Abstract
The discovery of the first microRNA (miRNA) over 20 years ago has ushered in a new era in molecular biology. There are now over 2000 miRNAs that have been discovered in humans and it is believed that they collectively regulate one third of the genes in the genome. miRNAs have been linked to many human diseases and are being pursued as clinical diagnostics and as therapeutic targets. This review presents an overview of the miRNA pathway, including biogenesis routes, biological roles, and clinical approaches.
Keywords: miRNA, microRNA, Dicer, Drosha, Argonaute, RNAi
Graphical Abstract
1. miRNAs are small noncoding regulatory RNAs
The first miRNA was discovered over 30 years ago in the nematode Caenorhabditis elegans with the identification of the developmental regulator lin-4[1]. Originally believed to be a conventional protein coding gene, the Ruvkun and Ambros labs made the startling discovery that lin-4 did not code for a protein but instead coded for a 22 nucleotide regulatory RNA[2, 3]. They demonstrated that the lin-4 RNA could base pair with the mRNA for another gene in the C. elegans developmental network, lin-14, and control the production of the LIN-14 protein[3]. This discovery would have had limited significance outside of the C. elegans research community had a second miRNA, let-7, not been discovered[4]. More importantly, let-7 is conserved across many organisms, including humans, suggesting that this class of small regulatory RNAs has a more general role in biology[5]. The other stunning development, occurring around the same time, was the discovery of the RNAi pathway; specifically, the ~21 nucleotide RNA triggers of the silencing machinery[6-9]. These two pathways have since been shown to be different arms of the same gene silencing pathway[10-12]. Subsequently, many thousands of miRNAs have been discovered in many organisms, and there are currently 2588 annotated miRNAs in the human genome[13]. Since each miRNA is predicted to regulate the expression of hundreds of target genes, the miRNA pathway as a whole is a critical mechanism for gene expression control[14].
2. The canonical miRNA biogenesis pathway
All miRNA families undergo a series of biogenesis steps that convert the primary miRNA transcript into the active, ~22 nucleotide mature miRNA (see Figure 1). The mature miRNA is loaded into the RNA induced silencing complex (RISC) where it directs the complex to target mRNAs, leading to translational repression and target mRNA degradation. This section will cover the canonical miRNA biogenesis pathway that is responsible for the maturation of most miRNA families. For simplicity we will focus on the mammalian enzyme components. Several exceptions to this canonical pathway have been described for unique miRNA families, and are discussed in a later section.
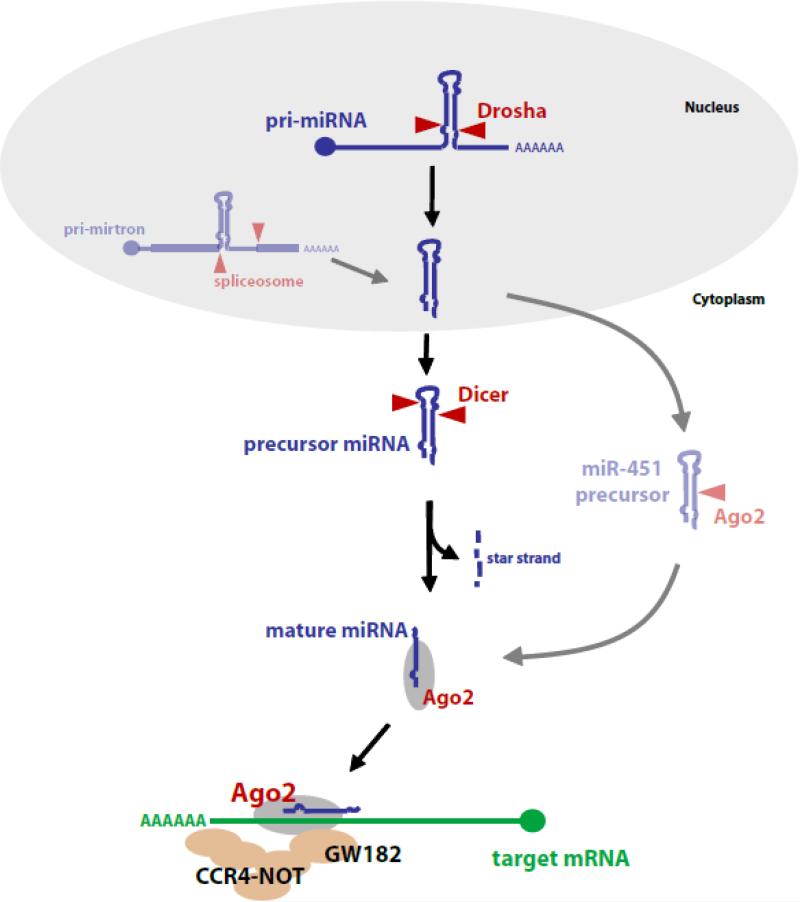
Figure 1. The miRNA biogenesis pathway.
The canonical miRNA biogenesis pathway is shown in the central part of the figure. miRNAs are transcribed by RNA polymerase II. The pri-miRNA transcript is cut by the enzyme Drosha yielding the precursor miRNA. After Dicer cleavage the mature miRNA is loaded into the effector complex RISC where it directs translational repression of mRNA targets. Two well characterized non-canonical biogenesis routes are shown in the semi-transparent graphic. mirtrons bypass the Drosha processing step; a splicing reaction produces the precursor. Subsequent biogenesis steps are the same. The second non-canonical route shown is specific for miR-451. This miRNA precursor is directly bound and cut by Argonaute 2 in a Dicer independent reaction. The remaining RNA is trimmed back to yield a mature miRNA.
2.1. Transcription
miRNA genes are located throughout the genome[15]. Many miRNA genes are noncoding genes whose sole transcriptional product is the miRNA. In other cases the miRNA is located within an intron or untranslated region (UTR) of a protein coding gene. Figure 2 exemplifies a cluster of three miRNAs located within an intron of the protein coding gene MCM7[16, 17]. The defining feature of all miRNA genes is the stem-loop precursor RNA structure, with one (or sometimes both) strands of the stem the source of the mature miRNA[18-20]. In the case of MCM7 there are three clustered miRNA stem loops, each leading to a distinct mature miRNA with a unique target set[21]. miRNAs are not generally located within coding exons, as excision of the miRNA would lead to loss of the protein coding transcript.
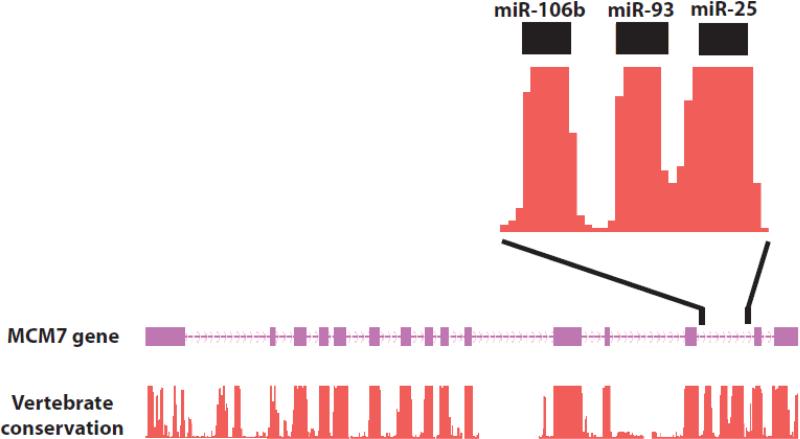
Figure 2. Anatomy of a miRNA poly-cistron.
The miRNA cluster containing miR-106b, miR-93, and miR-25 is shown in its genomic context. Three miRNAs are located within an intron of the protein coding gene MCM7. Boxes in the MCM7 gene model represent exons, and the arrow line represents introns. Sequence conservation is shown. Data were adapted from the UCSC genome browser.
Transcription of the miRNA host gene by RNA polymerase II leads to the pri-miRNA, or primary miRNA transcript[22, 23]. The pri-miRNA is typically spliced, capped, and polyadenylated in a similar manner to protein coding mRNAs[24]. While most miRNA promoters have not been characterized in detail, the few examples studied are similar in structure to protein coding gene promoters, including control elements and histone marks[25-33].
2.2. Processing by Drosha and Dicer
The pri-miRNA requires two endonuclease processing steps before it becomes a mature, active miRNA[34]. The first processing step occurs during or subsequent to transcription of the pri-miRNA by the enzyme Drosha[35-38]. The RNA binding protein DGCR8 is associated with Drosha and is required for cleavage of the pri-miRNA[39-41]. Drosha cleavage releases the stem-loop precursor from flanking pri-miRNA transcript sequences. The precursor is exported out of the nucleus in a Ran-GTPase dependent manner by Exportin5[42-44]. In the cytoplasm the second processing step occurs. The endonuclease Dicer cleaves the loop region of the precursor releasing the mature miRNA[10-12, 45]. Like Drosha, Dicer is also associated with an RNA binding protein, in this case TRBP[46-50]. The product of the Dicer reaction is a duplex RNA of approximately 21 nucleotides length (depending on the miRNA). One strand of the duplex is loaded into RISC as a mature miRNA. The other strand, termed the star strand, is typically degraded. An asterisk (star) is appended to the miRNA name to designate this star strand, e.g. miR-145*. However, for some miRNAs both strands may be loaded into RISC at similar frequencies. In this case, the strand from the 5′ end of the stem-loop is termed “5p” and the 3′ strand the “3p”. In fact, while RISC loading may strongly favor incorporation of one strand, recent next generation sequencing (NGS) efforts have demonstrated a small fraction of star strand loaded for essentially all miRNA families[51]. Further complicating the nomenclature, some miRNAs show different strand usage depending on cell type or biological state[52]. For these reasons, 5p/3p naming schemes are being increasingly used rather than the arbitrary mature/star nomenclature.
The specific nucleotide locations of Drosha and Dicer cleavage are usually narrowly defined, leading to mature miRNAs with clearly defined terminal ends. Some miRNAs, however, have heterogenous cleavage sites, leading to multiple “isomiRs” of the mature miRNA[53, 54]. This heterogeneity could lead to differential target repression and distinct biological activities. IsomiR preference can vary by cell type and isomiR switching has been detected in disease[52, 55, 56]. The molecular mechanism for isomiR switching is unclear.
2.3. RISC loading and target repression
The ultimate fate of the miRNA is to be incorporated into RISC (or miRISC). The exact composition of this protein complex is not clear but contains the essential protein Argonaute, of which four family members have been identified in humans (Ago1-4)[57, 58]. After Dicer cleavage, the miRNA duplex is loaded into RISC and the star strand is removed by one of several possible mechanisms. If the miRNA duplex has complementarity in the central region, the star strand can be cut by Ago2 and further degraded by the nuclease complex C3PO[59-62]. This is the mechanism for RISC loading for the related siRNA pathway. Most miRNA duplexes, however, lack central complementarity and therefore cannot participate in star strand cleavage. These miRNA duplexes rely on strand unwinding, and several helicases have been described to possess this activity[63-67].
Argonaute directly binds the mature miRNA and seeks target mRNAs that have complementarity to the miRNA. In particular, nucleotides 2-7 of the miRNA, termed the “seed” region, are important for target association[68-70]. The 3′ end of the miRNA can also contribute to target recognition, and centrally matched targets have been identified[71-74]. If complementarity exists in the central region of the miRNA (nucleotides 9-11) then the mRNA target can be cleaved via the endonuclease activity of Ago2[75, 76]. Most miRNA targets in humans, however, lack this central sequence match and are not directly cleaved by Ago2. Rather, Argonaute is recruited to a complex containing GW182 (TNRC6A/B/C) within cytoplasmic P bodies where translational repression occurs. The CCR4-NOT deadenylase complex is recruited to RISC and this facilitates removal of the poly(A) tail and eventual degradation of the mRNA target [77-79].
2.4. Alternative pathways
While the majority of identified miRNAs are produced by the canonical pathway there are several examples of alternative biogenesis. The most common variation is the mirtron class of miRNAs[80-83]. These miRNAs, located in introns at the exon junction site, bypass the Drosha step. Instead, the precursor ends are formed by the splicing cleavage events. The resultant precursor stem loop is processed by Dicer and loaded into RISC. Dicer independent miRNA biogenesis has also been described[84-86]. The best studied example is miR-451. After transcription and Drosha cleavage the precursor directly binds Ago2, instead of Dicer, and Ago2 cleaves the star strand (like it would cleave a target RNA). The remaining loop sequence is trimmed back to yield the pre-loaded, mature miR-451. Other miRNAs may follow this same pathway[87]. These non-canonical pathways are of particular relevance when interpreting data from Dicer or Drosha/DGCR8 knockout studies.
While the aforementioned RNAs are mature miRNAs that are generated through non-canonical routes, there are other classes of small RNAs that are not miRNAs, yet are produced by Drosha and/or Dicer activity and bound to RISC[88]. The best characterized class of these RNAs is the tRNA derived small RNA fragments (tRF or tsRNAs). These RNAs are a similar size to miRNAs and can be moderately abundant. Some tsRNAs are produced by Dicer cleavage of mature tRNAs and are loaded into RISC[89, 90]. Another class is derived from outside the mature tRNA and is generated by the tRNA splicing reaction[89, 90]. Examples of these RNAs are able to repress gene expression similar to conventional miRNAs, and have been shown to have essential cellular functions[90].
Further complicating the model, several studies have suggested that miRNAs can function independent of the RISC complex. A quantitative analysis of the total miRNA population in a cell, compared to the RISC protein copy number, suggested that a large fraction of miRNAs are not associated with RISC[91]. The functional roles of these RISC-free miRNAs are not known. However, one miRNA, miR-328, has been reported to be in a complex with the RNA binding protein hnRNP E2[92]. This miRNA functionally blocks association of the RNP with its target mRNA C/EBPα, enhancing translation of the C/EBP protein. It should be noted that these non-canonical pathways appear to be rare events. For example, miR-328 predominantly functions as a canonical miRNA within the RISC complex, directing translational repression of target mRNAs[93-97].
3. miRNA target identification
Defining target sets for miRNAs is important for several reasons. For biologists, defining the target set of a miRNA is key to understanding its biological role. For scientists developing miRNA therapeutics, validated targets provide the best biomarker(s) for determination of the efficacy of a miRNA mimic or inhibitor. The identification of miRNA targets has followed three general approaches: bioinformatic target prediction, biochemical isolation of miRNA/mRNA complexes, and transcriptomic/proteomic analysis. The three approaches will be briefly summarized.
3.1. Bioinformatic target prediction
Since miRNAs base pair with target mRNAs using standard Watson-Crick rules, this fact should make bioinformatic target identification straightforward. Unfortunately, the most important determinant of target binding is the seed sequence of the miRNA which is only 6 nucleotides in length. This will lead to a great number of candidate targets, many of which are false negatives. Therefore, all bioinformatic target prediction algorithms use additional factors to improve accuracy[98]. Since the best characterized targets are in mRNA 3′ untranslated regions, many algorithms limit target sites to this region. Other factors are employed, including sequence conservation, target site accessibility, flanking sequence determinants, and compensatory pairing outside the seed region. Current approaches have also used machine learning algorithms that incorporate validated targets in the learning sets[98]. A number of algorithms have been developed, with TargetScan, miRanda, and PicTar perhaps the most popular[69, 99-101]. In general, bioinformatic approaches are a good starting point for miRNA analysis and many research labs make use of them[102, 103].
3.2. Biochemical target identification
Several related approaches have used physical association of miRNA/RISC complexes with target mRNAs to isolate and identify targets. These are based on immunoprecipitation of RISC using anti-Argonaute antibodies, with or without prior RNA crosslinking, and definition of bound target RNAs by microarray or NGS profiling[104-107]. Crosslinking before cell lysis is generally preferred since artifactual RNA associations have been observed during cell lysis[108]. An alternative approach is isolation of specific biotinylated miRNAs followed by target identification[109, 110]. This has the advantage of capturing targets of a single known miRNA, but requires ectopic introduction of the biotinylated miRNA. While these physical approaches have been successfully used to define binding partners of miRNA complexes, not all bound mRNA targets may be biologically repressed targets. Studies have reported that some target sites, often in coding sequences of mRNAs, are bound to Argonaute but are not repressed targets[104]. Like all approaches, validation of individual targets is essential.
3.3. Omics -based strategies for target identification
The third general approach to target identification is a proteomic or transcriptomic analysis of cells/tissues in the presence and absence of a miRNA. Quantitative proteomic analysis directly measures the effect of a miRNA on protein production, and is possibly more reflective of the true target set, but is also technically challenging. Since most miRNA targets have reduced mRNA steady state levels, the simpler transcriptome analysis can be performed[111]. This can be done by microarray profiling and several analysis tools are available[112-115]. An example of this approach is the identification of targets of the neutrophil-specific miRNA miR-223[116]. Neutrophils were isolated from wild type and miR-223 knockout mice. Microarray and quantitative mass spectrometry were performed, and differences in mRNA and protein content was used to define targets of miR-223. It should be noted that the candidate target sets will also contain downstream secondary targets, requiring validation of individual targets.
4. Regulation of the miRNA pathway
Early work in the field focused on transcriptional regulation of miRNA host genes[25, 26, 117, 118]. For example, the cardiac-specific miRNA families miR-1 and miR-133 are transcriptionally regulated by well established cardiomyocyte transcription factor networks via SRF, MEF2, and MyoD[119-123]. Accordingly, enhancer elements for these factors have been identified in upstream regions of the miR-1/133 host genes. Similarly, miRNAs that are located in introns of protein coding genes are often regulated by the same transcription factor networks that control the protein coding mRNA. In general, approaches that have been used for mRNA promoter analysis will work similarly for miRNA promoters.
In addition to transcriptional regulation, miRNA production is often controlled during post-transcriptional biogenesis steps[34]. A well studied example of this is the regulation of the let-7 family during differentiation of embryonic cells. In stem cells the levels of mature let-7 are virtually undetectable. During differentiation, let-7 expression increases over a hundred fold, becoming one of the most abundant miRNA families in differentiated cells of a variety of cell types. Interestingly, transcription of the let-7 host genes does not increase significantly[124]. Rather, let-7 maturation is blocked in embryonic cells by the RNA binding protein Lin28[125-127]. This protein directly binds to the loop region of the pri-miRNA and precursor and inhibits Drosha and Dicer cleavage, respectively[128-130]. In a secondary repressive mechanism, Lin28 directs the uridyl transferase enzyme Tut4 to add an oligouridine tail to let-7 precursors, marking them for degradation[131-134]. During cellular differentiation, Lin28 expression is shut down allowing maturation of abundant lt-7 pri-miRNAs. More recently, other RNA binding proteins have been linked to the regulation of miRNA biogenesis, with activating and repressing roles described[135-138]. Regulated degradation of specific miRNAs has also been described[139].
5. Biological roles of miRNAs
The biological importance of miRNAs in mammalian development was first demonstrated with the generation of mouse models deficient for Dicer and DGCR8 (Drosha is involved in other RNA metabolic pathways while DGCR8 is specific for miRNA biogenesis, thus the DGCR8 knockout is more informative). Loss of either step in miRNA biogenesis results in embryonic lethality[140, 141]. Similarly, tissue-specific knockout of either gene leads to developmental defects in that tissue, underscoring the requirement for miRNA function in most tissue types[142]. Of course, knockout of these genes leads to loss of all miRNAs (except for the rare non-canonical miRNAs, described above); therefore, it is difficult to assign roles for specific miRNAs in the development of those tissues. This has been addressed by miRNA knockout mouse models[142, 143]. In general, individual miRNAs are not required for specification of individual tissues. For example, mice lacking the cardiac specific miRNA miR-208 still develop a heart[144, 145]. Rather, miRNAs are often required for maintaining tissue homeostasis. In the heart example, miR-208 deficient animals have defects in stress response and exhibit cardiac hypertrophy. miRNAs may be necessary for maintaining tissue differentiation state, since many tissue-restricted miRNAs are reduced in diseases, including cancer (see below).
Transgenic overexpression studies are another approach for the identification of the biological roles of individual miRNAs. These studies have the disadvantage of overexpression artifacts that could lead to misinterpretation of data. However, they have been used to uncover biological roles that have not been detected in knockout studies due to miRNA family redundancy. Furthermore, miRNA expression is often altered in disease, and overexpression studies can mimic this pathological condition (see below). The above-mentioned approaches, along with miRNA target network identification, have revealed roles for the miRNA pathway in the development and/or function of most tissues in the body. miRNAs have been linked to numerous aspects of neuronal function, including developmental remodeling, amygdala-dependent memory formation, dendritic spine development, and post-mitotic neuron cell survival [146-149]. miRNAs have been linked to the function of many hematopoietic lineages including neutrophils, lymphocytes, erythrocytes, megakaryocytes, an hematopoetic stem cells [17, 150-161]. These examples are a limited subset of the described functions of miRNAs in normal human biology.
6. Dysregulation of miRNAs in disease
Gene expression profiling studies have demonstrated alterations in miRNA expression in a wide range of human disease. In many cases, functional studies have linked miRNA dysregulation as a causal factor in disease progression. This section will summarize some of the disease states that have been linked to miRNA alterations.
6.1. Cancer
Alterations in miRNA expression have now been demonstrated in many cancer types. Early studies were performed using microarray, RT-PCR, and bead-based hybridization (Luminex) platforms, while more recent studies have used NGS-based profiling[117, 162-167]. miRNAs have been identified that are elevated in cancer, for example miR-21 and the miR-17-92 cluster, while other miRNA families are frequently downregulated in cancer, including the let-7 and miR-34 families[168]. Tissue specific miRNAs are often reduced in cancer: for example, the liver-specific miR-122 in hepatocellular carcinoma[169, 170]. This supports the hypothesis that many miRNA families reinforce cellular differentiation state, and reductions in these miRNAs may allow pathological reduction in differentiation or cell state. Functional studies have supported the role of some miRNAs in promoting or preventing cancer development and progression. A well established example is the miR-17-92 cluster. This cluster of six miRNAs is elevated in many cancer types, most notably in several lymphomas and leukemias[171]. Many studies have demonstrated that this miRNA cluster can promote cancer when ectopically expressed, often in cooperation with the oncogene c-Myc[172-175]. miR-34 is another notable cancer-associated miRNA family[176]. Comprised of three family members, these miRNAs are directly transactivated by p53 and function in parallel with p21 and Bcl2 family proteins to promote cell cycle arrest and apoptosis. Interestingly, this miRNA is at the focus of a frequent deletion in neuroblastoma. The functional importance of these miRNAs in cancer pathogenesis presents a strong case for their development as therapeutic targets[177].
Many other miRNA families are altered in cancers, and large scale profiling studies have uncovered signatures for cancer types and subtypes[178]. There is significant interest in developing miRNA signatures as clinical diagnostics.
6.2. Metabolic disease
miRNAs have been linked to several metabolic pathways, including cholesterol and fatty acid metabolism and transport, pancreatic islet function, and glucose metabolism[179]. miR-122 was introduced in an earlier section as a liver-specific miRNA, and is in fact the most abundantly expressed miRNA in hepatocytes. Targeted deletion of miR-122 in mouse models leads to reduced serum cholesterol and triglyceride levels[180, 181]. This effect was also observed in mouse and primate studies whereby miR-122 was inhibited by an antisense oligonucleotide[182, 183]. The effect on serum cholesterol is due in part to reduction in cholesterol biosynthetic enzyme expression in hepatocytes. miR-33 has been linked to serum high density lipoprotein (HDL) and triglyceride levels, though it is not clear if this is a hepatocyte-specific event or if expression in macrophages, among other cells, is controlling lipid metabolic regulatory networks[184-187]. Both of these miRNAs are being investigated as potential therapeutic targets.
Several miRNA families have been linked to glucose metabolism. miR-375 is highly expressed in pancreatic islet cells and regulates a large number of genes involved in islet cell proliferation and function[188, 189]. Let-7 has also been linked to glucose metabolism. Transgenic overexpression of let-7 in the pancreas leads to reduced glucose tolerance, while overexpression of the let-7 inhibitory protein Lin28 improves glucose uptake[190, 191]. The miR-103/107 family has also been functionally linked to glucose metabolism[192].
6.3. Viral pathogenesis
Several interesting connections have been made between miRNAs and virus biology. To date, 522 viral-encoded miRNAs have been discovered, most notably in the herpesvirus family[13]. While these miRNAs are not essential for virus replication, at least in the examples studied so far, there is evidence that miRNAs impact virus pathogenesis[193]. Functional studies have forged a link between viral miRNAs and the latent/lytic phase transition as well as host inflammatory response. This presents an interesting therapeutic option since these miRNAs are not in the human genome and interfering with their function should not have undesired side-effects.
Perhaps the most interesting example of a role for a miRNA in viral pathogenesis is the connection between miR-122 and hepatitis C virus (HCV). miR-122, highly expressed in hepatocytes, is required for efficient replication of the HCV virus[194]. Surprisingly, miR-122 enhances replication HCV by promoting stability of the viral genomic RNA (in contrast to the conventional role of miRNAs in reducing target mRNA stability). Mutation of either miR-122 binding site in the HCV genome results in decreased viral RNA stability and virus replication[195, 196]. The reason HCV evolved to use a host cell miRNA is not clear, but may involve host cell trophism. A miR-122 inhibitor is undergoing clinical trials as an HCV therapeutic[197].
7. miRNAs as diagnostic markers
The extensive alterations in miRNA expression in disease provide great potential for clinical diagnostics based on miRNA signatures. Furthermore, miRNAs are more stable than mRNAs and can be recovered from formalin fixed paraffin sections (FFPE) and other sources with low overall RNA quality[198-200]. Current focus is on developing miRNA signatures for disease diagnosis, identifying cancers of unknown primary, and predicting response to therapy and drug resistance[201-208]. As yet, no in vitro diagnostic (IVD) using miRNA signatures has received FDA approval. However, a number of companies are offering Laboratory Developed Tests (LDT) that use miRNA signatures for cancer diagnostics[209, 210].
7.1. miRNA detection methods
miRNA detection methods can be divided into two general categories[211]. Discovery methods are designed to profile the expression of many miRNAs at once, often with high throughput. These approaches are largely centered on microarray hybridization and NGS profiling. The latter is especially powerful since it is possible to characterize novel miRNAs, while most other methods are limited to detection of known miRNA sequences. There are several NGS platforms available but all begin with the construction of template libraries. RNA adapters are ligated to both ends of small RNA fractions and the targets are RT-PCR amplified with primers directed at the adapters. This process amplifies all RNAs in the desired size range. The libraries can then be sequenced on several instruments, with the Illumina platforms probably the most common. Alternatively, single molecule instruments are available that avoid the PCR amplification step altogether[212, 213]. Since current instruments are capable of 200 million or more reads per library run, it is possible to multiplex 48 libraries (or more) in a single run and still achieve adequate sequence read depth[211]. Sequence reads are counted and quantitative expression profiles are obtained. As stated above, novel miRNAs and other small RNA species can be identified. While NGS based profiling has clear advantages and is becoming the default technology, nucleotide biases incurred during ligation steps has been observed[214, 215]. As with any profiling method a validation step is essential.
While NGS platforms are capable of high throughput profiling of the entire miRNA population, most clinical diagnostic approaches are based on rapid analysis of a small gene signature set. Therefore, RT-PCR and Nanostring are the focus of current diagnostic platforms[216]. Nanostring is a single molecule hybridization method that allows quantitation of ~500 targets, either mRNA or miRNA, in a rapid experimental run[217]. An example of an LDT miRNA diagnostic is the pancreatic cancer test from Asuragen[218]. This RT-PCR based test uses a 7 miRNA signature to differentiate between pancreatic ductal adenocarcinoma and benign tissue.
7.2. miRNAs in extracellular fluids
Several years ago the startling discovery was made that miRNAs are present in human plasma[219, 220]. Due to the presence of RNase enzymes in plasma it was not expected that miRNAs would survive in this environment. It is now known that miRNAs are bound to protein, HDL, and/or encapsulated in lipid exosomes protecting them from degradation[221-227]. Originally thought to be cellular “debris” in plasma, possibly released from apoptotic cells, several studies have indicated that these extracellular miRNAs can be taken up by target cells and mediate target gene repression[228-230]. miRNA content in exosomes has distinct profiles from the parent cell, raising the possibility that miRNAs are sorted to exosomes in a controlled manner[231]. Thus, exosomes may function as extracellular messengers and may promote disease progression.
miRNAs have also been isolated from other extracellular fluids including saliva, urine, and stool[232]. Major efforts are underway to profile the miRNAs in these and other fluids, in normal subjects and in patients with disease[233]. Since many of these fluids can be easily obtained without invasive procedures they offer tremendous potential as a source material for clinical diagnostics. While numerous studies have reported extracellular miRNA signatures for diseases it remains to be determined whether these signatures have acceptable accuracy and sensitivity for the clinic.
8. miRNAs as therapeutic targets
As miRNAs become causally linked to increasing numbers of human diseases there are expanding efforts to develop therapeutics that directly target the miRNA[234-236]. These candidates are either miRNA mimics or antisense inhibitors, depending on the miRNA and underlying therapeutic goal. These drug candidates are based on modified nucleic acids and therefore present delivery challenges. The most promising candidate to date is the Regulus drug RG-101, currently in Phase II trials[197]. This drug is an antisense inhibitor of miR-122 that is being developed as a HCV therapeutic. A number of other miRNA therapeutic targets are being pursued by academia and industry, including targets for cardiovascular disease and cancer.
8.1. miRNA mimics and inhibitors
miRNA therapeutics fall under two general categories[234]. For miRNAs that are reduced in disease the therapeutic goal is to replace the miRNA. To achieve this a miRNA mimic is developed. These are typically dsRNA duplexes that directly load into RISC, though a stem-loop precursor can also be employed. In this case, the precursor is cleaved by Dicer and loaded into RISC. Since miRNA mimics are similar to siRNAs, the design strategies that have been developed for siRNA therapeutics have been adapted to miRNAs. Details of these designs are discussed in other sections of this special issue. Briefly, the duplex RNA requires backbone and ribose modifications to promote stability in vivo and improve pharmacodynamics. Some strategy to promote cellular uptake must be included. This may involve conjugation to sugar ligands or encapsulation in liposomes. Some therapeutic mimics that are under development are miR-34 for cancer (Phase I) and pre-clinical therapeutics including let-7 for cancer and miR-29 for fibrosis[237, 238].
miRNAs that are elevated in disease can be targeted by antisense oligonucleotide inhibitors[234]. These inhibitors bind to the miRNA, most likely in the RISC complex, and prevent miRNA binding to target mRNAs. As with mimics, miRNA inhibitors require modifications for stability, and in some cases conjugation to ligands or encapsulation. The best example of a miRNA inhibitor is the RG101 anti-miR-122 in Phase II trials for HCV infection[197].
8.2. Small molecule inhibitors of the miRNA pathway
An interesting alternative to inhibiting the disease-associated miRNA directly is to develop small molecule compounds that modulate production or function of the miRNA[239]. This would avoid the problems associated with oligonucleotide delivery in vivo. Several research groups have performed high throughput screens for compounds that inhibit the function of specific miRNAs[240-245]. The readout for these screens is often a cell-based reporter that is normally silenced by the miRNA. If a compound interferes with production or activity of the miRNA, the reporter will be desilenced and the compound scored a hit. As an example, compounds that inhibit the activity of the oncogenic miR-21 have been identified using this approach[241, 242, 244]. The disadvantage of these small molecule drugs is that, by definition, the mechanism of action is unknown. There is much more downstream work required to define the specificity and direct mode of action.
9. Genome editing approaches
The miRNA pathway is a subset of post-transcriptional gene silencing (PTGS) pathways. In these related biological pathways the mRNA is the target of the silencing machinery, hence “post-transcriptional” silencing. A complementary approach to silence gene expression is to target the genome itself. Several approaches have been developed to redirect an endonuclease to a desired target site in genomic DNA[246]. The resultant double strand break is repaired, usually by non-homologous end joining, generating a short deletion at the repair site. This “editing” event is permanent and often leads to loss of expression of the targeted gene.
Early strategies for genome editing were based on the FokI nuclease domain fused to DNA binding protein modules[246]. Zinc finger nuclease (ZFN) and transcription activator like effector (TALE) domains are protein domains that bind to nucleic acids with predictable specificity. By constructing tandem arrangements of these domain variants it is possible to generate a DNA targeting protein with the desired sequence specificity. This targeting protein, fused to FokI, can direct permanent genome editing. The major factor that limited the widespread use of ZFNs and TALENs is the technical difficulty in construction of the targeting module. Multiple cloning steps are required, and since genome editing success is not guaranteed, many laboratories were dissuaded from the approach. This changed with the discovery of the CRISPR/Cas system.
The CRISPR/Cas pathway originates in bacterial where it functions as a primitive immune system[247]. Clustered regularly interspaced short palindromic repeats (CRISPR) are repeat sequences in bacterial genomes that are derived from pathogenic viruses. Bacteria use guide RNAs derived from these sequences to direct the Cas9 endonuclease to invading viruses, cutting the viral genome and preventing viral attack. By modifying the sequence of this guide RNA, it is possible to target any desired DNA sequence (see Figure 3)[248-251]. The resultant DNA break is repaired by NHEJ, leading to a short deletion. If a homologous DNA template is supplied, the break is repaired by homologous recombination, allowing precise control of the editing event. This approach has received rapid acceptance in molecular biological studies and has been used for disease modeling in cultured cells and in the mouse[252]. The major limitation for the use of CRISPR/Cas system as a therapeutic is that the Cas9 nuclease protein is bacterial in origin and must be ectopically expressed in the target cell. This requires the use of viral gene therapy vectors and the associated delivery and safety challenges that have limited clinical progress in that field. There is much excitement in the field, however, that CRISPR/Cas is a potential therapeutic approach, and studies in the mouse have demonstrated incredible potential[253].
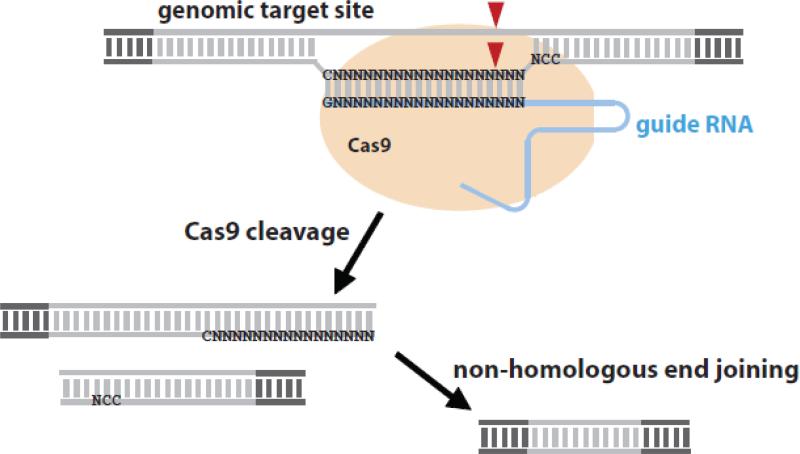
Figure 3. CRISPR/Cas as a genome editing tool.
The guide RNA directs the Cas9 endonuclease to target DNA. Modification of the guide RNA sequence allows any DNA sequence to be targeted. The only sequence requirement is the PAM motif NCC downstream of the target site, and a C at the 5′ end of the target site. The double strand break is repaired by non-homologous end joining, leading to a short deletion.
10. Conclusion
We are entering an exciting time in the field of miRNA research. A decade of strong basic science research has forged links between specific miRNA alterations and many human diseases. We are now entering a phase where miRNA diagnostics and therapeutics can be developed with confidence, and several commercial efforts are underway to bring these developments to the clinic.
Acknowledgments
Work performed in the author's lab was supported by funding from the NIH (R01GM070674, RC1HL100108) and the Department of Cell Biology and Physiology.
List of abbreviations
- RISC
RNA induced silencing complex
- MCM7
Minichromosome maintenance protein 7
- DGCR8
DiGeorge Syndrome critical region 8
- TRBP
TAR RNA binding protein
- NGS
Next generation sequencing
- C3PO
Component 3 promoter of RISC
- TNRC6
Trinucleotide repeat containing 6
- CCR4-NOT
carbon catabolite repression 4 negative on TATA-less complex
- tRF
tRNA derived fragment
- tsRNA
tRNA derived small RNA
- hnRNP
Heterogenous nuclear ribonuclear protein
- C/EBP
CCAAT/enhancer binding protein
- SRF
Serum response factor
- MEF2
Myocyte enhancer factor 2
- HCV
Hepatitis C virus
- FDA
Food and Drug Administration
- LDT
Laboratory developed test
- FFPE
Formalin fixed paraffin embedded
- IVD
In vitro diagnostic
- siRNA
small interfering RNA
- PTGS
Post-transcriptional gene silencing
- ZFN
Zinc finger nuclease
- TALEN
Transcription activator like effector nuclease
- CRISPR
Clustered regularly interspaced short palindromic repeats
- NHEJ
Non-homologous end joining
- PAM
Protospacer adjacent motif
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Horvitz HR, Sulston JE. Isolation and genetic characterization of cell-lineage mutants of the nematode Caenorhabditis elegans. Genetics. 1980;96:435–454. doi: 10.1093/genetics/96.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 3.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 4.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 5.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, Spring J, Srinivasan A, Fishman M, Finnerty J, Corbo J, Levine M, Leahy P, Davidson E, Ruvkun G. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 6.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 7.Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 8.Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP- dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 9.Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post- transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 10.Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 11.Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 12.Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 15.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poliseno L, Salmena L, Riccardi L, Fornari A, Song MS, Hobbs RM, Sportoletti P, Varmeh S, Egia A, Fedele G, Rameh L, Loda M, Pandolfi PP. Identification of the miR-106b~25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Sci Signal. 2010;3:ra29. doi: 10.1126/scisignal.2000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, Jaenisch R, Sharp PA, Jacks T. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 19.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 20.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 21.Papadopoulos GL, Reczko M, Simossis VA, Sethupathy P, Hatzigeorgiou AG. The database of experimentally supported targets: a functional update of TarBase. Nucleic Acids Res. 2009;37:D155–158. doi: 10.1093/nar/gkn809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bortolin-Cavaille ML, Dance M, Weber M, Cavaille J. C19MC microRNAs are processed from introns of large Pol-II, non-protein-coding transcripts. Nucleic Acids Res. 2009;37:3464–3473. doi: 10.1093/nar/gkp205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 26.Woods K, Thomson JM, Hammond SM. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J Biol Chem. 2007;282:2130–2134. doi: 10.1074/jbc.C600252200. [DOI] [PubMed] [Google Scholar]
- 27.Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, Calabrese JM, Dennis LM, Volkert TL, Gupta S, Love J, Hannett N, Sharp PA, Bartel DP, Jaenisch R, Young RA. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, Arking DE, Beer MA, Maitra A, Mendell JT. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, Oren M. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 30.He L, He X, Lim LP, de Stanchina E, Xuan Z, Yamakuchi, Liang W, Xue L, Zender J, Magnus D, Ridzon AL, Jackson PS, Linsley C, Chen SW, Lowe MA, Cleary GJ, Hannon A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Georgakilas G, Vlachos IS, Paraskevopoulou MD, Yang P, Zhang Y, Economides AN, Hatzigeorgiou AG. microTSS: accurate microRNA transcription start site identification reveals a significant number of divergent pri-miRNAs. Nat Commun. 2014;5:5700. doi: 10.1038/ncomms6700. [DOI] [PubMed] [Google Scholar]
- 32.Bhattacharyya M, Feuerbach L, Bhadra T, Lengauer T, Bandyopadhyay S. MicroRNA transcription start site prediction with multi-objective feature selection. Stat Appl Genet Mol Biol. 2012;11 doi: 10.2202/1544-6115.1743. Article 6. [DOI] [PubMed] [Google Scholar]
- 33.Ozsolak F, Poling LL, Wang Z, Liu H, Liu XS, Roeder RG, Zhang X, Song JS, Fisher DE. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008;22:3172–3183. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 35.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 36.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ballarino M, Pagano F, Girardi E, Morlando M, Cacchiarelli D, Marchioni M, Proudfoot NJ, Bozzoni I. Coupled RNA processing and transcription of intergenic primary microRNAs. Mol Cell Biol. 2009;29:5632–5638. doi: 10.1128/MCB.00664-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morlando M, Ballarino M, Gromak N, Pagano F, Bozzoni I, Proudfoot NJ. Primary microRNA transcripts are processed co-transcriptionally. Nat Struct Mol Biol. 2008;15:902–909. doi: 10.1038/nsmb.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 41.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 42.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 44.Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 46.Maniataki E, Mourelatos Z. A human, ATP-independent, RISC assembly machine fueled by pre miRNA. Genes Dev. 2005;19:2979–2990. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 48.Gatignol A, Laine S, Clerzius G. Dual role of TRBP in HIV replication and RNA interference: viral diversion of a cellular pathway or evasion from antiviral immunity? Retrovirology. 2005;2:65. doi: 10.1186/1742-4690-2-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haase AD, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A, Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang JS, Phillips MD, Betel D, Mu P, Ventura A, Siepel AC, Chen KC, Lai EC. Widespread regulatory activity of vertebrate microRNA* species. RNA. 2011;17:312–326. doi: 10.1261/rna.2537911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ohanian M, Humphreys DT, Anderson E, Preiss T, Fatkin D. A heterozygous variant in the human cardiac miR-133 gene, MIR133A2, alters miRNA duplex processing and strand abundance. BMC Genet. 2013;14:18. doi: 10.1186/1471-2156-14-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee LW, Zhang S, Etheridge A, Ma L, Martin D, Galas D, Wang K. Complexity of the microRNA repertoire revealed by next-generation sequencing. RNA. 2010;16:2170–2180. doi: 10.1261/rna.2225110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marti E, Pantano L, Banez-Coronel M, Llorens F, Minones-Moyano E, Porta S, Sumoy L, Ferrer I, Estivill X. A myriad of miRNA variants in control and Huntington's disease brain regions detected by massively parallel sequencing. Nucleic Acids Res. 2010;38:7219–7235. doi: 10.1093/nar/gkq575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baran-Gale J, Fannin EE, Kurtz CL, Sethupathy P. Beta Cell 5'-Shifted isomiRs Are Candidate Regulatory Hubs in Type 2 Diabetes. PLoS One. 2013;8:e73240. doi: 10.1371/journal.pone.0073240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vickers KC, Sethupathy P, Baran-Gale J, Remaley AT. Complexity of microRNA function and the role of isomiRs in lipid homeostasis. J Lipid Res. 2013;54:1182–1191. doi: 10.1194/jlr.R034801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- 58.Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- 59.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 60.Shin C. Cleavage of the star strand facilitates assembly of some microRNAs into Ago2-containing silencing complexes in mammals. Mol Cells. 2008;26:308–313. [PubMed] [Google Scholar]
- 61.Liu Y, Ye X, Jiang F, Liang C, Chen D, Peng J, Kinch LN, Grishin NV, Liu Q. C3PO, an endoribonuclease that promotes RNAi by facilitating RISC activation. Science. 2009;325:750–753. doi: 10.1126/science.1176325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ye X, Huang N, Liu Y, Paroo Z, Huerta C, Li P, Chen S, Liu Q, Zhang H. Structure of C3PO and mechanism of human RISC activation. Nat Struct Mol Biol. 2011;18:650–657. doi: 10.1038/nsmb.2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tomari Y, Du T, Haley B, Schwarz DS, Bennett R, Cook HA, Koppetsch BS, Theurkauf WE, Zamore PD. RISC assembly defects in the Drosophila RNAi mutant armitage. Cell. 2004;116:831–841. doi: 10.1016/s0092-8674(04)00218-1. [DOI] [PubMed] [Google Scholar]
- 64.Meister G, Landthaler M, Peters L, Chen PY, Urlaub H, Luhrmann R, Tuschl T. Identification of novel argonaute-associated proteins. Curr Biol. 2005;15:2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- 65.Nykanen A, Haley B, Zamore PD. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107:309–321. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- 66.Robb GB, Rana TM. RNA helicase A interacts with RISC in human cells and functions in RISC loading. Mol Cell. 2007;26:523–537. doi: 10.1016/j.molcel.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 67.Liu X, Jin DY, McManus MT, Mourelatos Z. Precursor microRNA-programmed silencing complex assembly pathways in mammals. Mol Cell. 2012;46:507–517. doi: 10.1016/j.molcel.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lai EC, Tam B, Rubin GM. Pervasive regulation of Drosophila Notch target genes by GY-box-, Brd-box-, and K-box-class microRNAs. Genes Dev. 2005;19:1067–1080. doi: 10.1101/gad.1291905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 70.Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 71.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shin C, Nam JW, Farh KK, Chiang HR, Shkumatava A, Bartel DP. Expanding the microRNA targeting code: functional sites with centered pairing. Mol Cell. 2010;38:789–802. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Helwak A, Kudla G, Dudnakova T, Tollervey D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013;153:654–665. doi: 10.1016/j.cell.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 75.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 76.Doench JG, Petersen CP, Sharp PA. siRNAs can function as miRNAs. Genes Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pfaff J, Meister G. Argonaute and GW182 proteins: an effective alliance in gene silencing. Biochem Soc Trans. 2013;41:855–860. doi: 10.1042/BST20130047. [DOI] [PubMed] [Google Scholar]
- 78.Braun JE, Huntzinger E, Izaurralde E. The role of GW182 proteins in miRNA-mediated gene silencing. Adv Exp Med Biol. 2013;768:147–163. doi: 10.1007/978-1-4614-5107-5_9. [DOI] [PubMed] [Google Scholar]
- 79.Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat Struct Mol Biol. 2012;19:586–593. doi: 10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- 80.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, Ma E, Mane S, Hannon GJ, Lawson ND, Wolfe SA, Giraldez AJ. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yang JS, Lai EC. Dicer-independent, Ago2-mediated microRNA biogenesis in vertebrates. Cell Cycle. 2010;9:4455–4460. doi: 10.4161/cc.9.22.13958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Diederichs S, Haber DA. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131:1097–1108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 88.Garcia-Silva MR, Cabrera-Cabrera F, Guida MC, Cayota A. Hints of tRNA-Derived Small RNAs Role in RNA Silencing Mechanisms. Genes (Basel) 2012;3:603–614. doi: 10.3390/genes3040603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16:673–695. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23:2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Janas MM, Wang B, Harris AS, Aguiar M, Shaffer JM, Subrahmanyam YV, Behlke MA, Wucherpfennig KW, Gygi SP, Gagnon E, Novina CD. Alternative RISC assembly: binding and repression of microRNA-mRNA duplexes by human Ago proteins. RNA. 2012;18:2041–2055. doi: 10.1261/rna.035675.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Eiring AM, Harb JG, Neviani P, Garton C, Oaks JJ, Spizzo R, Liu S, Schwind S, Santhanam R, Hickey CJ, Becker H, Chandler JC, Andino R, Cortes J, Hokland P, Huettner CS, Bhatia R, Roy DC, Liebhaber SA, Caligiuri MA, Marcucci G, Garzon R, Croce CM, Calin GA, Perrotti D. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell. 2010;140:652–665. doi: 10.1016/j.cell.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Paduano F, Dattilo V, Narciso D, Bilotta A, Gaudio E, Menniti M, Agosti V, Palmieri C, Perrotti N, Fusco A, Trapasso F, Iuliano R. Protein tyrosine phosphatase PTPRJ is negatively regulated by microRNA-328. FEBS J. 2013;280:401–412. doi: 10.1111/j.1742-4658.2012.08624.x. [DOI] [PubMed] [Google Scholar]
- 94.Turrini E, Haenisch S, Laechelt S, Diewock T, Bruhn O, Cascorbi I. MicroRNA profiling in K-562 cells under imatinib treatment: influence of miR-212 and miR-328 on ABCG2 expression. Pharmacogenet Genomics. 2012;22:198–205. doi: 10.1097/FPC.0b013e328350012b. [DOI] [PubMed] [Google Scholar]
- 95.Boissonneault V, Plante I, Rivest S, Provost P. MicroRNA-298 and microRNA-328 regulate expression of mouse beta-amyloid precursor protein-converting enzyme 1. J Biol Chem. 2009;284:1971–1981. doi: 10.1074/jbc.M807530200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang CH, Lee DY, Deng Z, Jeyapalan Z, Lee SC, Kahai S, Lu WY, Zhang Y, Yang BB. MicroRNA miR-328 regulates zonation morphogenesis by targeting CD44 expression. PLoS One. 2008;3:e2420. doi: 10.1371/journal.pone.0002420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen KC, Hsi E, Hu CY, Chou WW, Liang CL, Juo SH. MicroRNA-328 may influence myopia development by mediating the PAX6 gene. Invest Ophthalmol Vis Sci. 2012;53:2732–2739. doi: 10.1167/iovs.11-9272. [DOI] [PubMed] [Google Scholar]
- 98.Ekimler S, Sahin K. Computational Methods for MicroRNA Target Prediction. Genes (Basel) 2014;5:671–683. doi: 10.3390/genes5030671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Grun D, Wang YL, Langenberger D, Gunsalus KC, Rajewsky N. microRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLoS Comput Biol. 2005;1:e13. doi: 10.1371/journal.pcbi.0010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Thomas M, Lieberman J, Lal A. Desperately seeking microRNA targets. Nat Struct Mol Biol. 2010;17:1169–1174. doi: 10.1038/nsmb.1921. [DOI] [PubMed] [Google Scholar]
- 103.Huang Y, Zou Q, Song H, Song F, Wang L, Zhang G, Shen X. A study of miRNAs targets prediction and experimental validation. Protein Cell. 2010;1:979–986. doi: 10.1007/s13238-010-0129-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr., Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Karginov FV, Conaco C, Xuan Z, Schmidt BH, Parker JS, Mandel G, Hannon GJ. A biochemical approach to identifying microRNA targets. Proc Natl Acad Sci U S A. 2007;104:19291–19296. doi: 10.1073/pnas.0709971104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Leung AK, Young AG, Bhutkar A, Zheng GX, Bosson AD, Nielsen CB, Sharp PA. Genome-wide identification of Ago2 binding sites from mouse embryonic stem cells with and without mature microRNAs. Nat Struct Mol Biol. 2011;18:237–244. doi: 10.1038/nsmb.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Riley KJ, Yario TA, Steitz JA. Association of Argonaute proteins and microRNAs can occur after cell lysis. RNA. 2012;18:1581–1585. doi: 10.1261/rna.034934.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5'UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 110.Lal A, Thomas MP, Altschuler G, Navarro F, O'Day E, Li XL, Concepcion C, Han YC, Thiery J, Rajani DK, Deutsch A, Hofmann O, Ventura A, Hide W, Lieberman J. Capture of microRNA-bound mRNAs identifies the tumor suppressor miR-34a as a regulator of growth factor signaling. PLoS Genet. 2011;7:e1002363. doi: 10.1371/journal.pgen.1002363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Eichhorn SW, Guo H, McGeary SE, Rodriguez-Mias RA, Shin C, Baek D, Hsu SH, Ghoshal K, Villen J, Bartel DP. mRNA destabilization is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Mol Cell. 2014;56:104–115. doi: 10.1016/j.molcel.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.van Dongen S, Abreu-Goodger C, Enright AJ. Detecting microRNA binding and siRNA off- target effects from expression data. Nat Methods. 2008;5:1023–1025. doi: 10.1038/nmeth.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rasmussen SH, Jacobsen A, Krogh A. cWords - systematic microRNA regulatory motif discovery from mRNA expression data. Silence. 2013;4:2. doi: 10.1186/1758-907X-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Diao L, Marcais A, Norton S, Chen KC. MixMir: microRNA motif discovery from gene expression data using mixed linear models. Nucleic Acids Res. 2014;42:e135. doi: 10.1093/nar/gku672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sood P, Krek A, Zavolan M, Macino G, Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc Natl Acad Sci U S A. 2006;103:2746–2751. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sylvestre Y, De Guire V, Querido E, Mukhopadhyay UK, Bourdeau V, Major F, Ferbeyre G, Chartrand P. An E2F/miR-20a autoregulatory feedback loop. J Biol Chem. 2007;282:2135–2143. doi: 10.1074/jbc.M608939200. [DOI] [PubMed] [Google Scholar]
- 119.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Niu Z, Iyer D, Conway SJ, Martin JF, Ivey K, Srivastava D, Nordheim A, Schwartz RJ. Serum response factor orchestrates nascent sarcomerogenesis and silences the biomineralization gene program in the heart. Proc Natl Acad Sci U S A. 2008;105:17824–17829. doi: 10.1073/pnas.0805491105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 122.Liu N, Williams AH, Kim Y, McAnally J, Bezprozvannaya S, Sutherland LB, Richardson JA, Bassel-Duby R, Olson EN. An intragenic MEF2-dependent enhancer directs muscle-specific expression of microRNAs 1 and 133. Proc Natl Acad Sci U S A. 2007;104:20844–20849. doi: 10.1073/pnas.0710558105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sweetman D, Goljanek K, Rathjen T, Oustanina S, Braun T, Dalmay T, Munsterberg A. Specific requirements of MRFs for the expression of muscle specific microRNAs, miR-1, miR-206 and miR-133. Dev Biol. 2008;321:491–499. doi: 10.1016/j.ydbio.2008.06.019. [DOI] [PubMed] [Google Scholar]
- 124.Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 128.Nam Y, Chen C, Gregory RI, Chou JJ, Sliz P. Molecular basis for interaction of let-7 microRNAs with Lin28. Cell. 2011;147:1080–1091. doi: 10.1016/j.cell.2011.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ali PS, Ghoshdastider U, Hoffmann J, Brutschy B, Filipek S. Recognition of the let-7g miRNA precursor by human Lin28B. FEBS Lett. 2012;586:3986–3990. doi: 10.1016/j.febslet.2012.09.034. [DOI] [PubMed] [Google Scholar]
- 130.Lightfoot HL, Bugaut A, Armisen J, Lehrbach NJ, Miska EA, Balasubramanian S. A LIN28- dependent structural change in pre-let-7g directly inhibits dicer processing. Biochemistry. 2011;50:7514–7521. doi: 10.1021/bi200851d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 132.Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 134.Lehrbach NJ, Armisen J, Lightfoot HL, Murfitt KJ, Bugaut A, Balasubramanian S, Miska EA. LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans. Nat Struct Mol Biol. 2009;16:1016–1020. doi: 10.1038/nsmb.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Towbin H, Wenter P, Guennewig B, Imig J, Zagalak JA, Gerber AP, Hall J. Systematic screens of proteins binding to synthetic microRNA precursors. Nucleic Acids Res. 2013;41:e47. doi: 10.1093/nar/gks1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, Gherzi R, Rosenfeld MG. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Guil S, Caceres JF. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat Struct Mol Biol. 2007;14:591–596. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- 138.Newman MA, Hammond SM. Emerging paradigms of regulated microRNA processing. Genes Dev. 2010;24:1086–1092. doi: 10.1101/gad.1919710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 140.Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 141.Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Park CY, Choi YS, McManus MT. Analysis of microRNA knockouts in mice. Hum Mol Genet. 2010;19:R169–175. doi: 10.1093/hmg/ddq367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Vidigal JA, Ventura A. The biological functions of miRNAs: lessons from in vivo studies. Trends Cell Biol. 2014 doi: 10.1016/j.tcb.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress- dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 145.van Rooij E, Quiat D, Johnson BA, Sutherland LB, Qi X, Richardson JA, Kelm RJ, Jr., Olson EN. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell. 2009;17:662–673. doi: 10.1016/j.devcel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kole AJ, Swahari V, Hammond SM, Deshmukh M. miR-29b is activated during neuronal maturation and targets BH3-only genes to restrict apoptosis. Genes Dev. 2011;25:125–130. doi: 10.1101/gad.1975411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Somel M, Liu X, Tang L, Yan Z, Hu H, Guo S, Jiang X, Zhang X, Xu G, Xie G, Li N, Hu Y, Chen W, Paabo S, Khaitovich P. MicroRNA-driven developmental remodeling in the brain distinguishes humans from other primates. PLoS Biol. 2011;9:e1001214. doi: 10.1371/journal.pbio.1001214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 149.Griggs EM, Young EJ, Rumbaugh G, Miller CA. MicroRNA-182 regulates amygdala- dependent memory formation. J Neurosci. 2013;33:1734–1740. doi: 10.1523/JNEUROSCI.2873-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Fazi F, Rosa A, Fatica A, Gelmetti V, De Marchis ML, Nervi C, Bozzoni I. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 151.Zhou B, Wang S, Mayr C, Bartel DP, Lodish HF. miR-150, a microRNA expressed in mature B and T cells, blocks early B cell development when expressed prematurely. Proc Natl Acad Sci U S A. 2007;104:7080–7085. doi: 10.1073/pnas.0702409104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Vigorito E, Perks KL, Abreu-Goodger C, Bunting S, Xiang Z, Kohlhaas S, Das PP, Miska EA, Rodriguez A, Bradley A, Smith KG, Rada C, Enright AJ, Toellner KM, Maclennan IC, Turner M. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847–859. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, Schmidt-Supprian M, Rajewsky N, Yancopoulos G, Rao A, Rajewsky K. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 154.Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, Rajewsky N, Bender TP, Rajewsky K. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 155.Lu J, Guo S, Ebert BL, Zhang H, Peng X, Bosco J, Pretz J, Schlanger R, Wang JY, Mak RH, Dombkowski DM, Preffer FI, Scadden DT, Golub TR. MicroRNA-mediated control of cell fate in megakaryocyte-erythrocyte progenitors. Dev Cell. 2008;14:843–853. doi: 10.1016/j.devcel.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ben-Ami O, Pencovich N, Lotem J, Levanon D, Groner Y. A regulatory interplay between miR-27a and Runx1 during megakaryopoiesis. Proc Natl Acad Sci U S A. 2009;106:238–243. doi: 10.1073/pnas.0811466106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Dore LC, Amigo JD, Dos Santos CO, Zhang Z, Gai X, Tobias JW, Yu D, Klein AM, Dorman C, Wu W, Hardison RC, Paw BH, Weiss MJ. A GATA-1-regulated microRNA locus essential for erythropoiesis. Proc Natl Acad Sci U S A. 2008;105:3333–3338. doi: 10.1073/pnas.0712312105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Felli N, Fontana L, Pelosi E, Botta R, Bonci D, Facchiano F, Liuzzi F, Lulli V, Morsilli O, Santoro S, Valtieri M, Calin GA, Liu CG, Sorrentino A, Croce CM, Peschle C. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc Natl Acad Sci U S A. 2005;102:18081–18086. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, Brummelkamp TR, Fleming MD, Camargo FD. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- 160.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Garzon R, Pichiorri F, Palumbo T, Iuliano R, Cimmino A, Aqeilan R, Volinia S, Bhatt D, Alder H, Marcucci G, Calin GA, Liu CG, Bloomfield CD, Andreeff M, Croce CM. MicroRNA fingerprints during human megakaryocytopoiesis. Proc Natl Acad Sci U S A. 2006;103:5078–5083. doi: 10.1073/pnas.0600587103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Thomson JM, Parker J, Perou CM, Hammond SM. A custom microarray platform for analysis of microRNA gene expression. Nat Methods. 2004;1:47–53. doi: 10.1038/nmeth704. [DOI] [PubMed] [Google Scholar]
- 163.Liu CG, Calin GA, Meloon B, Gamliel N, Sevignani C, Ferracin M, Dumitru CD, Shimizu M, Zupo S, Dono M, Alder H, Bullrich F, Negrini M, Croce CM. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci U S A. 2004;101:9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 165.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.The Cancer Genome Atlas. http://cancergenome.nih.gov/
- 167.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 169.Kutay H, Bai S, Datta J, Motiwala T, Pogribny I, Frankel W, Jacob ST, Ghoshal K. Downregulation of miR-122 in the rodent and human hepatocellular carcinomas. J Cell Biochem. 2006;99:671–678. doi: 10.1002/jcb.20982. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 170.Girard M, Jacquemin E, Munnich A, Lyonnet S, Henrion-Caude A. miR-122, a paradigm for the role of microRNAs in the liver. J Hepatol. 2008;48:648–656. doi: 10.1016/j.jhep.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 171.Ota A, Tagawa H, Karnan S, Tsuzuki S, Karpas A, Kira S, Yoshida Y, Seto M. Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res. 2004;64:3087–3095. doi: 10.1158/0008-5472.can-03-3773. [DOI] [PubMed] [Google Scholar]
- 172.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Dews M, Homayouni A, Yu D, Murphy D, Sevignani C, Wentzel E, Furth EE, Lee WM, Enders GH, Mendell JT, Thomas-Tikhonenko A. Augmentation of tumor angiogenesis by a Myc- activated microRNA cluster. Nat Genet. 2006;38:1060–1065. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Lu Y, Thomson JM, Wong HY, Hammond SM, Hogan BL. Transgenic over-expression of the microRNA miR-17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev Biol. 2007;310:442–453. doi: 10.1016/j.ydbio.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Bonauer A, Dimmeler S. The microRNA-17-92 cluster: still a miRacle? Cell Cycle. 2009;8:3866–3873. doi: 10.4161/cc.8.23.9994. [DOI] [PubMed] [Google Scholar]
- 176.He L, He X, Lowe SW, Hannon GJ. microRNAs join the p53 network--another piece in the tumour-suppression puzzle. Nat Rev Cancer. 2007;7:819–822. doi: 10.1038/nrc2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Berindan-Neagoe I, Monroig Pdel C, Pasculli B, Calin GA. MicroRNAome genome: a treasure for cancer diagnosis and therapy. CA Cancer J Clin. 2014;64:311–336. doi: 10.3322/caac.21244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Barbarotto E, Schmittgen TD, Calin GA. MicroRNAs and cancer: profile, profile, profile. Int J Cancer. 2008;122:969–977. doi: 10.1002/ijc.23343. [DOI] [PubMed] [Google Scholar]
- 179.Fernandez-Hernando C, Ramirez CM, Goedeke L, Suarez Y. MicroRNAs in metabolic disease. Arterioscler Thromb Vasc Biol. 2013;33:178–185. doi: 10.1161/ATVBAHA.112.300144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Hsu SH, Wang B, Kota J, Yu J, Costinean S, Kutay H, Yu L, Bai S, La Perle K, Chivukula RR, Mao H, Wei M, Clark KR, Mendell JR, Caligiuri MA, Jacob ST, Mendell JT, Ghoshal K. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J Clin Invest. 2012;122:2871–2883. doi: 10.1172/JCI63539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Tsai WC, Hsu SD, Hsu CS, Lai TC, Chen SJ, Shen R, Huang Y, Chen HC, Lee CH, Tsai TF, Hsu MT, Wu JC, Huang HD, Shiao MS, Hsiao M, Tsou AP. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest. 2012;122:2884–2897. doi: 10.1172/JCI63455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, Subramaniam A, Propp S, Lollo BA, Freier S, Bennett CF, Bhanot S, Monia BP. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 183.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 184.Marquart TJ, Allen RM, Ory DS, Baldan A. miR-33 links SREBP-2 induction to repression of sterol transporters. Proc Natl Acad Sci U S A. 2010;107:12228–12232. doi: 10.1073/pnas.1005191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, Naar AM. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 2010;328:1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Rayner KJ, Suarez Y, Davalos A, Parathath S, Fitzgerald ML, Tamehiro N, Fisher EA, Moore KJ, Fernandez-Hernando C. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Horie T, Ono K, Horiguchi M, Nishi H, Nakamura T, Nagao K, Kinoshita M, Kuwabara Y, Marusawa H, Iwanaga Y, Hasegawa K, Yokode M, Kimura T, Kita T. MicroRNA-33 encoded by an intron of sterol regulatory element-binding protein 2 (Srebp2) regulates HDL in vivo. Proc Natl Acad Sci U S A. 2010;107:17321–17326. doi: 10.1073/pnas.1008499107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, Stoffel M. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 189.Poy MN, Hausser J, Trajkovski M, Braun M, Collins S, Rorsman P, Zavolan M, Stoffel M. miR-375 maintains normal pancreatic alpha- and beta-cell mass. Proc Natl Acad Sci U S A. 2009;106:5813–5818. doi: 10.1073/pnas.0810550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Frost RJ, Olson EN. Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proc Natl Acad Sci U S A. 2011;108:21075–21080. doi: 10.1073/pnas.1118922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Zhu H, Shyh-Chang N, Segre AV, Shinoda G, Shah SP, Einhorn WS, Takeuchi A, Engreitz JM, Hagan JP, Kharas MG, Urbach A, Thornton JE, Triboulet R, Gregory RI, Altshuler D, Daley GQ. The Lin28/let-7 axis regulates glucose metabolism. Cell. 2011;147:81–94. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Trajkovski M, Hausser J, Soutschek J, Bhat B, Akin A, Zavolan M, Heim MH, Stoffel M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;474:649–653. doi: 10.1038/nature10112. [DOI] [PubMed] [Google Scholar]
- 193.Cullen BR. Herpesvirus microRNAs: phenotypes and functions. Curr Opin Virol. 2011;1:211–215. doi: 10.1016/j.coviro.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 195.Jopling CL, Schutz S, Sarnow P. Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microbe. 2008;4:77–85. doi: 10.1016/j.chom.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Shimakami T, Yamane D, Welsch C, Hensley L, Jangra RK, Lemon SM. Base pairing between hepatitis C virus RNA and microRNA 122 3' of its seed sequence is essential for genome stabilization and production of infectious virus. J Virol. 2012;86:7372–7383. doi: 10.1128/JVI.00513-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. http://www.regulusrx.com/therapeutic-areas/rg-101/
- 198.Nelson PT, Baldwin DA, Kloosterman WP, Kauppinen S, Plasterk RH, Mourelatos Z. RAKE and LNA-ISH reveal microRNA expression and localization in archival human brain. RNA. 2006;12:187–191. doi: 10.1261/rna.2258506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Xi Y, Nakajima G, Gavin E, Morris CG, Kudo K, Hayashi K, Ju J. Systematic analysis of microRNA expression of RNA extracted from fresh frozen and formalin-fixed paraffin-embedded samples. RNA. 2007;13:1668–1674. doi: 10.1261/rna.642907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Lehmann U. MicroRNA-profiling in formalin-fixed paraffin-embedded specimens. Methods Mol Biol. 2010;667:113–125. doi: 10.1007/978-1-60761-811-9_8. [DOI] [PubMed] [Google Scholar]
- 201.Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20:460–469. doi: 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 202. http://www.rosettagenomics.com/pros-origin.
- 203.Li X, Lu Y, Chen Y, Lu W, Xie X. MicroRNA profile of paclitaxel-resistant serous ovarian carcinoma based on formalin-fixed paraffin-embedded samples. BMC Cancer. 2013;13:216. doi: 10.1186/1471-2407-13-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Vecchione A, Belletti B, Lovat F, Volinia S, Chiappetta G, Giglio S, Sonego M, Cirombella R, Onesti EC, Pellegrini P, Califano D, Pignata S, Losito S, Canzonieri V, Sorio R, Alder H, Wernicke D, Stoppacciaro A, Baldassarre G, Croce CM. A microRNA signature defines chemoresistance in ovarian cancer through modulation of angiogenesis. Proc Natl Acad Sci U S A. 2013;110:9845–9850. doi: 10.1073/pnas.1305472110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Dai Y, Xie CH, Neis JP, Fan CY, Vural E, Spring PM. MicroRNA expression profiles of head and neck squamous cell carcinoma with docetaxel-induced multidrug resistance. Head Neck. 2011;33:786–791. doi: 10.1002/hed.21540. [DOI] [PubMed] [Google Scholar]
- 206.Salter KH, Acharya CR, Walters KS, Redman R, Anguiano A, Garman KS, Anders CK, Mukherjee S, Dressman HK, Barry WT, Marcom KP, Olson J, Nevins JR, Potti A. An integrated approach to the prediction of chemotherapeutic response in patients with breast cancer. PLoS One. 2008;3:e1908. doi: 10.1371/journal.pone.0001908. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 207.Du L, Borkowski R, Zhao Z, Ma X, Yu X, Xie XJ, Pertsemlidis A. A high-throughput screen identifies miRNA inhibitors regulating lung cancer cell survival and response to paclitaxel. RNA Biol. 2013;10 doi: 10.4161/rna.26541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Du L, Subauste MC, DeSevo C, Zhao Z, Baker M, Borkowski R, Schageman JJ, Greer R, Yang CR, Suraokar M, Wistuba II, Gazdar AF, Minna JD, Pertsemlidis A. miR-337-3p and its targets STAT3 and RAP1A modulate taxane sensitivity in non-small cell lung cancers. PLoS One. 2012;7:e39167. doi: 10.1371/journal.pone.0039167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Asuragen [Google Scholar]
- 210.RosettaGenomics [Google Scholar]
- 211.Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet. 2012;13:358–369. doi: 10.1038/nrg3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Kapranov P, Ozsolak F, Milos PM. Profiling of short RNAs using Helicos single-molecule sequencing. Methods Mol Biol. 2012;822:219–232. doi: 10.1007/978-1-61779-427-8_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Gu LQ, Wanunu M, Wang MX, McReynolds L, Wang Y. Detection of miRNAs with a nanopore single-molecule counter. Expert Rev Mol Diagn. 2012;12:573–584. doi: 10.1586/erm.12.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Hafner M, Renwick N, Brown M, Mihailovic A, Holoch D, Lin C, Pena JT, Nusbaum JD, Morozov P, Ludwig J, Ojo T, Luo S, Schroth G, Tuschl T. RNA-ligase-dependent biases in miRNA representation in deep-sequenced small RNA cDNA libraries. RNA. 2011;17:1697–1712. doi: 10.1261/rna.2799511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Leshkowitz D, Horn-Saban S, Parmet Y, Feldmesser E. Differences in microRNA detection levels are technology and sequence dependent. RNA. 2013;19:527–538. doi: 10.1261/rna.036475.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Knutsen E, Fiskaa T, Ursvik A, Jorgensen TE, Perander M, Lund E, Seternes OM, Johansen SD, Andreassen M. Performance Comparison of Digital microRNA Profiling Technologies Applied on Human Breast Cancer Cell Lines. PLoS One. 2013;8:e75813. doi: 10.1371/journal.pone.0075813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. http://www.nanostring.com/products/nCounter.
- 218. http://asuragen.com/products-and-services/clinical-lab/mirinform-pancreas/
- 219.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 220.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Kharaziha P, Ceder S, Li Q, Panaretakis T. Tumor cell-derived exosomes: a message in a bottle. Biochim Biophys Acta. 2012;1826:103–111. doi: 10.1016/j.bbcan.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 222.Hannafon BN, Ding WQ. Intercellular Communication by Exosome-Derived microRNAs in Cancer. Int J Mol Sci. 2013;14:14240–14269. doi: 10.3390/ijms140714240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39:7223–7233. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Turchinovich A, Burwinkel B. Distinct AGO1 and AGO2 associated miRNA profiles in human cells and blood plasma. RNA Biol. 2012;9:1066–1075. doi: 10.4161/rna.21083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Kroh EM, Parkin RK, Mitchell PS, Tewari M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR) Methods. 2010;50:298–301. doi: 10.1016/j.ymeth.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, Tait JF, Tewari M. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Hergenreider E, Heydt S, Treguer K, Boettger T, Horrevoets AJ, Zeiher AM, Scheffer MP, Frangakis AS, Yin X, Mayr M, Braun T, Urbich C, Boon RA, Dimmeler S. Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat Cell Biol. 2012;14:249–256. doi: 10.1038/ncb2441. [DOI] [PubMed] [Google Scholar]
- 229.Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, de Gruijl TD, Wurdinger T, Middeldorp JM. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci U S A. 2010;107:6328–6333. doi: 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Mittelbrunn M, Gutierrez-Vazquez C, Villarroya-Beltri C, Gonzalez S, Sanchez-Cabo F, Gonzalez MA, Bernad A, Sanchez-Madrid F. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun. 2011;2:282. doi: 10.1038/ncomms1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Koppers-Lalic D, Hackenberg M, Bijnsdorp IV, van Eijndhoven MA, Sadek P, Sie D, Zini N, Middeldorp JM, Ylstra B, de Menezes RX, Wurdinger T, Meijer GA, Pegtel DM. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep. 2014;8:1649–1658. doi: 10.1016/j.celrep.2014.08.027. [DOI] [PubMed] [Google Scholar]
- 232.Azmi AS, Bao B, Sarkar FH. Exosomes in cancer development, metastasis, and drug resistance: a comprehensive review. Cancer Metastasis Rev. 2013 doi: 10.1007/s10555-013-9441-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. http://commonfund.nih.gov/Exrna/index.
- 234.van Rooij E, Kauppinen S. Development of microRNA therapeutics is coming of age. EMBO Mol Med. 2014;6:851–864. doi: 10.15252/emmm.201100899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Li Z, Rana TM. Therapeutic targeting of microRNAs: current status and future challenges. Nat Rev Drug Discov. 2014;13:622–638. doi: 10.1038/nrd4359. [DOI] [PubMed] [Google Scholar]
- 236.Chen Y, Gao D, Huang L. In vivo delivery of miRNAs for cancer therapy: Challenges and strategies. Adv Drug Deliv Rev. 2015;81C:128–141. doi: 10.1016/j.addr.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Bader AG. miR-34 - a microRNA replacement therapy is headed to the clinic. Front Genet. 2012;3:120. doi: 10.3389/fgene.2012.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Montgomery RL, Yu G, Latimer PA, Stack C, Robinson K, Dalby CM, Kaminski N, van Rooij E. MicroRNA mimicry blocks pulmonary fibrosis. EMBO Mol Med. 2014;6:1347–1356. doi: 10.15252/emmm.201303604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Monroig PD, Chen L, Zhang S, Calin GA. Small molecule compounds targeting miRNAs for cancer therapy. Adv Drug Deliv Rev. 2015;81C:104–116. doi: 10.1016/j.addr.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Xiao Z, Li CH, Chan SL, Xu F, Feng L, Wang Y, Jiang JD, Sung JJ, Cheng CH, Chen Y. A small-molecule modulator of the tumor-suppressor miR34a inhibits the growth of hepatocellular carcinoma. Cancer Res. 2014;74:6236–6247. doi: 10.1158/0008-5472.CAN-14-0855. [DOI] [PubMed] [Google Scholar]
- 241.Shi Z, Zhang J, Qian X, Han L, Zhang K, Chen L, Liu J, Ren Y, Yang M, Zhang A, Pu P, Kang C. AC1MMYR2, an inhibitor of dicer-mediated biogenesis of Oncomir miR-21, reverses epithelial-mesenchymal transition and suppresses tumor growth and progression. Cancer Res. 2013;73:5519–5531. doi: 10.1158/0008-5472.CAN-13-0280. [DOI] [PubMed] [Google Scholar]
- 242.Shum D, Bhinder B, Radu C, Calder P, Ramirez CN, Djaballah H. An image-based biosensor assay strategy to screen for modulators of the microRNA 21 biogenesis pathway. Comb Chem High Throughput Screen. 2012;15:529–541. doi: 10.2174/138620712801619131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Connelly CM, Deiters A. Identification of Inhibitors of MicroRNA Function from Small Molecule Screens. Methods Mol Biol. 2014;1095:147–156. doi: 10.1007/978-1-62703-703-7_12. [DOI] [PubMed] [Google Scholar]
- 244.Gumireddy K, Young DD, Xiong X, Hogenesch JB, Huang Q, Deiters A. Small-molecule inhibitors of microrna miR-21 function. Angew Chem Int Ed Engl. 2008;47:7482–7484. doi: 10.1002/anie.200801555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Young DD, Connelly CM, Grohmann C, Deiters A. Small molecule modifiers of microRNA miR-122 function for the treatment of hepatitis C virus infection and hepatocellular carcinoma. J Am Chem Soc. 2010;132:7976–7981. doi: 10.1021/ja910275u. [DOI] [PubMed] [Google Scholar]
- 246.Cai M, Yang Y. Targeted genome editing tools for disease modeling and gene therapy. Curr Gene Ther. 2014;14:2–9. doi: 10.2174/156652321402140318165450. [DOI] [PubMed] [Google Scholar]
- 247.Marraffini LA, Sontheimer EJ. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet. 2010;11:181–190. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual- RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA- guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 253.Long C, McAnally JR, Shelton JM, Mireault AA, Bassel-Duby R, Olson EN. Prevention of muscular dystrophy in mice by CRISPR/Cas9-mediated editing of germline DNA. Science. 2014;345:1184–1188. doi: 10.1126/science.1254445. [DOI] [PMC free article] [PubMed] [Google Scholar]