Abstract
Oral drug delivery is a route of choice for vaccine administration because of its noninvasive nature and thus efforts have focused on efficient delivery of vaccine antigens to mucosal sites. An effective oral vaccine delivery system must protect the antigen from degradation upon mucosal delivery, penetrate mucosal barriers, and control the release of the antigen and costimulatory and immunomodulatory agents to specific immune cells (i.e., APCs). In this paper, mannan-modified pH-responsive P(HEMA-co-MAA) nanogels were synthesized and assessed as carriers for oral vaccination. The nanogels showed pH-sensitive properties, entrapping and protecting the loaded cargo at low pH values, and triggered protein release after switching to intestinal pH values. Surface decoration with mannan as carbohydrate moieties resulted in enhanced internalization by macrophages as well as increasing the expression of relevant costimulatory molecules. These findings indicate that mannan-modified P(HEMA-co-MAA) nanogels are a promising approach to a more efficacious oral vaccination regimen.
INTRODUCTION
Up to date, most licensed vaccines are administered parenterally and result in robust systemic immune response but no mucosal immunity.1 In fact, mucosal delivery is the only vaccination route that induces both mucosal and systemic immune responses2 as well as immunization at distant mucosal sites.3 Oral drug delivery is a route of choice for vaccine administration because of its noninvasive nature, which improves patient compliance, facilitate administration and distribution, and reduces cost, in comparison to traditional administration methods (i.e., subcutaneous or intramuscular injections).2 However, the low mucosal permeability and lack of stability of most protein-based antigens in the gastrointestinal environment (i.e., gastric acidity and proteolytic enzymes) result in low bioavailability and lack of control on absorbed dose, which makes necessary the use of adjuvants4 and slows down products development.2,5 Efforts have focused on efficient delivery of vaccine antigens to mucosal sites, and one of the most promising strategies relies on the use of polymer nanoparticles.2
Several aspects must be considered in the rational design of oral-delivered nanovaccines. A primary goal is to protect the antigen from degradation upon mucosal delivery and provide controlled release at the targeted site (i.e., antigen-sampling M cells of immune-cell rich Peyer’s patches in the small intestine for oral delivery);6–8 the amount of bioactive antigen administrated can determine the Th1/Th2 bias in the immune response9 and, according to some authors, induce a specific oral tolerance mechanism upon repeated administration.7,10 Antigen should then be transported to organized mucosal-associated lymphoid tissues (MALT) and presented to antigen-presenting cells (APCs) such as macrophages and dendritic cells (DCs). APCs must be adequately activated in order to generate protective mucosal immune responses.5 Generating robust protection via cellular (T cell) or humoral (B cell) immune responses highly depends on the mechanisms by which antigen-containing nanoparticles are internalized.8 In addition to nanoparticle size,4,5 nanoparticle composition (i.e., surface chemistry and polymer architecture) plays a critical role in cell uptake and is of upmost importance in immune activation and modulation.1 Nanoparticle carriers can be designed to emulate pathogen associated molecular patterns (PAMPs), mimicking the entry of pathogens3 and targeting important pathogen recognition receptors (PRRs) on APCs.11
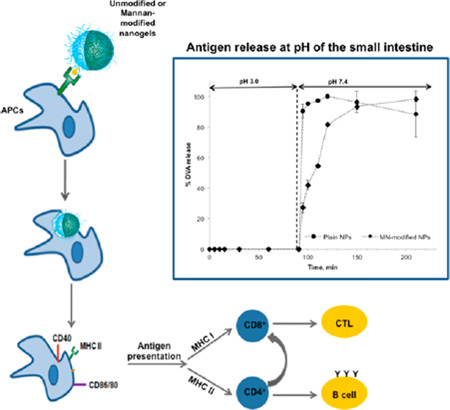
In this study, we design novel mannan (MN)-surface conjugated pH-responsive nanogels for targeting C-type lectin receptors (CLRs), which are PRRs with highly conserved carbohydrate-recognition domains and that are highly expressed in cells of the mucosal immune system,12,13 as a strategy to enhance and shape the immune response. Ligand recognition by CLRs leads to pathogen internalization via receptor mediated endocytosis and subsequent degradation and presentation of the pathogen as antigen to T cells by major histocompatibility complex class I or II (MHC I or MHC II), or both, promoting one or both humoral/cellular responses.14 Thus, surface conjugation of CLRs ligands (i.e., mannan) in combination with polymer-based strategies has proved to be a powerful approach for the rational design of targeted nanovaccines to induce robust immune responses.15
Our research team has largely worked on protein oral delivery by means of pH-responsive hydrogels.16 Extensive studies on methacrylic acid (MAA)-based hydrogel compositions such as poly(methacrylic acid –grafted-ethylene glycol (P(MAA-g-PEG))17 and poly(methacrylic acid-co-N-vinylpyrrolidone) (P(MAA-co-NVP))18 as delivery systems for protein therapeutics such as insulin,19 calcitonin,20 and interferon beta21 have been successfully carried out within our group. Furthermore, the tunable physicochemical characteristics of these polymer networks allow for modulation of biological activity through attachment of targeting ligands and mucoadhesion enhancers.22,23 Herein, we describe the synthesis of nanogels of poly(2-hydroxiethyl methacrylate-co-methacrylic acid), P(HEMA-co-MAA) and their in vitro evaluation as an oral vaccine delivery platform. The surface of these carriers is optimized by the covalent linkage of mannan to mimic carbohydrate moieties found on the surface of pathogens,24 providing “pathogen-like” properties to enhance M cell uptake3 and specifically target CLRs on APCs,15 with the additional benefit of potential complement activation.25 Results demonstrated the efficacy of the proposed surface-modified pH-responsive nanogels to efficiently release a model antigen in conditions similar to those at the small intestine (i.e., neutral pH) and to be uptaken by and activate APCs (i.e., macrophages).
MATERIALS AND METHODS
Chemicals
HEMA, MAA, ethylene glycol dimethacrylate (EGDMA), potassium persulfate (KPS), poly(vinyl alcohol) (PVA) MW 31000–50000 hydrolyzed 98–99%, ovalbumin from chicken egg (OVA), Mannan from Saccharomyces cerevisiae, 1-ethyl-3-(3-(dimethylamino)propyl) carbodiimide hydrochloride (EDC), N-hydroxisuccinimide (NHS), MES sodium salt, Dulbecco’s Modified Eagle Medium (DMEM; high glucose without l-glutamine), phenol red free DMEM, penicillin, streptomycin, sterile 1× phosphate buffer saline (PBS), Hank’s buffered salt solution without (HBSS) calcium and magnesium, chloroacetic acid 99% A.C.S. grade, and Alexa Fluor594 wheat germ-agglutinin (WGA) were purchased from Sigma-Aldrich (St. Louis, MO). Sodium hydroxide (NaOH), hydrochloric acid (HCl), 100% ethanol, potassium chloride (KCl), glacial acetic acid, nitric acid, acetone, basic alumina, sulfo-N-hydroxisuccinimide (suflo-NHS), ethylendiamine dihydrochloride, and paraformaldehyde were obtained from Thermo Fisher Scientific Inc. (Fairlawn, NJ). CellTiter 96 Aqueous Non-Radioactive Cell Proliferation Assay (MTS) was purchased from VWR International (Radnor, PA), and Pro-Long with DAPI was acquired from Molecular Probes Invitrogen (Grand Island, NY).
Methods
Synthesis of P(HEMA-co-MAA) Nanogels
Hydrogels were prepared by thermal-initiated surfactant-free emulsion polymerization, as described elsewhere26 with some modification. Prior to use, MAA was vacuum distilled23 to remove the inhibitor (hydroquinone). HEMA and EGDMA were passed through a basic alumina column for the same purpose and stored at 4 °C until use. For the preparation of particles, the stabilizer, poly(vinyl alcohol) (PVA) was dissolved in 50 mL of ultrapure water (Milli-Q Plus, Millipore) for the preparation of the continuous phase, at a concentration of 1, 1.5, or 3% (w/v). In the interim, monomers were mixed at a molar feed ratio of 3:1, 1:1, and 1:3 of HEMA/MAA and at 1 wt % to the continuous phase, to be assayed at each of the PVA concentrations prepared, yielding a total of nine different formulations. The cross-linker, EGDMA, was added to the monomers at 1.0 mol % of total monomers. Then, the monomers and cross-linker solution was added to the continuous phase and mixed in an ultrasonic bath (VWR Symphony Ultrasonic, VWR International) for an hour. Prior to polymerization, potassium persulfate (KPS, initiator) was added in the amount of 0.05 wt % of total monomers, and the mixture was purged with nitrogen for 20 min to remove oxygen. Polymerization was carried out under a nitrogen atmosphere and constant magnetic stirring at a temperature of 70 °C for 24 h. After polymerization was completed, particles were purified by washing with methanol and 0.5 N NaOH several times to remove the unreacted monomers. For this purpose, the particles were precipitated and collected by centrifugation (Centra-CL3R with ThermoIEC Rotor, Thermo Electron Corporation) at 25 °C, 3150 rcf for 1 h. The resulting pellets were resuspended in a 1:1 mixture of ethanol and 0.5 N NaOH and the process was repeated four more times. Following, the nanogels were resuspended in DI water and dialyzed in 100 kDa MWCO Biotech RC dialysis tubing (Spectra/Por, Rancho Dominguez, CA) and freeze-dried (FreeZone Cascade Benchtop Freeze-Drying System, LabConco).
Nanogels Characterization
The pH-responsive swelling behavior of the hydrogel nanoparticles was characterized by measuring the z-average diameters and zeta potential using a Malvern ZetaSizer nano ZS (Malvern Instruments Corp., Malvern, U.K.) equipped with a 633 nm laser source and MPT-2 Autotitrator. For this purpose, 2 mg of freeze-dried particles were suspended in 10 mL of 5 mM KCl and filtered through 1.2 µm pore size syringe filters (Whatman Int. Ltd., VWR International) to eliminate larger aggregates. In the measuring process, the autotitrator increased the pH to 11 and then stepwise decreased the pH while measuring the z-average diameter and zeta potential for each pH value. To evaluate particle morphology, nanogels were imaged using a Hitachi S-5500 Scanning Electron Microscope (SEM; 30 kV). For this purpose, ~3 mg of nanoparticles were spread on carbon tab previously stuck to aluminum stubs and dried overnight. Samples were sputter coated with a 10 nm gold layer using a Pelco Model 3 sputter coater (Pelco International, Redding, CA).
Surface Modification of Nanogels
Synthesis of Carboxymethyl Mannan (CMM)
Carboxymethyl mannan (CMM) was prepared through base-catalyzed methylation.27,28 Briefly, 100 mg of Mannan from Saccharomyces cerevisiae were dissolved in 1 mL of DI water and 1 mL of 1 M NaOH was added dropwise with constant stirring while the solution was kept in an ice bath. After 30 min, 1 mL of a 20% w/v chloroacetic acid solution were subsequently added dropwise, and then the mixture was allowed to react at 60 °C for 6 h with constant stirring. Finally, the mixture was cooled down to RT and neutralized with 1 M HCl. CMM was collected by precipitation in cold absolute ethanol, dialyzed overnight in DI water, and freeze-dried.
Mannan Attachment to Nanogels
Mannan was conjugated to the surface of P(HEMA-co-MAA) nanogels by an amine-carboxylic acid coupling reaction.15,29,30 In a previous step, ethylenediamine moieties were attached to the carboxylated mannan (AECM-MN) by incubating a solution of 50 mg of CMM in 1 mL of 0.1 M MES at pH 7.4 with 5 mg of 1-ethyl-3-(3-(dimethylamino)propyl) carbodiimide hydrochloride (EDC) and 5 mg of sulfo-N-hydroxysuccinimide (Sulfo-NHS), to activate the CMM’s carboxylic acid groups, and 160 mg of ethylenediamine dihydrochloride. The reaction mixture was incubated at RT for 12 h with constant agitation. After the incubation period, the AECM-MN was purified overnight by dialysis against DI water and freeze-dried. Conjugation of AECM-MN to the surface of nanogels was then performed by suspending the nanogels (2% w/v) in 0.1 M MES buffer following by the addition of 5 mg of Sulfo-NHS and 5 mg of EDC; mixture was allowed to react for 30 min at RT with constant end-to-end rotation. After the incubation period was completed, particles were centrifuged 3× at 8000 rpm for 5 min at 25 °C (Eppendorf MiniSpin Plus, Eppendorf), and the supernatant was removed to eliminate any remaining reagents. A new incubation was carried out by resuspending the particles in 500 µL of 0.1 MES with 0.1 mg of AECM-MN and incubating for 12 h at RT with constant rotation. Then, the particles were centrifuged 3× at 8000 rpm for 5 min at 25 °C, the supernatant was removed, and the pellet was resuspended once in ethanol and 2× in DI water. Finally, the particles were dried under vacuum for 2 h.
Characterization of Mannan-Modified Nanogels
The amount of mannan associated with the P(MAA-co-HEMA) nanogels was estimated by differences between the initial amount added and the amount quantified in the supernatants collected during the purification process. The amount of mannan in the initial solution and supernatants samples was estimated by a colorimetric phenol-sulfuric acid assay using a mannan standard curve as a reference. Additionally, an in vitro agglutination assay was performed to verify the conjugation of mannan to nanogels and to assess the availability of mannose ligand after conjugation. Briefly, mannan-modified nanogels were dispersed in water (1% w/v) and incubated with concanavalin A (Con-A). A time-dependent increase in turbidity at 550 nm was monitored spectrophotometrically for 3 h.31 Unmodified nanogels were also assayed with Con-A as control.
Ovalbumin (OVA) Loading
OVA loading was done by equilibrium partitioning. Initially, nanogels were incubated overnight in 5 mL of 10 mM phosphate buffer solution (PBS, pH 7.4) with constant end-to-end rotation to ensure wetting and swelling of the nanogels. A solution of OVA was prepared at a concentration of 2 mg/mL in PBS, pH 7.4. Before the addition of OVA to the particles, nanogels were allowed to set down for 2 min and a 200 µL sample of supernatant was taken and filtered using 0.22 µm PVDF filters (Millipore, Bedford, MA) and then replaced with equal amounts of 10 mM PBS, pH 7.4. Then, 5 mL of OVA solution were added to the particles suspension and incubated at room temperature (RT) with constant end-to-end rotation. Supernatant samples were taken at fixed time points; 1, 2, 4, 6, and 24 h. After 24 h of incubation, particles were collapsed using 0.1 N HCl until reaching pH 3.5. The supernatant was removed and its pH was adjusted to 7.4 with 0.1 N NaOH and stored at 4 °C until assayed. The particles were resuspended with 5 mL of 10 mM PBS at pH 3.5 and vortexed, followed by centrifugation at 300 rcf and 25 °C for 5 min. The resulting supernatant was again taken and its pH adjusted to 7.4 with 0.1 N NaOH, and stored at 4 °C until assayed. Finally, the particles pellet was resuspended in 5 mL of 10 mM PBS at pH 3.5 and freeze-dried. Determination of OVA concentration in the supernatant was carried out with a Micro BCA Protein Assay Kit (Thermo Scientific, Sugar Land, TX).
Loading efficiency was defined as follows:23
| (1) |
where C0 is the initial OVA concentration and Cf is the final OVA concentration remaining in solution.
OVA Release
Release studies aimed to simulate the pH changes the P(HEMA-co-MAA) nanogels would experience in vivo when passing through the stomach and into the small intestine (i.e., from an acidic pH to a neutral pH).23 For this experiment, 5 mg of OVA-loaded nanogels was suspended in low-adhesion microcentrifuge tubes by adding 2 mL of 10 mM PBS at pH 3.0 and vortexing. The suspensions were maintained at 37 °C with constant end-to-end rotation for a total of 90 min, and samples from the supernatant were taken at 0, 5, 10, 15, 30, 60, and 90 min of incubation and replaced with an equal volume of PBS buffer, pH 3.0. After 90 min, pH was raised to 7.0 with 0.1 N NaOH, and the suspensions were further incubated at 37 °C with constant end-to-end rotation for a total of 6 h, taking samples from the supernatant as described before, at 0, 5, 10, 20, 30, 60, 120, 240, and 360 min of incubation. Determination of OVA concentration in the supernatant was carried out with a Micro BCA Protein Assay Kit.
In Vitro Evaluation of Unmodified and Mannan-Modified P(HEMA-co-MAA) Nanogel’s Targeting Capabilities
Cell Culture
L929 fibroblasts and RAW 264.7 macrophages cell lines (American Type Culture Collection (ATCC, Rockwell, MD) were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% l-glutamine and 1% penicillin and streptomycin.32 Cultures were maintained in T-75 flasks (Corning, Corning, NY) at 37 °C in a humidified environment of 5% CO2 (Sheldon Signature HEPA Clean CO2 incubator, VWR). The medium was replaced every other day, and the cells were routinely passaged at 80% confluence. For experiments, cells were detached mechanically and adjusted to the required concentration of viable cells by counting in a hemocytometer.
In Vitro Nanogel Cytotoxicity
L929 fibroblasts and RAW 264.7 macrophages cells were seeded at 20000 cells per well in 96-well plates (Thermo Scientific Nunc Microwell Microplates, Waltham, MA) in phenol red free DMEM with 2% FBS and incubated at 37 °C, 5% CO2, for 24 h before testing. Cells were incubated with particle suspensions at concentrations of 1, 0.5, 0.25, and 0.125 mg/mL in phenol red free DMEM with 2% FBS. Lived control wells received only medium, and lysed control wells were included. Following 2 and 48 h of incubation, the particles were removed and the cells were rinsed 3× with sterile Hank’s balanced salt solution (HBSS). Cell viability was then assayed by adding the CellTiter Aqueous One Solution reagent (MTS, Promega) directly to the culture wells and incubating for 2 h. Finally, the absorbance at 690 nm (background) and 490 nm (MTS assay) was recorded using a microplate reader (Synergy HT, BioTek Instruments, Inc., Winooski, VT). 690 nm readings from each well were subtracted from absorbance (OD) values at 490 nm, and the viability fraction was defined as
| (2) |
Evaluation of Uptake by RAW 264.7 Macrophages Cell Line
In order to visualize nanogel uptake by confocal microscopy, unmodified and mannan-modified nanogels were conjugated to the CF488A amine fluorescence dye (Biotium, Hayward, CA). Briefly, 10 mg of nanogels were suspended in 0.1 M MES buffer at pH 4.7 (1% w/v) and vortexed. A total of 5 mg of Sulfo-NHS and 5 mg of EDC were added and suspensions were mixed gently by vortexing and incubated for 30 min at RT with constant end-to-end rotation. After initial activation of surface carboxylic acid groups, nanogels were centrifuged at 8000 rpm for 5 min, the supernatant was removed, and nanogels were resuspended in 500 µL of 0.1 M MES buffer (pH 4.7) by vortexing. This step was repeated one more time to ensure unreacted reagents were removed. After the final wash step, nanogels were resuspended in 500 µL of 0.1 M MES buffer (pH 4.7) and 5 µL of 1 mg/mL CF488A amine fluorescence dye was added. Nanogel suspensions were mixed by vortexing and incubated at RT for 2 h with constant end-to-end rotation. After the reaction time was completed, nanogels were washed twice, as previously described, once with 0.1 M MES buffer, pH 4.7, and a second time with DI water. Finally, nanogels were dried under vacuum for at least 2 h.
To study nanoparticle interactions with macrophages, RAW 264.7 macrophage cells were seeded in 12-well culture plates (Thermo Scientific Nunc Microwell Microplates, Waltham, MA) containing an acid washed 18 mm circular coverslip on each well (Thermo Fisher Scientific Inc.) at a density of 0.5 × 105 cells/well in supplemented DMEM and incubated overnight at 37 °C in humidified environment of 5% CO2. Next day, the medium was removed by aspiration and 1 mL of CF488A-labeled nanogels suspension in 10 mM sterile calcium and magnesium-free PBS were added to the cells to achieve a final concentration of 125 µg/mL. Cells were subsequently incubated for 2 h at 37 °C in humidified environment of 5% CO2. After incubation time, cells were washed 3× with sterile ice cold calcium and magnesium-free PBS to remove extracellular and nonadherent particles, and then fixed with 0.3 mL of 4% paraformaldehyde in PBS (pH 7.4) for 10 min at RT. Cultures were washed again 3× with HBSS and stained with 0.2 mL of Alexa Fluor594 wheat germ-agglutinin (WGA) at 5 µg/mL in HBSS for 10 min at RT in complete darkness. Cells were further washed 2× with HBSS and once with DI water, and the coverslips were finally mounted by placing them cell-side down on a drop of Pro-Long with DAPI mounting media (Invitrogen, Carlsbad, CA) on glass slides. The slides were cured overnight at RT and coverslips were stabilized in glass slides by using nail polish. Confocal microscopy was performed using a Leica SP2 AOBS laser scanning confocal microscope (Leica Microsystems, Buffalo Grove, IL). Final image preparation was performed utilizing particle-counting algorithms of ImageJ v1.36b software (NIH, Bethesda, MD).
Cell Surface Marker Expression Analysis by Flow Cytometry
For flow cytometry analysis of surface molecule expression a previous protocol described by Torres et al. was adapted.33 RAW 264.7 macrophages cells were cultured as previously described and seeded in 12-well culture plates at 1 × 106 cells/well in 2 mL of supplemented DMEM media. After overnight incubation at 37 °C in humidified environment of 5% CO2, unmodified and mannan-modified OVA-loaded nanogels suspended in culture medium were added to the macrophage cultures at a concentration of 125 µg/mL. Unstimulated macrophages and cells stimulated with lipopolysaccharide (LPS, 100 ng/mL) were used as negative and positive controls, respectively. Cultures were incubated for 24 h (37 °C, 5% CO2). After 24 h of stimulation, macrophages were harvested and washed with azide free and serum/protein free PBS, and then stained for cell viability with the fixable viability dye eFluor 780 (eBioscience, San Diego, CA) for 30 min at 4 °C. After viability staining was completed, cells were washed with fluorescence-activated cell sorting buffer (FACS, 0.1% w/v sodium azide and 0.1% w/v bovine serum albumin in phosphate buffer saline). A total of 0.1% v/v of unlabeled hamster IgG and 0.1% v/v/of antimouse CD16/CD32 antibody (eBioscience, San Diego, CA) were added to cultures in order to block Fcγ receptors and prevent nonspecific binding. After blocking for 20 min at 4 °C, cells were then incubated with the appropriate antibody or isotype control for 30 min at 4 °C. Antibodies used included allophycocyanin (APC) antimouse MHC II (I-Ad, clone AMS-32–1), PerCP-eFluor 710 antimouse CD40 (clone 1C10), and fluorescein isothiocyanate (FITC)- conjugated antimouse CD80 (B7–1, clone 16–10A1) purchased from eBioscience (San Diego, CA), and (PE)/Cy7 antimouse CD86 (clone GL-1) and (PE) antimouse CD206 (MMR, clone C068C2) purchased from BioLegend (San Diego, CA). Isotype-specific control antibodies included APC-conjugated mouse IgG2b κ, PerCP-eFluor 710 rat IgG2a κ, FITC-conjugated Armenian Hamster IgG purchased from eBioscience (San Diego, CA), and (PE)/Cy7 Rat IgG2a κ and (PE) rat IgG2a κ from BioLegend (San Diego, CA). Single color compensation controls were prepared using OneComp eBeads (eBioscience, San Diego, CA), as described by the manufacturer. Following staining, cells were washed in FACS buffer and fixed in 3% paraformaldehyde and stored at 4 °C until analysis. Analysis was performed on a BD LSRFortessa cell analyzer (BD Bioscience, San Jose, CA) and data were analyzed using FlowJo software (TreeStar, Inc., Ashland, OR).
Analysis of Released Cytokines by ELISA
Cell-free supernatants were collected from DCs cultured for 24 h in the presence of unmodified and mannan-modified OVA-loaded nanogels and stored at −20 °C until analysis. Pro-inflammatory cytokines IL-6 and TNF-α were measured using cytokine-specific ELISA kits from eBioscience (San Diego, CA).
Statistical Analysis
All the experiments were performed with n = 3–6, and data were analyzed with the statistical software IBM SPSS Statistics 20, one-way ANOVA, and Student–Newman–Keuls. Post Hoc tests were used to determine statistical significance among treatments, and p-values < 0.05 were considered significant.
RESULTS AND DISCUSSION
Synthesized P(HEMA-co-MAA) Nanogels Showed pH-Responsive Profile Required for Oral Delivery of Protein Antigens
The surfactant-free emulsion polymerization technique34–36 is a useful approach to overcome problems related with the presence of these additives and their purification processes, such as instrument calibration, pore size determination, and destabilization of polymer latex by coagulation or flocculation after surfactant removal.37 As a consequence of the lower total particle surface area that can be stabilized in the absence of surfactant, particle number is characteristically lower in surfactant-free emulsion polymerization processes, resulting in a higher particle size. However, this can be overcome by the addition of a nonionic stabilizer such as PVA.26 Stabilizers provide colloidal stabilization via steric interference with the van der Waals attraction between polymer particles.37 Several researchers reported the size dependence on the concentration of a stabilizer for various polymerization systems,38–40 describing an inversely proportional relationship between size and stabilizer concentration.
To the best of our knowledge, this is the first time that this technique is used to produce pH-responsive nanogels. In these pH-responsive systems, the polymer network contains weak acid or base groups that undergo complexation by hydrogen bonding, thus, exhibiting pH-dependent swelling properties with fast expansion and contraction.41 In the case of acrylic acid-based copolymers,17 the monomer MAA provides the weak acid groups that allow for swelling performance.41 In the proposed system, MAA is copolymerized with the monomer HEMA, which has many advantageous properties for biomedical use including high water content, low toxicity, and tissue compatibility.27 The presence of polar groups of hydroxyl and carboxyl and hydrophobic α-methyl groups42 allows for the interaction with proteins of different isoelectric points,43 and the selection of comonomers can provide further improvement of HEMA copolymer carrier’s performance for a given compound.44
In this study, different ratios of monomers HEMA/MAA (i.e., 3:1, 1:1, and 1:3) in combination with different concentrations of stabilizer, PVA (i.e., 1, 2, and 3 wt %) were assayed to obtain particles with an adequate pH-responsive behavior and size (i.e., nanosized particles). Aiming to obtain a nonvariable mesh size in all the formulations, not leading to significant differences in posterior assays, cross-linker concentration, EGDMA, was used in the constant amount of 1.0 mol % of total monomers for all the formulations, which has been a widely assayed cross-linker ratio for HEMA-MAA hydrogels.45,46
SEM
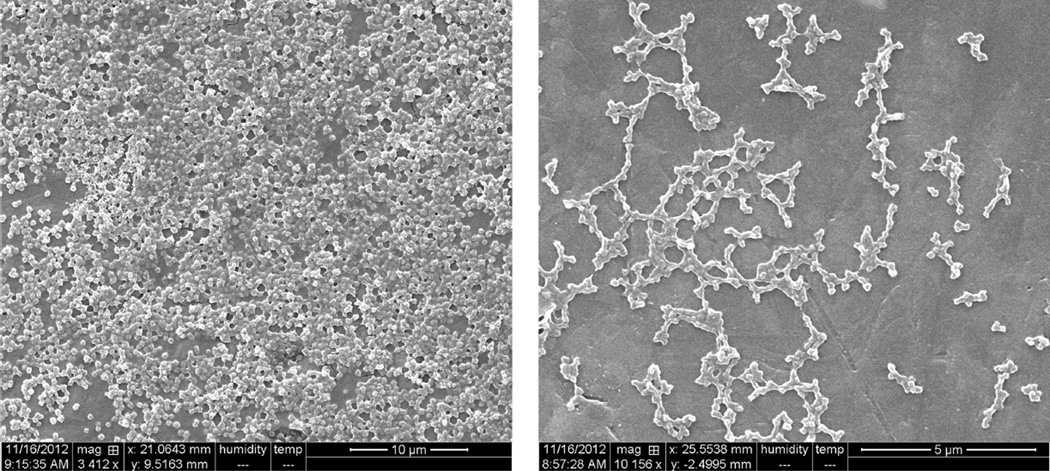
Freeze-dried particles were mounted on aluminum stubs and observed by SEM. Representative SEM microphotographs of 1:3 HEMA/MAA nanogels are shown Figure 1. Spherical particles with a size in the nanometer range were observed, which is in agreement with light scattering measurements. Some aggregation was noted in all the samples, which may be due to the dehydration process during freeze-drying of nanogels and sample preparation.
Figure 1.
SEM images of freeze-dried spherical polyanionic P(HEMA-co-MAA) nanogels (magnification 3412−10156×, Hitachi S-5500 Scanning Electron Microscope (SEM; 30 kV)).
Light Scattering
The elucidation of size was crucial in understanding the performance of the P(HEMA-co-MAA) delivery system and its potential behavior throughout human gastrointestinal (G.I.) tract. Monodisperse polyanionic nanoparticles were obtained for all the formulations assayed with size ranging from 292 nm to 1.5 µm, as determined by SEM (Figure 1) and corroborated by light scattering measurements. No significant differences in particle size were observed when PVA concentrations varied from 1 to 3 wt % (data not shown). This is probably due to the fact that the minimum concentration assayed was already enough to provide steric stability to the amount of particles produced and their surface in contact with the continuous phase. Thus, higher stabilizer concentrations did not provide further stability to the suspension, and formulations with 1 wt % were selected to continue with further studies.
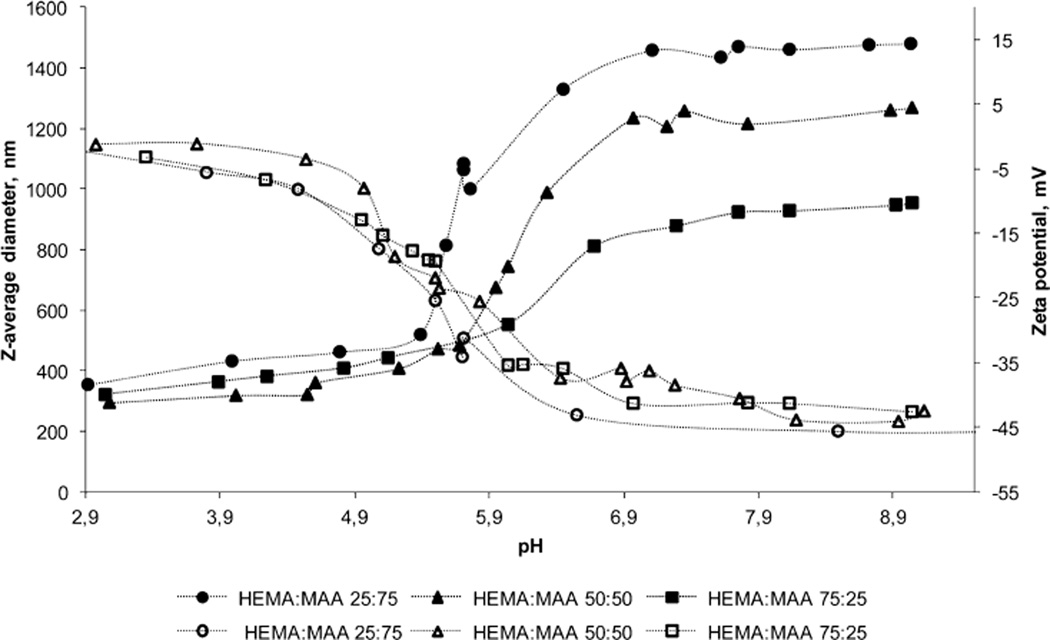
The pH-responsive swelling profile of the particles produced with 1 wt % PVA is pictured as of z-average diameter and zeta potential values in Figure 2. As it can be observed, all the formulations showed swelling/deswelling pH-responsive behavior due to hydrogen bonding with a clear transition in size and zeta potential measures between pH values of 5.8 and 7.0. This pH transition frame is attributed to MAA pKa value, which is 4.6 but has been reported to undergo significant modifications due to the change of chemical environment upon polymerization.47 Furthermore, a similar increase in pH transition values was previously reported for various copolymer pH-responsive networks containing MAA.40,48,49 Owing to this transition, the nanogel particles exhibited an increase in zeta potential and a decrease in z-average diameter as pH was decreased in the experiment; at high pH values, pendent MAA groups are ionized, showing zeta potential values of −45 mV and leading to the dissociation of the interpolymer network.20 The electrostatic repulsion causes the polymer complex to swell, and hence, z-average diameter values of 961.8 nm to 1.5 µm were observed, depending on the polymer composition (Figure 2). These divergences in particle size are probably due to the degree of MAA present in the network; higher contents of ionizable monomer led to higher repulsion and dissociation within the polymer network. Previous studies on MAA copolymerized with N-vinylpyrrolidone (P(MAA-co-NVP)) reported a higher degree of hydrogen bonding complexation at low pH values when higher concentrations of MAA were employed,49 which accounts for a larger z-average diameter transition in particles with higher contents of MAA. Furthermore, similar results were previously obtained with P(MAA-g-PEG) networks,17 where a higher MAA feed ratio correlated with the more dynamic change in size on gel nanospheres as the pH was increased. Therefore, larger swollen particles were obtained for 1:3 HEMA/MAA nanoparticles relative to 1:1 HEMA/MAA particles, which were subsequently larger than 3:1 HEMA/MAA particles. As pH was progressively decreased, hydrogen bonding minimized the electrostatic repulsion leading to deswelling of particles, which reached similar sizes of about 300 nm for all the formulations. These size values are also in accordance with published studies on pH-responsive acrylic-based polymer nanospheres.17 Zeta potential values increased as well to near 0 mV due to surface charge neutralization, which led to poor colloidal stability of the suspension and difficulty for the measure of particles32 at pH values lower than 3 (data not shown).
Figure 2.
Synthesized P(HEMA-co-MAA) nanogels showed pH-responsive profile required for oral delivery of antigens. pH-responsive swelling of polyanionic P(HEMA-co-MAA) nanoparticles prepared with 1:3; 1:1, and 3:1 HEMA/MAA ratio, 1 mol % EGDMA, and 1 wt % PVA. z-Average diameter (filled icons) and zeta potential values (empty icons) were measured using dynamic light scattering with Malvern ZetaSizer Nano ZS Instrument equipped with MPT-2 Autotitrator. Formulations were not colloidally stable at low pH values; measures at pH values lower than 3 are not shown.
Hence, particle size at the swelled state could be modulated based on the initial monomer feed ratio employed, which is highly advantageous since nanoparticle size plays a critical role in cell uptake,5 amount of delivered antigen,1 and bias of immune response,8 as previously exposed. In addition, formulation parameters such as initiator molar ratio and monomers and cross-linker concentration in the system could be further explored in terms of particle size modulation. Since the obtained results were optimal for the purpose of this study, and higher swelling potentially leads to higher release of compounds within the delivery system, 1:3 HEMA/MAA particles were selected to continue with further studies.
Mannan-Attachment to Nanogels Surface was Accomplished by Amine–Carboxylic Acid Coupling
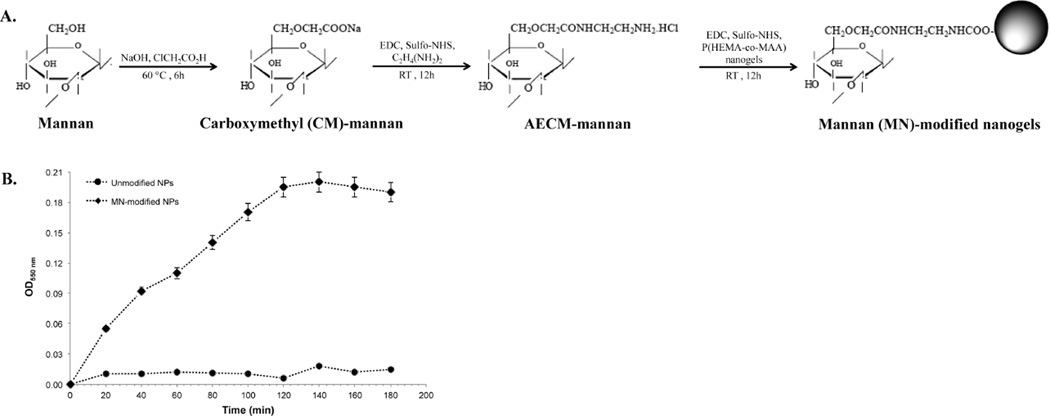
Mannan, an intended ligand to CLR on APCs, was attached to nanogels in three steps, which are depicted in Figure 3A. In the first step, carboxymethylation of mannan (CM-mannan) was achieved by a reaction of mannan with sodium hydroxide and chloroacetic acid. Infrared spectroscopy indicated the presence of a carbonyl group at 1740 cm, as evidence of carboxymethylation, which was corroborated by an acid–base titration procedure (data not shown). In the second step, CM-mannan was reacted with ethylenediamine hydrochloride in the presence of EDC and Sulfo-NHS as coupling agent to form aminoethylcarbomethylmannan (AECM-mannan). CHN elemental analysis indicated the substitution degree of between 7 and 9%.
Figure 3.
Mannan-attachment to nanogels surface was accomplished by amine-carboxylic acid coupling. (A) Molecular structures of mannan and intermediates products, as well as mannan-attached to nanogels surface. (B) In vitro ligand agglutination of unmodified and MN-modified nanogels with Con-A. Optical density was measured at 550 nm. Values represent mean ± SD (n = 3).
In the final step, AECM-mannan was conjugated on the surface of P(HEMA-co-MAA) nanogels by an amine-carboxylic acid coupling reaction. Specifically, the amine groups introduced to mannan after the reaction with ethylenediamine were reacted with carboxylic acid residues from MAA. To corroborate the attachment of mannan moieties to the surface of the nanogels, a colorimetric phenol-sulfuric acid assay was employed. Higher absorbance at 490 nm, were observed for mannan-modified P(HEMA-co-MAA) nanogels when compared with unmodified nanogels (data not shown). By utilizing a standard curve generated with AECM-mannan and unmodified nanogels as controls to account for polymer interference with the assay, the mannan content was found to be about 20 µg per mg of nanogels. Mannan presence, integrity, and activity after attachment to P(HEMA-co-MAA) nanogels was also confirmed by the agglutination test with Con-A, which binds specifically with saccharides such as mannose, fructose, and glucose residues. As shown in Figure 3B, the measured absorbance at 550 nm increased when Con-A is added to MN-modified nanogels suspension, while the unmodified formulation shows no significant increase in measured absorbance. The results clearly indicate that mannose residues from mannan are capable of interacting with the lectin receptors, which may result in efficient internalization by cells. The increase in absorbance is due to the increase in turbidity caused by agglutination of Con-A on interaction with mannose.
OVA Antigen was Efficiently Loaded into P(HEMA-co-MAA) Nanogels
An OVA loading optimization experiment was performed to determine the appropriate amount of time for loading of OVA into the nanogels.23 From this study, it was concluded that maximum loading efficiency was achieved at the longest incubation time tested, that is, 24 h, with a loading efficiency value of 24 and 12 wt % of loaded OVA relative to particle mass. Loading efficiencies were also determined after the collapsing of particles with the addition of HCl and posterior washing of remaining amounts of OVA not encapsulated, as registered in Table 1. After the addition of acid, loading efficiency increased to 57%, which is most probably due to the entrapment of the protein within the collapsed polymer network. It should be noted that loading efficiencies are usually reported to decrease after acid addition, due to the collapse of the network forcing some of the protein out of the polymer.23 In comparison, loading efficiency of OVA into the particles proved to increase after HCl addition, which could be attributed to the isoelectric point (IEP) of the protein that determines a net charge at low pH values. Hence, the protein would lose molecular mobility with the decrease of pH, favoring the entrapment within the polymer complex. In addition, the loading efficiency value experimented a slight decrease after washing the particles by centrifugation, which is probably due to certain amounts of OVA adsorbed on particles surface in the collapsing step that were eliminated after washing.
Table 1.
OVA Loading Efficiencies for P(HEMA-co-MAA) Nanoparticles after 24 h Incubation and Both before and after Acid Collapsing
| loading efficiency (%) ± SD |
wt (%) loaded (mg OVA/mg particles) ± SD |
|
|---|---|---|
| before collapsing | 24 ± 9.6 | 12 ± 4.2 |
| after collapsing | 57 ± 3.6 | 29 ± 0.5 |
| after washing | 51 ± 2.0 | 26 ± 0.6 |
Both MN-Modified and Unmodified P(HEMA-co-MAA) Nanogels Release Antigen at pH of the Small Intestine
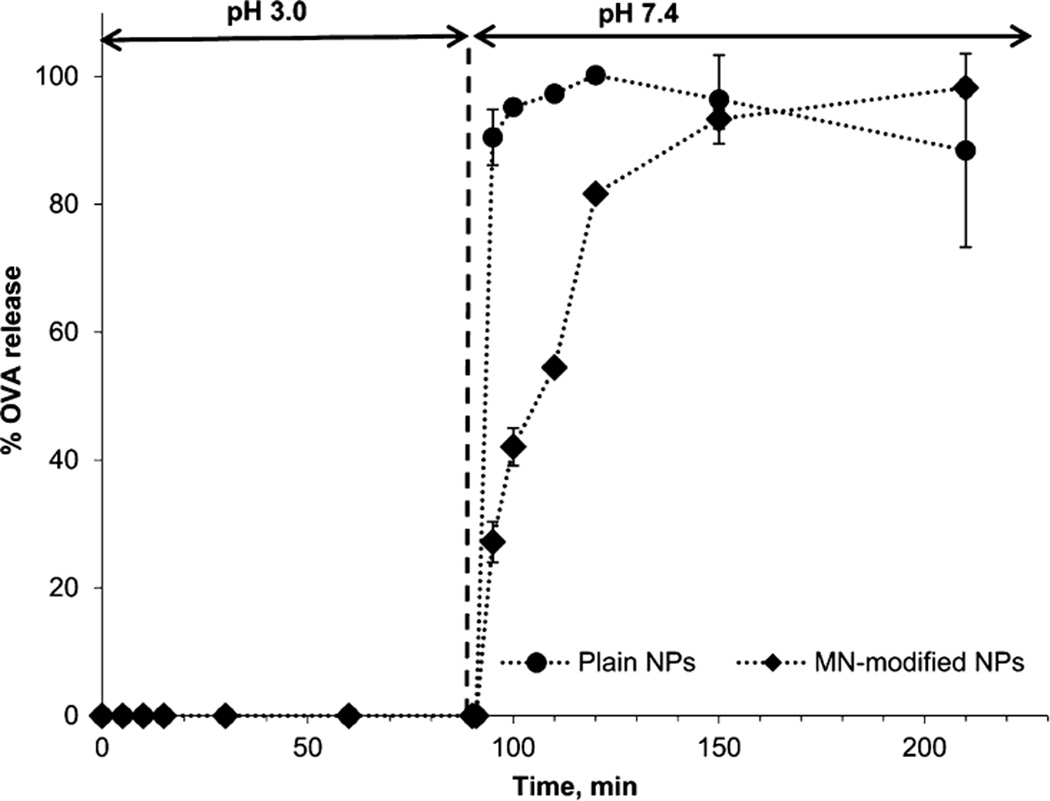
OVA release studies aimed to simulate pH changes that the P(HEMA-co-MAA) nanogels would experience in vivo when passing through the stomach and into the small intestine (i.e., from an acidic pH to a neutral pH). Figure 4 shows the results of OVA release from P(HEMA-co-MAA) and mannan-modified P(HEMA-co-MAA) (MN-modified NPs) nanogels. At low pH, both unmodified and MN-modified nanogels limited the release of OVA from the particles. After 90 min, the release media pH was increased and the protein was rapidly released from the particles, which is in agreement with the previous experiments. P(HEMA-co-MAA) carriers released the maximum amount of protein after 30 min at pH 7.4. MN-modified P(HEMA-co-MAA) carriers also showed a fast initial release but reached 81.65% of total OVA at that time point, showing a lower release rate in a second stage and reaching the maximum amount of protein release at 210 min. This slower release profile could be attributed to electrostatic interactions between the released protein and mannan molecules on the particles surface. Similar results were reported in the literature for calcitonin-loaded and insulin-loaded poly(methacrylic acid-grafted-ethylene glycol) (P(MAA-g-EG).20,23 Another possibility to explain the slower released profile observed from mannan-modified particles could be a restricted swelling potential due to surface cross-linking by the presence of mannan molecules. More detailed characterization is needed to corroborate any of these hypothesis but those studies are out of the scope of the present work. This study demonstrated that OVA is quickly released from both unmodified and mannan-modified P(HEMA-co-MAA) carriers after the pH is increased above the pKa of MAA. Furthermore, release results indicate that the change in pH between stomach and the small intestine can be used as a physiologic trigger to release OVA from these nanogels. As the particles pass into the small intestine, they would recognize a pH shift to ~7, which ionize MAA pendent groups triggering decomplexation of the hydrogel and quick release of the compound at the targeted site. Although the fast release profile observed at pH 7.4 for both unmodified and mannan-modified P(HEMA-co-MAA) nanogels may translate to premature release of the antigen before nanogels can reached the Payer’s patches for M-cell uptake; these nanogels would meet the criteria of antigen protection from degradation upon mucosal delivery,6 initially stated in this work.
Figure 4.
Both MN-modified and unmodified P(HEMA-co-MAA) nanogels release antigen at pH of the small intestine. OVA release profiles from pH-responsive P(HEMA-co-MAA) and MN-P(HEMA-co-MAA) NPs incubated in 10 mM PBS undergoing a shift in pH from 3.0 to 7.4 at 90 min.
In Vitro Evaluation of Unmodified and Mannan-Modified P(HEMA-co-MAA) Nanogel’s Targeting Capabilities
In Vitro Biocompatibility of Unmodified and Mannan-Modified P(HEMA-co-MAA) Nanogels was Demonstrated in Fibroblast (L929) and Macrophage (RAW 264.7) Cell Cultures
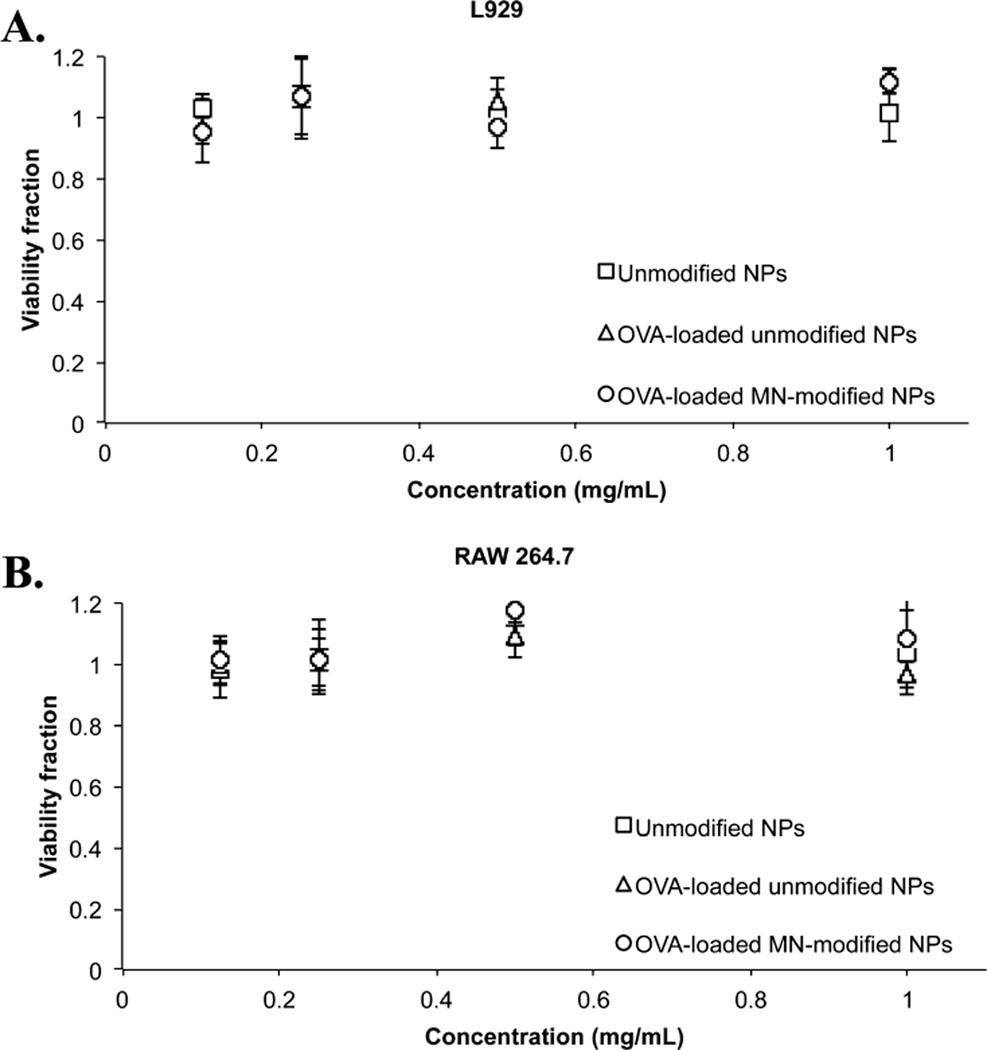
Fibroblast (L929) and macrophage (RAW 264.7) cells were used to determine biocompatibility of nonloaded and OVA-loaded and unmodified and MN-modified P(HEMA-co-MAA) nanogels. Results are shown in Figure 5. Cell viability was determined using MTS assay after 2 and 48 h of incubation with the particles. Both cell lines presented similar responses when exposed to the different carrier formulations, showing no cytotoxic effect of neither of the formulations assayed. Post hoc tests were applied to cell viability data with IBM SPSS Statistics 20 software to analyze if there were statistically significant differences between values. Student–Newman–Keuls test showed no statistically significant differences between the treatments and provided an only statistical subgroup (p = 0.053).
Figure 5.
In vitro biocompatibility of unmodified and mannan-modified P(HEMA-co-MAA) nanogels was demonstrated in fibroblast (L929) and macrophage (RAW 264.7) cell cultures. (A) Fibroblasts L929 and (B) RAW 264.7 cell viability fraction assayed with MTS after 48 h incubation with unmodified and OVA-loaded unmodified and OVA-loaded MN-modified P(HEMA-co-MAA) nanogels. No significant cytotoxicity was observed at the concentrations tested with either cell line; n = 3−6 ± SD.
Both Unmodified and Mannan-Modified P(HEMA-co-MAA) Nanogels were Internalized Effectively by Macrophages, but Higher Internalization Rates were Observed for MN-Modified Nanogels
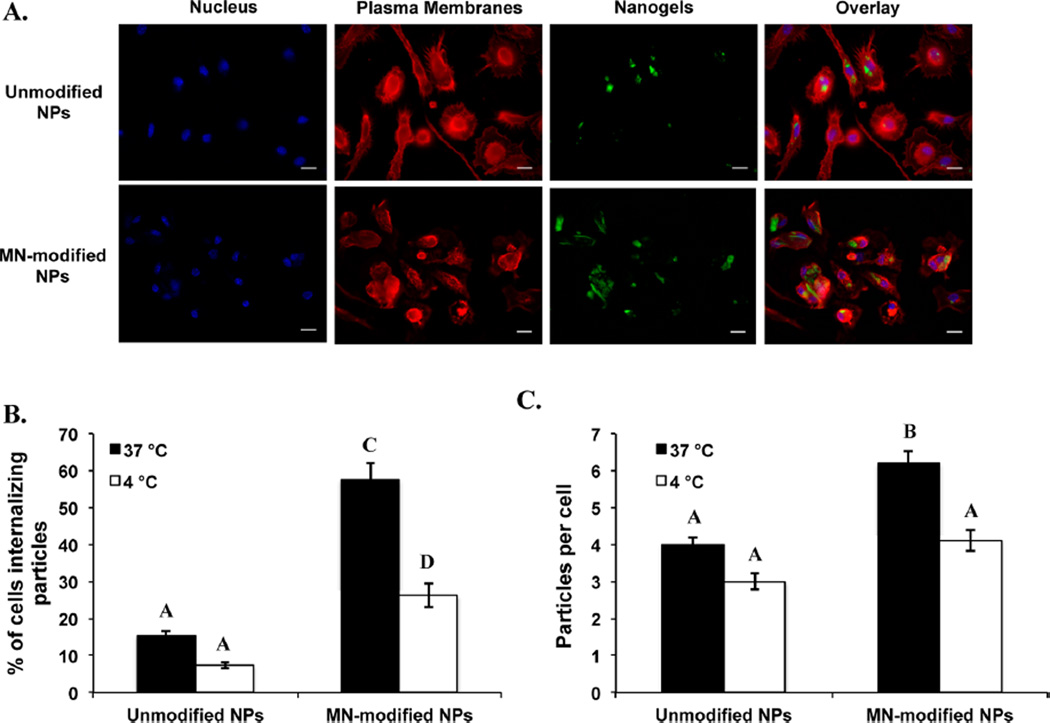
To determine the effect of mannan-attachment to the surface of P(HEMA-co-MAA) nanogels in the internalization of nanogels by macrophages, microscopic analyses were performed. As shown in Figure 6A, both unmodified and mannan-modified P(HEMA-co-MAA) nanogels were efficiently internalized by macrophages. These qualitative findings were corroborated by applying particle-counting algorithms, results of this quantitative analysis are summarize in Figure 6B,C. Higher percentage of cells internalizing nanogels (Figure 6B) as well as average number of nanogels internalized per cell (Figure 6C) resulted when macrophages were incubated with mannan-modified P(HEMA-co-MAA) nanogels when compared with unmodified nanogels. Together, this data shows that surface functionalization of P(HEMA-co-MAA) nanogels with mannan significantly improved their internalization by macrophages. The enhanced uptake of mannan-modified nanogels increases the likelihood that more macrophages would be available to present antigen to T cells following in vivo administration of these particles.
Figure 6.
Both unmodified and mannan-modified P(HEMA-co-MAA) nanogels were internalized effectively by macrophages but higher internalization rates were observed for MN-modified nanogels. (A) Chemical attachment of amine-containing fluorescent dye CF 488A (green) to the surface of P(HEMA-co-MAA) nanogels allow for imaging using confocal microscopy. Cell membrane (red) and nucleus (blue) were stained after 2 h of incubation of cells with the unmodified or mannan-modified nanogels. (B) Percentage of cells internalizing unmodified or mannan-modified nanogels. (C) Average number of unmodified or mannan-modified nanogels per cell. Data are expressed as the mean ± SEM of three independent experiments. Different letters indicate statistically significant differences between the groups at p < 0.05.
These results also suggest that mannan-modified nanogels are internalized by a different mechanism than unmodified nanogels. While mannan-modified nanogels may interact directly with CLRs present on macrophages driving a receptor-mediated endocytosis mechanism,12,50 a nonendocytotic or energy-independent pathway may be involved in the internalization of unmodified nanogels.51 To corroborate this hypothesis, internalization studies were performed at 4 °C, which inhibits all energy-dependent pathways.51,52 As shown in Figure 6B, during incubation, the percentage of cells internalizing unmodified P(HEMA-co-MAA) nanogels did not decrease significantly when incubation was performed at 4 °C; the internalization of mannan-modified P(HEMA-co-MAA) nanogels was significantly decreased (from ~60 to 25%) at this temperature. These results suggests that endocytotic or energy-dependent mechanisms are involved in the cellular uptake of mannan-modified P(HEMA-co-MAA) nanogels, although more detailed studies around this observation are needed to establish a clear mechanism for internalization of both unmodified and MN-modified P(HEMA-co-MAA) nanogels.
Mannan-Modified P(HEMA-co-MAA) Nanogels Enhanced the Expression of Costimulatory Molecules on Macrophages
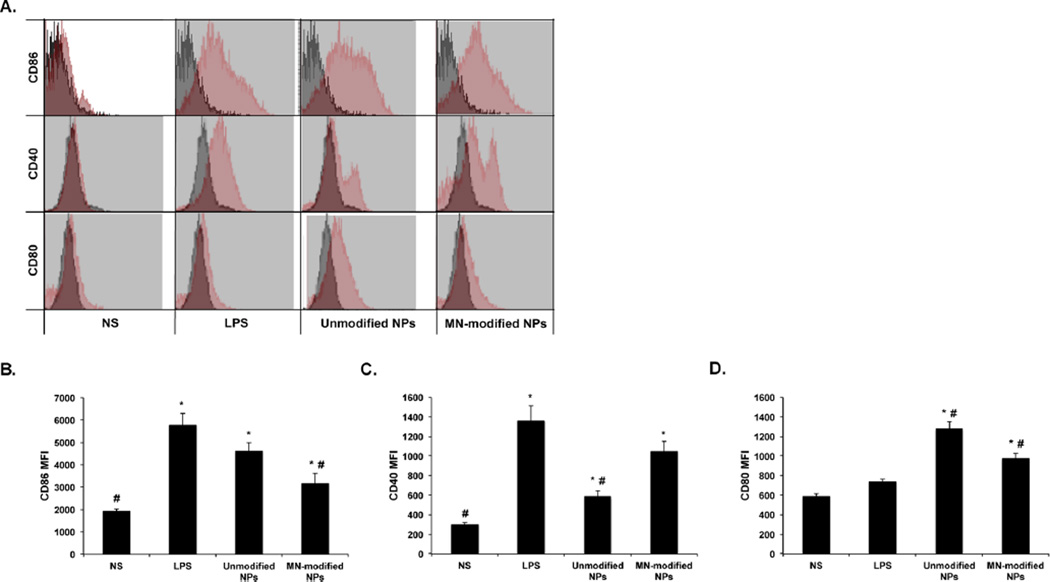
Flow cytometry was utilized to evaluate the stimulating effects of unmodified and mannan-modified P(HEMA-co-MAA) nanogels in comparison with LPS, a pathogen associated molecular pattern (PAMP) known to enhance the expression of cell surface markers on APCs by interacting with specific PRRs.53 Stimulation of RAW 264.7 macrophages with unmodified or MN-modified P(HEMA-co-MAA) nanogels enhanced the expression of the T cell costimulatory molecules CD86 (Figure 7B), CD40 (Figure 7C), and CD80 (Figure 7D) over nonstimulated cells. Moreover, the expression of these cell surface marker molecules was, in most of the cases, comparable or at higher levels than the positive control (i.e., LPS). This is also noted in the histograms by the increased shift from the isotype control (Figure 7A). The expression of these costimulatory molecules on APCs is of great relevance since they play an important role in further antigen presentation and activation of T cells, which may lead to the generation of a most robust immune response.54 The enhanced stimulation of costimulatory molecules by unmodified P(HEMA-co-MAA) nanogels may be due to the interaction of macrophages with the hydrophobic surface of these nanogels, which may result in endogenous danger signals, which are known to stimulate robust immune responses in the absence of inflammatory response.55 Consistent with this statement, the secretion of pro-inflammatory cytokines (i.e., IL-6 and TNF-α) by macrophages was not increased after stimulation with unmodified P(HEMA-co-MAA) nanogels when compared with nonstimulated controls (data not shown).
Figure 7.
Mannan-modified P(HEMA-co-MAA) nanogels enhanced the expression of costimulatory molecules on macrophages. Expression of cell surface molecules on the surface of macrophages (RAW 264.7) cells was studied by flow cytometry after incubation of cells with unmodified and MN-modified P(HEMA-co-MAA) nanogels for 24 h. (A) Represented histograms comparing between treatments in red and the isotype nonspecific control in gray. Treatments represented in the histograms include unmodified P(HEMA-co-MAA) nanogels, MN-modified P(HEMA-co-MAA) nanogels, and unstimulated cells and lipopolysaccharide (LPS)-stimulated cells were used as negative and positive controls, respectively. Below the histograms is the complete set of results for mean fluorescence intensity (MFI) expression of RAW 264.7 macrophages cell surface markers: (B) CD86, (C) CD40, and (D) CD80. Data are graphed as mean ± SEM of a minimum of three replicates per stimulation group. * and # represent groups that are significantly different (p < 0.05) from the nonstimulated or LPS-treated DCs, respectively.
As shown in Figure 7C, cells incubated with mannan-modified P(HEMA-co-MAA) nanogels showed the higher expression of CD40 surface molecules. The expression of this molecule on APCs has been directly correlated to increase in particle internalization,56 which is consistent with the results previously discussed in which MN-modified nanogels were internalized at a higher rate than unmodified nanogels (Figure 6).
Previous published work showed that dimannose-functionalized polymer nanoparticles enhance the expression of DC activation markers (i.e., MHC II, CD40, and CD86) by targeting the mannose receptor (MMR, CD206).15 The stimulating effect of mannan-modified P(HEMA-co-MAA) nanogels may, therefore, be attributed to the direct targeting of CLRs (i.e., MMR) on macrophages with the mannan ligands attached on the surface of nanogels. To corroborate this hypothesis, the expression of the macrophage mannose receptor (i.e., CD206) was assessed after stimulation with MN-modified P(HEMA-co-MAA) nanogels. Results did not show an increase in the expression of MMR compared to unmodified nanogels (data not shown). These findings may indicate that the mannan-ligands presence on the surface of P(HEMA-co-MAA) nanogels do not interact directly with the mannose receptor on macrophages. However, the observed increase in internalization and enhancement in the expression of costimulatory molecules may be attributed to targeting of other CLRs, such as the DC-SIGN receptor. It has been previously suggested that DC-SIGN recognized and interact more specifically with high-order mannose structures,12,14 which is the case of the mannan-ligand utilized in the design of these novel nanogel carriers.
The observed efficient internalization of both unmodified and mannan-modified nanogels by macrophages and the ability of both carrier formulations to induce the expression of costimulatory molecules indicate the potential of both systems for efficient antigen delivery to APCs. The need of taking an extra bioconjugation step (i.e., mannan functionalization of nanogels) will depend on the specific targeting capabilities that could be obtained with the incorporation of surface ligands and the influence that direct targeting will have in modulating the induced immune response, which needs to be addressed mainly in in vivo systems.
CONCLUSIONS
Oral drug delivery is a route of choice for vaccine administration because of its noninvasive nature. However, most bioactive drugs such as peptides and proteins result in low bioavailability lack of control on absorbed dose and need of adjuvants. An effective oral vaccine delivery system that overcomes these problems must protect the antigen from degradation upon mucosal delivery, penetrate mucosal barriers and control the release of the antigen and costimulatory and immunomodulatory agents to specific immune cells (i.e., APCs). In this paper, mannan-modified pH-responsive P-(HEMA-co-MAA) nanogels were synthesized and their effectiveness as carriers for oral vaccination was assessed. The nanogels showed pH sensitive properties, entrapping and protecting the loaded cargo at low pH values, and triggered protein release after pH switching to intestinal pH values. Unmodified P(HEMA-co-MAA) nanogels showed intrinsic adjuvant properties by being efficiently internalized by macrophages and induced the expression of costimulatory molecules. Surface decoration with mannan as carbohydrate moieties resulted in enhanced internalization by macrophages as well as increasing the expression of relevant costimulatory molecules (i.e., CD40) at higher levels to those observed for unmodified nanogels. Collectively, these findings indicate that both unmodified and mannan functionalized P(HEMA-co-MAA) nanogels are a viable strategy for enhancing antigen delivery to APCs and, in turn, these delivery platforms may facilitate a more efficacious oral vaccination regimen.
ACKNOWLEDGMENTS
M.D.-L. thanks Junta de Andalucía for financial support and is especially grateful to Jennifer Knipe for her assistance with SEM imaging.
Footnotes
The authors declare no competing financial interest.
REFERENCES
- 1.Woodrow KA, Bennett KM, Lo DD. Annu. Rev. Biomed. Eng. 2012;14:17–46. doi: 10.1146/annurev-bioeng-071811-150054. [DOI] [PubMed] [Google Scholar]
- 2.des Rieux A, Fievez V, Garinot M, Schneider Y, Preat V. J. Controlled Release. 2006;116:1–27. doi: 10.1016/j.jconrel.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 3.Azizi A, Kumar A, Diaz-Mitoma F, Mestecky J. PLoS Pathog. 2010;6:e1001147. doi: 10.1371/journal.ppat.1001147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson-Welder JH, Torres MP, Kipper MJ, Mallapragada SK, Wannemuehler MJ, Narasimhan B. J. Pharm. Sci. 2009;98:1278–1316. doi: 10.1002/jps.21523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark MA, Jepson MA, Hirst BH. Adv. Drug Delivery Rev. 2001;50:81–106. doi: 10.1016/s0169-409x(01)00149-1. [DOI] [PubMed] [Google Scholar]
- 6.Storni T, Kundig TM, Senti G, Johansen P. Adv. Drug Delivery Rev. 2005;57:333–355. doi: 10.1016/j.addr.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 7.Garside P, Mowat AM. Semin. Immunol. 2001;13:177–185. doi: 10.1006/smim.2001.0310. [DOI] [PubMed] [Google Scholar]
- 8.Mann JFS, Shakir E, Carter KC, Mullen AB, Alexander J, Ferro VA. Vaccine. 2009;27:3643–3649. doi: 10.1016/j.vaccine.2009.03.040. [DOI] [PubMed] [Google Scholar]
- 9.Guery JC, Galbiati F, Smiroldo S, Adorini L. J. Exp. Med. 1996;183:485–497. doi: 10.1084/jem.183.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mowat AM. Nat. Rev. Immunol. 2003;3:331–341. doi: 10.1038/nri1057. [DOI] [PubMed] [Google Scholar]
- 11.Pashine A, Valiante NM, Ulmer JB. Nat. Med. 2005;11:S63–S68. doi: 10.1038/nm1210. [DOI] [PubMed] [Google Scholar]
- 12.Figdor CG, van Kooyk Y, Adema GJ. Nat. Rev. Immunol. 2002;2:77–84. doi: 10.1038/nri723. [DOI] [PubMed] [Google Scholar]
- 13.Geijtenbeek TBH, Gringhuis SI. Nat. Rev. Immunol. 2009;9:465–479. doi: 10.1038/nri2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Kooyk Y, Rabinovich GA. Nat. Immunol. 2008;9:593–601. doi: 10.1038/ni.f.203. [DOI] [PubMed] [Google Scholar]
- 15.Carrillo-Conde B, Song E, Chavez-Santoscoy A, Phanse Y, Ramer-Tait AE, Pohl NLB, Wannemuehler MJ, Bellaire BH, Narasimhan B. Mol. Pharmaceutics. 2011;8:1877–1886. doi: 10.1021/mp200213r. [DOI] [PubMed] [Google Scholar]
- 16.Liechty WB, Peppas NA. Eur. J. Pharm. Biopharm. 2012;80:241–246. doi: 10.1016/j.ejpb.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foss AC, Goto T, Morishita M, Peppas NA. Eur. J. Pharm. Biopharm. 2004;57:163–169. doi: 10.1016/S0939-6411(03)00145-0. [DOI] [PubMed] [Google Scholar]
- 18.Carr DA, Peppas NA. J. Biomed. Mater. Res., Part A. 2010;92A:504–512. doi: 10.1002/jbm.a.32395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lowman AM, Morishita M, Kajita M, Nagai T, Peppas NA. J. Pharm. Sci. 1999;88:933–937. doi: 10.1021/js980337n. [DOI] [PubMed] [Google Scholar]
- 20.Torres-Lugo M, Peppas NA. Macromolecules. 1999;32:6646–6651. [Google Scholar]
- 21.Kamei N, Morishita M, Chiba H, Kavimandan NJ, Peppas NA, Takayama K. J. Controlled Release. 2009;134:98–102. doi: 10.1016/j.jconrel.2008.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wood KM, Stone GM, Peppas NA. Acta Biomater. 2010;6:48–56. doi: 10.1016/j.actbio.2009.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wood KM, Stone GM, Peppas NA. Biomacromolecules. 2008;9:1293–1298. doi: 10.1021/bm701274p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernandez N, Alonso S, Valera I, Vigo AG, Renedo M, Barbolla L, Crespo MS. J. Immunol. 2005;174:8154–8162. doi: 10.4049/jimmunol.174.12.8154. [DOI] [PubMed] [Google Scholar]
- 25.Reddy ST, van der Vlies AJ, Simeoni E, Angeli V, Randolph GJ, O’Neill CP, Lee LK, Swartz MA, Hubbell JA. Nat. Biotechnol. 2007;25:1159–1164. doi: 10.1038/nbt1332. [DOI] [PubMed] [Google Scholar]
- 26.Oeztuerk N, Bereli N, Akgoel S, Denizli A. Colloids Surf., B. 2008;67:14–19. doi: 10.1016/j.colsurfb.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 27.Karabinos JV, Hindert M. Adv. Carbohydr. Chem. 1954;9:285–302. doi: 10.1016/s0096-5332(08)60378-1. [DOI] [PubMed] [Google Scholar]
- 28.An NT, Dong NT, Le Dung P, Thien DT. Carbohydr. Polym. 2011;83:645–652. [Google Scholar]
- 29.Sheehan JC, Boshart GL, Cruickshank PA. J. Org. Chem. 1961;26:2525–&. [Google Scholar]
- 30.Staros JV, Wright RW, Swingle DM. Anal. Biochem. 1986;156:220–222. doi: 10.1016/0003-2697(86)90176-4. [DOI] [PubMed] [Google Scholar]
- 31.Copland MJ, Baird MA, Rades T, McKenzie JL, Becker B, Reck F, Tyler PC, Davies NM. Vaccine. 2003;21:883–890. doi: 10.1016/s0264-410x(02)00536-4. [DOI] [PubMed] [Google Scholar]
- 32.Forbes DC, Creixell M, Frizzell H, Peppas NA. Eur. J. Pharm. Biopharm. 2013;84:472–478. doi: 10.1016/j.ejpb.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 33.Torres MP, Wilson-Welder JH, Lopac SK, Phanse Y, Carrillo-Conde B, Ramer-Tait AE, Bellaire BH, Wannemuehler MJ, Narasimhan B. Acta Biomater. 2011;7:2857–2864. doi: 10.1016/j.actbio.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chainey M, Hearn J, Wilkinson MC. J. Polym. Sci., Part A: Polym. Chem. 1987;25:505–518. [Google Scholar]
- 35.Li JQ, Salovey R. J. Polym. Sci., Part A: Polym. Chem. 2000;38:3181–3187. [Google Scholar]
- 36.Ni HM, Du YZ, Ma GH, Nagai M, Omi S. Macromolecules. 2001;34:6577–6585. [Google Scholar]
- 37.Odian G. Principles of Polymerization. New York: Wiley-Interscience; 2004. [Google Scholar]
- 38.Bulmus V, Tuncel A, Piskin E. J. Appl. Polym. Sci. 1996;60:697–704. [Google Scholar]
- 39.Tuncel A, Kahraman R, Piskin E. J. Appl. Polym. Sci. 1994;51:1485–1498. [Google Scholar]
- 40.Torres-Lugo M, Peppas NA. J. Nanopart. Res. 2002;4:73–81. [Google Scholar]
- 41.Caldorera-Moore M, Peppas NA. Adv. Drug Delivery Rev. 2009;61:1391–1401. doi: 10.1016/j.addr.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chou KF, Han CC, Lee S. Polym. Eng. Sci. 2000;40:1004–1014. [Google Scholar]
- 43.Michalek J, Pradny M, Artyukhov A, Slouf M, Smetana K. J. Mater. Sci.: Mater. Med. 2005;16:783–786. doi: 10.1007/s10856-005-2617-2. [DOI] [PubMed] [Google Scholar]
- 44.Moustafa AB, Sobh RA, Rabie AM, Nasr HE, Ayoub MMH. J. Appl. Polym. Sci. 2013;129:853–865. [Google Scholar]
- 45.Ende MTA, Peppas NA. J. Appl. Polym. Sci. 1996;59:673–685. [Google Scholar]
- 46.Kotsmar C, Sells T, Taylor N, Liu DE, Prausnitz JM, Radke CJ. Macromolecules. 2012;45:9177–9187. [Google Scholar]
- 47.Dong H, Du H, Qian X. J. Phys. Chem. A. 2008;112:12687–12694. doi: 10.1021/jp807315p. [DOI] [PubMed] [Google Scholar]
- 48.Lowman AM, Peppas NA. Macromolecules. 1997;30:4959–4965. [Google Scholar]
- 49.Carr DA, Peppas NA. Macromol. Biosci. 2009;9:497–505. doi: 10.1002/mabi.200800235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adams EW, Ratner DM, Seeberger PH, Hacohen N. ChemBioChem. 2008;9:294–303. doi: 10.1002/cbic.200700310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Madani F, Lindberg S, Langel U, Futaki S, Graslund A. J. Biophys. 2011;2011:414729–414729. doi: 10.1155/2011/414729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Langel U. Cell-Penetrating Peptides: Processes and Applications. Boca Raton, FL: CRS Press; 2006. [Google Scholar]
- 53.Steinman RM. J. Invest. Dermatol. 2000;114:209–209. [Google Scholar]
- 54.Janewey CA, Travers P, Shlomichik MJ, Walport M. Immunobiology. New York: Garland Science; 2005. The immune system in health and disease; pp. 1–134. [Google Scholar]
- 55.Seong SY, Matzinger P. Nat. Rev. Immunol. 2004;4:469–478. doi: 10.1038/nri1372. [DOI] [PubMed] [Google Scholar]
- 56.Petersen LK, Ramer-Tait AE, Broderick SR, Kong C, Ulery BD, Rajan K, Wannemuehler MJ, Narasimhan B. Biomaterials. 2011;32:6815–6822. doi: 10.1016/j.biomaterials.2011.05.063. [DOI] [PubMed] [Google Scholar]