis biofilms. The Journal of antimicrobial chemotherapy. 2010;65:1955–8. doi: 10.1093/jac/dkq257. [DOI] [PubMed] [Google Scholar]
- SQUIRES TM, MASON TG. Fluid Mechanics of Microrheology. Annual Review of Fluid Mechanics. 2010;42:413–438. [Google Scholar]
- STAAL M, BORISOV SM, RICKELT LF, KLIMANT I, KUHL M. Ultrabright planar optodes for luminescence life-time based microscopic imaging of O-2 dynamics in biofilms. Journal of Microbiological Methods. 2011;85:67–74. doi: 10.1016/j.mimet.2011.01.021. [DOI] [PubMed] [Google Scholar]
- STEWART PS. Theoretical aspects of antibiotic diffusion into microbial biofilms. Antimicrobial agents and chemotherapy. 1996;40:2517–22. doi: 10.1128/aac.40.11.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STEWART PS. A review of experimental measurements of effective diffusive permeabilities and effective diffusion coefficients in biofilms. Biotechnology and Bioengineering. 1998;59:261–272. doi: 10.1002/(sici)1097-0290(19980805)59:3<261::aid-bit1>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- STEWART PS. Mechanisms of antibiotic resistance in bacterial biofilms. International journal of medical microbiology: IJMM. 2002;292:107–13. doi: 10.1078/1438-4221-00196. [DOI] [PubMed] [Google Scholar]
- STEWART PS. Diffusion in biofilms. Journal of bacteriology. 2003;185:1485–91. doi: 10.1128/JB.185.5.1485-1491.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STEWART PS. Mini-review: convection around biofilms. Biofouling. 2012;28:187–98. doi: 10.1080/08927014.2012.662641. [DOI] [PubMed] [Google Scholar]
- STEWART PS, COSTERTON JW. Antibiotic resistance of bacteria in biofilms. Lancet. 2001;358:135–8. doi: 10.1016/s0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- STEWART PS, FRANKLIN MJ. Physiological heterogeneity in biofilms. Nature reviews. Microbiology. 2008;6:199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- STONE G, WOOD P, DIXON L, KEYHAN M, MATIN A. Tetracycline rapidly reaches all the constituent cells of uropathogenic Escherichia coli biofilms. Antimicrobial agents and chemotherapy. 2002;46:2458–61. doi: 10.1128/AAC.46.8.2458-2461.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STOODLEY P, CARGO R, RUPP CJ, WILSON S, KLAPPER I. Biofilm material properties as related to shear-induced deformation and detachment phenomena. Journal of Industrial Microbiology & Biotechnology. 2002;29:361–367. doi: 10.1038/sj.jim.7000282. [DOI] [PubMed] [Google Scholar]
- STOODLEY P, DEBEER D, LEWANDOWSKI Z. Liquid Flow in Biofilm Systems. Applied and Environmental Microbiology. 1994;60:2711–2716. doi: 10.1128/aem.60.8.2711-2716.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STOODLEY P, LEWANDOWSKI Z, BOYLE JD, LAPPIN-SCOTT HM. Structural deformation of bacterial biofilms caused by short-term fluctuations in fluid shear: An in situ investigation of biofilm rheology. Biotechnology and Bioengineering. 1999;65:83–92. [PubMed] [Google Scholar]
- STOODLEY P, YANG SN, LAPPINSCOTT H, LEWANDOWSKI Z. Relationship between mass transfer coefficient and liquid flow velocity in heterogenous biofilms using microelectrodes and confocal microscopy. Biotechnology and Bioengineering. 1997;56:681–688. doi: 10.1002/(SICI)1097-0290(19971220)56:6<681::AID-BIT11>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- SUTHERLAND I. Biofilm exopolysaccharides: a strong and sticky framework. Microbiology. 2001a;147:3–9. doi: 10.1099/00221287-147-1-3. [DOI] [PubMed] [Google Scholar]
- SUTHERLAND IW. The biofilm matrix–an immobilized but dynamic microbial environment. Trends in microbiology. 2001b;9:222–7. doi: 10.1016/s0966-842x(01)02012-1. [DOI] [PubMed] [Google Scholar]
- SVENSSON E, HANBERGER H, NILSSON M, NILSSON LE. Factors affecting development of rifampicin resistance in biofilm-producing Staphylococcus epidermidis. The Journal of antimicrobial chemotherapy. 1997;39:817–20. doi: 10.1093/jac/39.6.817. [DOI] [PubMed] [Google Scholar]
- THURNHEER T, GMUR R, SHAPIRO S, GUGGENHEIM B. Mass transport of macromolecules within an in vitro model of supragingival plaque. Applied and Environmental Microbiology. 2003;69:1702–9. doi: 10.1128/AEM.69.3.1702-1709.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TOWLER BW, RUPP CJ, CUNNINGHAM AB, STOODLEY P. Viscoelastic properties of a mixed culture biofilm from rheometer creep analysis. Biofouling. 2003;19:279–285. doi: 10.1080/0892701031000152470. [DOI] [PubMed] [Google Scholar]
- TSENG BS, ZHANG W, HARRISON JJ, QUACH TP, SONG JL, PENTERMAN J, SINGH PK, CHOPP DL, PACKMAN AI, PARSEK MR. The extracellular matrix protects Pseudomonas aeruginosa biofilms by limiting the penetration of tobramycin. Environmental Microbiology. 2013;15:2865–78. doi: 10.1111/1462-2920.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VOGT M, FLEMMING HC, VEEMAN WS. Diffusion in Pseudomonas aeruginosa biofilms: a pulsed field gradient NMR study. Journal of Biotechnology. 2000;77:137–146. doi: 10.1016/s0168-1656(99)00213-8. [DOI] [PubMed] [Google Scholar]
- VU B, CHEN M, CRAWFORD RJ, IVANOVA EP. Bacterial extracellular polysaccharides involved in biofilm formation. Molecules. 2009;14:2535–54. doi: 10.3390/molecules14072535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WAHARTE F, STEENKESTE K, BRIANDET R, FONTAINE-AUPART MP. Diffusion measurements inside biofilms by image-based fluorescence recovery after photobleaching (FRAP) analysis with a commercial confocal laser scanning microscope. Applied and environmental microbiology. 2010;76:5860–9. doi: 10.1128/AEM.00754-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WAIGH TA. Microrheology of complex fluids. Reports on Progress in Physics. 2005;68:685–742. doi: 10.1088/0034-4885/79/7/074601. [DOI] [PubMed] [Google Scholar]
- WALKER TS, TOMLIN KL, WORTHEN GS, POCH KR, LIEBER JG, SAAVEDRA MT, FESSLER MB, MALCOLM KC, VASIL ML, NICK JA. Enhanced Pseudomonas aeruginosa biofilm development mediated by human neutrophils. Infection and immunity. 2005;73:3693–701. doi: 10.1128/IAI.73.6.3693-3701.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WALTERS MC, 3RD, ROE F, BUGNICOURT A, FRANKLIN MJ, STEWART PS. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrobial agents and chemotherapy. 2003;47:317–23. doi: 10.1128/AAC.47.1.317-323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHALEN WJ, BUNGAY HR, SANDERS WM. MICROELECTRODE DETERMINATION OF OXYGEN PROFILES IN MICROBIAL SLIME SYSTEMS. Environmental Science & Technology. 1969;3:1297–8. [Google Scholar]
- WHITE J, STELZER E. Photobleaching GFP reveals protein dynamics inside live cells. Trends in cell biology. 1999;9:61–5. doi: 10.1016/s0962-8924(98)01433-0. [DOI] [PubMed] [Google Scholar]
- WILKING JN, ZABURDAEV V, DE VOLDER M, LOSICK R, BRENNER MP, WEITZ DA. Liquid transport facilitated by channels in Bacillus subtilis biofilms. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:848–52. doi: 10.1073/pnas.1216376110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WIRTZ D. Particle-Tracking Microrheology of Living Cells: Principles and Applications. Annual Review of Biophysics. 2009;38:301–326. doi: 10.1146/annurev.biophys.050708.133724. [DOI] [PubMed] [Google Scholar]
- WOOL RP. Self-healing materials: a review. Soft Matter. 2008;4:400–418. doi: 10.1039/b711716g. [DOI] [PubMed] [Google Scholar]
- WRANGSTADH M, SZEWZYK U, OSTLING J, KJELLEBERG S. Starvation-specific formation of a peripheral exopolysaccharide by a marine Pseudomonas sp., strain S9. Applied and Environmental Microbiology. 1990;56:2065–72. doi: 10.1128/aem.56.7.2065-2072.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YANG S, LEWANDOWSKI Z. Measurement of local mass transfer coefficient in biofilms. Biotechnology and Bioengineering. 1995;48:737–44. doi: 10.1002/bit.260480623. [DOI] [PubMed] [Google Scholar]
- YU T, BISHOP PL. Stratification and oxidation-reduction potential change in an aerobic and sulfate-reducing biofilm studied using microelectrodes. Water Environ Res. 2001;73:368–73. doi: 10.2175/106143001x139399. [DOI] [PubMed] [Google Scholar]
- YU T, DE LA ROSA C, LU R. Microsensor measurement of oxygen concentration in biofilms: from one dimension to three dimensions. Water Science and Technology. 2004;49:353–8. [PubMed] [Google Scholar]
- YURT N, BEYENAL H, SEARS J, LEWANDOWSKI Z. Quantifying selected growth parameters of Leptothrix discophora SP-6 in biofilms from oxygen concentration profiles. Chemical Engineering Science. 2003;58:4557–4566. [Google Scholar]
- ZHANG Z, NADEZHINA E, WILKINSON KJ. Quantifying diffusion in a biofilm of Streptococcus mutans. Antimicrobial agents and chemotherapy. 2011;55:1075–81. doi: 10.1128/AAC.01329-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHU XY, KUNDUKAD B, VAN DER MAAREL JRC. Viscoelasticity of entangled lambda-phage DNA solutions. Journal of Chemical Physics. 2008;129 doi: 10.1063/1.3009249. [DOI] [PubMed] [Google Scholar]
- ZUSTIAK SP, BOUKARI H, LEACH JB. Solute diffusion and interactions in cross-linked poly(ethylene glycol) hydrogels studied by Fluorescence Correlation Spectroscopy. Soft Matter. 2010;6 doi: 10.1039/C0SM00111B. [DOI] [PMC free article] [PubMed] [Google Scholar]


