Abstract
Stimulating or maintaining the proliferative capacity of postnatal mammalian cardiomyocytes is a major challenge to cardiac regeneration. Previously, we found that fetal cardiac extracellular matrix (ECM) could promote neonatal rat cardiomyocyte proliferation in vitro better than neonatal or adult ECM. We hypothesized that partial digestion of adult ECM (PD-ECM), would liberate less crosslinked components that promote cardiomyocyte proliferation, similar to fetal ECM. Neonatal rat cardiac cells were seeded onto substrates coated with adult rat cardiac ECM that had been solubilized in pepsin-HCl for 1, 3, 6, 12, 24, or 48 hr. Cardiomyocyte proliferation and fold-change in numbers from 1 to 5 days were highest on 1hr and 3hr PD-ECM compared to other conditions. Sarcomeres tended to mature on 24hr and 48hr PD-ECM where low proliferation was observed. 3hr PD-ECM was primarily composed of Fibrillin-1, Fibrinogen, and Laminins while 48hr PD-ECM was dominated by Collagen I. Our results suggest that adult ECM retains regenerative cues that may be masked by more abundant, mature ECM components. PD-ECM provides a simple yet powerful approach to promoting cardiomyocyte proliferation.
Keywords: cardiomyocyte, cell proliferation, ECM (extracellular matrix), image analysis, integrin
1. Introduction
Congenital heart defects (CHDs) are a leading cause of mortality in young children and cardiovascular disease is the leading cause of death in adults.[1] Although medical and surgical interventions have significantly improved survival over the years, many patients inevitably develop heart failure.[2] Current therapies to treat heart failure slow down its progression but do not restore the contractile function of the myocardium.[3] Thus, the only successful long-term option is either heart transplantation or the use of ventricular assist devices.[2] The prognosis for pediatric patients with end stage heart failure is particularly poor due to limited donor organs and few devices that are designed for children.[4–6] Thus, there is a need to develop new strategies to either maintain or restore the function of the failing heart, especially for young patients.
As cardiomyocytes are responsible for the contractile function of the heart, a major goal of cardiac regeneration and cell-based therapies is to restore the numbers of functional cardiomyocytes that have been lost. Although cardiomyocytes are highly proliferative during fetal development,[7] they undergo a switch from hyperplastic to hypertrophic growth after birth.[8, 9] The extracellular matrix (ECM) appears to play an important role in regulating cardiomyocyte proliferation and maturation. ECM proteins synthesized by embryonic cardiac fibroblasts enhance the proliferative response of embryonic cardiomyocytes to growth factors.[10] Integrins, which are necessary for cell adhesion and response to the ECM, change throughout heart development and maturation,[11–15] particularly during the switch from hyperplasia to hypertrophy.[16] Indeed, neonatal and adult cardiomyocytes have preferential adhesion for different types of ECM[17] and the ECM can significantly influence cardiomyocyte response.[10, 18] An ECM-based approach to stimulating or maintaining the proliferative capacity of postnatal mammalian cardiomyocytes could be a powerful tool for cardiac regeneration.
Solubilized cardiac ECM shows much promise as an injectable therapeutic biomaterial that can promote vascularization and improve cardiac function in animal models of myocardial infarction.[2, 19, 20] In our previous work, we found that neonatal rat cardiomyocytes were more proliferative when cultured on fetal cardiac ECM compared to neonatal or adult ECM.[21] However, the use of fetal ECM as a biomaterial for cardiac regeneration is unrealistic due to ethical concerns (in regards to human-derived tissues) and low yields of material compared to the adult heart. We noted in our previous study that fetal ECM solubilized in pepsin within 1 hr while adult ECM required 24–48 hr to fully solubilize.[21] Fetal ECM tends to be provisional and less crosslinked than mature adult ECM,[22] which likely allows it to solubilize more rapidly. We hypothesized that shorter digestions of adult cardiac ECM, or “partially digested ECM” (PD-ECM), would liberate more soluble or less crosslinked components such as those present in the fetal ECM, and thus promote cardiomyocyte proliferation. In this work, we studied the effect of adult rat PD-ECM on proliferation and sarcomere structure in neonatal rat cardiomyocytes. We also determined the complex composition of the PD-ECM and identified candidate peptides that may play a role in the proliferative response. We anticipate that the PD-ECM approach will be a beneficial therapy in future regenerative medicine strategies.
2. Results
2.1. Cardiomyocyte Morphology
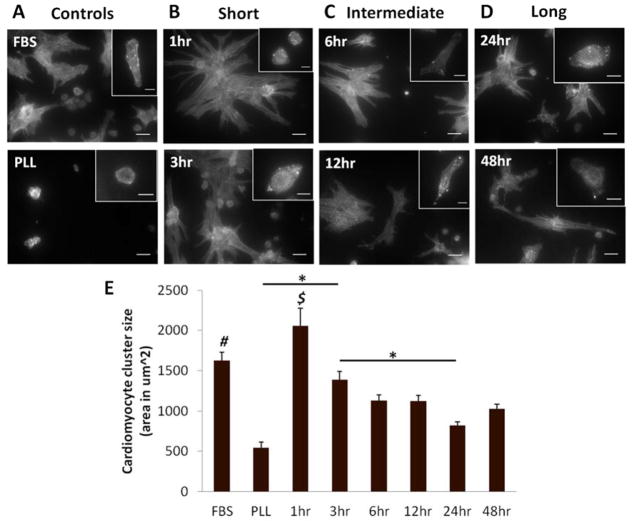
Cardiomyocyte morphology was assessed at 1 and 5 days in culture. After 1 day, myocytes on PLL cultured with FBS demonstrated good initial adhesion and spreading. Cells on PLL with serum-free media were rounded, indicating poor adhesion. At 5 days, FBS controls had small clusters of well-spread myocytes while the serum-free PLL controls remained rounded (Figure 1A). There were also striking differences among the PD-ECM conditions. Cardiomyocytes on “short” PD-ECM (1 hr, 3 hr) were generally small and rounded but with observable sarcomeres at 1 day. After 5 days, large clusters or “colonies” of well-spread myocytes were observed (Figure 1B). Cardiomyocytes had the best initial adhesion and spreading on “intermediate” PD-ECM (6 hr, 12 hr) and also formed clusters after 5 days, but these appeared smaller than on the short PD-ECM (Figure 1C). Myocytes were able to spread reasonably well on “long” PD-ECM (24 hr, 48 hr) although after 5 days they tended to remain as individual cells or very small clusters (Figure 1D). Measurements of cardiomyocyte cluster size showed that the largest clusters formed on 1hr PD-ECM, followed by FBS and 3hr PD-ECM (Figure 1E). Given that our conditions were serum free, we generally did not observe contraction of the cardiomyocytes on PD-ECM; contraction was observed in FBS controls.
Figure 1. Cardiomyocyte morphology on PD-ECM.
Representative images of cardiac α-actinin stain showing cardiomyocytes at 1 day (insets) and 5 days (full image). (A) FBS-treated controls on PLL (“FBS”) show well spread cardiomyocytes even at 1 day, with small clusters observed at 5 days. PLL controls without serum (“PLL”) had rounded cardiomyocytes at both time points. (B) “Short” digestions of 1hr and 3hr PD-ECM had some spreading at 1 day and large clusters of cardiomyocytes at 5 days. (C) “Intermediate” digestions of 6hr and 12hr PD-ECM had well-spread myocytes at 1 day and small clusters or individual well-spread cells at 5 days. (D) “Long” digestions of 24hr and 48hr PD-ECM resulted in some spreading at 1 day and small clusters and many individual cardiomyocytes at 5 days. Scale bars for insets = 10 μm; full images = 20 μm. (E) Measurements of cardiomyocyte cluster size show significantly larger clusters for FBS and 1hr PD-ECM compared to all other conditions. $ = significant difference vs. all conditions except FBS; # = significant difference vs. PLL, 6hr, 12hr, 24hr, and 48hr PD-ECM. Graph shows mean ± S.E.M.
2.2. Cardiac Cell Density and Myocyte Populations
To quantitatively assess initial cell adhesion and changes in cardiac cell populations over time, we determined cell density and cardiomyocyte populations at 1 day and 5 days. Total cell and cardiomyocyte density were similar among conditions at 1 day, with the exception of FBS, where significantly more cells were observed (Figure 2A & B, blue). Total cell density increased significantly after 5 days for all conditions except PLL serum-free. Cell density was highest for FBS controls and 1hr PD-ECM, and lowest for PLL controls and 24hr and 48hr PD-ECM (Figure 2A, brown). Cardiomyocyte density increased significantly for FBS controls and 1 hr, 3 hr, and 12 hr PD-ECM after 5 days in culture. There were significantly more myocytes for FBS, 1 hr and 3 hr PD-ECM vs. all other conditions after 5 days (Figure 2A & B, brown).
Figure 2. Cardiac cell adhesion at 1 day and changes in cell populations over time.
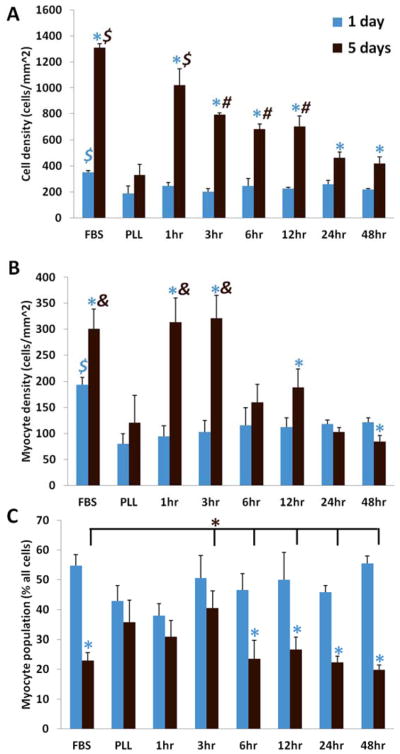
(A) Cell density measurements at 1 day (blue bars) and 5 days (brown bars). (B) Cardiomyocyte density measurements at 1 day and 5 days. Note that cardiomyocyte numbers were highest for FBS controls and “short” (1hr and 3hr) PD-ECM. (C) Myocytes as a percentage of the total cardiac cell population. Cardiomyocyte populations dropped significantly for all conditions except PLL and short PD-ECM. Blue * = significant change compared to 1 day time point; $ = significant difference vs. all other conditions at corresponding time point; # = significantly greater vs. PLL, 24hr and 48hr PD-ECM at corresponding time point; & = significant difference vs. PLL, 6hr, 12hr, 24hr, and 48hr PD-ECM at corresponding time point. Graphs show mean ± S.D.
Typically in culture, neonatal rat cardiomyocytes become an increasingly smaller percentage of the population of mixed cardiac cells as they are overtaken by the more proliferative “fibroblasts” (non-myocytes). At 1 day, the cardiomyocyte population was 40–55% of the total cardiac cell population across the various substrates (Figure 2C, blue). After 5 days, the myocyte population decreased significantly for all conditions except for PLL, 1hr and 3hr PD-ECM compared to their respective 1 day time points. These conditions maintained the highest percentage of cardiomyocytes after 5 days (~30–40%) compared to all others (~20–25%) (Figure 2C, brown).
2.3. Proliferation on PD-ECM
To determine whether cardiomyocyte proliferation may contribute to increased cardiomyocyte density on short PD-ECM, we assessed fold-changes in cell numbers and the proliferation marker Ki67 (Figure 3A). Ki67 is present in cell nuclei during all active phases of the cell cycle (G1, S, G2, and mitosis), but is not expressed in quiescent cells (G0 phase).[23] At 5 days, the percentage of proliferating cells that were cardiomyocytes was highest for “short” PD-ECM (1hr and 3hr, 38–48% of Ki67+ cells), and lowest for PLL and “long” PD-ECM (24hr and 48hr – only 10–16% of Ki67+ cells) (Figure 3B).
Figure 3. Proliferation of cells on PD-ECM.
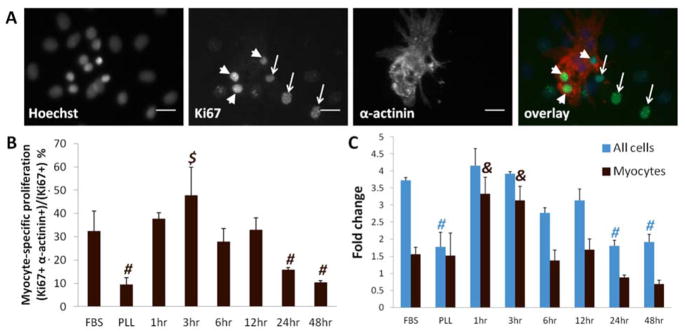
(A) Example images showing all nuclei stained with Hoechst, Ki67 staining of myocyte nuclei (arrowheads) and non-myocyte nuclei (arrows), and cardiac α-actinin staining to label myocytes. Overlay shows proliferating cardiomyocytes and non-myocytes. Scale bar = 20 μm. (B) Cardiomyocyte-specific proliferation (the percentage of Ki67+ cells that are myocytes) is highest on 3hr PD-ECM compared to all other conditions except FBS and 1hr PD-ECM. (C) Fold-changes in cell populations calculated from 1 day to 5 days show greatest increases in cell numbers for FBS and “short” (1hr and 3hr) PD-ECM. Cardiomyocyte fold-change was highest on short PD-ECM. $ = significant difference vs. PLL, 6hr, 12hr, 24hr, and 48hr PD-ECM at corresponding time point; # = significantly lower compared to FBS, 1hr, 3hr, 6hr, and 12hr PD-ECM; & = significant difference vs. FBS, PLL, 6hr, 12hr, 24hr, and 48hr PD-ECM. Graphs show mean ± S.D.
Total cell numbers had the greatest increases for FBS and “short” PD-ECM (~3.5–4-fold) and were lowest for PLL and “long” PD-ECM (~2-fold) (Figure 3C, blue). Cardiomyocyte numbers increased most on “short” PD-ECM (~3-fold), had modest increases for FBS, PLL, and “intermediate” PD-ECM (~1.5-fold), and did not increase on “long” PD-ECM (Figure 3C, brown). Interestingly, myocyte-specific proliferation roughly followed the trends observed in fold-changes in myocyte numbers.
2.4. Sarcomere Maturation on PD-ECM
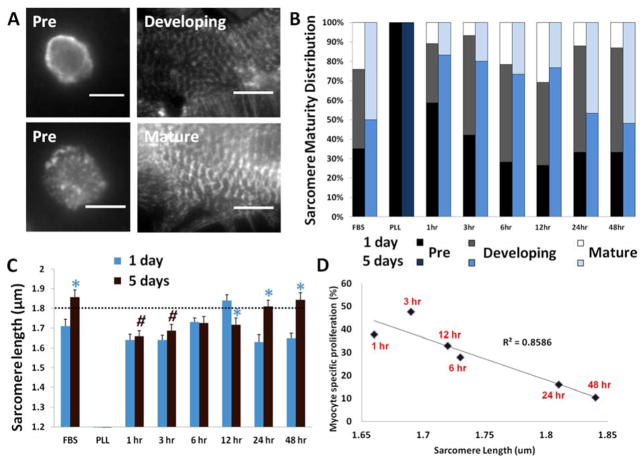
To determine cardiomyocyte maturation on PD-ECM, we analyzed sarcomeres. Sarcomeres were categorized based on organization and length (Figure 4A). At 1 day, a number of cardiomyocytes with no or ill-defined sarcomeric structures were observed. These cells were most abundant on PLL and “short” PD-ECM (Figure 4B, black). Developing sarcomeres were observed for all conditions except PLL (Figure 4B, gray). Mature sarcomeres were mostly observed for FBS and “intermediate” PD-ECM (Figure 4B, white). After 5 days, a shift in the sarcomere maturity distribution was observed for all conditions except PLL, on which cells failed to develop sarcomeric structures (Figure 4B, dark blue). A shift to a greater abundance of mature sarcomeres was observed for FBS and “long” PD-ECM (Figure 4B, light blue). “Short” and “intermediate” PD-ECM were characterized by developing sarcomeres (Figure 4B, blue).
Figure 4. Sarcomere development and correlation with proliferation.
(A) Examples of sarcomere categorization by α-actinin staining. Pre-developed sarcomeres are non-existent or highly disorganized. Developing sarcomeres are small (< 1.8 μm) and disorganized. Mature sarcomeres are large (> 1.8 μm) and organized into fibrils. Scale bars = 10 μm. (B) Sarcomere maturity distribution changes from 1 day to 5 days. (C) Average sarcomere length measurements show that sarcomeres lengthened or matured for FBS and long PD-ECM but remained small or immature for short PD-ECM. Dotted line indicates threshold for maturation. (D) Myocyte-specific proliferation at 5 days plotted against average sarcomere length at 5 days shows inverse correlation with proliferation and sarcomere size on PD-ECM. Blue * = significant change compared to 1 day time point; # = significantly lower compared to FBS and 48hr PD-ECM at corresponding time point. Graph in C shows mean ± S.E.M.
Quantification of sarcomere length mirrors the maturity distribution data. At 1 day, average sarcomere size was largest on FBS and 12hr PD-ECM (Figure 4C, blue). After 5 days, the largest sarcomeres were observed on FBS and “long” PD-ECM, which crossed the threshold for mature sarcomere size (indicated by the dotted line). Sarcomere size increased significantly from 1 day to 5 days for these conditions as well, but cardiomyocytes tended to maintain smaller, developing sarcomeres on “short” and “intermediate” PD-ECM (Figure 4C, brown).
As cardiomyocytes undergo a dramatic switch from hyperplastic to hypertrophic growth after birth,[8] cardiomyocyte maturation is generally considered to be inversely correlated with proliferative capacity. We found that cardiomyocyte-specific proliferation on PD-ECM had a strong inverse correlation with sarcomere size at 5 days (R2 = 0.86) (Figure 4D). This data suggests that “short” PD-ECM maintains cardiomyocytes in a less mature state that may help promote or maintain their ability to proliferate.
2.5. Cardiac Cell Markers and Integrin Gene Expression
To determine the effects of PD-ECM on gene expression, we analyzed a panel of cardiac markers and integrins by quantitative PCR after 5 days in culture (full list of genes in Supplemental Table 1). For these studies, we focused on a select set of conditions: 3hr PD-ECM (high proliferation), 12hr PD-ECM (intermediate proliferation), and 48hr PD-ECM (low proliferation). We found that transcription factors and contractile genes were not significantly affected by PD-ECM at the time point studied, although there was generally a trend in decreased expression on 48hr PD-ECM (Supplemental Figure 1A, B). The expression levels of most integrin genes were also unaffected (not shown). The only significant differences were found in Kit (stem cell/non-terminally differentiated cardiomyocyte marker[24–27]) and Itga6 (integrin subunit α6), which were increased on 3hr PD-ECM compared to 12hr and 48hr PD-ECM (Supplemental Figure 1C).
2.6. Composition of PD-ECM
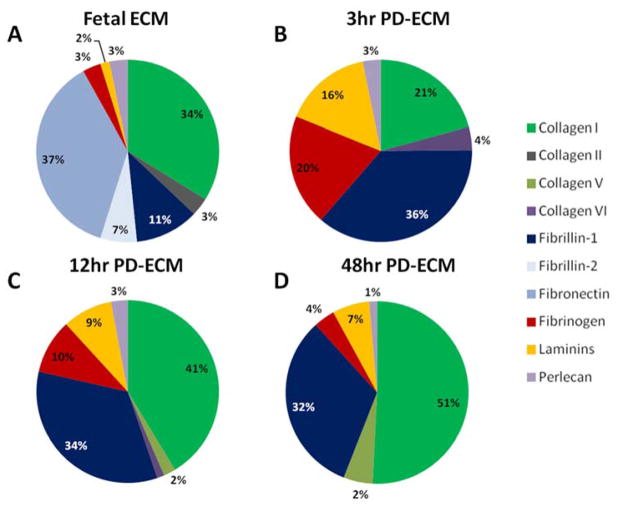
The composition of the adult PD-ECM for 3hr, 12hr, and 48hr digestions, as well as embryonic day 18 (E18) fetal ECM, was determined from spectrum count data obtained by LC-MS/MS. The most abundant protein in fetal ECM was Fibronectin (37%), similar to our previous findings,[21] followed by Collagen I (34%), Fibrillin-1 (11%), and Fibrillin-2 (7%) (Figure 5A). Of note, Fibronectin was not detected in adult PD-ECM. The most abundant protein identified in 3hr PD-ECM (which had the highest cardiomyocyte proliferation), was Fibrillin-1 (36%), followed by Collagen I (21%), Fibrinogen (20%), and Laminins (16%) (Figure 5B). With longer digestions (12hr and 48hr), Collagen I became more and more dominant in the relative composition (41% and 51%) while the percentage of Fibrillin-1 stayed the same (32–34%). Fibrinogen and Laminins also decreased in relative abundance for 12hr and 48hr PD-ECM (Figure 5C, D). This data suggests that either (1) the ECM proteins/peptides which promote cardiomyocyte proliferation are not the same in fetal ECM and adult “short” PD-ECM or (2) the proliferation-promoting peptides are the same but are not the most abundant proteins present.
Figure 5. Composition of PD-ECM.
Composition of select PD-ECM and fetal ECM was determined from LC-MS/MS data. (A) Fetal ECM was mostly composed of Fibronectin and Collagen I. (B) 3hr PD-ECM was mostly composed of Fibrillin-1, Collagen I, Fibrinogen, and Laminins. Collagen I content increased with digestion and was the major component of (C) 12hr and (D) 48hr PD-ECM. Note that Fibronectin was not detected in adult PD-ECM.
2.7. Peptide sequences in PD-ECM
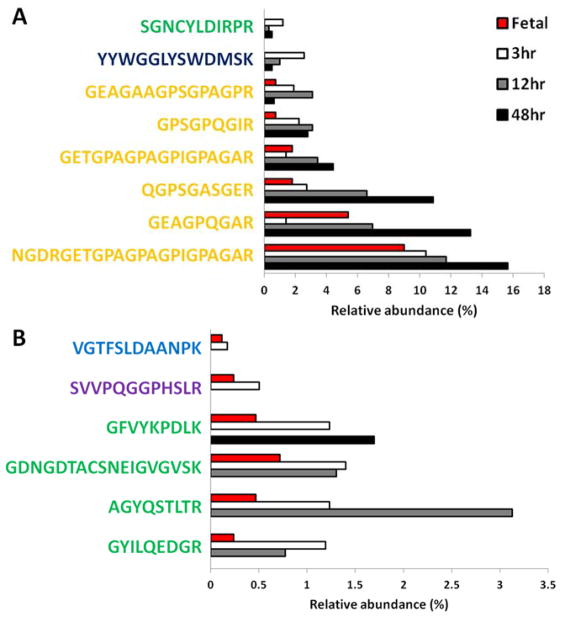
To identify potential candidate sequences in the proliferative response, we further studied the peptide results from LC-MS/MS. Of note, a greater variety of unique peptide sequences were found in 3hr PD-ECM (44), followed by 12hr (36) and then 48hr PD-ECM (26). There were 8 peptide sequences ubiquitous in all PD-ECM, but to varying degrees of relative abundance (Figure 6A). Of these, six were derived from Collagen I; one was derived from Fibrinogen (YYWGGLYSWDMSK), and one from Fibrillin-1 (SGNCYLDIRPR). 48hr PD-ECM tended to have higher abundance of ubiquitous peptides, particularly 3 of the Collagen I-derived peptides. Fetal ECM also contained the Collagen I-derived peptides. Given that both fetal ECM[21] and 3hr PD-ECM promote cardiomyocyte proliferation, we compared their compositions to determine if there were any shared peptides (Figure 6B). There were few peptides common to both and these were also in low abundance, likely due to the high concentration of Fibronectin in fetal ECM that was absent in adult PD-ECM. However, of the six common peptides found, four were derived from Fibrillin-1, while the remaining two were from Perlecan and Laminin α5. Three of these peptides were also found in 12hr PD-ECM, and only one in 48hr PD-ECM.
Figure 6. Common peptides in PD-ECM.
(A) Ubiquitous peptides found in all PD-ECM and (B) peptides common to fetal ECM and 3hr PD-ECM. Note that ubiquitous peptides tend to be more abundant in 48hr PD-ECM. Fetal ECM and 3hr PD-ECM share some peptides but these are in relatively low abundance. Collagen I derived peptides shown in yellow; Fibrillin-1 in green; Fibrinogen in dark blue; Laminin in light blue; Perlecan in purple.
We identified the peptides that were exclusive to or characteristic of each PD-ECM. 3hr PD-ECM was characterized by a variety of peptides derived from Fibrillin-1, Fibrinogen, and Laminin (Figure 7A). Some of these peptides were also found in 12hr PD-ECM in lower abundance but were absent in 48hr PD-ECM. Interestingly, 12hr PD-ECM contained no unique peptides, but was characterized by two sub-sets of peptide groups that were also found in 3hr PD-ECM and 48hr PD-ECM (Figure 7B). 48hr PD-ECM had the fewest characteristic peptides, with only one exclusive Collagen V-derived peptide sequence not found in 3hr and 12hr PD-ECM. The remaining three characteristic peptides were also found to some degree in 12hr PD-ECM but not in 3hr PD-ECM (Figure 7C).
Figure 7. Exclusive and characteristic peptides in PD-ECM.
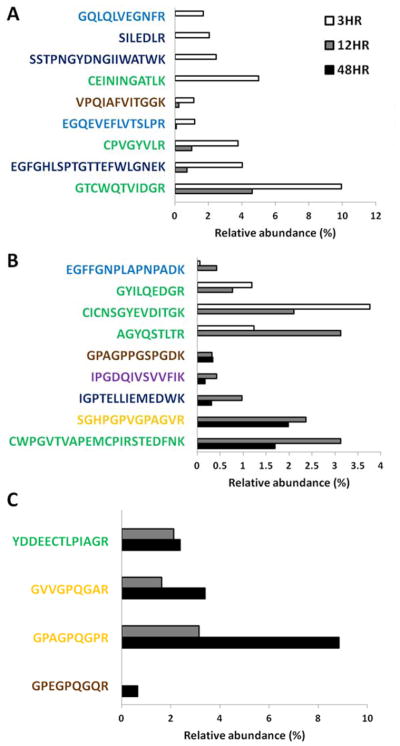
(A) Peptides found in 3hr PD-ECM were mostly derived from Fibrillin-1, Fibrinogen, and Laminin. (B) No exclusive peptides in 12hr PD-ECM were found, but two groups of peptides were identified as those characteristic of 12hr PD-ECM but also detected in 3hr or 48hr PD-ECM. (C) Far fewer exclusive or characteristic peptides were found in 48hr PD-ECM. Collagen I derived peptides shown in yellow; Fibrillin-1 in green; Fibrinogen in dark blue; Laminin in light blue; Perlecan in purple; Collagens V and VI in brown.
3. Discussion
Current cardiac regeneration strategies are limited by the inability of cardiomyocytes to proliferate significantly after birth.[28] Although neonates have a greater capacity for regeneration compared to adults,[29, 30] this capability is quickly lost. Much effort has focused on autologous stem cell-based therapies for cardiac repair, such as mesenchymal stem cells or cardiac progenitors.[31–33] However, these cells have poor differentiation efficiency toward the cardiomyocyte lineage and most benefits are attributed to paracrine signaling.[34–38] Although pluripotent stem cells can generate significant numbers of cardiomyocytes, there still exist ethical and safety concerns regarding the use of these cells in humans.[39–41] An alternative approach would be to use biomaterials that can promote cardiac repair or regeneration. Indeed, adult porcine cardiac ECM shows considerable promise as a potential injectable therapeutic for treating myocardial infarction.[2, 19, 42–44] However, we have found that fully digested adult cardiac ECM does not promote cardiomyocyte proliferation as well as younger ECM, such as that derived from fetal animals.[21] Fetal and adult cardiac ECM share many of the same individual ECM protein components, but in differing ratios.[21, 45] This raised an interesting question as to whether the adult ECM retains “regenerative” cues that are masked by the more abundant “mature” cues, and was a major motivator for our current work.
Our rationale for using the partial digestion approach was our observation in previous work that fetal cardiac ECM fully digested in about 1 hr while adult ECM required 24–48 hr.[21] Thus, we hypothesized that short digestion of adult ECM would release less crosslinked components that may be similar to the fetal ECM and that these “short” PD-ECM would promote cardiomyocyte proliferation better than “long” PD-ECM that fully solubilized the ECM. We found that cardiomyocyte numbers increased most significantly on “short” (1hr and 3hr) PD-ECM and myocyte-specific Ki67 staining was higher on 3hr PD-ECM. In addition, cardiomyocytes were found in large clusters on “short” PD-ECM while they tended to remain as individual cells on “long” PD-ECM. Cardiomyocyte clusters are suggestive of colonies undergoing expansion. Taken together, the data indicates that significant expansion of the cardiomyocyte population occurs on “short” PD-ECM via cardiomyocyte proliferation, similar to what we found previously for fetal cardiac ECM.
Cardiomyocytes undergo a switch from hyperplasia to hypertrophy after birth, in which growth ceases to be primarily via proliferation and changes to increases in cell size. In the rat, this transition occurs rapidly around postnatal day 3 (P3).[8] It is generally accepted that mammalian cell maturity and proliferative capacity are inversely correlated: the more mature the cell, the less proliferative it is. In cardiomyocytes, it has been shown that there is a correlation between sarcomere structure and proliferation. The immature cardiomyocytes in the developing heart are highly proliferative and have small, disorganized sarcomeres.[8] When proliferation has been observed in adult cardiomyocytes, it is typically in association with “sarcomere disassembly”, in which the large, highly organized sarcomeres become smaller and more disorganized, reminiscent of an immature myocyte.[46–48] In line with previous literature, we found that sarcomere size and cardiomyocyte-specific proliferation on PD-ECM were strongly inversely correlated. Sarcomeres tended to remain small over time in culture on “short” PD-ECM where the highest proliferation was observed, while sarcomeres lengthened on “long” PD-ECM in which cardiomyocyte proliferation was low. Thus, our data suggest that cardiomyocytes were maintained in a less mature state that may have facilitated proliferation on “short” PD-ECM while cells matured on “long” PD-ECM and lost their proliferative capacity.
To further investigate the effects of PD-ECM on the cardiac cell population, we studied various cell markers and integrins via quantitative PCR for select conditions. The most notable was significant up-regulation of the kit gene on 3hr PD-ECM compared to 12hr and 48hr PD-ECM. C-kit is generally considered a cardiac progenitor cell marker;[26, 49, 50] however, it is also transiently expressed in rodent cardiomyocytes soon after birth.[24, 25] Loss of c-kit expression in cardiomyocytes coincides with terminal differentiation.[24] Thus, the increased expression of kit on 3hr PD-ECM with respect to other conditions suggests that cardiomyocyte terminal differentiation may be inhibited, although more rigorous follow-up studies that focus exclusively on the cardiomyocyte population will be needed to confirm this hypothesis.
Integrins are important for mediating cell adhesion and response to the ECM.[51] We found Itga6 (α6 subunit) was 3–4-fold higher on 3hr PD-ECM vs. 12hr and 48hr PD-ECM. Integrin α6β1 is expressed in the early developing heart and is also transiently up-regulated in the neonatal heart.[16] In the context of our ECM composition data, it is well known that α6 integrin binds Laminins,[52, 53] which was a major component of 3hr PD-ECM that promoted cardiomyocyte proliferation. The other major component that characterized 3hr PD-ECM was Fibrillin-1, which is known to bind integrins αvβ3, αvβ6, and α5β1.[54–56] We did not find any differences in these integrin subunits on PD-ECM (data not shown). The adhesion of cardiomyocytes to Fibrillin-1 has not been well studied; thus, it may be possible that novel Fibrillin-1 peptides which bind other integrins have yet to be discovered. Collagen I, a stable, mature protein that is characteristic of the adult heart,[21, 45] became more and more dominant with longer digestions, while other components decreased in their relative abundance. In our experience, Collagen I does not significantly promote cardiomyocyte proliferation (unpublished), and thus its signaling likely masked the “proliferative cues” that may have still been present in longer digestions.
The finding that 3hr PD-ECM and fetal ECM had very different compositions, yet both promote cardiomyocyte proliferation, was intriguing. The major component of fetal ECM is Fibronectin, which is known to mediate embryonic cardiomyocyte proliferation.[10] Interestingly, Fibronectin was not detected in adult PD-ECM, even at short digestions. It is possible that a protein or peptide in low abundance which is present in both fetal and 3hr PD-ECM promotes proliferation. Indeed, we did find peptide sequences that were common to both fetal ECM and 3hr PD-ECM; these peptide sequences were mostly derived from Fibrillin-1. However, it is also probable that fetal ECM and adult 3hr PD-ECM promote cardiomyocyte proliferation through different mechanisms. 3hr PD-ECM contained nine “characteristic” sequences that were mostly derived from Fibrillin-1, Laminin, and Fibrinogen. These peptides were different than the common peptides that were also identified in fetal ECM. 48hr PD-ECM had few unique characteristic peptides and contained many ubiquitous Collagen I-derived peptides that were found in 3hr and 12hr PD-ECM as well. It was also interesting to note that 3hr PD-ECM contained a greater variety of unique peptides compared to longer digestions. The diversity of the 3hr PD-ECM composition may be important in cardiomyocyte proliferation. Further study of the candidate peptides identified in this paper, both individually and in combinations, will be crucial in our future work and may lead to the design of a synthetic “regenerative” ECM.
Elucidating the regenerative mechanisms of fetal ECM and adult PD-ECM will be critical in the context of treating children vs. adults. For example, immature (fetal, early neonatal) myocytes adhere well to Fibronectin, while adult myocytes do not; on the other hand, both immature and adult cardiomyocytes can adhere to Laminins[10, 17] which are present in the developing and mature cardiac ECM. A fetal ECM-based approach may not be appropriate for adult cardiac regeneration if adult myocytes cannot respond to its components. Indeed, immature myocytes can proliferate in response to Fibronectin[10] which is a major component of fetal ECM, while it has been reported that Fibronectin does not stimulate adult cardiomyocyte proliferation.[57] PD-ECM could address this challenge by providing adult ECM-derived cues. It will be intriguing to determine the proliferative response of cardiomyocytes at different developmental ages to PD-ECM. It could be expected that fetal ECM would promote a greater response in fetal myocytes compared to neonatal, while the response in older myocytes may be blunted. However, if PD-ECM does indeed work via different mechanisms, then perhaps older myocytes could be stimulated to proliferate. In addition, given that solubilized pig cardiac ECM is making considerable progress towards clinical translation to treat myocardial infarction,[2] other immediate goals will include evaluating the regenerative potential of PD-ECM in adult animal models of heart disease.
Finally, we note several limitations to our current study. We did not attempt to purify the cardiomyocytes via pre-plating or centrifugation techniques. As such, there was a significant fibroblast population in our cell cultures. It is possible that cardiomyocytes had an indirect response to PD-ECM via the fibroblasts, which are likely releasing paracrine signals and depositing new ECM. The presence of fibroblasts also makes gene data difficult to interpret, since the cardiomyocyte specific population cannot be analyzed. For example, we cannot determine if up-regulation of Itga6 is attributed to the cardiomyocytes or the fibroblasts. Working with a purified cardiomyocyte population, particularly one derived from human pluripotent stem cells, will yield a greater understanding of the mechanisms and therapeutic potential of PD-ECM.
4. Conclusion
In this work, we have demonstrated that short partial digestion of adult cardiac ECM, or “PD-ECM”, can promote neonatal cardiomyocyte proliferation in vitro. Cardiomyocyte numbers increased 3-fold from 1 to 5 days in culture on “short” PD-ECM in serum-free conditions but only increased ~1.5-fold or less when cultured with FBS or on “intermediate” and “long” PD-ECM. Cardiomyocytes on “short” PD-ECM tended to remain immature, as determined by sarcomere length, while cardiomyocytes on “long” PD-ECM underwent lengthening of sarcomeres over time in culture. 3hr PD-ECM was mostly composed of Fibrillin-1, Fibrinogen, Collagen I, and Laminins, while longer digestions led to increasing abundance of Collagen I. A unique set of peptides characteristic of 3hr PD-ECM was also identified. This work suggests that adult ECM contains regenerative cues that may be masked by more abundant, mature proteins. PD-ECM offers a simple but powerful approach to promoting cardiomyocyte proliferation, and the method will likely be adaptable to the regeneration of other cells and tissues as well.
5. Experimental Section
Partial Digestion of Adult Cardiac ECM
All animal procedures were performed in accordance with the Institutional Animal Care and Use Committee at Tufts University and the NIH Guide for the Care and Use of Laboratory Animals. Hearts were harvested from euthanized adult female Sprague Dawley rats (Charles River Laboratories). The ventricular tissue was minced and submerged in 1% (wt/vol) sodium dodecyl sulfate (SDS) in deionized water with gentle shaking at room temperature (RT) until decellularization was complete (about 48 hr). The ECM was transferred to a 1% TritonX-100 (vol/vol) solution for 4 hr to remove SDS and then washed with deionized water at least 3 times for 12 hr per wash. The ECM was frozen at −20°C, lyophilized for 24 hours, and solubilized at a concentration of 10 mg/ml (from initial dry weight, assuming fully digested) in 1 mg/ml pepsin dissolved in 0.1 M HCl. To obtain PD-ECM, solubilization was carried out for 1 hr, 3 hr, 6 hr, 12 hr, 24 hr, and 48 hr at RT. At the end of the designated digestion time, the cardiac ECM solution was neutralized to pH 7 with 1 N NaOH. Any remaining intact tissue was discarded and the supernatant was collected. The concentrations of the various PD-ECM were determined using a NanoDrop 2000 Spectrophotometer (Thermo Scientific).
Culture substrates were prepared the day before cell seeding as described previously.[21] PD-ECM was coated onto 48-well tissue culture plates or 9 × 9 mm glass cover slips at 50 μg/cm2 and allowed to air dry overnight in a sterile tissue culture cabinet. Control surfaces were coated with poly-L-lysine (PLL) (0.01% solution, Sigma) according to the manufacturer’s instructions.
Cell Isolation and Culture
Cardiac cells were isolated from P2–P3 neonatal rat pups according to previously described methods.[58] Pups were euthanized by conscious decapitation and the hearts were collected in ice-cold sterile PBS supplemented with 20 mM glucose. Ventricular tissue was gently minced and underwent 7 × 7 min digestions in collagenase type 2 (Worthington Biochemical Corp, Lakewood, NJ). Cells were counted with a hemocytometer and seeded at a density of 100,000 cells/cm2 on the PLL- and PD-ECM-coated substrates in serum-free media (50/50 mixture of DMEM and Ham’s F12 Nutrient Mix (Invitrogen), 0.2% (wt/vol) bovine serum albumin (BSA) (Sigma), 0.5% insulin–transferrin–selenium-X (Invitrogen) and 1% penicillin–streptomycin (Invitrogen), with 0.1 mM L-ascorbic acid (Sigma) added fresh at every feeding).[21, 59] As a positive control for cell proliferation, a set of cells on PLL were cultured with DMEM containing 15% FBS. Fresh media was given on days 1 and 3 of culture, and samples were analyzed at days 1 and 5, as described below.
Immunocytochemistry and Imaging
Samples were fixed for immunostaining at days 1 and 5 in 4% paraformaldehyde (Alfa Aesar) for 10 min at 4°C and then permeabilized with 0.1% (v/v) TritonX-100 in PBS for 10 min at RT. Non-specific binding was blocked with 1% BSA and 5% donkey serum for 1 hr at RT. To label nuclei, proliferating cells, and cardiomyocytes, samples were incubated for 1 hr with Hoechst 33258 (Invitrogen), rabbit polyclonal Ki67 antibody (1:500 dilution, Abcam), and mouse monoclonal sarcomeric α-actinin antibody (1:500 dilution, Sigma), respectively. Incubation with secondary antibodies Alexa Fluor 488 donkey anti-rabbit IgG and Alexa Fluor 555 donkey anti-mouse IgG (Invitrogen) was carried out for 45 min at RT. In between incubations, samples were washed 3 × 5 min with PBST (PBS with 0.1% Tween20). Images were acquired on an Olympus IX8I microscope using Metamorph Basic software (version 7.7.4.0, Molecular Devices).
Image Analysis
Ten random fields of each sample were imaged using a 20× or 40× objective lens to study cardiomyocyte populations, cell proliferation, and sarcomere maturation. Analysis of cardiac cell populations and cardiomyocyte proliferation was similar to previously described methods.[21] Briefly, a custom-designed pipeline was used in CellProfiler (release 11710, The Broad Institute) to identify total cell nuclei or Ki67+ cell nuclei. The identified nuclei were then “masked” with the corresponding image of sarcomeric α-actinin stain to determine cardiomyocyte-specific nuclei and proliferation, respectively.
Total cell and cardiomyocyte density was determined using the total imaged area (converted to mm2) for each sample. Cardiomyocyte population was determined as the percentage of the total number of nuclei that were contained within the sarcomeric α-actinin “mask”. Cardiomyocyte-specific proliferation was measured as the percentage of proliferating cells (Ki67+ nuclei) that were also positive for sarcomeric α-actinin. As an additional measure of proliferation, fold-changes in total cells and cardiomyocytes were determined from changes in cell density from 24 hr to 5 days.
Sarcomere length has been used as a measure of cardiomyocyte maturation.[60] To assess sarcomeres, images of sarcomeric α-actinin staining at 40× magnification were acquired. Analysis was performed using ImageJ software (NIH, version 1.45s). When organized sarcomeres were observed, a line was manually drawn across multiple sarcomeres perpendicular to alignment. The “Plot Profile” function was used to display the staining intensity across the line. The number of sarcomeres in a given length were counted and average sarcomere length determined. Sarcomeres were also categorized according to the following definitions: “pre-myofibril” (no sarcomeres or highly disorganized sarcomeres were observed); “developing” (sarcomeres measuring < 1.8 μm in length); and “mature” (sarcomeres measuring > 1.8 μm in length).
Gene Expression
Gene expression profiles for various cardiac cell markers and integrins were determined at 5 days using custom-designed PCR array plates from SA Biosciences as well as individual primers from Life Technologies. RNA was isolated using the RNeasy Mini Kit (Qiagen) per the manufacturer’s instructions and stored at −80°C. RNA concentration was determined using a NanoDrop. Equal amounts of RNA from each sample underwent genomic DNA elimination and reverse transcription using the RT2 First Strand Kit (Qiagen). The resulting cDNA was stored at −20°C until use. Quantitative PCR was carried out in the custom array plates per the manufacturer’s instructions using a Stratagene Mx3000P qPCR System (Agilent Technologies). Genes of interest on the custom array plates (Kit, Gata4, Tnnt3, integrins) were normalized to Actb and transcription factors Nkx2.5, Mef2c, Tbx5, and Isl1 were normalized to Gapdh. Cardiac contractile proteins, which should be exclusively expressed in cardiomyocytes, were normalized to Calsq, which has been used as a control for normalizing gene expression specific to the cardiomyocyte population.[61] (see Supplemental Table 1). The comparative CT method (ΔΔCT) was used to determine fold-changes in gene expression on select PD-ECM (3hr, 12hr, and 48hr) and normalized to 48hr PD-ECM. Genes with approximately 2-fold change or greater were identified and statistical analysis was performed as described below (N = 3 for each condition).
Proteomics Analysis of ECM
To determine the composition of select conditions of PD-ECM (3hr, 12hr, and 48hr), solubilized protein was precipitated with acetone and analyzed via liquid chromatography tandem mass spectrometry (LC-MS/MS) at the Beth Israel Deaconess Medical Center Mass Spectrometry Core Facility. Fetal cardiac ECM from E18 pups was included for comparison. Spectrum count data was used to determine relative amounts of ECM components within each group. Individual ECM components are reported as a percentage of the total ECM spectra identified.
Peptide report data was used to identify specific peptides in PD-ECM. The percentage of the total spectra for each unique peptide sequence was used to determine relative abundance within each sample. Ubiquitous peptides were identified as those found in all PD-ECM conditions studied (3hr, 12hr, and 48hr). Exclusive peptides were identified as those found only within a certain condition (e.g., only within 3hr PD-ECM). Characteristic peptides were those which were found in lower abundance in another condition but typically higher in the PD-ECM of interest. Common peptides in fetal ECM and 3hr PD-ECM were also identified. Relative abundances of the peptide sequences of interest were plotted.
Statistical Analysis
Statistical significance was determined using the appropriate dimension of analysis of variance and Tukey’s posthoc test in SigmaPlot 12.0 software. The Student’s t-test was used for pair-wise comparisons. Differences were considered statistically significant for p < 0.05.
Supplementary Material
Supplemental Table 1. Summary of genes analyzed.
Supplemental Figure 1. Gene expression at 5 days. (A) Cardiac transcription factors show a trend in decreasing expression for 48hr PD-ECM but no significant difference. (B) The expression of cardiac contractile markers was not affected by PD-ECM. (C) Kit and Itga6 were significantly higher on 3hr PD-ECM compared to 12hr and 48hr PD-ECM. Graphs show mean ± S.D.
Acknowledgments
This work was supported by the NRSA individual postdoctoral fellowship F32 HL112538 (C.W.), the AHA predoctoral fellowship 14PRE19960001 (K.S.), the NIH Pathway to Independence Award R00 HL093358 (L.D.B.), NIH-NHLBI Award R21 HL115570 (L.D.B.), and NSF CAREER Award NSF1351241 (L.D.B.). The authors gratefully acknowledge Professor John Asara and the Mass Spectrometry Core Laboratory (Beth Israel Deaconess Medical Center) for providing LC-MS/MS data.
References
- 1.Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, Borden WB, Bravata DM, Dai S, Ford ES, Fox CS, Franco S, Fullerton HJ, Gillespie C, Hailpern SM, Heit JA, Howard VJ, Huffman MD, Kissela BM, Kittner SJ, Lackland DT, Lichtman JH, Lisabeth LD, Magid D, Marcus GM, Marelli A, Matchar DB, McGuire DK, Mohler ER, Moy CS, Mussolino ME, Nichol G, Paynter NP, Schreiner PJ, Sorlie PD, Stein J, Turan TN, Virani SS, Wong ND, Woo D, Turner MB. Circulation. 2013;127:e6. doi: 10.1161/CIR.0b013e31828124ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seif-Naraghi SB, Singelyn JM, Salvatore MA, Osborn KG, Wang JJ, Sampat U, Kwan OL, Strachan GM, Wong J, Schup-Magoffin PJ, Braden RL, Bartels K, DeQuach JA, Preul M, Kinsey AM, DeMaria AN, Dib N, Christman KL. Science Transl Med. 2013;5:173ra25. doi: 10.1126/scitranslmed.3005503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ye KY, Black LD. J Cardiovasc Trans Res. 2011;4:575. doi: 10.1007/s12265-011-9303-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schure AY, Kussman BD. Pediatr Anesth. 2011;21:594. doi: 10.1111/j.1460-9592.2010.03418.x. [DOI] [PubMed] [Google Scholar]
- 5.Almond CSD, Thiagarajan RR, Piercey GE, Gauvreau K, Blume ED, Bastardi HJ, Fynn-Thompson F, Singh TP. Circulation. 2009;119:717. doi: 10.1161/CIRCULATIONAHA.108.815712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Almond CS, Morales DL, Blackstone EH, Turrentine MW, Imamura M, Massicotte MP, Jordan LC, Devaney EJ, Ravishankar C, Kanter KR, Holman W, Kroslowitz R, Tjossem C, Thuita L, et al. Circulation. 2013;127:1702. doi: 10.1161/CIRCULATIONAHA.112.000685. [DOI] [PubMed] [Google Scholar]
- 7.Soonpaa MH, Field LJ. Circ Res. 1998;83:15. doi: 10.1161/01.res.83.1.15. [DOI] [PubMed] [Google Scholar]
- 8.Li F, Wang X, Capasso JM, Gerdes AM. J Mol Cell Cardiol. 1996;28:1737. doi: 10.1006/jmcc.1996.0163. [DOI] [PubMed] [Google Scholar]
- 9.Mollova M, Bersell K, Walsh S, Savla J, Das LT, Park S-Y, Silberstein LE, Remedios CGd, Graham D, Colan S, Kuhn B. Proc Natl Acad Sci. 2013;110:1446. doi: 10.1073/pnas.1214608110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ieda M, Tsuchihashi T, Ivey KN, Ross RS, Hong TT, Shaw RM, Srivastava D. Dev Cell. 2009;16:233. doi: 10.1016/j.devcel.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ross RS, Borg TK. Circ Res. 2011;88:1112. doi: 10.1161/hh1101.091862. [DOI] [PubMed] [Google Scholar]
- 12.Terracio L, Rubin K, Gullberg D, Balog E, Carver W, Jyring R, Borg TK. Circ Res. 1991;68:734. doi: 10.1161/01.res.68.3.734. [DOI] [PubMed] [Google Scholar]
- 13.Carver W, Price RL, Raso DS, Terracio L, Borg TK. J Histochem Cytochem. 1994;42:167. doi: 10.1177/42.2.8288862. [DOI] [PubMed] [Google Scholar]
- 14.Collo G, Domanico SZ, Klier G, Quaranta V. Cell Adhes Commun. 1995;3:101. doi: 10.3109/15419069509081280. [DOI] [PubMed] [Google Scholar]
- 15.Pow CST, Hendrickx AG. Anat Rec. 1995;243:241. doi: 10.1002/ar.1092430211. [DOI] [PubMed] [Google Scholar]
- 16.Maitra N, Flink IL, Bahl JJ, Morkin E. Cardiovasc Res. 2000;47:715. doi: 10.1016/s0008-6363(00)00140-1. [DOI] [PubMed] [Google Scholar]
- 17.Borg TK, Rubin K, Lundgren E, Borg K, Obrink B. Dev Biol. 1984;104:86. doi: 10.1016/0012-1606(84)90038-1. [DOI] [PubMed] [Google Scholar]
- 18.Shanker AJ, Yamada K, Green KG, Yamada KA, Saffitz JE. Circ Res. 2005;96:558. doi: 10.1161/01.RES.0000158964.42008.a2. [DOI] [PubMed] [Google Scholar]
- 19.Singelyn JM, DeQuach JA, Seif-Naraghi SB, Littlefield RB, Schup-Magoffin PJ, Christman KL. Biomaterials. 2009;30:5409. doi: 10.1016/j.biomaterials.2009.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singelyn JM, Sundaramurthy P, Johnson TD, Schup-Magoffin PJ, Hu DP, Faulk DM, Wang J, Mayle KM, Bartels K, Salvatore M, Kinsey AM, DeMaria AN, Dib N, Christman KL. J Am Coll Cardiol. 2012;59:751. doi: 10.1016/j.jacc.2011.10.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams C, Quinn KP, Georgakoudi I, Black LD. Acta Biomater. 2014;10:194. doi: 10.1016/j.actbio.2013.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nandadasa S, Foulcer S, Apte SS. Matrix Biol. 2014;35:34. doi: 10.1016/j.matbio.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheri A, Dowsett M. Ann Oncol. 2012;23:x219. doi: 10.1093/annonc/mds307. [DOI] [PubMed] [Google Scholar]
- 24.Li M, Naqvi N, Yahiro E, Liu K, Powell PC, Bradley WE, Martin DIK, Graham RM, Dell’Italia LJ, Husain A. Circ Res. 2008;102:677. doi: 10.1161/CIRCRESAHA.107.161737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naqvi N, Li M, Yahiro E, Graham RM, Husain A. Pediatr Cardiol. 2009;30:651. doi: 10.1007/s00246-008-9366-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tallini Y, Greene KS, Craven M, Spealman A, Breitbach M, Smith J, Fisher PJ, Steffey M, Hesse M, Doran RM, Woods A, Singh B, Yen A, Fleischmann BK, Kotlikoff MI. Proc Natl Acad Sci. 2009;106:1808. doi: 10.1073/pnas.0808920106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He JQ, Vu DM, Hunt G, Chugh A, Bhatnagar A, Bolli R. PLoS One. 2011;6:e27719. doi: 10.1371/journal.pone.0027719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zimmermann WH, Cesnjevar R. Pediatr Cardiol. 2009;30:716. doi: 10.1007/s00246-009-9405-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simpson DL, Mishra R, Sharma S, Goh SK, Deshmukh S, Kaushal S. Circulation. 2012;126(suppl 1):S46. doi: 10.1161/CIRCULATIONAHA.111.084699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, Sadek HA. Science. 2011;331:1078. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bolli R, Chugh AR, D’Amario D, Loughran JH, Stoddard MF, Ikram S, Beache GM, Wagner SG, Leri A, Hosoda T, Sanada F, Elmore JB, Goichberg P, Cappetta D, Salankhi NK, Fahsah I, Rokosh DG, Slaughter MS, Kajstura J, Anversa P. The Lancet. 2011;378:1847. doi: 10.1016/S0140-6736(11)61590-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Makkar RR, Smith RR, Cheng K, Malliaras K, Thomson LEJ, Berman D, Czer LSC, Marban L, Mendizabal A, Johnston PV, Russell SD, Scheleri KH, Lardo AC, Gerstenblith G, Marban E. Lancet. 2012;379:895. doi: 10.1016/S0140-6736(12)60195-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Williams AR, Hare JM. Circ Res. 2011;109:923. doi: 10.1161/CIRCRESAHA.111.243147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van_Berlo JH, Kanisicak O, Maillet M, Vagnozzi RJ, Karch J, Lin S-CJ, Middleton RC, Marban E, Molkentin JD. Nature. 2014;509:337. doi: 10.1038/nature13309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kawaguchi N, Smith AJ, Waring CD, Hasan MK, Miyamoto S, Matsuoka R, Ellison GM. PLoS One. 2010;5:e14297. doi: 10.1371/journal.pone.0014297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uemura R, Xu M, Ahmad N, Ashraf M. Circ Res. 2006;98:1414. doi: 10.1161/01.RES.0000225952.61196.39. [DOI] [PubMed] [Google Scholar]
- 37.Gnecchi M, Zhang Z, Ni A, Dzau VJ. Circ Res. 2008;103:1204. doi: 10.1161/CIRCRESAHA.108.176826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hong KU, Guo Y, Li QH, Cao P, Al-Maqtari T, Vajravelu BN, Du J, Book MJ, Zhu X, Nong Y, Bhatnagar A, Bolli R. PLoS One. 2014;9:e96725. doi: 10.1371/journal.pone.0096725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mayshar Y, Ben-David U, Lavon N, Biancotti JC, Yakir B, Clark AT, Plath K, Lowry WE, Benvenisty N. Cell Stem Cell. 2010;7:521. doi: 10.1016/j.stem.2010.07.017. [DOI] [PubMed] [Google Scholar]
- 40.Ma H, Morey R, O’Neil RC, He Y, Daughtry B, Schultz MD, Hariharan M, Nery JR, Castanon R, Sabatini K, Thiagarajan RD, Tachibana M, Kang E, Tippner-Hedges R, Ahmed R, Gutierrez NM, Van_Dyken C, Polat A, Sugawara A, Sparman M, Gokhale S, Amato P, Wolf DP, Ecker JR, Laurent LC, Mitalipov S. Nature. 2014;511:177. doi: 10.1038/nature13551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miura K, Okada Y, Aoi T, Okada A, Takahashi K, Okita K, Nakagawa M, Koyanagi M, Tanabe K, Ohnuki M, Ogawa D, Ikeda E, Okano H, Yamanaka S. Nature Biotechnol. 2009;27:743. doi: 10.1038/nbt.1554. [DOI] [PubMed] [Google Scholar]
- 42.Singelyn JM, Christman KL. J Cardiovasc Trans Res. 2010;3:478. doi: 10.1007/s12265-010-9202-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seif-Naraghi SB, Salvatore MA, Schup-Magoffin PJ, Hu DP, Christman KL. Tissue Eng Part A. 2010;16:2017. doi: 10.1089/ten.tea.2009.0768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seif-Naraghi SB, Horn D, Schup-Magoffin PJ, Christman KL. Acta Biomaterialia. 2012;8:3695. doi: 10.1016/j.actbio.2012.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gershlak JR, Resnikoff JI, Sullivan KE, Williams C, Wang RM, Black LD. Biochem Biophys Res Comm. 2013;439:161. doi: 10.1016/j.bbrc.2013.08.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jopling C, Sleep E, Raya M, Marti M, Raya A, Belmonte JCI. Nature. 2010;464:606. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bersell K, Arab S, Haring B, Kuhn B. Cell. 2009;138:257. doi: 10.1016/j.cell.2009.04.060. [DOI] [PubMed] [Google Scholar]
- 48.Zhang Y, Li TS, Lee ST, Wawrowsky KA, Cheng K, Galang G, Malliaras K, Abraham MR, Wang C, Marban E. PLoS One. 2010;5:e12559. doi: 10.1371/journal.pone.0012559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miyamoto S, Kawaguchi N, Ellison GM, Matsuoka R, Shin’oka T, Kurosawa H. Stem Cells Dev. 2010;19:105. doi: 10.1089/scd.2009.0041. [DOI] [PubMed] [Google Scholar]
- 50.Hosoda T. Am J Cardiovasc Dis. 2012;2:58. [PMC free article] [PubMed] [Google Scholar]
- 51.Bokel C, Brown NH. Dev Cell. 2002;3:311. doi: 10.1016/s1534-5807(02)00265-4. [DOI] [PubMed] [Google Scholar]
- 52.Davis GE, Senger DR. Circ Res. 2005;97:1093. doi: 10.1161/01.RES.0000191547.64391.e3. [DOI] [PubMed] [Google Scholar]
- 53.Belkin AM, Stepp MA. Microsc Res Tech. 2000;51:280. doi: 10.1002/1097-0029(20001101)51:3<280::AID-JEMT7>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 54.Bax DV, Bernard SE, Lomas A, Morgan A, Humphries J, Shuttleworth CA, Humphries MJ, Kielty CM. J Biol Chem. 2003;278:34605. doi: 10.1074/jbc.M303159200. [DOI] [PubMed] [Google Scholar]
- 55.Pfaff M, Reinhardt DP, Sakai LY, Timpl R. FEBS Lett. 1996;384:247. doi: 10.1016/0014-5793(96)00325-0. [DOI] [PubMed] [Google Scholar]
- 56.Jovanovic J, Iqbal S, Jensen S, Mardon H, Handford P. Biochem Soc Trans. 2008;36:257. doi: 10.1042/BST0360257. [DOI] [PubMed] [Google Scholar]
- 57.Kuhn B, Monte Fd, Hajjar RJ, Chang Y-S, Lebeche D, Arab S, Keating MT. Nature Medicine. 2007;13:962. doi: 10.1038/nm1619. [DOI] [PubMed] [Google Scholar]
- 58.Ye KY, Sullivan KE, Black LD. J Vis Exp. 2011;19 doi: 10.3791/3251. pii: 3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Engel FB, Schebesta M, Duong MT, Lu G, Ren S, Madwed JB, Jiang H, Wang Y, Keating MT. Genes Dev. 2005;19:1175. doi: 10.1101/gad.1306705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Young JL, Engler AJ. Biomaterials. 2011;32:1002. doi: 10.1016/j.biomaterials.2010.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hullin R, Asmus F, Ludwig A, Hersel J, Boekstegers P. Circulation. 1999;100:155. doi: 10.1161/01.cir.100.2.155. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1. Summary of genes analyzed.
Supplemental Figure 1. Gene expression at 5 days. (A) Cardiac transcription factors show a trend in decreasing expression for 48hr PD-ECM but no significant difference. (B) The expression of cardiac contractile markers was not affected by PD-ECM. (C) Kit and Itga6 were significantly higher on 3hr PD-ECM compared to 12hr and 48hr PD-ECM. Graphs show mean ± S.D.