Abstract
Type 2 diabetes mellitus (T2DM) is a complex disease that is caused by an impairment in the secretion of β-cell insulin and by a peripheral resistance to insulin. Most patients suffering from T2DM and from obesity exhibit insulin resistance in the muscles, liver, and fat, resulting in a reduced response of these tissues to insulin. In healthy individuals, pancreatic islet β-cells secrete insulin to regulate the increase in blood glucose levels. Once these β-cells fail to function, T2DM develops. Despite the progress achieved in this field in recent years, the genetic causes for insulin resistance and for T2DM have not yet been fully discovered. The present study aims to characterize T2DM by comparing its gene expression with that of normal controls, as well as to identify biomarkers for early T2DM. Gene expression profiles were downloaded from the Gene Expression Omnibus, and differentially expressed genes (DEGs) were identified for type 2 diabetes. Furthermore, functional analyses were conducted for the gene ontology and for the pathway enrichment. In total, 781 DEGs were identified in the T2DM samples relative to healthy controls. These genes were found to be involved in several biological processes, including cell communication, cell proliferation, cell shape, and apoptosis. We constructed a protein–protein interaction (PPI) network, and the clusters in the PPI were analyzed by using ClusterONE. Six functional genes that may play important roles in the initiation of T2DM were identified within the network.
Introduction
Improvements in living standards and lifestyle changes have brought about an increase in the morbidity of type 2 diabetes mellitus (T2DM), a disease that seriously endangers people's lives.1,2 According to the International Diabetes Federation, 100 million patients suffered from diabetes in the end of the 20th Century, and this number has grown rapidly to 382 million as of 2013. In 2035, the world is expected to have 592 million people affected by T2DM.3
T2DM is a complex multifactorial disorder that is characterized by insulin resistance and impaired β-cell functions.4–6 Numerous multifactorial mechanisms are involved in the development of insulin resistance and in the impairment of insulin secretion, including genetic and environmental factors that lead to a decline in insulin activity, followed by a chronic pancreatic β-cell dysfunction.7,8 However, insulin resistance alone may not be a decisive pathogenic factor of T2DM because insulin resistance often exists in many people without diabetes, and, in fact, the pancreatic islets of the Langerhans play a more important role in the development of T2DM.9 Prediabetes insulin resistance does not always lead to diabetes because a considerable proportion of individuals with prediabetes do not develop T2DM when the β-cells compensate for the insulin deficiency by secreting insulin to maintain glucose homeostasis. When insulin secretion by the β-cells cannot keep pace with the increase in insulin resistance, high blood glucose levels will occur throughout the body, and T2DM will develop.7,8,10,11 The increase in blood glucose level in diabetes patients leads to a wide variety of long-term complications, including retinopathy, nephropathy, and cardiovascular disease. The complications associated with diabetes result in chronic tissue damage—especially of important organs of the central nervous system, the kidneys, and the heart—as well as in chronic damage to other tissues, seriously jeopardizing the normal functions of the human body.
In this study, the differentially expressed genes (DEGs) between those with diabetes and normal controls were identified, and the biomarkers of diabetes were screened based on a protein–protein interaction (PPI) network. Our results may contribute to the further understanding of diabetes.
Materials and Methods
DNA microarray data
GSE25724 microarray data were downloaded from the Gene Expression Omnibus database,12 and a microarray analysis was performed to evaluate the differences in the transcriptome of human T2DM islets samples in comparison with those of samples from subjects without diabetes. Collagenase digestion, followed by density gradient purification, was used to isolate human islets from the pancreas of organ donors, and the islets were then hand-picked and cultured for 2 days in M199 culture medium. A GeneChip® human genome U133A array (Affymetrix, Santa Clara, CA) was used to process a total of 13 arrays that were available for further analysis, including six arrays of T2DM and seven without diabetes. Pancreata were obtained with the approval of the local Ethics Committee and consent by donors' relatives.
Human islets were isolated from seven organ donors without diabetes (age, 58±17 years; gender, four males/three females; body mass index, 24.8±2.5 kg/m2) and six organ donors with T2DM (age, 71±9 years; gender, three males/three females; body mass index, 26.0±2.2 kg/m2) by collagenase digestion followed by density gradient purification as previously reported.
Identification of DEGs and methylated sites
We use the Qlucore Omics Explorer version 3.0 software program (Qlucore AB, Lund, Sweden) to analyze the data from the microarray. Qlucore Omics Explorer is a next-generation bioinformatics software that can analyze and explore datasets, including those obtained from gene expression arrays, micro RNA arrays, protein arrays, and DNA methylation arrays. Qlucore Omics Explorer uses state-of-the-art mathematical and statistical methods to provide new methods to perform data analysis and data mining.
Principal component analyses and hierarchical clustering analysis of DEGs
A hierarchical clustering analysis was carried out for the DEGS to identify the differences in gene expression in a more intuitive manner. The Qlucore Omics Explorer version 3.0 software was used to generate the principal component analyses, hierarchical clustering, and a heat map.
Function and pathway enrichment analysis of DEGs
The Database for Annotation, Visualization and Integrated Discovery (DAVID) of bioinformatics resources consists of an integrated biological knowledge base and analytical tools aimed at systematically extracting biological meaning from large lists of genes or proteins.13 DAVID was applied in order to conduct the gene ontology analysis, and the Kyoto Encyclopedia of Genes and Genomes was used to carry out the pathway enrichment analysis. A false discovery rate of <0.05 and a count of >2 were chosen as the cutoff criteria.
PPI data
In order to obtain more reliable relationships, duplicate edges and interactions with self-loops were deleted. Finally, in total, 5,233 PPIs were obtained for further investigation. The PPI data were downloaded from the Biological General Repository for Interaction Datasets (BioGRID), the Molecular INTeraction database (MINT), the IntAct molecular interaction database (IntAct), and the iRefIndex database. BioGRID houses and distributes collections of protein and genetic interactions from major model organism species. As of August 1, 2013, BioGRID (a general repository for interaction datasets) contains 703,150 interactions from 32 different species, as derived from 41,099 high-throughput and conventional focused studies. At present, the MINT team focuses on the physical interactions between proteins, storing information on molecular interactions in a structured format in the MINT database by extracting experimental details from studies published in journals. The data in the current version of MINT include 241,458 experimentally verified interactions. IntAct (an open source molecular and open interaction database) is a comprehensive, open resource database as well as an analysis system for protein interactions that can promote consistency in the interaction datasets by using the same infrastructure and annotation system. The iRefIndex database (a consolidated protein interaction database with provenance) provides an index of protein interactions available in several primary interaction databases, including BIND, BioGRID, CORUM, DIP, HPRD, IntAct, MINT, MPact, MPPI, and OPHID. These data include multiple interaction types, including physical and genetic interactions (mapped to their corresponding protein products) that are determined through a multitude of methods.
PPI network analysis
The PPI information data were imported into Cytoscape (an open source software platform), and a PPI network was easily constructed. Clustering with overlap neighborhood expansion (ClusterONE) was used in the Cytoscape network in order to discover densely connected regions in the network with possible overlap. A quality function can guide ClusterONE by growing dense regions out of small seeds. The modules were identified to have a minimum density of >0.05 and a degree of >6. A cluster with a P value of <0.001 was determined to be a new network. Several parameters from the nodes in the network were obtained by using the Network Analyzer in Cytoscape, which can be used to help us determine the hub nodes for the network.
Results
Identification of DEGs and methylated sites
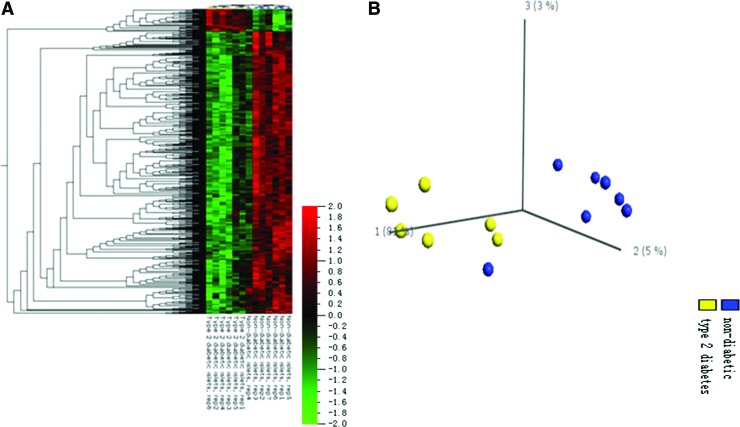
The DEGs of the GSE25724 data for human T2DM islets and those from subjects without diabetes were identified using Qlucore Omics Explorer version 3.0. As shown in Figure 1, in total, 781 genes were screened, including 741 down-regulated genes, leaving 40 genes remaining up-regulated (P=0.005, q=0.0037, fold change ≥2 times).
FIG. 1.
(A) Cluster analysis of differentially expressed genes. The colors from green to black and to red represent the expression values of the differentially expressed genes that have increased. (B) Principal cluster analysis plots. The yellow samples represent type 2 diabetes mellitus islets, and the blue samples represent islets from subjects without diabetes. Color graphics are available online at www.liebertonline.com/dia.
Hierarchical clustering analysis
As shown in Figure 1, the expression value for most DEGs for islets from subjects without diabetes was higher (deeper) than that for T2DM islets, suggesting that almost all the DEGs in islets from subjects without diabetes were down-regulated. These observations were consistent with the fact that DEG analysis indicated 741 out of 781 genes were down-regulated.
Function and pathway annotation
In Table 1, in total, 10 functions were enriched; the most significant function was that of intracellular transport (P=3.21E-16). In Table 2, the most significant pathway was the proteasome signaling pathway (P=2.16E-07). The dysregulated pathways in T2DM included pathways associated with signal transduction, such as the proteasome pathway, citrate cycle (tricarboxylic acid cycle) pathway, and the ubiquitin-mediated proteolysis pathway.
Table 1.
Classification of Genes in the Functional Modules According to Gene Ontology Terms with P<0.05 (Top 10)
| Term ID | Description | Count | P value |
|---|---|---|---|
| 0046907 | Intracellular transport | 82 | 3.24E-16 |
| 0006511 | Ubiquitin-dependent protein catabolic process | 41 | 1.78E-12 |
| 0032268 | Regulation of cellular protein metabolic process | 51 | 4.26E-08 |
| 0044265 | Cellular macromolecule catabolic process | 78 | 4.87E-12 |
| 0051438 | Regulation of ubiquitin-protein ligase activity | 22 | 3.17E-11 |
| 0006084 | Acetyl-coenzyme A metabolic process | 9 | 6.22E-05 |
| 0051340 | Regulation of ligase activity | 22 | 6.94E-11 |
| 0008135 | Translation factor activity, nucleic acid binding | 17 | 7.02E-06 |
| 0031396 | Regulation of protein ubiquitination | 23 | 7.61E-10 |
| 0005739 | Mitochondrion | 87 | 8.04E-08 |
Table 2.
Classification of Genes in Functional Modules According to Kyoto Encyclopedia of Gene and Genomes Terms with False Discovery Rate of <0.05 (Top 10)
| Term | Count | P value |
|---|---|---|
| hsa03050:Proteasome | 15 | 2.16E-07 |
| hsa03010:Ribosome | 16 | 1.17E-04 |
| hsa00020:Citrate cycle (tricarboxylic acid cycle) | 9 | 2.85E-04 |
| hsa04120:Ubiquitin-mediated proteolysis | 20 | 3.04E-04 |
| hsa04114:Oocyte meiosis | 15 | 0.004212 |
| hsa03060:Protein export | 4 | 0.0087 |
| hsa03040:Spliceosome | 15 | 0.013725 |
| hsa05012:Parkinson's disease | 15 | 0.015611 |
| hsa00510:N-Glycan biosynthesis | 8 | 0.015655 |
| hsa00071:Fatty acid metabolism | 7 | 0.026233 |
| hsa00620:Pyruvate metabolism | 7 | 0.026233 |
| hsa04110:Cell cycle | 14 | 0.028226 |
| hsa00640:Propanoate metabolism | 6 | 0.03522 |
| hsa03430:Mismatch repair | 5 | 0.041016 |
| hsa03022:Basal | 6 | 0.04936 |
Construction of PPI network and network clustering
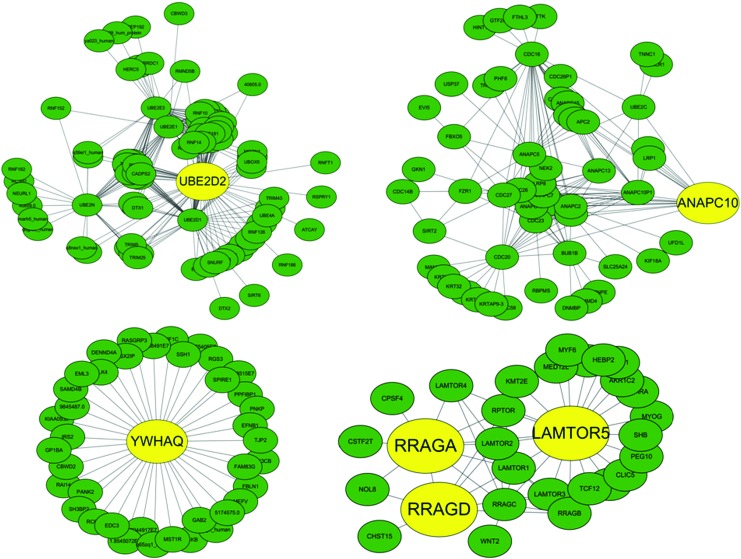
The PPI network was constructed with 8,731 nodes and 35,456 edges, and the DEGs were mapped into PPIs of the network. The clustering analysis was conducted in the PPI network by ClusterONE plugin in Cytoscape. The cluster with a significant value of 0.001 remained in order to construct a new network containing 295 nodes. The hub nodes with betweenness centrality and closeness centrality more than average, and a degree of more than twofold on average was kept for further analysis after the network analysis. Six of these are DEGs: ANAPC10, LAMTPR5, RRAGA, RRAGD, UBE2D2, and YWHAQ (Fig. 2).
FIG. 2.
The significant functional modules in the protein–protein interaction network. The yellow node represents differentially expressed genes, and the green node represents normal genes. The edge represents the interaction relationship among proteins. Color graphics are available online at www.liebertonline.com/dia.
Discussion
In the past few decades, the development of bioinformatics has enabled for targeted genetic and molecular treatments to become an important part of disease therapy.14,15 In this study, we analyzed the gene expression profiles to select DEGs in normal subjects and T2DM patients at different stages. In total, 781 genes were identified between normal subjects and T2DM patients. Of these, 740 genes were down-regulated, and 41 were up-regulated. DAVID was used to conduct function and pathway enrichment. In addition, the interaction network was constructed for the DEGs, and the degrees of hub genes were calculated.16
The anaphase-promoting complex (APC) is a cell cycle-regulated ubiquitin ligase (also referred to as a cyclosome) that is composed of at least 11 subunits. ANAPC10 is a core subunit of the APC, and the C-terminus of APC10 binds to CDC27/AP3, playing a role in controlling progression through mitosis and the G1 phase of the cell cycle. 17 The subunit ANANC10/APC10 may have a function in ubiquitination reactions due to the conserved core (residues of 22–161 amino acids of all 185) in its structure that is homologous to other founded domains in putative ubiquitin ligases. The low expression of this gene in human β-cells may reduce the secretion of glucagon, which results in the occurrence of T2DM.18
The hepatitis B X-interacting protein (HBXIP/LAMTOR5) is located at the human chromosome 1p13.3 and encodes a 9.6-kDa protein. It was originally identified as a result of its interaction with the C-terminus of the hepatitis B virus X protein, which negatively regulates the activity of hepatitis B virus X protein.19 In hepatoma cells, hepatitis B virus X protein plays a role in suppressing apoptosis initiated via the mitochondrial/cytochrome c pathway. LAMTOR5 promotes the proliferation of hepatoma cancer cells via the phosphatidylinositol 3-kinase/Akt pathway.20–22 In our study, the low expression of this gene in T2DM patients may promote apoptosis in the pancreas islets. However, its potential role in the development of T2DM should also be investigated.
The YWHAQ(14-3-3) family of proteins, including the seven isoforms β, ɛ, η, γ, σ, ζ, and τ in mammals, belongs to a family of highly conserved 30-kDa molecules23,24 that can bind to target proteins depending on the phosphorylated Ser or Thr residues mainly within the two 14-3-3 binding motifs, RSXpSXP (mode I) and RXXXpSXP (mode II), of the target protein (pS represents phosphoserine).25,26 The 14-3-3 proteins have a wide range of potential functions and pathological relevance,27–33 regulating the intercellular signal processes for differentiation, development, and growth, and binding to many important target molecules, including transcription cofactors. In addition, they can prevent or mediate apoptosis and survival by controlling the localization of the potential signaling molecules between the nucleus or the mitochondrion and the cytoplasm. In our study, 14-3-3 was down-regulated in T2DM patients, and we could conclude that 14-3-3 plays an important role in the development of T2DM.
The ubiquitination of proteins is mediated by a cascade of enzymes, including E1 (ubiquitin activating), E2 (ubiquitin conjugating), and E3 (ubiquitin ligases) enzymes. UBE2D2 encodes a member of the E2 enzyme family. Also, the tumor suppressor protein p53 and the peroxisomal biogenesis factor 5 are the substrates of this enzyme. UBE2D2 is an essential protein in the degradation of many regulatory and abnormal proteins,34 and UbcH5A and UbcH5C together form one of the most active classes of E2 enzymes related to UBE2D2.35 However, these could be associated with the degradation of several important human transcription factors, such as p53,36 nuclear factor κB, and c-Fos.37 Recently, it has been noticed that the ubiquitination of proteins could not be restricted to protein degradation only, but could also contain other regulatory functions in, for example, signal transduction, transcription regulation, chromatin remodeling, and DNA repair.38 In our study, the low expression of this gene in T2MD patients may result in the degradation of insulin.
RRAG A/Rag A is a functional homolog to yeast Gtr1p and is a member of the Ras superfamily of small G proteins. Moreover, while polymerase chain reaction methods were used to isolate RRAGA/Rag A/FIP-1 during the search for novel G proteins, RRAGA/RagA was observed to develop a complex with other G proteins, RRAG C/RagC and RRAG D/Rag D.39 RRAGD is a monomeric guanine nucleotide-binding protein, or G protein, that acts as a molecular switch in numerous cell processes and signaling pathways while binding to GTP or GDP, which are small G proteins. Previous studies indicated that RRAGA is closely related to RRAGD and that both participate in the mammalian target of rapamycin signaling pathway related to the insulin signaling pathway. This suggests that RagD has a role in mTORC1 activation and that a molecular function for the FLCN tumor suppressor could potentially exist. In our study, we can conclude that the down-regulated expression of RRAGA and RRAGD is a potential biomaker for early-onset T2DM.
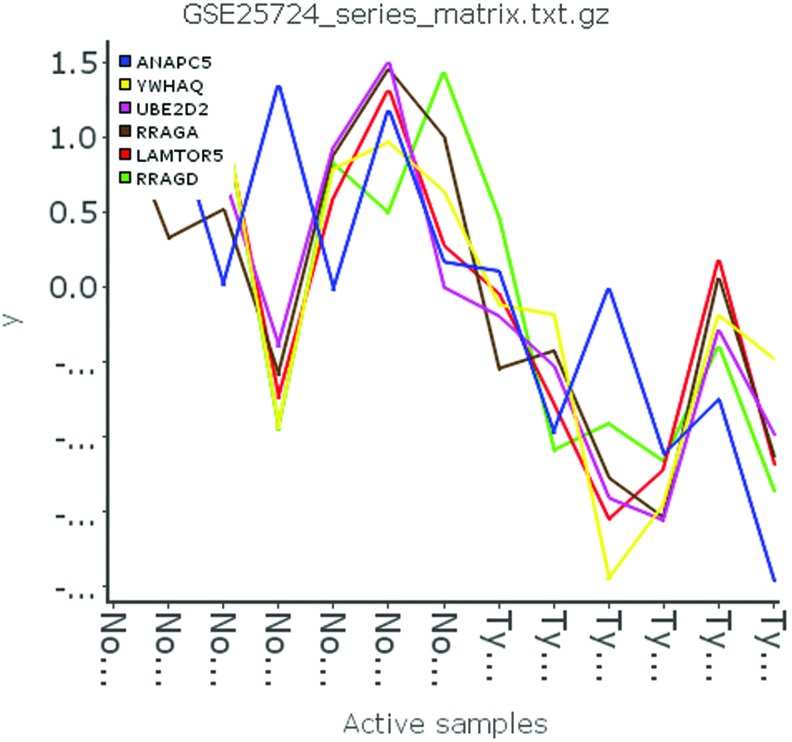
From the above discussion, we could draw the conclusion that the six genes (ANAPC10, LAMTPR5, RRAGA, RRAGD, UBE2D2, and YWHAQ) are identified as differentially expressed and are all down-regulated in T2DM patients (Fig. 3). Meanwhile, these six DEGs are hub nodes in the PPI network (Table 3). Thus, we predict that all these six genes could be potential biomarkers of early T2DM. Furthermore, Figure 2 indicates that RRAGA, RRAGD, and LAMTOR5 are closely related. All in all, we can regard the six genes to be biomarkers for T2DM.
FIG. 3.
The line map of GSE25724 shows six genes that are all down-regulated in type 2 diabetes mellitus. Color graphics are available online at www.liebertonline.com/dia.
Table 3.
The Six Differentially Expressed Genes and Their 30 Hub Nodes
| Hub node | Closeness centrality | Degree | Betweenness centrality |
|---|---|---|---|
| ANAPC1 | 0.28416779 | 15 | 0.00161029 |
| ANAPC10 | 0.36020583 | 18 | 0.08334712 |
| ANAPC10P1 | 0.35897436 | 16 | 0.07459775 |
| ANAPC13 | 0.33386328 | 15 | 0.02966666 |
| ANAPC16 | 0.28340081 | 13 | 5.98E-04 |
| ANAPC2 | 0.28074866 | 11 | 0.00957107 |
| ANAPC4 | 0.28416779 | 16 | 0.01794462 |
| ANAPC5 | 0.34201954 | 24 | 0.07845842 |
| ANAPC7 | 0.28340081 | 14 | 0.00149308 |
| BUB1B | 0.26785714 | 14 | 0.03080296 |
| CDC16 | 0.34768212 | 31 | 0.1242791 |
| CDC20 | 0.2872777 | 22 | 0.07109287 |
| CDC23 | 0.28493894 | 16 | 0.0116858 |
| CDC26 | 0.28532609 | 16 | 0.00281479 |
| CDC27 | 0.28649386 | 19 | 0.01848354 |
| CTNNB1 | 0.40540541 | 42 | 0.34130418 |
| LAMTOR1 | 0.59574468 | 9 | 0.03143739 |
| LAMTOR5 | 0.8 | 22 | 0.77270723 |
| MTUS2 | 1 | 14 | 1 |
| RCHY1 | 0.328125 | 27 | 0.24110276 |
| RRAGA | 0.60869565 | 10 | 0.21375661 |
| RRAGC | 0.57142857 | 8 | 0.079806 |
| RRAGD | 0.44444444 | 8 | 0.08906526 |
| UBE2C | 0.32061069 | 8 | 0.02287318 |
| UBE2D1 | 0.42424242 | 64 | 0.33524711 |
| UBE2D2 | 0.43298969 | 69 | 0.45291098 |
| UBE2E1 | 0.33653846 | 46 | 0.04686355 |
| UBE2E3 | 0.328125 | 41 | 0.05150276 |
| UBE2N | 0.3030303 | 32 | 0.06705781 |
| YWHAQ | 1 | 39 | 1 |
Conclusions
Overall, we have constructed a PPI network of T2MD and have identified six functional modules that may play important roles in the initiation of early-onset T2MD. The hub nodes of these six modules and the genes ANAPC10, LAMTOR5, RRAGA, RRAGD, UBE2D2, and YWHAQ may serve as biomarkers of early-onset T2MD and have the potential to be targets for therapeutic intervention. However, further studies are still needed in order to confirm our findings.
Acknowledgments
This research was supported by the Institute of Genetic Engineering at the Southern Medical University, Guangzhou, Guangdong, China. This study was supported by the National Natural Science Foundation of China (grant 39880032) and the Guangdong Foundation for Leading Talented Scientists (grant C1030925).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Danaei G, Finucane MM, Lu Y, et al. : National, regional, and global trends in fasting plasma glucose and diabetes prevalence since 1980: systematic analysis of health examination surveys and epidemiological studies with 370 country-years and 2·7 million participants. Lancet 2011;378:31–40 [DOI] [PubMed] [Google Scholar]
- 2.Wild S, Roglic G, Green A, et al. Global prevalence of diabetes: estimates for the year 2000 and projections for 2030. Diabetes Care 2004;27:1047–1053 [DOI] [PubMed] [Google Scholar]
- 3.International Diabetes Federation: Diabetes Atlas, 3rd ed. Brussels: International Diabetes Federation, 2007 [Google Scholar]
- 4.Dayeh T, Volkov P, Salö S, et al. : Genome-wide DNA methylation analysis of human pancreatic islets from type 2 diabetic and non-diabetic donors identifies candidate genes that influence insulin secretion. PLoS Genet 2014;10:e1004160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunetti A, Chiefari E, Foti D: Recent advances in the molecular genetics of type 2 diabetes mellitus. World J Diabetes 2014;5:128–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stumvoll M, Goldstein BJ, van Haeften TW: Type 2 diabetes: principles of pathogenesis and therapy. Lancet 2005;365:1333–1346 [DOI] [PubMed] [Google Scholar]
- 7.Donath MY, Shoelson SE: Type 2 diabetes as an inflammatory disease. Nat Rev Immunol 2011;11:98–107 [DOI] [PubMed] [Google Scholar]
- 8.Gupta D, Krueger CB, Lastra G: Over-nutrition, obesity and insulin resistance in the development of β-cell dysfunction. Curr Diabetes Rev 2012;8:76–83 [DOI] [PubMed] [Google Scholar]
- 9.Jain P, Vig S, Datta M, et al. : Systems biology approach reveals genome to phenome correlation in type 2 diabetes. PLoS One 2013;8:e53522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kolb H, Eizirik DL: Resistance to type 2 diabetes mellitus: a matter of hormesis? Nat Rev Endocrinol 2011;8:183–192 [DOI] [PubMed] [Google Scholar]
- 11.Lusis AJ, Attie AD, Reue K: Metabolic syndrome: from epidemiology to systems biology. Nat Rev Genet 2008;9:819–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dominguez V, Raimondi C, Somanath S, et al. : Class II phosphoinositide 3-kinase regulates exocytosis of insulin granules in pancreatic beta cells. J Biol Chem 2011;286:4216–4225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eisen MB, Spellman PT, Brown PO, et al. : Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A 1998;95:14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Da Huang W, Sherman BT, Lempicki RA: Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44–57 [DOI] [PubMed] [Google Scholar]
- 15.Dinh P, Sotiriou C, Piccart MJ: The evolution of treatment strategies: aiming at the target. Breast 2007;16(Suppl 2):10–16 [DOI] [PubMed] [Google Scholar]
- 16.He H, Mao L, Xu P, et al. : Ossification of the posterior longitudinal ligament related genes identification using microarray gene expression profiling and bioinformatics analysis. Gene 2014;533:515–519 [DOI] [PubMed] [Google Scholar]
- 17.Kurasawa Y, Todokoro K: Identification of human APC10/Doc1 as a subunit of anaphase promoting complex. Oncogene 1999;18:5131–5137 [DOI] [PubMed] [Google Scholar]
- 18.Wendt KS, Vodermaier HC, Jacob U, et al. : Crystal structure of the APC10/DOC1 subunit of the human anaphase promoting complex. Nature 2001;8:784–788 [DOI] [PubMed] [Google Scholar]
- 19.Melegari M, Scaglioni PP, Wands JR: Cloning and characterization of a novel hepatitis B virus X binding protein that inhibits viral replication. J Virol 1998;72:1737–1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marusawa H, Matsuzawa S, Welsh K, et al. : HBXIP functions as a cofactor of survivin in apoptosis suppression. EMBO J 2003;22:2729–2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang FZ, Fei HR, Lian LH, et al. : Hepatitis B x-interacting protein induces HepG2 cell proliferation through activation of the phosphatidylinositol 3-kinase/Akt pathway. Exp Biol Med (Maywood) 2011;236:62–69 [DOI] [PubMed] [Google Scholar]
- 22.Wang F-Z, Sha L, Zhang W-Y, et al. : Involvement of hepatitis B X-interacting protein (HBXIP) in proliferation regulation of cells. Acta Pharmacol Sin 2007;28:431–438 [DOI] [PubMed] [Google Scholar]
- 23.Aitken A: 14-3-3 proteins: a historic overview. Semin Cancer Biol 2006;16:162–172 [DOI] [PubMed] [Google Scholar]
- 24.Aitken A, Collinge DB, van Heusden BP, et al. : 14-3-3 proteins: a highly conserved, widespread family of eukaryotic proteins. Trends Biochem Sci 1992;17:498–501 [DOI] [PubMed] [Google Scholar]
- 25.Yaffe MB, Rittinger K, Volinia S, et al. : The structural basis for 14-3-3:phosphopeptide binding specificity. Cell 1997;91:961–971 [DOI] [PubMed] [Google Scholar]
- 26.Berg D, Holzmann C, Riess O: 14-3-3 proteins in the nervous system. Nat Rev Neurosci 2003;4:752–762 [DOI] [PubMed] [Google Scholar]
- 27.Dougherty MK, Morrison DK: Unlocking the code of 14-3-3. J Cell Sci 2004;117:1875–1884 [DOI] [PubMed] [Google Scholar]
- 28.Mhawech P: 14-3-3 proteins—an update. Cell Res 2005;15:228–236 [DOI] [PubMed] [Google Scholar]
- 29.Muslin AJ, Lau JM: Differential functions of 14-3-3 isoforms in vertebrate development. Curr Top Dev Biol 2005;65:211–228 [DOI] [PubMed] [Google Scholar]
- 30.Skoulakis EM, Davis RL: 14-3-3 proteins in neuronal development and function. Mol Neurobiol 1998;16:269–284 [DOI] [PubMed] [Google Scholar]
- 31.Takahashi Y: The 14-3-3 proteins: gene, gene expression, and function. Neurochem Res 2003;28:1265–1273 [DOI] [PubMed] [Google Scholar]
- 32.Taya S, Shinoda T, Tsuboi D, et al. : Disc1 regulates the transport of the nudel/lis1/14-3-3epsilon complex through kinesin-1. J Neurosci 2007;27:15–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tzivion G, Shen YH, Zhu J: 14-3-3 proteins; bringing new definitions to scaffolding. Oncogene 2001;20:6331–6338 [DOI] [PubMed] [Google Scholar]
- 34.Seufert W, Jentsch S: Ubiquitin-conjugating enzymes UBC4 and UBC5 mediate selective degradation of short-lived and abnormal proteins. EMBO J 1990;9:543–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scheffner M, Huibregtse JM, Howley PM: Identification of a human ubiquitin-conjugating enzyme that mediates the E6-AP-dependent ubiquitination of p53. Proc Natl Acad Sci U S A 1994;91:8797–8801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gonen H, Bercovich B, Orian A, et al. : Identification of the ubiquitin carrier proteins, E2s, involved in signal-induced conjugation and subsequent degradation of IκBα. J Biol Chem 1999;274:14823–14830 [DOI] [PubMed] [Google Scholar]
- 37.Stancovski I, Gonen H, Orian A, et al. : Degradation of the protooncogene product c-Fos by the ubiquitin proteolytic system in vivo and in vitro: identification and characterization of the conjugating enzymes. Mol Cell Biol 1995;15:7106–7116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aguilar RC, Wendland B: Ubiquitin: not just for proteasomes anymore. Curr Opin Cell Biol 2003;15:184–190 [DOI] [PubMed] [Google Scholar]
- 39.Sekiguchi T, Hirose E, Nakashima N, et al. : Novel G proteins, Rag C and Rag D, interact with GTP-binding proteins, Rag A and Rag B. J Biol Chem 2001;276:7246–7257 [DOI] [PubMed] [Google Scholar]