Abstract
Coordinated serotonin (5-HT) synthesis and reuptake depends on coexpression of Tph2, Aadc (Ddc) and Sert (Slc6a4) in brain 5-HT neurons. However, other gene products play critical roles in brain 5-HT synthesis and transport. For example, 5-HT synthesis depends on coexpression of genes encoding the enzymatic machinery necessary for the production and regeneration of tetrahydrobiopterin (BH4). In addition, the organic cation transporter 3 (Oct3, Slc22a3) functions as a low affinity, high capacity 5-HT reuptake protein in 5-HT neurons. The regulatory strategies controlling BH4 and Oct3 gene expression in 5-HT neurons have not been investigated. Our previous studies showed that Pet-1 is a critical transcription factor in a regulatory program that controls coexpression of Tph2, Aadc and Sert in 5-HT neurons. Here, we investigate whether a common regulatory program determines global 5-HT synthesis and reuptake through coordinate transcriptional control. We show with comparative microarray profiling of flow sorted YFP+ Pet-1−/− and wild type 5-HT neurons that Pet-1 regulates BH4 pathway genes, Gch1, Gchfr, and Qdpr. Thus, Pet-1 coordinates expression of all rate-limiting enzymatic (Tph2, Gch1) and post-translational regulatory (Gchfr) steps that determine the level of mammalian brain 5-HT synthesis. Moreover, Pet-1 globally controls acquisition of 5-HT reuptake in dorsal raphe 5-HT neurons by coordinating expression of Slc6a4 and Slc22a3. In situ hybridizations revealed that virtually all 5-HT neurons in the dorsal raphe depend on Pet-1 for Slc22a3 expression; similar results were obtained for Htr1a. Therefore, few if any 5-HT neurons in the dorsal raphe are resistant to loss of Pet-1 for their full neuron-type identity.
Keywords: Serotonin, tetrahydrobiopterin, Organic cation transporter 3, Pet-1, GTP cyclohydrolase I, GTP cyclohydrolase I feedback regulator, Quinoid dihydropteridine reductase
Coexpression of unique gene combinations encoding numerous kinds of neuron-type and pan neuronal characteristics establishes the identity of different neurons (1). However, the gene regulatory mechanisms controlling the acquisition of neuron-type identities are poorly understood. One key and obvious identity feature that distinguishes different neuron types is transmitter identity. Transmitter identity is commonly defined by the presence of a particular transmitter together with the coexpression of genes required for its synthesis, reuptake, and vesicular transport in specific neuron-types (2). In the case of 5-HT neurons, the gene products that typically define serotonergic transmitter identity are tryptophan hydroxylase 2 (Tph2), aromatic amino acid decarboxylase (Aadc, gene symbol Ddc), serotonin transporter (Sert, gene symbol Slc6a4), vesicular monoaminergic transporter 2 (Vmat2, gene symbol Slc18a2) and the 5-HT1a (gene symbol Htr1a) and 5-HT1b (gene symbol Htr1b) autoreceptors.
A serotonergic gene regulatory network, comprising multiple interacting transcription factors, has been identified that coordinates expression of Tph2, Aadc, Sert, Vmat2, 5-HT1a, and 5-HT1b in brain 5-HT neurons (3). Transcription factors Ascl1, Nkx2.2, and Foxa2 are required for specification of serotonergic progenitors in the ventral hindbrain. These factors subsequently activate a downstream transcription factor network comprising, Gata-2, Insm1, Gata3, Lmx1b, Engrailed1/2, and Pet-1, which acts in postmitotic serotonergic precursors to initiate 5-HT neuron-type differentiation (4–9). Germ line targeting of each of these factors results in aborted differentiation to varying extents depending on which factor is missing (3). For example, the Pet-1 ETS factor is required for coordinate expression of Tph2, Ddc, Slc6a4, Slc18a2, Htr1a, and Htr1b in postmitotic serotonergic precursors as expression of each of these 5-HT identity genes is severely reduced in Pet-1−/− 5-HT neurons (7, 10, 11). In vivo chromatin immunoprecipitation and in vitro DNA binding assays have demonstrated that Pet-1 coordinates expression of these serotonergic genes through direct binding to a common conserved ETS DNA binding site in their promoter regions (11, 12). Although Pet-1 is expressed in what appears to be all brain 5-HT neurons, Tph2 continues to be expressed, albeit at reduced levels, in a subset of Pet-1−/− 5-HT neurons suggesting the presence of a Pet-1 resistant subpopulation of 5-HT neurons (7, 10, 13).
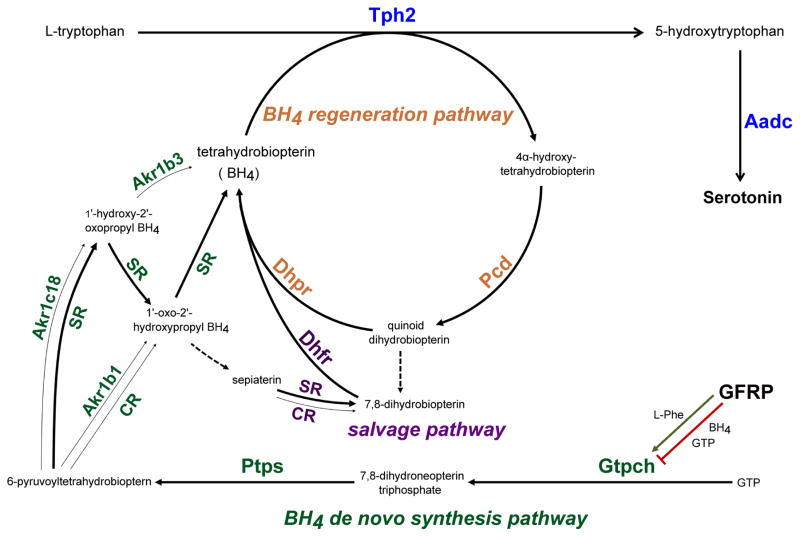
In addition to the genes described above, other gene products play critical roles in 5-HT synthesis and transport and therefore are necessary for 5-HT to function as a transmitter. For example, in addition to Tph2 and Aadc, 5-HT synthesis depends on coordinate expression of the enzymatic machinery catalyzing the production and regeneration of 6R-L-erythro-5,6,7,8-tetrahydrobiopterin (BH4), an obligatory cofactor for Tph2 enzymatic activity as well as the enzymatic activity of other monoaminergic monooxygenases, nitric oxide synthases, and alkylglycerol monooxygenase (14–17). BH4 is synthesized de novo from the precursor guanosine triphosphate (GTP) in four or five enzymatic steps (Figure 1) catalyzed by GTP cyclohydrolase I (Gtpch, gene symbol Gch1), 6-pyruvoyltetrahydropterin synthase (Ptps, gene symbol Pts), and Sepiapterin reductase (SR, gene symbol Spr). The enzymatic steps catalyzed by SR, however, can be alternatively catalyzed by aldo-keto-reductase family 1 member 3 (mouse ortholog Akr1c18), aldo-keto-reductase family 1 B1 (mouse ortholog Akr1b3), and carbonyl reductase (CR, gene symbol Cbr1) in various combinations (Figure 1) (14). Although, it is commonly accepted that Tph2 is a rate-limiting step for the production of 5-HT, Gtpch activity is rate limiting for BH4 synthesis and therefore control of its expression level is a critical determinant of 5-HT synthesis. Gtpch enzymatic activity is also controlled post-translationally through negative feedback regulation by GTP cyclohydrolase I feedback regulatory protein (Gfrp, gene symbol Gchfr) (18, 19). The allosteric binding of BH4 to Gtpch stimulates the formation of a multimeric Gfrp:Gtpch complex in which Gtpch activity is inhibited (19, 20). In contrast, L-phenylalanine can stimulate BH4 biosynthesis in a Gfrp-dependent manner (21). After its enzymatic conversion to 4α-hydroxy-tetrahydrobiopterin in a monooxygenase or synthase reaction, BH4 can be regenerated in a two-step pathway catalyzed by pterin-4α-carbinolamine dehydratase (PCD, gene symbol Pcbd1 and Pcbd2) and dihydropteridine reductase (Dhpr, gene symbol Qdpr, quinoid dihydropteridine reductase) (15). A large number of rare mutations, causing BH4 and monoamine deficiency, have been identified in human GCH1, PTS, SPR, PCBD1, and QDPR genes and are responsible for several neurological motor control disorders such as dopa-responsive dystonia or Segawa disease (22). The 5-HT transporter, Sert (gene symbol Slc6a4), is responsible for high affinity reuptake of 5-HT (23, 24). However, other transporters are now recognized as playing important roles in clearing 5-HT from the synaptic cleft and extrasynaptic sites (25). For example, Oct3 (gene symbol Slc22a3) has been shown to function as a low affinity, high capacity 5-HT transporter (26). Oct3 expression is abundant in brain 5-HT neurons and importantly it is a critical determinant of SSRI efficacy (27). However, the regulatory mechanisms that control expression of BH4 and Slc22a3 genes in 5-HT neurons have not been investigated.
Figure 1.
Schematic of BH4 de novo synthesis, salvage, and regeneration pathways and its role in 5-HT synthesis. Tph2, Tryptophan hydroxylase 2; Aadc (Ddc), aromatic L-amino acid decarboxylase; Gfrp (Gchfr), GTP cyclohydrolase I feedback regulator; Gtpch (Gch1), GTP cyclohydrolase; Ptps (Pts), 6-pyruvoyl-tetrahydropterin synthase; SR (Spr), sepiapterin reductase; Akr1c3, aldo-keto-reductase family 1 member 3; Akr1b1, aldo-keto-reductase family 1 B1; CR (Cbr1), carbonyl reductase; Dhfr dihydrofolate reductase; Dhpr (Qdpr), dihydropteridine reductase; Pcd (Pcbd1, Pcbd2), pterin-4 alpha-carbinolamine dehydratase. Dashed lines indicate non-enzymatic steps.
A possibility is that a regulatory network distinct from that controlling Tph2, Ddc, Slc6a4, Slc18a2, Htr1a, and Htr1b, controls these additional key serotonergic genes. This possibility seems plausible as BH4 enzymatic pathways are expressed in numerous neural and non-neural cell types of the brain and periphery and are required for other cellular functions in addition to 5-HT synthesis (14, 15). Similarly, Oct3 is widely expressed in many different neuron-types and glia of the adult rodent brain unlike Tph2, Sert, and Vmat2 (28, 29). Alternatively, as for other neural expressed genes (30, 31) the complex expression patterns of Oct3 and BH4 genes might be generated through separate cis regulatory control modules one of which positively responds to the same transcription factors that control other serotonergic genes such as Tph2.
To begin to understand the regulatory mechanisms that control Oct3 expression and BH4 production in mouse 5-HT neurons, we investigated whether their serotonergic expression is controlled by Pet-1. We previously reported a microarray method for transcriptome studies of flow sorted yellow fluorescent protein (YFP) -expressing fetal 5-HT neurons obtained from the ePet-EYFP transgenic mouse line (32, 33). Here, we present a protocol for flow sorting of ePet-EYFP-marked Pet-1−/− 5-HT neurons from the fetal rostral hindbrain. We used this new protocol for comparative microarray analyses of Slc22a3 and BH4 gene expression in wild type and Pet-1−/− 5-HT neurons. In addition, we investigated the relative Pet-1 dependency of serotonergic genes in the adult dorsal raphe.
Results and Discussion
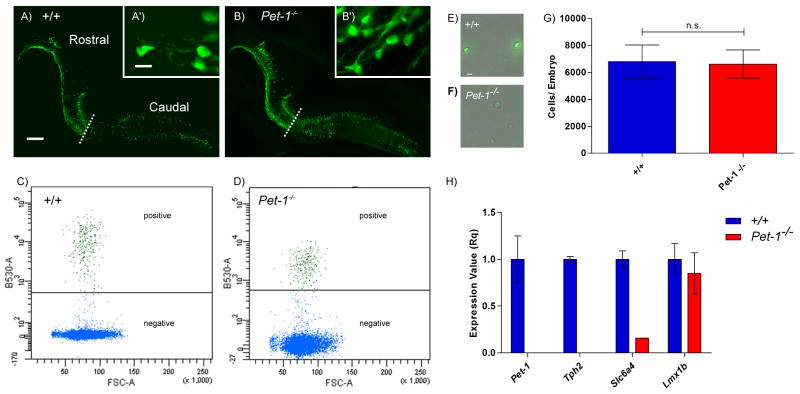
In previous histochemical studies, we found comparable numbers of Pet-1−/− 5-HT neuron cells bodies and wild type 5-HT neuron cell bodies in the midbrain dorsal raphe (34). Here, we crossed ePet-EYFP+/+ and ePet-EYFPPet-1−/− mice to generate ePet-EYFPPet+/− offspring. These offspring were then interbred to generate ePet-EYFP+/+ and ePet-EYFPPet-1−/− littermate embryos. Anti-YFP immunostaining of fetal 5-HT neurons in the ePet-EYFP+/+ and ePet-EYFPPet-1−/− brain revealed comparable levels of YFP expression and similar numbers of wild type and mutant neurons (Figure 2A, B). These findings suggested we should be able to sort sufficient numbers of Pet-1−/− 5-HT neurons for microarray gene expression profiling. Embryonic (E) 12.5 YFP+ rostral hindbrain domains were dissected, dissociated and purified by flow cytometry as previously described (33). Because Pet-1 may regulate the rostral and caudal 5-HT system differently, we only used tissue form the rostral 5-HT system which gives rise to the dorsal and median raphe and B9 nuclei.
Figure 2.
Isolation of ePet-EYFP+/+ and ePet-EYFPPet-1−/− 5-HT neurons. (A–B) Sagittal sections of E12.5 embryonic hindbrain. Dashed line indicates division between rostral and caudal 5-HT neurons. (C–D) Flow cytometry data of sorted YFP+ cells. X-axis Forward Scatter Area (FSC-A). Y-axis 488nm fluorescent intensity. (E–F) Sorted 5-HT neurons. (G) Number of YFP+ cells per embryo collected from either +/+ or Pet-1−/− animals (p=1.00). (H) RT-qPCR validation of selected 5-HT genes in +/+ vs Pet-1−/− samples. Scale, 200μm; zoom, 10μm.
Both +/+ and Pet-1−/− 5-HT neurons were readily sorted (Figure 2C–F) and comparable numbers were obtained (Figure 2G). RT-qPCR revealed, as expected, a complete lack of Pet-1 expression in YFP+ RNA isolated from Pet-1−/− rostral hindbrain (Figure 2H). Moreover, Tph2 and Slc6a4 RNA levels were also dramatically reduced in Pet-1−/− embryos compared to control levels as expected (7). Importantly, the expression of Lmx1b a serotonergic transcription factor whose expression is independent of Pet-1 at fetal stages was unchanged (5). Thus, flow sorted Pet-1−/− YFP+ 5-HT neurons can be used to determine the impact of Pet-1 loss of function on Slc22a3 and BH4 gene expression.
Having established a protocol for flow cytometry of Pet-1 mutant 5-HT neurons we set up crosses to generate sufficient numbers of embryos to perform four microarray biological replicates for both Pet-1−/− and +/+ 5-HT neurons. Based on our previous studies (33) we collected between 20 and 30 thousand cells per replicate. This yielded sufficient RNA to generate labeled cDNA probes, which were then hybridized to the GeneChiP Mouse Gene 1.0 ST array.
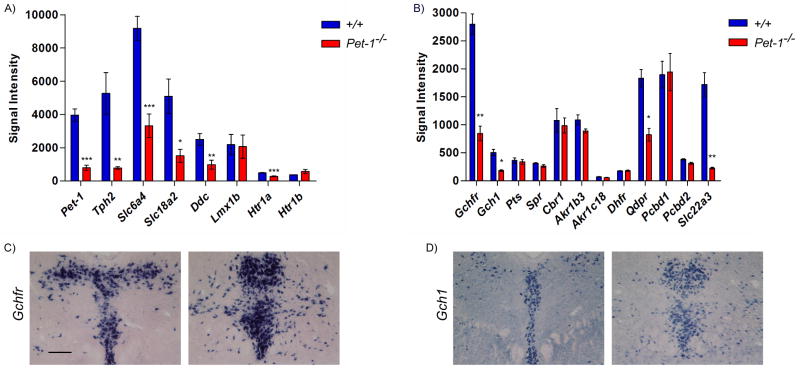
As a validation of our microarray approach, we first examined Pet-1 probe intensities in arrays hybridized with +/+ and Pet-1−/− cDNA (Figure 3A). Analysis of probe intensities revealed background levels of Pet-1 expression. As a further test of our approach we examined expression levels of several known Pet-1 downstream targets. Expression levels of Tph2, Ddc, Slc6a4, Slc18a2, and Htr1a were all significantly reduced in Pet-1−/− vs. +/+ arrays (Figure 3A) to extents that correlate well with previous histochemical studies of these genes in Pet-1−/− mice. We also note that expression of Lmx1b, a gene not regulated by Pet-1 in fetal 5-HT neurons (5, 11), was not altered in the present microarray analysis of its expression in mutant 5-HT neurons. Our array findings indicated very low expression of Hrt1a and Hrtr1b, which is consistent with our earlier findings that expression of these two autoreceptor genes is not strongly induced until after E14 (11). Thus, the very low, near background levels, of these genes at E12.5 likely precluded detection of Htr1b’s Pet-1 dependency although Htr1a did show significantly reduced expression in sorted Pet-1−/− 5-HT neurons (Figure 3B). Importantly, our array findings for Tph2, Ddc, Slc6a4, Slc18a2, Htr1a, and Lmx1b were perfectly consistent with our previously published studies of their Pet-1 dependency (35).
Figure 3.
Microarray analyses. (A) Signal intensities of 5-HT neuron-type identity genes. (B) Signal intensities of BH4 pathway genes and Slc22a3. (C) In situ hybridization of Gchfr and (D) Gch1 probes at two rostrocaudal levels of the dorsal raphe of 3 week old mice. Scale, 200μm. Genes were analyze by a Student’s t-test followed by a Bonferroni correction. Corrected p-value: *p<0.05, **p<0.01, ***p<0.001.
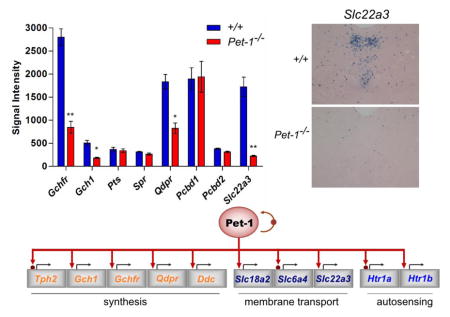
Having demonstrated the validity of our array approach for reproducible and accurate detection of gene expression changes in response to loss of Pet-1 we analyzed our datasets for expression of Slc22a3 and BH4 genes (Figure 3B). Examination of the +/+ arrays revealed that in comparison to Pet-1 and other 5-HT genes, Slc22a3, Gchfr, Qdpr, and Pcbd1 are robustly expressed at E12.5. Gchfr was the most abundantly expressed BH4 gene in our dataset, which is consistent with previous studies showing strong expression of this feedback regulator gene in adult rat 5-HT neurons (21) and as shown here it is expressed strongly in a serotonergic pattern in the adult mouse dorsal raphe (Figure 3C). In contrast, its expression is undetectable in other brain monoaminergic neuron types. This suggests that BH4 production in 5-HT neurons is especially sensitive to Gfrp-dependent BH4 negative feedback and Gfrp-dependent L-Phe stimulation of BH4 biosynthesis (21). One possibility is that this provides for rapid and precise adjustments in 5-HT synthetic rates in response to changing behavioral and metabolic states. The Pcbd1 paralog, Pcbd2, has a relatively low expression level (Pcbd1 average probe intensity: 1897±239, Pcbd2: 384±13) suggesting Pcbd1 plays the major role in BH4 regeneration in 5-HT neurons. Although, Gchfr, Qdpr, and Pcbd1 were robustly expressed at E12.5, Gch1, Pts and Spr expression was relatively low (Figure 3B). Yet, the level of Gch1, Pts, and Spr expression must be adequate to support evident abundant synthesis of 5-HT at this fetal stage. ISH confirmed that by adulthood, Gch1 is indeed strongly expressed in the mouse dorsal raphe (Figure 3D).
Besides SR, several other enzymes can catalyze the last three steps of the de novo BH4 synthesis pathway. Of these, enzymes encoded by Akr1b3, and Cbr1 but not Akr1c18 are robustly expressed at E12.5 suggesting multiple BH4 synthesis pathways may be involved in 5-HT synthesis in immature 5-HT neurons (Figure 3B, Figure 1)). Finally, the regeneration/salvage gene, DHFR, is weakly expressed in immature 5-HT neurons (Figure 3B).
Interestingly, our array experiments revealed a severe loss of Slc22a3 expression in Pet-1−/− 5-HT neurons thus demonstrating that Pet-1 controls not simply high affinity, low capacity 5-HT transport (Sert) but also low affinity, high capacity transport (Oct3) (Figure 3B). Our findings further revealed substantially reduced expression of Gchfr, and Gch1 (Figure 3B). Therefore, Pet-1 is a major transcriptional regulator of the rate limiting enzymatic and post-translational regulatory steps for BH4 synthesis. Given the substantial loss of expression of Gchfr and Gch1 expression in Pet-1−/− 5-HT neurons, it is somewhat surprising that the expression levels of the other BH4 synthetic genes, Spr and Pts were unchanged. Perhaps the very low expression of Pts and Spr near background levels detected for these genes at E12.5 precluded detection of Pet-1 dependency as was likely the case for Htr1b. Further studies of Pet-1 dependency for these two genes at later stages of life will be required. The array results also indicated a role for Pet-1 in regulation of the BH4 regeneration pathway as Qdpr levels were significantly reduced in Pet-1−/− 5HT neurons (Figure 3B). However, loss of Pet-1 had no effect on Pcbd1, Pcbd2, Cbr1, Akr1b3, Akr1c18, and Dhfr expression level. Given the strong and comparable levels of expression of Pcbd1, Cbr1, and Akr1b3 in the wild type and mutant arrays we conclude that Pet-1 is not necessary for their expression at least at E12.5. One possibility is that Lmx1b or Gata-3 is required for expression of these other BH4 synthesis, regeneration, and salvage pathway genes. Alternatively, all three transcription factors may play compensatory roles in the expression of these genes so that removal of a single regulatory partner has little or no effect on expression of these genes. A final possibility is that the more broadly expressed broadly functional BH4 genes such as Akr1b3 are controlled by another gene regulatory network.
Together these findings reveal an extended battery of serotonergic genes under the control of Pet-1. Slc22a3 and the BH4 pathway genes are expressed in many cell types in brain and periphery while Pet-1 expression in the brain is restricted to the 5-HT lineage. Therefore, it seems likely that the cis regulatory regions of the Slc22a3 and BH4 genes possess a Pet-1 binding module that functions specifically to direct their expression to 5-HT neurons. Either Lmx1b and/or Gata-3 or other unknown factors may also control expression of Gchfr, Gch1, Qdpr and may account for the residual expression of these BH4 pathway genes in Pet-1−/− 5-HT neurons.
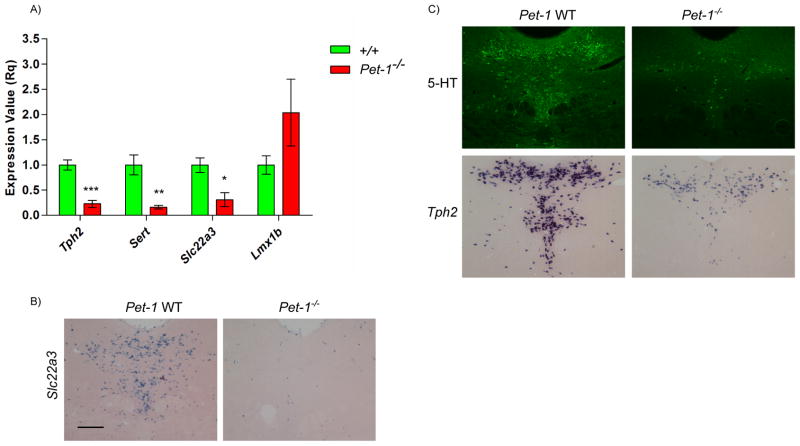
Although we obtained highly concordant array results among our biological replicates for Slc22a3, Gch1, Gchfr and Qdpr dependence on Pet-1, we selected Slc22a3 for RT-qPCR verification using dissected E12.5 neural tubes (Figure 4A). Consistent with the array results, we detected significantly reduced expression of Tph2, Slc6a4, and Slc22a3 but not Lmx1b in Pet-1−/− neural tubes (Figure 4A).
Figure 4.
Slc22a3 expression. (A) RT-qPCR analyses of 5-HT genes with RNA obtained from unsorted hindbrain dissections of E12.5 Pet-1+/− or Pet-1−/− embryos. (B) Comparative in situ hybridization of Slc22a3 probe in +/+ and Pet-1−/− mice. (C) 5-HT immunohistochemistry and Tph2 in situ hybridization in +/+ and Pet-1−/− adult mice. Scale, 200μm. Genes were analyze by a Student’s t-test. p-value: *p<0.05, **p<0.01, ***p<0.001.
We next investigated adult Slc22a3 expression with in situ hybridization (ISH). Consistent with the Allen Brain Atlas ISH analysis of Slc22a3, we found a highly restricted serotonergic raphe pattern of expression for this gene in the midbrain and pons (Figure 4B, left panel and data not shown) suggesting a highly selective serotonergic role for this gene in these brain regions.
Analysis of Slc22a3 expression in in adult Pet-1−/− mice revealed a striking nearly complete loss of its expression (Figure 4B, right panel). This finding was surprising given that expression of other serotonergic identity features such as Tph2 and 5-HT itself are not absolutely dependent on Pet-1 for their full expression in about 20–30% of 5-HT neurons (7, 13), which is shown here in Figure 4C for comparison to the Slc22a3 dependency on Pet-1. Thus, our Slc22a3 findings suggest that whether or not some 5-HT neurons are resistant to loss of Pet-1 depends on specific identity features expressed in these neurons. We note, however, that the residual expression of Tph2 in individual Pet-1−/− 5-HT neurons (Figure 4C, lower panels) appears to be substantially weaker than its expression level in individual wild type 5-HT neurons suggesting Pet-1 controls Tph2 expression in all 5-HT neurons.
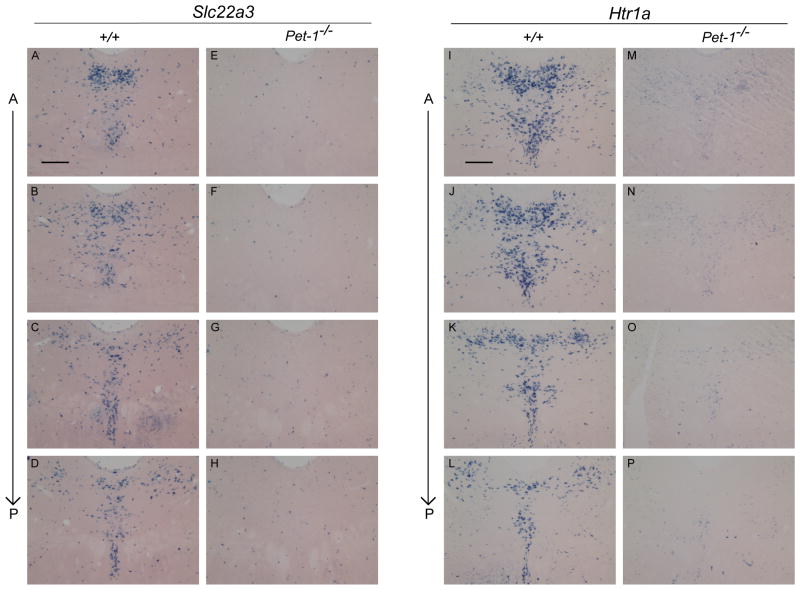
To further investigate Slc22a3 dependency on Pet-1, we systematically investigated its expression throughout the entire dorsal raphe nucleus of Pet-1−/− adults. ISH was performed on every other 25μm section spanning the entire dorsal raphe. As shown in Figure 5A–H, we found a nearly complete elimination of Slc22a3 expression at all levels of the dorsal raphe in Pet-1−/− mice. Residual expression, if any, was confined to scattered cells in numbers far lower than the number of mutant neurons still expressing Tph2 or 5-HT in the Pet-1−/− brain (Figure 4C). These findings indicate that in the case of the Slc22a3 identity feature its expression is completely dependent on Pet-1 as virtually no 5-HT neurons are resistant to loss of Pet-1. Thus, few if any 5-HT neurons acquire their complete adult identity in the absence of Pet-1 and the number of Pet-1 resistant 5-HT neurons that exist is far fewer than previously thought (7, 13).
Figure 5.
Slc22a3 and Htr1a expression in the adult dorsal raphe. 25μm coronal brain sections processed by in situ hybridization and developed for 15hrs using an (A–H) Slc22a3 or (I–P) Htr1a probe on either wild type or Pet-1−/− tissue sections. Alternate sections through the entire dorsal raphe were used for each probe. (A) anterior; (P) posterior. Scale, 200μm.
To extend this analysis to an additional key identity gene we investigated Htr1a’s dependency on Pet-1. ISH throughout the dorsal raphe indicated a similar severe loss of Htr1a expression in Pet-1−/− mice (Figure 5I–P). Few if any cells in the mutant dorsal raphe expressed Htr1a at levels comparable to levels of Htr1a in wild type dorsal raphe or levels of residual Tph2 expression in cells of the Pet-1−/− dorsal raphe (Figure 4C). Instead, a low uniform level of blue precipitate was present in cells of the mutant dorsal raphe, which might be background ISH signal, low residual expression in mutant 5-HT neurons or normal expression in non-serotonergic cells. Thus, in the case of the Htr1a identity feature far fewer 5-HT neurons are resistant to loss of Pet-1 compared to the number of Pet-1 resistant 5-HT neurons in the case of the Tph2 gene. Further gene expression studies are likely to reveal other serotonergic identity features whose expression is dependent on Pet-1 in all brain 5-HT neurons.
In summary, our findings provide additional insight into the regulatory strategies that enable 5-HT synthesis and reuptake in the brain. We show that Pet-1 globally controls acquisition of the brain’s capacity for 5-HT synthesis by coordinating coexpression of Tph2, Ddc, and genes required for BH4 cofactor synthesis, regeneration, and post-translational negative feedback (Figure 6). Thus, Pet-1 controls all of the known rate-limiting enzymatic (Tph2, Gch1) and post-translational regulatory (Gfrp) steps that determine the level of mammalian brain 5-HT synthesis. In addition, we show that Pet-1 globally controls acquisition of 5-HT reuptake in dorsal raphe 5-HT neurons by coordinating expression of high affinity, low capacity transport via Sert and low affinity, high capacity transport via Oct3 (Figure 6). We show that few, if any 5-HT neurons in the dorsal raphe are resistant to loss of Pet-1 for expression of Slc22a3. Similarly, Pet-1 is indispensable for expression of Htr1a in what appears to be all or nearly all 5-HT neurons of the dorsal raphe that normally express this gene. These findings indicate that while some 5-HT neurons are not absolutely dependent on Pet-1 for expression of Tph2 virtually all dorsal raphe 5-HT neurons require Pet-1 for expression of other key identity genes. Therefore, few if any 5-HT neurons in the dorsal raphe are truly resistant to loss of Pet-1 for their neurochemical differentiation.
Figure 6.
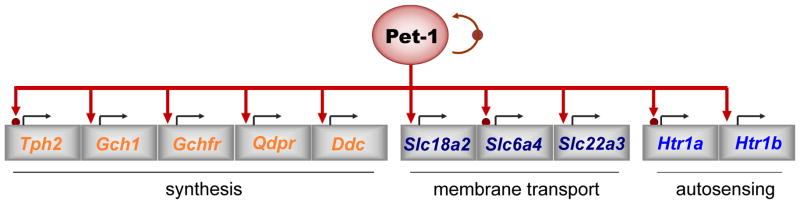
Pet-1 control of the 5-HT neuron-type gene battery. Pet-1 coordinates expression of genes required for 5-HT synthesis (orange), vesicular transport and reuptake (purple) and autoreceptor function (blue). Filled red circles indicate direct control of genes as determined by chromatin immunoprecipitation, reporter assays and in vitro mobility shift assays. Other genes in the schematic may also be direct targets of Pet-1 but have not yet been investigated.
Methods
Animals
Pet-1−/− mice are described in Hendricks et al. 2003. All in situ hybridizations were performed on age and sex matched adult mice 6–8 weeks of age unless otherwise stated. The National Institutes of Health guide was followed for the care and use of laboratory animals. All experiments were approved by the CWRU School of Medicine Institutional Animal Care.
Perfusion and Sectioning
Mice were anesthetized with Avertin (0.5 g tribromoethanol/39.5 ml H2O + 0.31ml tert-amyl alcohol) and transcardially perfused with saline followed by cold 4% paraformaldehyde (PFA). Brains were extracted and post-fixed for 2 hrs and incubated overnight (O/N) in 30% sucrose-PBS solution at 4°C. Floating 25μm coronal brain sections of the dorsal raphe serotonin system were taken, mounted on SuperFrost Plus slides (Fisher Scientific), and dried in a vacuum chamber for at least 1hr before use.
In Situ Hybridization (ISH)
Digoxigenin (Roche Diagnostics, Indianapolis, Indiana) labeled antisense RNA probes (~600bp) were synthesized using cDNA fragments of Slc22a3, Htr1a, Gch1, Gchfr or Tph2 that were PCR amplified with reverse primers containing bacteriophage T3 promoter sequences at their 5′ ends. A previously published in situ hybridization protocol was followed (10).
Quantitative Real-time PCR (RT-qPCR)
RNA was isolated from E12.5 Pet-1+/− and Pet-1−/− animals from the rostral serotonin system using PureLinkTM RNA Mini Kit (Ambion by Life Technologies, Carlsbad, California). Purified RNA was converted to cDNA by PCR using the Transcripter FirstStrand cDNA Synthesis Kit (Roche Diagnostics, Indianapolis, Indiana) and stored at −20°C. qPCR was performed in triplicate using Fast StartTM Universal Syber Green ROX Master solution (Roche Diagnostics, Indianapolis, Indiana). Samples were normalized to β-actin.
Immunohistochemistry (IHC)
Fluorescent immunohistochemistry was performed using a polyclonal primary rabbit antibody against 5-HT (1:1000, ImmunoStar, Hudson, WI) or a primary chicken antibody against GFP (1:1000, Abcam, San Francisco, Ca) O/N at 4°C and an anti-rabbit or anti-chicken Alexa Fluor®488 secondary antibody (1:500, Invitrogen by Life Technologies, Carlsbad, California) for 1hr at RT in the dark. A standard IHC protocol was used (6). Fluorescent images were taken using SPOT RT color digital camera (Diagnostic Instruments, Sterling Heights, MI) using an Olympus Optical BX51 microscope (Center Valley, PA).
FACS of YFP+ cells
The rostral hindbrain domain from the mesencephalic flexure to the pontine flexure of either ePet-EYFP+/+ and ePet-EYFPPet-1−/− embryos was dissected and then treated with 0.25% trypsin-EDTA (Life Technologies) to dissociate cells as described previously (33). Cells were filtered through a 40μm filter and sorted using a Becton Dickinson FACS Aria digital cell sorter with an argon laser (200mW @ 488nm). Cells were sorted directly into Trizol (Invitrogen) for RNA extraction. Approximately 7000 YFP+ cells were isolated from +/+ or Pet-1−/− embryos. Each of the 4 biological replicates (4 +/+,4 Pet-1−/−) consisted of between 20–30 thousand cells from independent litters.
Microarray
Total RNA was isolated after the addition of 20μg of glycogen (Invitrogen) using phenol chloroform extraction. RNA amplification and cDNA libraries were prepared using the Ambion®WT Expression Kit (Life Technologies) according to the manufacturer’s protocol. 5.5μg of single-stranded DNA was fragmented and labeled using Affymetrix GeneChip® WT Terminal Labeling Kit. Probes were hybridized overnight at 45°C to a GeneChip Mouse Gene 1.0 ST Array (Affymetrix). After hybridization, chips were washed in a Genechip Fluidics Station (Affymetrix) and scanned at high resolution using an Affymetrix High Density GeneChip Scanner 3000. The.CEL files from the 8 chips were normalized using the Robust MultiChip Averaging (RMA) using Affymetrix® Expression ConsoleTM Software version 1.1.
Acknowledgments
Funding
This research was supported by NIH grants P50 MH096972, RO1 MH062723, and T32 NS067431.
We thank Katherine Lobur for assistance with mouse genotyping and breeding.
Abbreviations
- 5-HT
serotonin
- BH4
6R-L-erythro-5,6,7,8-tetrahydrobiopterin
- Tph2
Tryptophan hydroxylase 2
- Sert
serotonin transporter
- Oct3
Organic cation transporter 3
- Gch1
GTP cyclohydrolase I
- Gchfr
GTP cyclohydrolase feedback regulator
- Qdpr
quinoid dihydropteridine reductase
- ISH
in situ hybridization
Footnotes
Author contributions
SCW, LJD, ED conceived and designed the experiments. SCW, LJD, MY performed the experiments. SCW, LJD, ED analyzed the data and wrote the manuscript.
References
- 1.Hobert O, Carrera I, Stefanakis N. The molecular and gene regulatory signature of a neuron. Trends in neurosciences. 2010;33:435–445. doi: 10.1016/j.tins.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deneris ES, Hobert O. Maintenance of postmitotic neuronal cell identity. Nat Neurosci. 2014 doi: 10.1038/nn.3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deneris ES, Wyler SC. Serotonergic transcriptional networks and potential importance to mental health. Nature neuroscience. 2012 doi: 10.1038/nn.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Craven SE, Lim KC, Ye W, Engel JD, De Sauvage F, Rosenthal A. Gata2 specifies serotonergic neurons downstream of sonic hedgehog. Development. 2004;131:1165–1173. doi: 10.1242/dev.01024. [DOI] [PubMed] [Google Scholar]
- 5.Ding YQ, Marklund U, Yuan W, Yin J, Wegman L, Ericson J, Deneris E, Johnson RL, Chen ZF. Lmx1b is essential for the development of serotonergic neurons. Nat Neurosci. 2003;6:933–938. doi: 10.1038/nn1104. [DOI] [PubMed] [Google Scholar]
- 6.Fox SR, Deneris ES. Engrailed is required in maturing serotonin neurons to regulate the cytoarchitecture and survival of the dorsal raphe nucleus. J Neurosci. 2012;32:7832–7842. doi: 10.1523/JNEUROSCI.5829-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hendricks TJ, Fyodorov DV, Wegman LJ, Lelutiu NB, Pehek EA, Yamamoto B, Silver J, Weeber EJ, Sweatt JD, Deneris ES. Pet-1 ETS gene plays a critical role in 5-HT neuron development and is required for normal anxiety-like and aggressive behavior. Neuron. 2003;37:233–247. doi: 10.1016/s0896-6273(02)01167-4. [DOI] [PubMed] [Google Scholar]
- 8.Jacob J, Storm R, Castro DS, Milton C, Pla P, Guillemot F, Birchmeier C, Briscoe J. Insm1 (IA-1) is an essential component of the regulatory network that specifies monoaminergic neuronal phenotypes in the vertebrate hindbrain. Development. 2009;136:2477–2485. doi: 10.1242/dev.034546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pattyn A, Simplicio N, Van Doorninck JH, Goridis C, Guillemot F, Brunet JF. Ascl1/Mash1 is required for the development of central serotonergic neurons. Nat Neurosci. 2004;7:589–595. doi: 10.1038/nn1247. [DOI] [PubMed] [Google Scholar]
- 10.Hendricks T, Francis N, Fyodorov D, Deneris E. The ETS Domain Factor Pet-1 is an Early and Precise Marker of Central 5-HT Neurons and Interacts with a Conserved Element in Serotonergic Genes. J Neurosci. 1999;19:10348–10356. doi: 10.1523/JNEUROSCI.19-23-10348.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu C, Maejima T, Wyler SC, Casadesus G, Herlitze S, Deneris ES. Pet-1 is required across different stages of life to regulate serotonergic function. Nat Neurosci. 2010;13:1190–1198. doi: 10.1038/nn.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacobsen KX, Czesak M, Deria M, Le Francois B, Albert PR. Region-specific regulation of 5-HT1A receptor expression by Pet-1-dependent mechanisms in vivo. J Neurochem. 2011 doi: 10.1111/j.1471-4159.2010.07161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kiyasova V, Fernandez SP, Laine J, Stankovski L, Muzerelle A, Doly S, Gaspar P. A Genetically Defined Morphologically and Functionally Unique Subset of 5-HT Neurons in the Mouse Raphe Nuclei. J Neurosci. 2011;31:2756–2768. doi: 10.1523/JNEUROSCI.4080-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kapatos G. The neurobiology of tetrahydrobiopterin biosynthesis: a model for regulation of GTP cyclohydrolase I gene transcription within nigrostriatal dopamine neurons. IUBMB life. 2013;65:323–333. doi: 10.1002/iub.1140. [DOI] [PubMed] [Google Scholar]
- 15.Werner ER, Blau N, Thony B. Tetrahydrobiopterin: biochemistry and pathophysiology. Biochem J. 2011;438:397–414. doi: 10.1042/BJ20110293. [DOI] [PubMed] [Google Scholar]
- 16.Tietz A, Lindberg M, Kennedy EP. A New Pteridine-Requiring Enzyme System for the Oxidation of Glyceryl Ethers. J Biol Chem. 1964;239:4081–4090. [PubMed] [Google Scholar]
- 17.Watschinger K, Keller MA, Golderer G, Hermann M, Maglione M, Sarg B, Lindner HH, Hermetter A, Werner-Felmayer G, Konrat R, Hulo N, Werner ER. Identification of the gene encoding alkylglycerol monooxygenase defines a third class of tetrahydrobiopterin-dependent enzymes. Proc Natl Acad Sci U S A. 2010;107:13672–13677. doi: 10.1073/pnas.1002404107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harada T, Kagamiyama H, Hatakeyama K. Feedback regulation mechanisms for the control of GTP cyclohydrolase I activity. Science. 1993;260:1507–1510. doi: 10.1126/science.8502995. [DOI] [PubMed] [Google Scholar]
- 19.Yoneyama T, Hatakeyama K. Decameric GTP cyclohydrolase I forms complexes with two pentameric GTP cyclohydrolase I feedback regulatory proteins in the presence of phenylalanine or of a combination of tetrahydrobiopterin and GTP. J Biol Chem. 1998;273:20102–20108. doi: 10.1074/jbc.273.32.20102. [DOI] [PubMed] [Google Scholar]
- 20.Maita N, Hatakeyama K, Okada K, Hakoshima T. Structural basis of biopterin-induced inhibition of GTP cyclohydrolase I by GFRP, its feedback regulatory protein. J Biol Chem. 2004;279:51534–51540. doi: 10.1074/jbc.M409440200. [DOI] [PubMed] [Google Scholar]
- 21.Kapatos G, Hirayama K, Shimoji M, Milstien S. GTP cyclohydrolase I feedback regulatory protein is expressed in serotonin neurons and regulates tetrahydrobiopterin biosynthesis. J Neurochem. 1999;72:669–675. doi: 10.1046/j.1471-4159.1999.0720669.x. [DOI] [PubMed] [Google Scholar]
- 22.Thony B, Blau N. Mutations in the BH4-metabolizing genes GTP cyclohydrolase I, 6-pyruvoyl-tetrahydropterin synthase, sepiapterin reductase, carbinolamine-4a-dehydratase, and dihydropteridine reductase. Human mutation. 2006;27:870–878. doi: 10.1002/humu.20366. [DOI] [PubMed] [Google Scholar]
- 23.Murphy DL, Lesch KP. Targeting the murine serotonin transporter: insights into human neurobiology. Nat Rev Neurosci. 2008;9:85–96. doi: 10.1038/nrn2284. [DOI] [PubMed] [Google Scholar]
- 24.Blakely RD, Edwards RH. Vesicular and plasma membrane transporters for neurotransmitters. Cold Spring Harbor perspectives in biology. 2012;4 doi: 10.1101/cshperspect.a005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Daws LC. Unfaithful neurotransmitter transporters: focus on serotonin uptake and implications for antidepressant efficacy. Pharmacology & therapeutics. 2009;121:89–99. doi: 10.1016/j.pharmthera.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baganz NL, Horton RE, Calderon AS, Owens WA, Munn JL, Watts LT, Koldzic-Zivanovic N, Jeske NA, Koek W, Toney GM, Daws LC. Organic cation transporter 3: Keeping the brake on extracellular serotonin in serotonin-transporter-deficient mice. Proc Natl Acad Sci U S A. 2008;105:18976–18981. doi: 10.1073/pnas.0800466105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horton RE, Apple DM, Owens WA, Baganz NL, Cano S, Mitchell NC, Vitela M, Gould GG, Koek W, Daws LC. Decynium-22 enhances SSRI-induced antidepressant-like effects in mice: uncovering novel targets to treat depression. J Neurosci. 2013;33:10534–10543. doi: 10.1523/JNEUROSCI.5687-11.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vialou V, Balasse L, Callebert J, Launay JM, Giros B, Gautron S. Altered aminergic neurotransmission in the brain of organic cation transporter 3-deficient mice. J Neurochem. 2008;106:1471–1482. doi: 10.1111/j.1471-4159.2008.05506.x. [DOI] [PubMed] [Google Scholar]
- 29.Gasser PJ, Orchinik M, Raju I, Lowry CA. Distribution of organic cation transporter 3, a corticosterone-sensitive monoamine transporter, in the rat brain. J Comp Neurol. 2009;512:529–555. doi: 10.1002/cne.21921. [DOI] [PubMed] [Google Scholar]
- 30.Serrano-Saiz E, Poole RJ, Felton T, Zhang F, De La Cruz ED, Hobert O. Modular control of glutamatergic neuronal identity in C. elegans by distinct homeodomain proteins. Cell. 2013;155:659–673. doi: 10.1016/j.cell.2013.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wenick AS, Hobert O. Genomic cis-regulatory architecture and trans-acting regulators of a single interneuron-specific gene battery in C. elegans. Dev Cell. 2004;6:757–770. doi: 10.1016/j.devcel.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 32.Scott MM, Krueger KC, Deneris ES. A differentially autoregulated Pet-1 enhancer region is a critical target of the transcriptional cascade that governs serotonin neuron development. J Neurosci. 2005;25:2628–2636. doi: 10.1523/JNEUROSCI.4979-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wylie CJ, Hendricks TJ, Zhang B, Wang L, Lu P, Leahy P, Fox S, Maeno H, Deneris ES. Distinct transcriptomes define rostral and caudal serotonin neurons. J Neurosci. 2010;30:670–684. doi: 10.1523/JNEUROSCI.4656-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krueger KC, Deneris ES. Serotonergic transcription of human FEV reveals direct GATA factor interactions and fate of Pet-1-deficient serotonin neuron precursors. J Neurosci. 2008;28:12748–12758. doi: 10.1523/JNEUROSCI.4349-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deneris ES. Molecular genetics of mouse serotonin neurons across the lifespan. Neuroscience. 2011 doi: 10.1016/j.neuroscience.2011.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]