Abstract
Purpose
In a recent phase II study of onartuzumab (MetMAb), patients whose non–small cell lung cancer (NSCLC) tissue scored as positive for MET protein by immunohistochemistry (IHC) experienced a significant benefit with onartuzumab plus erlotinib (O+E) versus erlotinib. We describe development and validation of a standardized MET IHC assay and, retrospectively, evaluate multiple biomarkers as predictors of patient benefit.
Experimental Design
Biomarkers related to MET and/or EGF receptor (EGFR) signaling were measured by IHC, FISH, quantitative reverse transcription PCR, mutation detection techniques, and ELISA.
Results
A positive correlation between IHC, Western blotting, and MET mRNA expression was observed in NSCLC cell lines/tissues. An IHC scoring system of MET expression taking proportional and intensity-based thresholds into consideration was applied in an analysis of the phase II study and resulted in the best differentiation of outcomes. Further analyses revealed a nonsignificant overall survival (OS) improvement with O+E in patients with high MET copy number (mean ≥5 copies/cell by FISH); however, benefit was maintained in “MET IHC-positive”/MET FISH-negative patients (HR, 0.37; P = 0.01). MET, EGFR, amphiregulin, epiregulin, or HGF mRNA expression did not predict a significant benefit with onartuzumab; a nonsignificant OS improvement was observed in patients with high tumor MET mRNA levels (HR, 0.59; P = 0.23). Patients with low baseline plasma hepatocyte growth factor (HGF) exhibited an HR for OS of 0.519 (P = 0.09) in favor of onartuzumab treatment.
Conclusions
MET IHC remains the most robust predictor of OS and progression-free survival benefit from O+E relative to all examined exploratory markers.
Introduction
MET, a receptor tyrosine kinase (RTK) that binds hepatocyte growth factor (HGF) is frequently overexpressed in a variety of human malignancies. MET activation has been implicated in tumorigenesis, and MET signaling can be dysregulated through a variety of genetic or epigenetic mechanisms in cancer (1, 2). In non–small cell lung cancer (NSCLC), tumor MET receptor protein expression, HGF protein expression, and high MET gene copy number are indicative of poor prognosis (3–6). Although focal amplification of the MET gene is rare in primary lung tumors (~1%–7%; ref. 4), it is associated with oncogenic addiction, and with sensitivity, in preclinical models, to small-molecule inhibitors (SMI) targeting MET (7, 8). No activating mutations have been identified in the kinase domain of MET in NSCLC; however, somatic variants causing exon 14 skipping, can result in an alternatively spliced MET receptor lacking the juxtamembrane domain that sustains enhanced ligand-dependent MET signaling (9). Finally, genetic polymorphisms have been linked to enhanced MET signaling (R970C, T990I; ref. 10), as well as to lower HGF-binding affinity (N375S; ref. 11).
A growing body of evidence has emerged to support a link between the MET and EGF receptor (EGFR) signaling pathways. These RTKs are often coexpressed in tumors, and evidence exists for functional transactivation that may amplify downstream signals (12). For example, activation of EGFR may occur through MET amplification or HGF-mediated induction of EGFR ligands (13). MET activation has been associated with resistance to EGFR inhibitors both preclinically and clinically (14–16). Collectively, these findings support the rationale for dual inhibition of MET and EGFR signaling.
Onartuzumab (MetMAb) is a recombinant, humanized, monovalent monoclonal antibody targeting MET (17). A phase II study (OAM4558g) evaluated onartuzumab plus erlotinib (O+E) versus placebo plus erlotinib (p+E) in patients with second-/third-line NSCLC therapy (18). Patient tumor samples were evaluated for MET expression by immunohistochemistry (IHC) and were classified as MET-positive or MET-negative, after randomization, but before unblinding the treatment assignment. There was neither a progression-free survival (PFS; HR, 1.09; P = 0.69), nor overall survival (OS) benefit (HR, 0.80; P = 0.34), in the intent-to-treat (ITT) population. However, the combination of O+E in MET-positive disease resulted in improved PFS and OS (HR, 0.53; P = 0.04; HR, 0.37; P = 0.002, respectively; ref. 18).
In this article, we describe the development and validation of the specific IHC assay and the corresponding scoring system that was used to assess MET protein expression in the OAM4558g clinical trial. In addition, we carry out retrospective analyses to further assess the diagnostic cutoff point and evaluate additional biomarkers related to the MET and/or EGFR pathways, as predictors of benefit from O+E.
Materials and Methods
Patients
Patients ages ≥ 18 years with measurable and previously treated (up to two prior regimens) stage IIIB/IV NSCLC were eligible. Submission of tumor tissue (archival permitted), as either a tissue block or unstained serial slides, was required. Written informed consent was obtained before any study-specific screening procedures. A total of 137 patients were randomized; 68 to p+E and 69 to O+E. Demographic and baseline characteristics were generally well-balanced across the treatment arms in the ITT and MET diagnostic subgroups, with few exceptions noted. A similar pattern in these characteristics was also observed across the treatment arms in the subgroups of FISH evaluable patients and quantitative reverse transcription PCR (qRT-PCR) evaluable patients (data not shown). Prioritization of specific biomarker analyses and number of tissues examined for the given analysis was as follows: MET IHC (n = 128), EGFR/KRAS mutation analysis (n = 112), MET/EGFR FISH (n = 96), MET exon14 (n = 87) and N375S genotyping (n = 113) and finally, qRT-PCR (n = 67). Plasma HGF levels were evaluated from 96 patients. The overlap in biomarker analyses is shown in Supplementary Table S1.
Study design
This was a phase II double-blinded, multicenter, international randomized clinical trial (OAM4558g). Patients were randomly assigned in a 1:1 ratio to receive either onartuzumab (15 mg/kg) or placebo (both administered by intravenous infusion every 3 weeks), plus erlotinib (oral 150 mg daily). The coprimary endpoints were PFS in the ITT- and MET-positive populations. Approximately, 120 patients would be needed to estimate the PFS benefit in both these populations. Further details of the study design have been published previously (18).
MET expression by IHC
MET expression levels were evaluated using CONFIRM anti-total MET (SP44) rabbit monoclonal primary antibody (Ventana Medical Systems, Inc.; cat no. 790-4430), and a composite scoring system was devised to determine the status of MET. The IHC cutoff point for positivity was prospectively defined in OAM4558g. The scoring of OAM4558g specimens was independently reviewed by a second pathologist, with an 88.3% concordance in calling MET-positive between pathologists. We did not observe a difference in outcome based upon the interpretation of pathologist #2 of the data. Further details are included in the Supplementary Methods section.
Western blot analysis
Cell lines used for Western blotting were obtained from the American Type Culture Collection (ATCC), National Cancer Institute Division of Cancer Treatment and Diagnosis Tumor Repository (Bethesda, MD); or Japan Health Sciences Foundation (Tokyo, Japan) and were authenticated. For authentication, short tandem repeat (STR) profiles were determined for each line using the Promega PowerPlex 16 System. This was performed once and compared with external STR profiles of cell lines (when available) to determine cell line ancestry. Sixteen loci (fifteen STR loci and Amelogenin for gender identification) were analyzed, including D3S1358, TH01, D21S11, D18S51, Penta E, D5S818, D13S317, D7S820, D16S539, CSF1PO, Penta D, AMEL, vWA, D8S1179, and TPOX. NSCLC cell lines were lysed in T-PER tissue extraction reagent (Thermo Scientific) supplemented with protease and phosphatase inhibitors. Proteins resolved by SDS–PAGE were electrophoretically transferred to polyvinylidene difluoride membrane and Western blots were carried out using standard techniques. The primary antibodies used in this study were: MET clone SP44 (Ventana Medical Systems), MET clone L41G3 (Cell Signaling Technology), MET clone 5D5 (HB-11895; ATCC), and actin (cat. no. sc-8432; Santa Cruz Biotechnology). A polyclonal antibody raised against MST1R was a kind gift from Dr Amitabha Chaudhuri (Genentech, Inc.).
Molecular assessments
MET and EGFR gene copy numbers were evaluated by FISH; a CEP7 centromere probe (Abbott Molecular) was used as a control. High-level MET amplification was defined as tight gene clusters of ≥ 15 copies in ≥ 10% of tumor cells, or a MET: CEP7 ratio of ≥2. A cutoff of ≥5 copies of MET/ cell was predefined as the criterion for FISH-positive status (FISH+), based on prior prognostic data supporting this cutoff in NSCLC (4). Tumors were considered EGFR FISH+ based on a scoring system used in multiple clinical studies (19). Further details are included in the Supplementary Methods section.
DNA and RNA were isolated from macrodissected tissue to enrich for tumor content. EGFR and KRAS mutations were evaluated using the DxS Genotyping Kit. MET exon 14 variants were evaluated by Surveyor nuclease digestion and detection by WAVE analysis (Transgenomics, Inc.). A polymorphism at position N375S of MET was evaluated by pyrosequencing on the Pyromark Q24 (11). Expression of MET, HGF, EGFR, amphiregulin (AREG), and epiregulin (EREG) mRNA transcripts was evaluated by qRT-PCR on the Biomark platform (Fluidigm). The primer/probes used for profiling are shown in Supplementary Table S2 and further details are included in the Supplementary Methods section.
HGF quantification by ELISA
Plasma HGF levels were measured by ELISA, as described previously (20).
Statistical analysis
PFS was defined as the time from randomization to the first occurrence of disease progression or death on study from any cause, or to the last tumor assessment date if patients were progression free. OS was determined from randomization to the date of death, or the last known patient contact. For each treatment arm, the median PFS and OS were estimated from Kaplan–Meier curves. The PFS and OS treatment comparisons were based on a log-rank test at the 0.05 level of significance (two-sided). Estimated treatment effects for O+E relative to p+E were expressed as HRs with 95% confidence intervals (CI), derived from an unstratified Cox model. Survival analyses were performed in biomarker subgroups for PFS and OS.
Results
Development of the SP44 IHC test
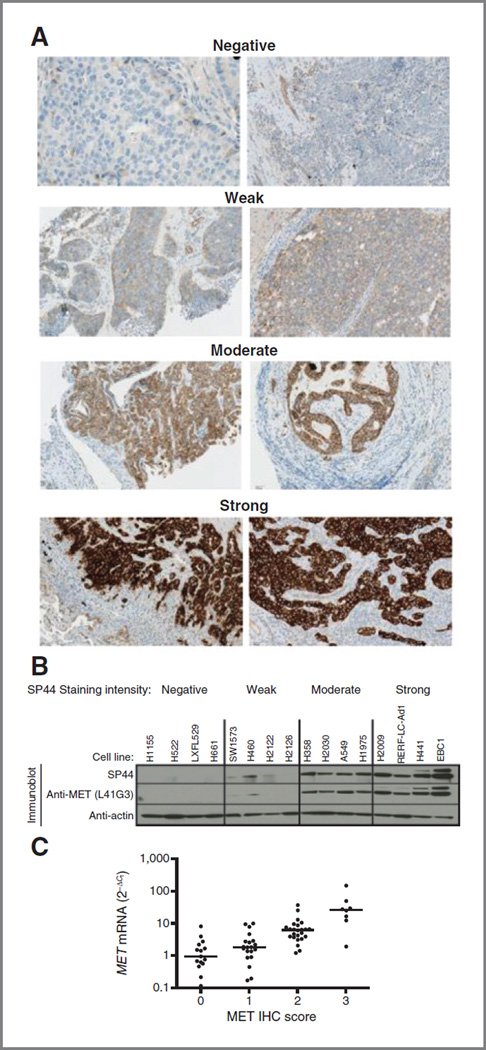
To identify antibodies that would be suitable for detecting MET protein expression levels in formalin-fixed paraffin-embedded tissue specimens, we initially tested 16 commercially or internally available antibodies for their specificity and staining intensity on control tissues and cell lines (data not shown). The CONFIRM anti-total cMET (SP44) rabbit monoclonal antibody (mAb), generated against an intracellular epitope of MET, was chosen for further analysis based upon its initial reactivity pattern in NSCLC cell lines engineered to knockdown or ectopically express the MET gene (Supplementary Fig. S1). Furthermore, SP44 did not cross-react with the Ron receptor (MST1R), the closest homolog of MET (Supplementary Fig. S1C). IHC staining of NSCLC tissues (Fig. 1A) and cell lines (Supplementary Fig. S2A) revealed a range of staining intensities characterized as no staining (negative), weak, moderate, or strong, with a pattern that was primarily membranous with a variably strong cytoplasmic component. In NSCLC cell lines, the SP44 staining intensities associated with the degree of MET protein expression, as determined by Western blot analysis using both SP44, as well as an alternative anti-MET antibody (L41G3; Fig. 1B). SP44 staining intensities also corresponded with the levels of MET detected on the surface of NSCLC cell lines using flow cytometry when probed with the bivalent form of the parent mAb to onartuzumab (5D5) and no IHC staining was observed with SP44 in cell lines in which 5D5 did not bind (Supplementary Fig. S2B and S2C). Finally, a statistically significant association was observed between the intensity of IHC staining with SP44 and MET mRNA levels across a larger panel of NSCLC cell lines (Supplementary Fig. S3) further supporting the specificity of this reagent.
Figure 1.
Characterization of SP44. A, representative images depicting the range of SP44 staining intensities (negative, weak, moderate, and strong staining) in malignant NSCLC tissues. B, relationship of SP44 immunohistochemical staining intensity and MET protein levels by Western blotting in NSCLC cell lines using two anti-MET antibodies; four representative cell lines and staining intensity are shown. C, correlation of IHC score with MET mRNA levels in NSCLC tissues determined by qRT-PCR.
In NSCLC tissues, heterogeneity in the intensity was frequently observed within individual NSCLC tumors (Supplementary Fig. S4). To account for this heterogeneity, a comprehensive clinical scoring system was developed that evaluated both membranous and cytoplasmic staining intensity and the percentage of cells staining at a given intensity level (Supplementary Table S3). A proportional cutoff of ≥50% was selected to ensure that a majority of the cells within a given specimen expressed MET at either a weak (clinical score = 1 +), moderate (clinical score = 2+), or strong (clinical score = 3+) intensity level. Specimens with no or equivocal staining in tumor cells or <50% of tumor cells staining at any given intensity were considered negative (clinical score = 0). Using these IHC scoring criteria, we continued to observe a statistically significant (Jonckheere–Terpstra P < 0.0001) relationship with MET mRNA expression in tumor specimens (Fig. 1C). NSCLC tumors expressing moderate or strong levels of MET in ≥50% of cells (clinical IHC score 2+ or 3+) were classified as MET-positive (MET IHC diagnostic positive). To understand how intratumoral heterogeneity may affect this clinical diagnostic score, we evaluated MET expression in 10 patients for whom more than one paraffin block had been submitted for analysis (Supplementary Table S4). Although variation was observed in the proportion of cells staining at a given intensity, there was complete concordance in the final IHC score for the multiple tissue specimens in these patients. Finally, using these scoring criteria, we determined that 58% (±6%) of nonsquamous and 26% (±11%) of squamous cell tumor specimens were MET-positive across three separate series of NSCLC tumor specimens (Table 1).
Table 1.
Prevalence of MET expression by SP44 IHC assay in NSCLC tumor specimens
| Frequency (%) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Study | Group | Subgroup | Total (n) | Dx-negative | Dx-positive | 0 | 1 | 2 | 3 |
| NSCLC TMA 1 | Histology | SCC | 36 | 64 | 36 | 33 | 31 | 33 | 3 |
| Non-SCC | 31 | 39 | 61 | 23 | 16 | 55 | 6 | ||
| OAM4558g | Histology | SCC | 36 | 72 | 28 | 11 | 61 | 22 | 6 |
| Non-SCC | 92 | 39 | 61 | 16 | 23 | 47 | 14 | ||
| NSCLC TMA 2 | Histology | SCC | 155 | 86 | 14 | 22 | 65 | 13 | 1 |
| Non-SCC | 291 | 49 | 51 | 20 | 29 | 44 | 7 | ||
| Final pathologic | I | 253 | 60 | 40 | 21 | 39 | 35 | 5 | |
| Stage (mountain) | II | 74 | 74 | 26 | 24 | 50 | 24 | 1 | |
| III | 96 | 60 | 40 | 15 | 46 | 34 | 5 | ||
| IV | 23 | 48 | 52 | 22 | 26 | 44 | 9 | ||
| Final pathologic | I | 229 | 59 | 41 | 22 | 37 | 36 | 5 | |
| Stage (IASLC) | II | 106 | 74 | 26 | 19 | 55 | 25 | 2 | |
| III | 96 | 59 | 41 | 19 | 41 | 35 | 5 | ||
| IV | 15 | 47 | 53 | 20 | 27 | 40 | 13 | ||
| Race | African American | 24 | 75 | 25 | 21 | 54 | 21 | 4 | |
| Asian | 6 | 50 | 50 | 33 | 17 | 50 | 0 | ||
| Caucasian | 402 | 62 | 38 | 20 | 42 | 33 | 5 | ||
| Hispanic | 14 | 36 | 64 | 14 | 21 | 50 | 14 | ||
Abbreviations: Dx, diagnostic; IASLC, International Association for the Study of Lung Cancer; SCC, squamous cell carcinoma; TMA, tissue microarray.
Evaluation of alternative scoring systems
We also evaluated the H-score method of semiquantitatively assessing MET expression as an alternative to take into account the intensity and heterogeneity of MET staining (21). Both membranous and cytoplasmic staining were combined into a single intensity score. We observed a range of H-scores from 0 to 300, with a median H-score of 160 in the set of tissue samples collected from patients in OAM4558g. Most specimens within each clinical score category (0, 1+, 2+, and 3+) were distributed around the H-scores of 0, 100, 200, and 300, respectively (Supplementary Fig. S5 and S6), as would be expected if staining was completely homogeneous. When heterogeneity in staining intensity was present (n = 64 specimens exhibiting tumor cell populations with multiple staining intensity levels), positivity was primarily distributed between two different staining intensity levels (n = 48). Only 13% of specimens (n = 16) exhibited a significant degree of heterogeneity in which staining intensities were noted across three different levels. There were four specimens below the median H-score that were considered MET-positive when applying the clinical scoring algorithm; conversely, there was one specimen at the median H-score that was MET-negative by the clinical scoring system.
Defining the best cutoff for the MET IHC assay
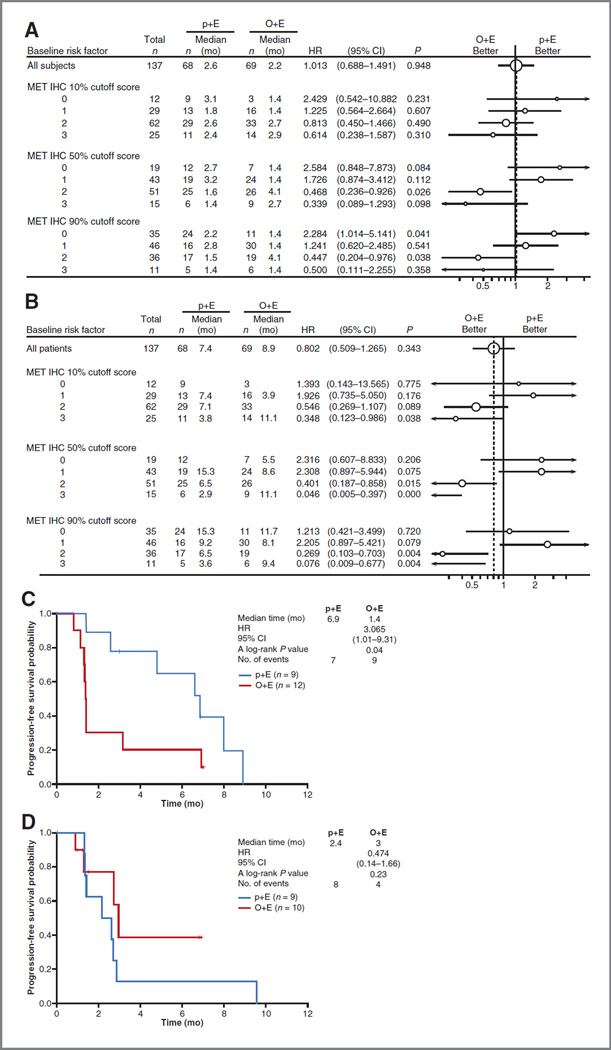
The IHC cutoff point for positivity was prospectively defined in OAM4558g. An analysis evaluating the appropriateness of this cutoff point revealed that alternative proportional cutoff points to define MET positivity of ≥10% or ≥90% of tumor cells staining moderately or strongly did not result in a better differentiation of patient outcomes (18). To evaluate alternative cutoff points in more detail, we examined PFS (Fig. 2A) and OS (Fig. 2B) based upon individual staining intensities using a 10%, 50%, or 90% proportional cutoff. Consistent with prior results, the best differentiation of patient outcomes was observed at the 50% cutoff and the differentiation was not greatly altered by applying different cutoffs to different staining intensities. More importantly, PFS was also analyzed in two subsets of patients whose IHC scores bordered the diagnostic cutoff: group 1, patients categorized as MET-positive using a 10% cutoff but as MET-negative using 50% cutoff (patients whose tumors exhibited moderate to strong staining in 10%–49% of tumor cells; Fig. 2C); and group 2, patients categorized as MET-positive using 50% cutoff, but as MET-negative using a 90% cutoff (patients whose tumors exhibited moderate to strong staining in 50%–89% of tumor cells; Fig. 2D). In group 1 (n = 21), patients treated with O+E had shorter PFS than patients treated with p+E (HR, 3.065; P = 0.04), suggesting that using a 10% cutoff may incorporate patients who may not benefit from the study drug. Outcomes in group 2 patients were not statistically different between treatment arms; however, there was a trend toward benefit in those who received O+E (n = 19; HR, 0.47; P = 0.23). Similar findings were observed for OS (data not shown).
Figure 2.
Evaluation of patient outcomes with O+E treatment based upon alternative IHC cutoffs. A, the forest plot for PFS from OAM4558g according to different IHC intensities and different proportional cutoffs (10%, 50%, and 90%); B, the forest plot for OS in OAM4558g according to different IHC intensities and different proportional cutoffs (10%, 50%, and 90%). Kaplan–Meier estimates for PFS in patients with IHC scores bordering the diagnostic cutoff; C, patients defined as MET-positive using a 10% cutoff and MET-negative using a 50% cutoff; D, patients defined as MET-positive using a 50% cutoff and MET-negative using a 90% cutoff.
Evaluation of alternative predictive markers of onartuzumab + erlotinib activity
Pathway mutations
EGFR and KRAS genotyping data were obtained from 112 patients, mutations were identified in 13 (12%) and 26 (23%) samples, respectively, and were mutually exclusive. Nine of 13 EGFRmut cases and 13 of the 26 KRASmut cases were MET-positive. EGFR mutations were identified in 6 of the 7 patients with objective responses (Supplementary Table S5 and S6); there were no responses in patients with KRASmut tumors. Exon 14, encoding the juxtamembrane domain of MET, was genotyped in 87 patients. A splice site deletion was detected in only one individual and alternative splicing was confirmed at the mRNA level (data not shown). Two additional patients (both on p+E) had tumors harboring exon 14 polymorphisms R970C and T990I. Polymorphisms in MET N375S were found in 12 of 113 (11%) tumors tested; because of an imbalance between treatment subgroups, no further analysis was carried out.
MET/EGFR copy number
MET and EGFR copy number were evaluated by FISH for 96 patients. The median MET copy number was 3.44 copies per cell (range, 1.6–25.0), with 19 (20%) evaluable tumors being MET FISH+, including eight (8%) harboring high-level MET amplification. The relationship between MET copy number and MET IHC clinical score is shown in Supplementary Fig. S7, with a statistically significant association of FISH positivity with IHC 3+ cases observed (χ2, P < 0.0001). No high-level MET gene amplifications were identified in patients who had objective responses to therapy (Supplementary Table S6); however, a high-level MET amplification was identified in an EGFRmut patient who did not respond to O+E.
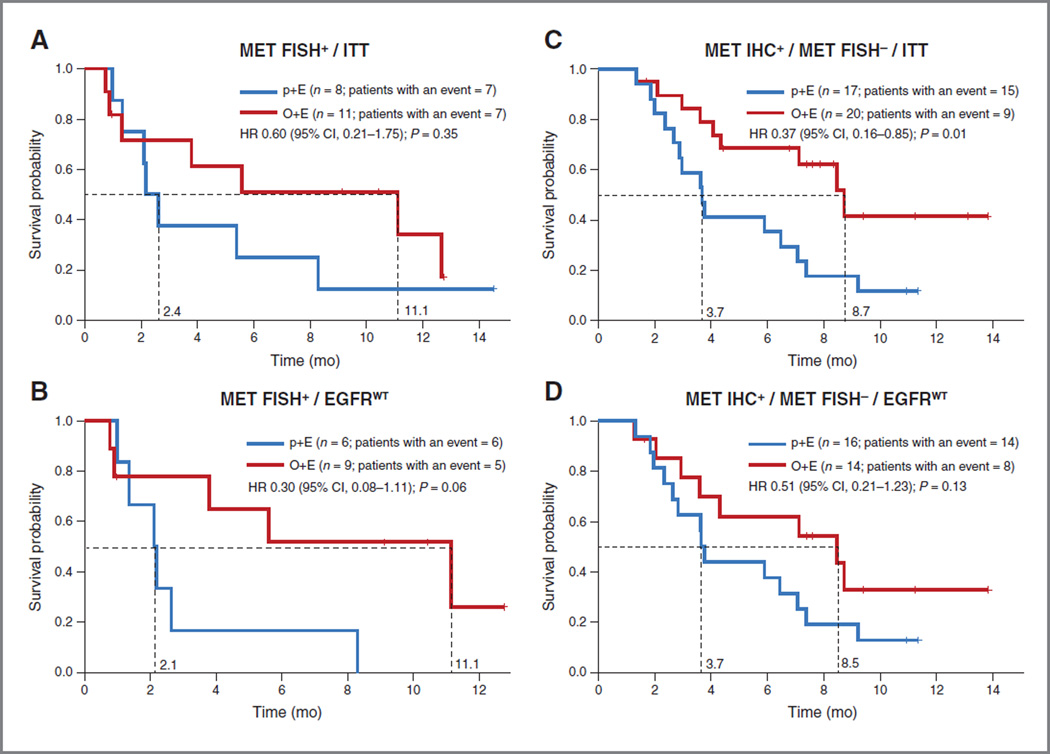
The estimated HR for OS for patients who were MET FISH+ was 0.60 (P = 0.35) in favor of O+E (Fig. 3A). No improvement in OS was observed using lower cutoffs for MET copy number (HR, 0.89; P = 0.79 for ≥4 copies and HR, 0.75; P = 0.37 for ≥3 copies; Supplementary Fig. S8A and S8B). To evaluate the impact of EGFR mutations on patient outcomes, we examined outcomes in the known EGFRwt population: OS HR for patients with MET FISH+ tumors receiving O+E was 0.30 (P = 0.06); PFS HRwas 0.58 (P = 0.38; Fig. 3B). As observed in the ITT population, no improvement in OS was seen in the EGFRwt populations further defined by ≥4 (HR, 0.76; P = 0.55) or ≥ 3 (HR, 0.75; P = 0.40) copies of MET/cell (Supplementary Fig. S8C and S8D, respectively).
Figure 3.
Evaluation of patient outcomes with O+E based upon MET gene copy number. Kaplan–Meier estimates of OS according to MET FISH positivity (≥5 copies of MET/cell) in the ITT (A; n = 19) and EGFRwt (B; n = 15) populations. Kaplan–Meier estimates of OS in patients with MET-positive and MET FISH-negative (<5 copies of MET/cell) tumors in the ITT (C) and EGFRwt (D) populations.
To determine whether the MET FISH+ patients influenced the benefit observed in the MET-positive subgroup, OS and PFS analyses were performed in both the MET IHC+/FISH− subgroups of the ITT and EGFRwt populations. An OS HR of 0.37 (P = 0.01) and a PFS HR of 0.24 (P = 0.003) were observed in the MET IHC+/FISH− ITT population (Fig. 3C). An OS HR of 0.51 (P = 0.13) and a PFS HR of 0.44 (P = 0.10) was observed in the MET IHC+/FISH− and EGFRwt patients (Fig. 3D).
Because of chromosome 7 polyploidy, gains in MET gene copy number have been shown to associate with high EGFR copy number gains (4). To ensure an unbiased analysis, EGFR and MET copy number variations were evaluated using identical criteria, as previously adopted for use in EGFR FISH testing (19). A significant association between EGFR and MET copy number was observed (Fisher exact, P = 2.2 × 10−5), with 24% (n = 23) of evaluable cases being positive for gains in both EGFR and MET copy number (data not shown). However, no clinical benefit was observed in this subpopulation (Supplementary Fig. S8E): OS HR, 1.36 (P = 0.54); PFS HR, 1.0 (P = 1.0).
MET/EGFR pathway transcript expression
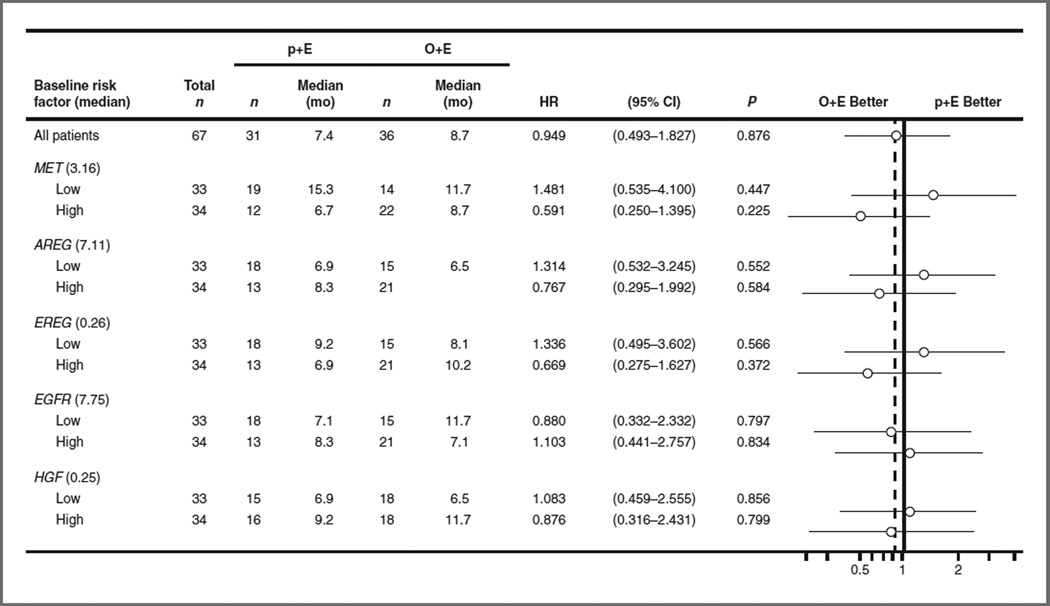
Expression of mRNA transcripts for MET, HGF, and EGFR, and the EGFR ligands, AREG and EREG, was evaluated by qRT-PCR in tumor biopsies from 67 patients. In this biomarker sub-population, a nonsignificant improvement in OS (HR, 0.49; P = 0.11) and PFS (HR, 0.57; P = 0.22) was observed in IHC MET–positive patients. In patients with high tumor MET mRNA levels (median ≥3.16), an OS HR of 0.59 (P = 0.23), and a PFS HR of 0.76 (P = 0.51) was observed (Fig. 4; Supplementary Table S7). No similar improvements in outcome were seen in any other mRNA subgroups for patients receiving O+E.
Figure 4.
Evaluation of patient outcomes with O+E based upon tumor mRNA expression. The forest plot showing association of MET, AREG, EREG, EGFR, and HGF mRNA expression as determined by quantitative PCR with OS. High, mRNA levels ≥ median; low, mRNA levels < median.
Plasma HGF
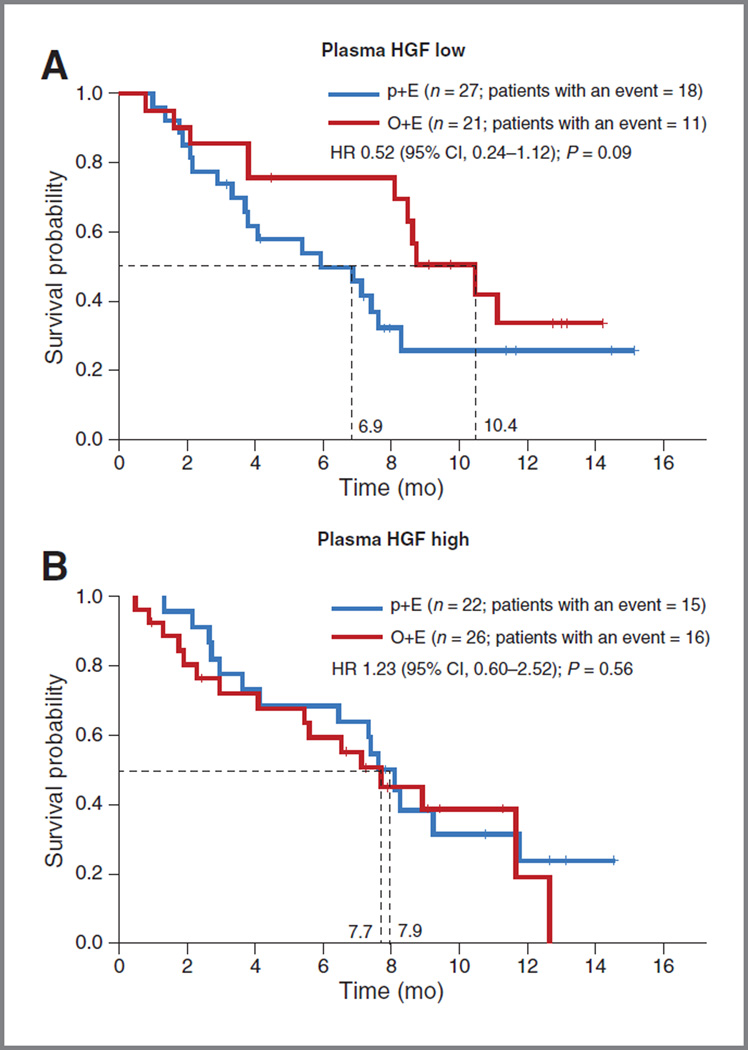
Pretreatment plasma protein levels of HGF were evaluated in 96 patients. The median level of HGF was 480.2 pg/mL (range, 203–11,375 pg/mL), with no significant difference in distribution between MET IHC diagnostic subgroups (Supplementary Fig. S9). No correlation of plasma HGF with tumor HGF mRNA levels was observed (data not shown). In this subpopulation of patients with evaluable plasma HGF data, both OS and PFS were consistent with what was observed in the general population: HR, 0.39 (P = 0.006) in OS and HR, 0.52 (P = 0.06) in PFS, respectively, in MET-positive patients. In patients who had baseline HGF protein below the median, the OS HR was 0.52 (P = 0.09) and PFS HR was 0.56 (P = 0.10) favoring O+E (Fig. 5A). In patients with baseline HGF protein ≥ median, the OS HR was 1.23 (P = 0.56; Fig. 5B). In addition, a Cox proportional hazard regression model revealed no statistically significant association between OS and HGF level (HR, 0.81; 95% CI, 0.52–1.25; P = 0.339) or between OS and HGF level and treatment (HR, 1.68; 95% CI, 0.91–3.10; P = 0.096).
Figure 5.
Evaluation of patient outcomes with O+E based upon plasma HGF levels. Kaplan–Meier estimates of OS in patients with low (<median) baseline HGF levels (A) and with high (≥median) baseline HGF levels (B).
Discussion
In the OAM4558g study, the addition of onartuzumab to erlotinib resulted in improvements in both PFS and OS in patients whose tumor specimens were determined to be MET-positive by IHC (18). This benefit was not seen in the overall population, and patients with MET-negative tumors who received O+E had a worse outcome versus those treated with erlotinib alone. Therefore, the data indicate that the MET IHC assay may be a reliable diagnostic test for identifying NSCLC patients most likely to benefit from O+E; and as such, this test is currently being developed as a companion diagnostic for onartuzumab in combination with erlotinib (clinicaltrials.gov identifier: NCT01456325; ref. 22). In this report, we describe the development and performance of this IHC test and compare its performance relative to alternative biomarkers.
Our investigation has identified that the SP44 mAb binds MET with high specificity and generates a range of staining intensities that correlate well with MET mRNA levels and MET protein levels determined by alternative means. As with many histologic tests, we observed intratumoral heterogeneity in MET protein expression levels in NSCLC tissues. To account for such heterogeneity, we examined various scoring systems that incorporate both staining intensity and the percentage of malignant cell positivity. We reasoned that a cutoff that captures the MET expression signal in the majority of the malignant cell population in a specimen (i.e., ≥50%) would likely produce more reproducible findings. When using a cutoff of 50% of malignant cells in a biopsy sample with moderate and/or strong staining intensity, we observed that our call for positivity did not change when evaluating intratumoral heterogeneity in a small set of cases for which sections representing more than one area of the same resection specimen were available for analysis (Supplementary Table S4). More importantly, we demonstrated that this 50% cutoff was the criterion that best differentiated patient outcomes in the phase II OAM4558g trial. A less stringent cutoff of 10% of malignant cells staining at moderate and/or strong intensity resulted in the inclusion of a subset of patients who did not benefit from O+E therapy in this trial, as shown by worse outcomes (PFS HR, 3.065; P = 0.04). Conversely, implementation of a more stringent cutoff of 90% of malignant cells staining at moderate and/or strong intensity would result in the loss of a subset of patients who trended toward improved outcomes with O+E, although this did not achieve statistical significance (PFS HR, 0.464; P = 0.23). Additional evaluations of outcome based on different staining intensities supported these proposed criteria. Overall, these data strongly suggest that the diagnostic cutoff that was prospectively defined represents a suitable scoring system for future clinical studies evaluating O+E in NSCLC. Other scoring criteria, such as H-score, were considered. However, exploratory analysis did not show that H-score could alter the interpretation of the outcomes from previous analyses (data not shown).
To determine whether MET IHC represented the best predictor of patient benefit in this study, additional biomarkers related to the MET and/or EGFR signaling pathways were measured. EGFR-activating mutations represent the strongest predictor of response to EGFR-targeted therapies in NSCLC. As expected in a primarily Caucasian population, approximately 12% of patients harbored EGFR mutations in this study. Of the 7 patients who reported an objective response, all but one had an EGFRmut tumor. One EGFRwt patient had an objective response on O+E; however, it was not possible to associate this response to any other biomarker examined, including EGFR or MET gene amplification, MET mutation or high tumor/serum HGF levels. Most importantly, EGFR mutations did not drive the benefit observed in the MET-positive population, as the magnitude of the treatment benefit was maintained in MET-positive, EGFRwt patients (OS HR, 0.46; P = 0.03; ref. 18).
KRAS is a downstream effector of both EGFR and MET signaling and mutations in this gene provide a critical predictive marker for efficacy of EGFR-targeted antibodies in colorectal cancer. Although KRAS mutations predict for a lack of radiologic response to EGFR-targeted SMIs in NSCLC, any similar impact on PFS and OS remains unresolved (23, 24). In this study, the incidence of KRAS mutations was mutually exclusive from EGFR mutations, and did not significantly affect outcomes in patients receiving O+E in the ITT, MET-positive or MET-negative populations. However, given limitation in sample size, it remains to be determined whether this subgroup would indeed benefit from a MET inhibitor.
The role of high-level MET gene amplification has gained considerable attention, as it predicts for enhanced sensitivity to MET-targeted SMIs in preclinical models (7, 8), has recently been linked to anecdotal responses on MET-targeted agents in the clinic (25), and, importantly, represents a mechanism of acquired resistance to EGFR SMIs in patients with EGFRmut NSCLC (14–16). In addition, general increases in MET copy number have been reported in many, but not all studies, to represent a negative prognostic factor in patients with NSCLC and may be relevant in the context of EGFR-targeted combination therapy, given that EGFR and MET copy number gains can be associated because of chromosome 7 ploidy. In this study, the frequency of high-level MET gene amplification and high MET copy number was 8% and 20%, respectively, which was in accordance (albeit at the high end) with previous studies. The association of MET FISH positivity with the MET IHC 3+ subgroup suggests that high MET gene copy number leads to high protein expression in this subgroup. Although the MET IHC 2+ score is also represented within the MET-positive subgroup, there was no corresponding association with high copy number gains. Nonsignificant improvements in PFS and OS were observed in the MET FISH+ ITT and EGFRwt populations. However, this benefit was not maintained when lower copy number cutoffs were evaluated. This cutoff for MET FISH positivity (≥5 copies of MET/cell) has been linked with shortened OS and advanced-stage disease (4) and, hence, may identify a poor prognostic population who could benefit from O+E. However, a statistically significant benefit was also observed in the MET-positive/FISH-negative population. These results would suggest that MET IHC status, rather than MET FISH status, performs better for predicting benefit, and that FISH status alone would miss a large population who may benefit from O+E. No improvement in outcome was observed in patients harboring gains in both MET and EGFR copy number, further suggesting that the benefit to this MET and EGFR-targeted combination therapy was not driven by the cases of general chromosome 7 ploidy.
Tumor mRNA expression of key MET and EGFR pathway genes was additionally evaluated as alternative predictive biomarkers. This not only included expression of MET/HGF, but expression levels of two EGFR ligands that can be transcriptionally regulated by MET activation, AREG and EREG. MET was the only mRNA biomarker examined that showed a trend toward improvement in OS and PFS, albeit this did not achieve statistical significance. This could likely be attributed to the small sample size, as only approximately 1/2 of the patients (n = 67) had material remaining following the prioritization of other biomarker assessments for transcriptional analysis. Regardless, evaluation of MET mRNA levels as an alternative predictive biomarker may warrant further investigation in future studies.
High serum or plasma HGF levels have been associated with poor prognosis in several cancers, including NSCLC (26–28). In a study of patients with NSCLC treated with EGFR SMIs, high-serum HGF was associated with worse outcome (tumor response, PFS, and OS; ref. 26). Despite our expectations that the addition of onartuzumab to erlotinib could improve outcomes versus erlotinib alone in patients with high-plasma HGF at baseline, we did not observe a statistically significant association. In fact, a nonsignificant trend toward benefit was observed in patients with low baseline HGF. The study cited from Kasahara and colleagues evaluated serum HGF, whereas our study evaluated plasma HGF. However, this would unlikely explain the difference, as strong correlations between plasma and serum HGF levels have been observed (data not shown). High-plasma HGF also failed to show a prognostic link in the p+E arm in our study (OS HR, 0.75; P = 0.41), suggesting that the prognostic significance of circulating HGF may be influenced by differences in the population tested. It remains to be elucidated how HGF levels in circulation relate to intratumoral levels of HGF. Although elevated circulating HGF levels have been observed in subsets of patients with cancer, elevated circulating HGF levels have additionally been described in other settings, including certain viral or bacterial infections, graft versus host disease, and surgical procedures (29–31).
In conclusion, MET IHC was a reliable and accurate assay for identifying patients with NSCLC most likely to benefit from O+E, and outperformed the other examined exploratory markers. The retrospective, exploratory nature of these analyses, combined with small sample size, limit the scope of definitive conclusions to be drawn from these alternative biomarkers. Further investigations of these biomarkers using larger sample sizes are necessary to determine whether any would be useful in conjunction with MET IHC to better define subpopulations most likely to benefit from O+E. In addition, it will be imperative to determine whether such biomarkers will be useful in determining benefit to other onartuzumab-based therapies and/or other antagonists of the MET signaling pathway. The anti-HGF antibody, rilotumumab, in combination with standard-of-care chemotherapy exhibited a benefit in both PFS and OS compared with chemotherapy alone (OS HR, 0.29; P = 0.012) in patients with gastric or gastroesophageal cancer with METHIGH tumors by IHC (32). Interestingly, similar to the OAM4558g NSCLC trial with onartuzumab, patients with METLOW tumors receiving HGF-targeted therapy seemed to do worse compared with patients receiving chemotherapy only (HR, 1.84; P value not reported). However, tumor MET expression did not predict for benefit on rilotumumab in a phase II study in castration-resistant prostate cancer (33). Hence, the role of MET IHC in predicting benefit to onartuzumab and/or other MET-targeting therapies will likely need to be carefully evaluated within a given indication. Currently, the test reported here is being developed as a companion diagnostic for onartuzumab and is being used to screen and enroll only MET-positive patients in the phase III study comparing onartuzumab in combination with erlotinib versus placebo plus erlotinib (clinicaltrials.gov identifier: NCT01456325; ref. 22).
Supplementary Material
Translational Relevance.
A hallmark of the successful clinical development of targeted therapies is the identification of the patient subgroups most likely to benefit from these agents. Recent clinical studies evaluating molecules targeting the MET signaling pathway, including onartuzumab and rilotumumab, have demonstrated benefit in patients whose tumors express high levels of MET protein. We describe the development, performance, and scoring criteria of a MET IHC test that differentiated patient outcomes for onartuzumab plus erlotinib versus erloti-nib alone in a recent phase II study in non-small cell lung cancer. In a retrospective analysis of this study, multiple biomarkers related to MET and/or EGFR signaling were additionally examined. It was determined that the IHC test to detect tumor MET levels represented the best predictor of benefit. These results have guided the phase III clinical development strategy for onartuzumab and provided further support for development of the MET IHC test as a potential companion diagnostic.
Acknowledgments
The authors thank F. Hoffmann-La Roche Ltd. for third-party writing assistance for this article. The authors thank the OAM4558g investigators on this study, including Drs. D.R. Spigel, T.J. Ervin, R. Ramlau, D.B. Daniel, J.H. Goldschmidt Jr, G.R. Blumenschein Jr, M.J. Krzakowski, G. Robinet, B. Godbert, F. Barlesi, R. Govindan, T. Patel, S.V. Orlov, and M.S. Wertheim; as well as the patients enrolled on OAM4558g and their families for supporting this work.
Grant Support
This work was supported by Genentech, Inc. (South San Francisco, CA). Financial support for third-party writing assistance for this article was provided by F. Hoffmann-La Roche Ltd. I. Wistuba was supported by the UT Lung Specialized Programs of Research Excellence grant (P50CA70907) and the MD Anderson Cancer Center Support grant CA016672.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
H. Koeppen has ownership interest in Roche. L. Fu is an employee of Roche. T. Januario, M. Lazarov, and L.C. Amler are employees of Genentech. I.I. Wistuba reports receiving a commercial research grant from Genentech/ Roche and is a consultant/advisory board member for Genentech/Roche and Ventana. S.C. Phan is an employee of and has ownership interest (including patents) in Genentech. No potential conflicts of interest were disclosed by the other authors.
Authors’ Contributions
Conception and design: W. Yu, J. Zha, A. Pandita, E. Filvaroff, M. Lazarov, L. Amler, A. Peterson, R.L. Yauch
Development of methodology: W. Yu, J. Zha, A. Pandita, L. Rangell, S. Mohan, R. Patel, L. Fu, S.G. Louie, E. Filvaroff, J. Huang, R.L. Yauch
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): H. Koeppen, J. Zha, A. Pandita, L. Rangell, S. Mohan, R. Patel, L. Fu, A. Do, V. Parab, X. Xia, T. Januario, E. Filvaroff, D.S. Shames, I. Wistuba, A. Peterson, R.L. Yauch
Analysis and interpretation of data (e.g., statistical analysis, biosta-tistics, computational analysis): H. Koeppen, W. Yu, J. Zha, A. Pandita, E. Penuel, S. Mohan, V. Parab, T. Januario,E.Filvaroff, D.S. Shames,J. Huang, M. Lazarov, V. Ramakrishnan, L. Amler, S.-C. Phan, P. Patel, A. Peterson, R.L. Yauch
Writing, review, and/or revision of the manuscript: H. Koeppen, W. Yu, J. Zha, A. Pandita, L. Fu, V. Parab, T. Januario, I. Wistuba, J. Huang, M. Lazarov, V. Ramakrishnan, L. Amler, S.-C. Phan, P. Patel, A. Peterson, R.L. Yauch
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): W. Yu, A. Pandita, L. Rangell, R. Raja, E. Filvaroff, M. Lipkind, J. Huang, V. Ramakrishnan, A. Peterson Study supervision: J. Zha, A. Pandita, E. Filvaroff, L. Amler, P. Patel, A. Peterson, R.L. Yauch
Developed biomarker assays and methodologies, performed the assays and data analysis: R. Raja
Mutation analysis and interpretation of data: R. Desai
References
- 1.Trusolino L, Bertotti A, Comoglio PM. MET signalling: principles and functions in development, organ regeneration, and cancer. Nat Rev Mol Cell Biol. 2010;11:834–848. doi: 10.1038/nrm3012. [DOI] [PubMed] [Google Scholar]
- 2.Gherardi E, Birchmeier W, Birchmeier C, Woude GV. Targeting MET in cancer: rationale and progress. Nat Rev Cancer. 2012;12:89–103. doi: 10.1038/nrc3205. [DOI] [PubMed] [Google Scholar]
- 3.Masuya D, Huang C, Liu D, Nakashima T, Kameyama K, Haba R, et al. The tumour-stromal interaction between intratumoral c-Met and stromal hepatocyte growth factor associated with tumour growth and prognosis in non-small cell lung cancer patients. Br J Cancer. 2004;90:1555–1562. doi: 10.1038/sj.bjc.6601718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cappuzzo F, Marchetti A, Skokan M, Rossi E, Gajapathy S, Felicioni L, et al. Increased MET gene copy number negatively affects survival of surgically resected non-small cell lung cancer patients. J Clin Oncol. 2009;27:1667–1674. doi: 10.1200/JCO.2008.19.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beau-Faller M, Ruppert A, Voegeli A. MET gene copy number in non- small cell lung cancer: molecular analysis in a targeted tyrosine kinase inhibitor naive cohort. J Thorac Oncol. 2008;3:331–339. doi: 10.1097/JTO.0b013e318168d9d4. [DOI] [PubMed] [Google Scholar]
- 6.Siegfried JM, Weissfeld LA, Luketich JD, Weyant RJ, Gubish CT, Landreneau RJ. The clinical significance of hepatocyte growth factor for non-small cell lung cancer. Ann Thorac Surg. 1998;66:1915–1918. doi: 10.1016/s0003-4975(98)01165-5. [DOI] [PubMed] [Google Scholar]
- 7.Smolen G, Sordella R, Muir B, Mohapatra G, Barmettler A, Archibald H, et al. Amplification of MET may identify a subset of cancers with extreme sensitivity to the selective tyrosine kinase inhibitor PHA-665752. Proc Natl Acad Sci U S A. 2006;103:2316–2321. doi: 10.1073/pnas.0508776103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lutterbach B, Zeng Q, Davis LJ, Hatch H, Hang G, Kohl NE, et al. Lung cancer cell lines harboring MET gene amplification are dependent on Met for growth and survival. Cancer Res. 2007;67:2081–2088. doi: 10.1158/0008-5472.CAN-06-3495. [DOI] [PubMed] [Google Scholar]
- 9.Kong-Beltran M, Seshagiri S, Zha J, Zhu W, Bhawe K, Mendoza N, et al. Somatic mutations lead to an oncogenic deletion of Met in lung cancer. Cancer Res. 2006;66:283–289. doi: 10.1158/0008-5472.CAN-05-2749. [DOI] [PubMed] [Google Scholar]
- 10.Ma PC, Kijima T, Maulik G, Fox EA, Sattler M, Griffin JD, et al. c-MET mutational analysis in small cell lung cancer: novel juxtamembrane domain mutations regulating cytoskeletal functions. Cancer Res. 2003;63:6272–6281. [PubMed] [Google Scholar]
- 11.Krishnaswamy S, Kanteti R, Duke-Cohan JS, Loganathan S, Liu W, Ma PC, et al. Ethnic differences and functional analysis of MET mutations in lung cancer. Clin Cancer Res. 2009;15:5714–5723. doi: 10.1158/1078-0432.CCR-09-0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lai AZ, Abella JV, Park M. Crosstalk in Met receptor oncogenesis. Trends Cell Biol. 2009;19:542–551. doi: 10.1016/j.tcb.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 13.Reznik TE, Sang Y, Ma Y, Abounader R, Rosen EM, Xia S, et al. Transcription-dependent epidermal growth factor receptor activation by hepatocyte growth factor. Mol Cancer Res. 2008;6:139–150. doi: 10.1158/1541-7786.MCR-07-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 15.Bean J, Brennan C, Shih J-Y, Riely G, Viale A, Wang L, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci U S A. 2007;104:20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turke AB, Zejnullahu K, Wu Y-L, Song Y, Dias-Santagata D, Lifshits E, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell. 2010;17:77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Surati M, Patel P, Peterson A, Salgia R. Role of MetMAb (OA-5D5) in c-MET active lung malignancies. Expert Opin Biol Ther. 2011;11:1655–1662. doi: 10.1517/14712598.2011.626762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spigel D, Ervin TJ, Ramlau RA, Daniel DB, Goldschmidt JH, Jr, Blumenschein GR, Jr, et al. Randomized phase II trial of onartuzumab in combination with erlotinib in patients with advanced non-small cell lung cancer. J Clin Oncol. 2013;31:4105–4114. doi: 10.1200/JCO.2012.47.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Varella-Garcia M, Diebold J, Eberhard DA, Geenen K, Hirschmann A, Kockx M, et al. EGFR fluorescence in situ hybridisation assay: guidelines for application to non-small cell lung cancer. J Clin Pathol. 2009;62:970–977. doi: 10.1136/jcp.2009.066548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Catenacci DVT, Henderson L, Xiao SY, Patel P, Yauch RL, Hegde P, et al. Durable complete response of metastatic gastric cancer with anti-Met therapy followed by resistance at recurrence. Cancer Discov. 2011;1:573–579. doi: 10.1158/2159-8290.CD-11-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Detre S, Jotti GS, Dowsett M. A “quickscore” method for immunohis-tochemical semiquantitation: validation for oestrogen receptor in breast carcinomas. J Clin Path. 1995;48:876–878. doi: 10.1136/jcp.48.9.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spigel DR, Edelman MJ, Mok T, O’Byrne K, Paz-Ares L, Yu W, et al. Treatment rationale study design for the MetLung trial: a randomized, double-blind phase III study of onartuzumab (MetMAb) in combination with erlotinib versus erlotinib alone in patients who have received standard chemotherapy for stage IIIB or IV Met-positive non-small cell lung cancer. Clin Lung Cancer. 2012;13:500–504. doi: 10.1016/j.cllc.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 23.Zhu CQ, da Cunha Santos G, Ding K, Sakurada A, Cutz JC, Liu N, et al. Role of KRAS and EGFR as biomarkers of response to erlotinib in National Cancer Institute of Canada Clinical Trials Group Study BR.21. J Clin Oncol. 2008;26:4268–4275. doi: 10.1200/JCO.2007.14.8924. [DOI] [PubMed] [Google Scholar]
- 24.Jackman DM, Miller VA, Cioffredi LA, Yeap BY, Janne PA, Riely GJ, et al. Impact of epidermal growth factor receptor and KRAS mutations on clinical outcomes in previously untreated non-small cell lung cancer patients: results of an online tumor registry of clinical trials. Clin Cancer Res. 2009;15:5267–5273. doi: 10.1158/1078-0432.CCR-09-0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chi AS, Batchelor TT, Kwak EL, Clark JW, Wang DL, Wilner KD, et al. Rapid radiographic and clinical improvement after treatment of a MET-amplified recurrent glioblastoma with a mesenchymal-epithelial transition inhibitor. J Clin Oncol. 2012;30:e30–e33. doi: 10.1200/JCO.2011.38.4586. [DOI] [PubMed] [Google Scholar]
- 26.Kasahara K, Arao T, Sakai K, Matsumoto K, Sakai A, Kimura H, et al. Impact of serum hepatocyte growth factor on treatment response to epidermal growth factor receptor tyrosine kinase inhibitors in patients with non-small cell lung adenocarcinoma. Clin Cancer Res. 2010;16:4616–4624. doi: 10.1158/1078-0432.CCR-10-0383. [DOI] [PubMed] [Google Scholar]
- 27.Toiyama Y, Miki C, Inoue Y, Okugawa Y, Tanaka K, Kusunoki M. Serum hepatocyte growth factor as a prognostic marker for stage II or III colorectal cancer patients. Int J Cancer. 2009;125:1657–1662. doi: 10.1002/ijc.24554. [DOI] [PubMed] [Google Scholar]
- 28.Aune G, Lian A-M, Tingulstad S, Torp SH, Forsmo S, Reseland JE, et al. Increased circulating hepatocyte growth factor (HGF): a marker of epithelial ovarian cancer and an indicator of poor prognosis. Gynecol Oncol. 2011;121:402–406. doi: 10.1016/j.ygyno.2010.12.355. [DOI] [PubMed] [Google Scholar]
- 29.Barreiros AP, Sprinzl M, Rosset S, Höhler T, Otto G, Theobald M, et al. EGF and HGF levels are increased during active HBV infection and enhance survival signaling through extracellular matrix interactions in primary human hepatocytes. Int J Cancer. 2009;124:120–129. doi: 10.1002/ijc.23921. [DOI] [PubMed] [Google Scholar]
- 30.Okamoto T, Takatsuka H, Fujimori Y, Wada H, Iwasaki T, Kakishita E. Increased hepatocyte growth factor in serum in acute graft-versus-host disease. Bone Marrow Transplant. 2001;28:197–200. doi: 10.1038/sj.bmt.1703095. [DOI] [PubMed] [Google Scholar]
- 31.Yamada T, Hisanaga M, Nakajima Y, Kanehiro H, Watanabe A, Ohyama T, et al. Serum interleukin-6, interleukin-8, hepatocyte growth factor, and nitric oxide changes during thoracic surgery. World J Surg. 1998;22:783–790. doi: 10.1007/s002689900470. [DOI] [PubMed] [Google Scholar]
- 32.Oliner KS, Tang R, Anderson A, Lan Y, Iveson T, Donehower RC, et al. Evaluation of MET pathway biomarkers in a phase II study of rilotu-mumab (R, AMG 102) or placebo (P) in combination with epirubicin, cisplatin, and capecitabine (ECX) in patients (pts) with locally advanced or metastatic gastric (G) or esophagogastric junction (EGJ) cancer. J Clin Oncol. 2012;30:4005. [Google Scholar]
- 33.Ryan CJ, Rosenthal M, Ng S, Alumkal J, Picus J, Gravis G, et al. Targeted MET inhibition in castration-resistant prostate cancer: a randomized phase II study and biomarker analysis with rilotumumab plus mitoxantrone and prednisone. Clin Can Res. 2013;19:215–224. doi: 10.1158/1078-0432.CCR-12-2605. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.