Abstract
AIM: To investigate the expression and clinical significance of DEK, cyclin D1, insulin-like growth factor II (IGF-II), glypican 3 (GPC3), ribosomal phosphoprotein 0 (rpP0) mRNA in hepatocellular carcinoma (HCC) and its paraneoplastic tissues.
METHODS: The expression of mRNAs of DEK, cyclin D1, IGF-II, GPC3 and rpP0 mRNA was detected in HCC and its paraneoplastic tissues by multiplex RT-PCR.
RESULTS: By the simplex RT-PCR, the overexpression of mRNAs of DEK, cyclin D1, IGF-II, GPC3, rpP0 mRNA in HCC and its paraneoplastic tissues was 78.1%, 87.5%, 87.5%, 75.0%, 81.3% and 15.6%, 40.6%, 37.5%, 21.9%, 31.3% respectively (P < 0.05). By the multiplex RT-PCR, at least one of the mRNAs was detected in all HCC samples and in 75.0% of paraneoplastic samples (P > 0.05). However, all these five mRNAs were found in 68.8% of HCC samples, but only in 9.4% of paraneoplastic tissues (P < 0.05). The positive expression of mRNAs of DEK, cyclin D1, IGF-II, GPC3, rpP0 in well- and poorly-differentiated HCC was 89.0%, 66.7%, 66.7%, 66.7%, 77.8% and 73.9%, 95.7%, 95.7%, 95.7%, 82.6%, respectively (P > 0.05). The expression of these genes in HCCs with α-feto protein (AFP) negative and positive was 90.0%, 80.0%, 90.0%, 90.0%, 90.0% and 72.7%, 86.3%, 77.3%, 90.9%, 68.2% respectively (P > 0.05).
CONCLUSION: The expression of DEK, cyclin D1, IGF-II, GPC3, rpP0 mRNA in HCC is much higher in HCC than in its paraneoplastic tissues. Multiplex RT-PCR assay is an effective, sensitive, accurate, and cost-effective diagnostic method of HCC.
Keywords: HCC, DEK, Cyclin D1, IGF-II, GPC3, rpP0
INTRODUCTION
Hepatocellular carcinoma (HCC) is an aggressive malignancy with poor prognosis and one of the most common tumors in humans. The development of HCC is a chronic process and involves many factors, including infection of hepatitis virus and contamination with aflatoxin B1[1]. Recent advances in molecular genetics indicate that some tumor suppressor genes, oncogenes, and growth factors may play an important role in hepatocarcinogenesis[2].
Several methods such as DDRT-PCR, cDNA screening[3] are used to identify differential expression of mRNA in tumor and non-tumor tissues. It is reported that some genes, such as DEK, rpP0[4], cyclin D1[5], IGF-II[6] , and GPC3[7], are overexpressed in HCC tissue. The expression of DEK, IGF-II and rpP0 is higher in HCC than non-tumor tissues in our previous study.
RT-PCR is widely used to analyze gene expression in HCC and paraneoplastic liver tissue. However, the overexpressed genes are only relatively higher in HCC than in paraneoplastic liver tissue and the positive detection rate is relatively low. Moreover, RT-PCR can detect only a single gene once in the past, and the results obtained are fluctuant and less useful in clinics. In order to find a method to enhance the specificity and positive rate, multiplex PCR was used to detect several genes, such as DEK, cyclin D1, IGF-II, GPC3, rpP0.
MATERIALS AND METHODS
Tissue samples and patients
HCC and corresponding paraneoplastic tissues were obtained with informed consent from 32 patients who underwent hepatectomy at the First Affiliated Hospital of Guangxi Medical University and Guangxi Tumor Hospital. The profiles were obtained from medical records of 20 male and 12 female patients with an average age of 42.5 years. Twenty-two patients were positive for α-feto protein (AFP). HCC and paraneoplastic tissues were enucleated separately and immediately frozen in liquid nitrogen. Histological classification was performed according to the Edmondson’s grading criteria.
Multiplex RT-PCR
Total RNA was isolated from 100 mg of frozen tissue according to the manufacturer’s instructions using TRIzol kit (Sagon Company, Shanghai, China), and then dissolved in water that was treated by DEPC. Four micrograms of total RNA was used to produce cDNA using oligo(dT) primer and MuLV reverse transcription (MBI Company) in a final volume of 20 µL at 42°C for 1 h. The reaction was terminated by incubation at 75°C for 10 min. One microgram of products was PCR amplified with multiple primer sets (0.5 µmol/L, Sagon Company, Shanghai, China), 0.75 units of Taq DNA polymerase (MBI), 0.5 µL 10 mmol/L dNTPs, 2.5 µL 10 × buffer. The primers were as follows: DEK: 5’-AGG CAC TGT GTC CTC ATT AA and 5’-TCT GAC AGA ATT TCA GGA CA (332 bp); cyclin D1: 5’-TAT TTG CAT AAC CCT GAG CG and 5’-GTG ACT ACA TGC ATA TGA GC (350 bp); IGF: 5’-AGG AGC TCC TGG ATA ATT TC and 5’-AAT ATT TCA CGT GAC AGA AC (421 bp); GPC3: 5’-TGG ACA TCA ATG AGT GCC TC and 5’-CAC ATT CTG GTG AGC ATT CG (204 bp); rpP0: 5’-ATG TGA AGT CAC TGT GCC AG and 5’-CTT GGC TTC AAC CTT AGC TG (549 bp). GAPDH was used as control, the primers were 5’-TGA GTA CGT CGT GGA GTC CA and 5’-CAA AGT TGT CAT GGA TGA CC (230 bp). The conditions were initial denaturation at 94°C for 5 min, 30 cycles of amplification, each cycle consisting of denaturation at 94°C for 45 s, annealing at 58°C for 30 s, extension at 72°C for 30 s, final extension at 72°C for 7 min. Because of the limitation by the length of PCR products, the primers of cyclin D1 and GPC3 were used together in a single reaction and the primers of DEK, IGF-II, and rpP0 were used together in a single reaction.
Analysis by electrophoresis
The amplified products were electrophoresed on 1.2% agarose gel to detect the expression of the genes in HCC and paraneoplastic tissues. The images were analyzed by Quantity One software.
Statistical analysis
Results were analyzed by χ2 test to compare the differences between the groups. P < 0.05 was considered statistically significant.
RESULTS
Expression of GAPDH mRNA
The expression of GAPDH mRNA was detected in all HCC and non-HCC tissues. There was no difference between the two groups (Figure 1A).
Figure 1.
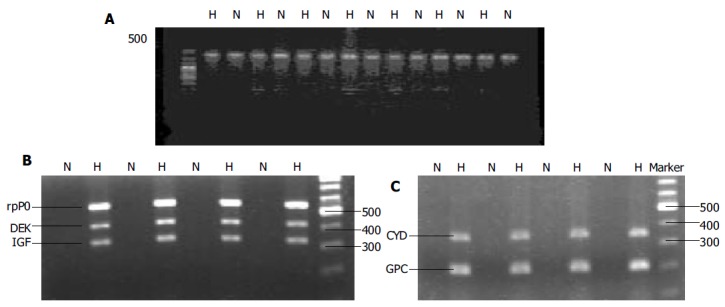
Expression of GAPDH (A), mRNA of rpP0, DEK, and IGF-II (B), mRNA of cyclin D1 and GPC3 (C) in HCC tissue (H) and adjacent nontumorous tissue (N).
Expression of DEK, cyclin D1, IGF-II, GPC3, rpP0 mRNAs in HCC and adjacent noncancerous liver tissues
The expression of DEK, cyclin D1, IGF-II, GPC3, rpP0 mRNAs in HCC and adjacent noncancerous liver tissues was 78.1%, 87.5%, 87.5%, 75.0%, 81.3%, and 15.6%, 40.6%, 37.5%, 21.9%, 31.3%, respectively (P < 0.05, Table 1), which were significantly higher in HCC tissues than in adjacent nontumorous tissue (Figures 1B and C). The density of the bands was also higher in HCC than in adjacent noncancerous liver tissues.
Table 1.
Expression of DEK, cyclin D1, IGF-II, GPC3, rpP0 mRNAs in HCC and adjacent noncancerous liver tissues
|
Cyclin D1a |
GPC3c |
DEKe |
IGFg |
rpP0i |
||||||
| T | N | T | N | T | N | T | N | T | N | |
| Positive | 28 | 13 | 24 | 7 | 25 | 5 | 28 | 12 | 26 | 10 |
| Negative | 4 | 19 | 8 | 25 | 7 | 27 | 4 | 20 | 6 | 22 |
T, HCC tumor tissue; N, adjacent nontumorous tissue. χ2-test:
P < 0.05,
P < 0.05,
P < 0.05,
P < 0.05,
P < 0.05 vs others.
The expression of DEK, cyclin D1, IGF-II, GPC3, rpP0 mRNAs in well- and poorly-differentiated HCC was 89.0%, 66.7%, 66.7%, 66.7%, 77.8% and 73.9%, 95.7%, 95.7%, 95.7%, 82.6%, respectively (P > 0.05, Table 2).
Table 2.
Expression of DEK, cyclin D1, IGF-II, GPC3, rpP0 mRNAs in well- and poorly-differentiated HCC
|
Cyclin D1 |
GPC3 |
DEK |
IGF-II |
rpP0 |
||||||
| + | - | + | - | + | - | + | - | + | - | |
| Well-differentiated HCC | 6 | 3 | 6 | 3 | 8 | 1 | 6 | 3 | 7 | 2 |
| Poor-differentiated HCC | 22 | 1 | 18 | 5 | 17 | 6 | 22 | 1 | 19 | 4 |
| Total | 28 | 4 | 24 | 8 | 25 | 7 | 28 | 4 | 26 | 6 |
The expression of DEK, cyclin D1, IGF-II, GPC3, rpP0 mRNAs in HCC with AFP negative and positive was 90.0%, 80.0%, 90.0%, 90.0%, 90.0% and 72.7%, 86.3%, 77.3%, 90.9%, 68.2%, respectively (P > 0.05, Table 3).
Table 3.
Expression of DEK, cyclin D1, IGF-II, GPC3, rpP0 mRNAs in HCC with AFP negative and positive HCC tissue
| n | Cyclin D1+ | GPC3+ | DEK+ | IGF-II+ | rpP0+ | |
| AFP(-) | 10 | 8 | 9 | 9 | 9 | 9 |
| AFP(+) | 22 | 20 | 15 | 16 | 19 | 17 |
| Total | 32 | 28 | 24 | 25 | 28 | 26 |
χ2 test, P > 0.05, the group of APF(+) vs the group of AFP(-).
Expression of DEK, cyclin D1, IGF-II, GPC3, rpP0 mRNAs in HCC and adjacent noncancerous liver tissues shown by multiplex RT-PCR
By multiplex RT-PCR, at least one of the mRNAs could be detected in all HCC tissues and in 75.0% of paraneoplastic tissues (P > 0.05) (Table 4). However, all these five mRNAs were found in 68.8% of HCC tissue, but only in 9.4% of paraneoplastic tissues (P < 0.05, Table 4).
Table 4.
Expression of DEK, cyclin D1, IGF-II, GPC3, rpP0 mRNAs in HCC and adjacent noncancerous liver tissues by multiplex RT-PCR Groups
| n |
Positive reaction |
Negative reaction |
|||
| Anyone of the five mRNAs (%) | All of the five mRNAs (%) | Anyone of the five mRNAs (%) | All of the five mRNAs (%) | ||
| T | 32 | 32 (100) | 22 (68.8) | 10 (31.3) | 0 (0) |
| N | 32 | 24 (75.0) | 3 (9.4) | 29 (90.6) | 8 (25.0) |
| P > 0.05 | P > 0.05 | P > 0.05 | P > 0.05 | ||
DISCUSSION
The growth of cells depends on the regulation by many factors, including oncogenes, tumor suppressor genes, growth factors, signal transduction factors, and apoptosis factors, etc. The origin of tumor is related to the modification of these genes. Genes such as DEK, cyclin D1, IGF-II, GPC3, rpP0, are involved in the initiation and development of HCC, and the possible markers for the diagnosis of HCC in clinic. To identify whether these genes were generally involved in hepatocarcinogenesis, multiplex PCR was used in the present study. We found that these genes had an upregulated expression in HCC and multiplex PCR could enhance the detective positive rate.
Cyclin D1
Cyclin, cyclin-dependent kinases, and tumor suppressor gene products interact and regulate the normal cell cycle. Cyclin D1 and cyclin-dependent kinases are required for completion of the G1/S transition in normal mammalian cells[8]. Cyclin D1 is located on chromosome 11q13 and exhibits many characteristics of cellular oncogenes. Overexpression of cyclin D1 may be associated with actual gene amplification or transcriptional dysregulation in cancer. Cyclin D1 is overexpressed in hyperplastic lesions, such as endometrioid adenocarcinoma[9], mantle cell lymphoma[10], and ovarian carcinoma[11]. The results in our study showed that the expression of cyclin D1 was significantly higher in HCC than in adjacent nontumorous tissue. The mechanism of cyclin D1 dysregulation in HCC is not clear, but it is likely that the dysregulation contributes to increasing the proportion of cells in transition from G1 to S phase. The overexpression of cyclin D1 may be one of the several mechanisms involved in hyperplasia of liver cells.
DEK
DEK is a 43-ku phosphoprotein that was first isolated as part of a fusion protein expressed in a subtype of acute myeloid leukemias with t(6;9) chromosomal translocations[12]. DEK was lately identified as an autoimmune antigen in patients with pauciarticular onset juvenile rheumatoid arthritis, systemic lupus erythematosus[13], and other autoimmune diseases. In recent study, it was demonstrated that DEK was a site-specific DNA binding protein that was involved likely in transcriptional regulation and signal transduction and it had implications not only for HIV-2 transcription[14] but also for multiple cellular processes involving with DEK. Recent data demonstrated that the major portion of DEK is associated with chromatin in vivo and suggested that it might play a role in chromatin architecture[15]. Our present experiment shows that the percentage of overexpression of mRNA of DEK (78.1%) in HCC was higher than in adjacent nontumorous tissues (15.6%). It indicates the overexpression of DEK may be involved in the transformation from normal liver tissue to HCC, perhaps by activating the oncogenes.
GPC3
GPC3 gene is located at Xq26, and mutated in the Simpson-Golabi-Behmel syndrome[16]. It may be regulated by methylation of the inactive X in expressing tissues, and encodes a developmentally regulated heparin sulfate proteoglycan that is bound to the cell surface through a glycosylphosphati-dylinositol anchor. Based on their localization on the cell surface, such glypicans are thought to regulate interactions between growth factor and their receptors. It is associated with apoptosis and cell signal transduction. It is reported that GPC3 is a tissue-specific gene in breast tumor[17], ovarian tumor[18], and malignant mesothelioma[19] in which it is downr-egulated by aberrant methylation of the GPC3 promoter region, and upregulated in HCC. In the present study, GPC3 was overexpressed in HCC tissue and the positive expression rate was 75.0%, which was higher than that in adjacent nontumorous tissue (21.9%). Although the role of overexpression of GPC3 in the development of HCC is not known, it may break the balance between cell growth and death.
IGF-II
It was reported that IGF-II is positive in some benign neoplastic nodules and HCC[6]. In the present study, the levels of IGF-II was higher in HCC than in adjacent nontumorous tissue, suggesting that the growth factor may act as an autocrine regulation of cell proliferation, GPC3 may act as a positive regulator of IGF-II, although we have not detected a direct interaction between GPC3 and IGF-II. It is possible that GPC3 positively regulates IGF-II activity by interacting with the components of its signaling system.
rpP0
Ribosome acts as a place for protein synthesis. It is composed of rRNA and ribosomal phosphoproteins. In the family of ribosomal phosphoprotein, there are five members: P0, P1α, P1β, P2α, and P2β. Three functional domains can be defined in the rpP0: one involved in binding to rRNA, one connected to P1/P2 protein interaction, and one associated with elongation factors[20]. It was reported that rpP0 expression increases in colon carcinoma cells[21]. In the present study, rpP0 was overexpressed in HCC tumor tissue, suggesting that upregulation of rpP0 is associated with HCC, which may be a signal for increasing protein synthesis.
Multiplex RT-PCR and HCC
In this study, we used multiplex RT-PCR to detect the expression of mRNAs of DEK, cyclin D1, IGF-II, GPC3, rpP0 in HCC and adjacent nontumorous liver tissue. We found that at least one of the mRNAs could be detected in all HCC tissues and 68.8% of HCC tissues expressed all these five mRNAs, while only 9.4% of paraneoplastic tissues expressed all of them (P < 0.05), suggesting that multiplex RT-PCR enhances the detective sensitivity and specificity when combining several specific primers. The expression of anyone of these mRNAs in liver tissue could be regarded as a risk factor for HCC. The higher the expression of these mRNAs the greater the risk. When all these mRNAs are negative in tissues, the possibility of HCC is lower. Multiplex RT-PCR provides an easy and quick method to detect the expression of these genes in liver tissues.
ACKNOWLEDGMENTS
The authors thank Dr. Dang-Rong Li for practical and productive technical advice regarding the RT-PCR technique and Dr. Le-Qun Li for providing useful tissues.
Footnotes
Supported by the National Natural Science Foundation of China, No. 39860079
Science Editor Wang XL and Guo SY Language Editor Elsevier HK
References
- 1.Park US, Su JJ, Ban KC, Qin L, Lee EH, Lee YI. Mutations in the p53 tumor suppressor gene in tree shrew hepatocellular carcinoma associated with hepatitis B virus infection and intake of aflatoxin B1. Gene. 2000;251:73–80. doi: 10.1016/s0378-1119(00)00183-9. [DOI] [PubMed] [Google Scholar]
- 2.Park DY, Sol MY, Suh KS, Shin EC, Kim CH. Expressions of transforming growth factor (TGF)-beta1 and TGF-beta type II receptor and their relationship with apoptosis during chemical hepatocarcinogenesis in rats. Hepatol Res. 2003;27:205–213. doi: 10.1016/s1386-6346(03)00264-x. [DOI] [PubMed] [Google Scholar]
- 3.Sell S. Mouse models to study the interaction of risk factors for human liver cancer. Cancer Res. 2003;63:7553–7562. [PubMed] [Google Scholar]
- 4.Kondoh N, Wakatsuki T, Ryo A, Hada A, Aihara T, Horiuchi S, Goseki N, Matsubara O, Takenaka K, Shichita M, et al. Identification and characterization of genes associated with human hepatocellular carcinogenesis. Cancer Res. 1999;59:4990–4996. [PubMed] [Google Scholar]
- 5.Azechi H, Nishida N, Fukuda Y, Nishimura T, Minata M, Katsuma H, Kuno M, Ito T, Komeda T, Kita R, et al. Disruption of the p16/cyclin D1/retinoblastoma protein pathway in the majority of human hepatocellular carcinomas. Oncology. 2001;60:346–354. doi: 10.1159/000058531. [DOI] [PubMed] [Google Scholar]
- 6.Tsai JF, Jeng JE, Chuang LY, You HL, Ho MS, Lai CS, Wang LY, Hsieh MY, Chen SC, Chuang WL, et al. Serum insulin-like growth factor-II and alpha-fetoprotein as tumor markers of hepatocellular carcinoma. Tumour Biol. 2003;24:291–298. doi: 10.1159/000076461. [DOI] [PubMed] [Google Scholar]
- 7.Sung YK, Hwang SY, Park MK, Farooq M, Han IS, Bae HI, Kim JC, Kim M. Glypican-3 is overexpressed in human hepatocellular carcinoma. Cancer Sci. 2003;94:259–262. doi: 10.1111/j.1349-7006.2003.tb01430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jirström K, Ringberg A, Fernö M, Anagnostaki L, Landberg G. Tissue microarray analyses of G1/S-regulatory proteins in ductal carcinoma in situ of the breast indicate that low cyclin D1 is associated with local recurrence. Br J Cancer. 2003;89:1920–1926. doi: 10.1038/sj.bjc.6601398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruhul Quddus M, Latkovich P, Castellani WJ, James Sung C, Steinhoff MM, Briggs RC, Miranda RN. Expression of cyclin D1 in normal, metaplastic, hyperplastic endometrium and endometrioid carcinoma suggests a role in endometrial carcinogenesis. Arch Pathol Lab Med. 2002;126:459–463. doi: 10.5858/2002-126-0459-EOCDIN. [DOI] [PubMed] [Google Scholar]
- 10.Howe JG, Crouch J, Cooper D, Smith BR. Real-time quantitative reverse transcription-PCR for cyclin D1 mRNA in blood, marrow, and tissue specimens for diagnosis of mantle cell lymphoma. Clin Chem. 2004;50:80–87. doi: 10.1373/clinchem.2003.024695. [DOI] [PubMed] [Google Scholar]
- 11.Raju U, Nakata E, Mason KA, Ang KK, Milas L. Flavopiridol, a cyclin-dependent kinase inhibitor, enhances radiosensitivity of ovarian carcinoma cells. Cancer Res. 2003;63:3263–3267. [PubMed] [Google Scholar]
- 12.Maeda T, Kosugi S, Ujiie H, Osumi K, Fukui T, Yoshida H, Kashiwagi H, Ishikawa J, Tomiyama Y, Matsuzawa Y. Localized relapse in bone marrow in a posttransplantation patient with t(6; 9) acute myeloid leukemia. Int J Hematol. 2003;77:522–525. doi: 10.1007/BF02986623. [DOI] [PubMed] [Google Scholar]
- 13.Wichmann I, Respaldiza N, Garcia-Lozano JR, Montes M, Sanchez-Roman J, Nuñez-Roldan A. Autoantibodies to DEK oncoprotein in systemic lupus erythematosus (SLE) Clin Exp Immunol. 2000;119:530–532. doi: 10.1046/j.1365-2249.2000.01154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faulkner NE, Hilfinger JM, Markovitz DM. Protein phosphatase 2A activates the HIV-2 promoter through enhancer elements that include the pets site. J Biol Chem. 2001;276:25804–25812. doi: 10.1074/jbc.M006454200. [DOI] [PubMed] [Google Scholar]
- 15.Kappes F, Burger K, Baack M, Fackelmayer FO, Gruss C. Subcellular localization of the human proto-oncogene protein DEK. J Biol Chem. 2001;276:26317–26323. doi: 10.1074/jbc.M100162200. [DOI] [PubMed] [Google Scholar]
- 16.Veugelers M, Cat BD, Muyldermans SY, Reekmans G, Delande N, Frints S, Legius E, Fryns JP, Schrander-Stumpel C, Weidle B, et al. Mutational analysis of the GPC3/GPC4 glypican gene cluster on Xq26 in patients with Simpson-Golabi-Behmel syndrome: identification of loss-of-function mutations in the GPC3 gene. Hum Mol Genet. 2000;9:1321–1328. doi: 10.1093/hmg/9.9.1321. [DOI] [PubMed] [Google Scholar]
- 17.Xiang YY, Ladeda V, Filmus J. Glypican-3 expression is silenced in human breast cancer. Oncogene. 2001;20:7408–7412. doi: 10.1038/sj.onc.1204925. [DOI] [PubMed] [Google Scholar]
- 18.Lin H, Huber R, Schlessinger D, Morin PJ. Frequent silencing of the GPC3 gene in ovarian cancer cell lines. Cancer Res. 1999;59:807–810. [PubMed] [Google Scholar]
- 19.Murthy SS, Shen T, De Rienzo A, Lee WC, Ferriola PC, Jhanwar SC, Mossman BT, Filmus J, Testa JR. Expression of GPC3, an X-linked recessive overgrowth gene, is silenced in malignant mesothelioma. Oncogene. 2000;19:410–416. doi: 10.1038/sj.onc.1203322. [DOI] [PubMed] [Google Scholar]
- 20.Rodríguez-Gabriel MA, Remacha M, Ballesta JP. The RNA interacting domain but not the protein interacting domain is highly conserved in ribosomal protein P0. J Biol Chem. 2000;275:2130–2136. doi: 10.1074/jbc.275.3.2130. [DOI] [PubMed] [Google Scholar]
- 21.Barnard GF, Staniunas RJ, Bao S, Mafune K, Steele GD, Gollan JL, Chen LB. Increased expression of human ribosomal phosphoprotein P0 messenger RNA in hepatocellular carcinoma and colon carcinoma. Cancer Res. 1992;52:3067–3072. [PubMed] [Google Scholar]


