Abstract
AIM: Fourteen urinary nucleosides, primary degradation products of tRNA, were evaluated to know the potential as biological markers for patients with colorectal cancer.
METHODS: The concentrations of 14 kinds of urinary nucleosides from 52 patients with colorectal cancer, 10 patients with intestinal villous adenoma and 60 healthy adults were determined by column switching high performance liquid chromatography method.
RESULTS: The mean levels of 12 kinds of urinary nucleosides (except uridine and guanosine) in the patients with colorectal cancer were significantly higher than those in patients with intestinal villous adenoma or the healthy adults. Using the levels of 14 kinds of urinary nucleosides as the data vectors for principal component analysis, 71% (37/52) patients with colorectal cancer were correctly classified from healthy adults, in which the identification rate was much higher than that of CEA method (29%). Only 10% (1/10) of patients with intestinal villous adenoma were indistinguishable from patients with colorectal cancer. The levels of m1G, Pseu and m1A were positively related with tumor size and Duke’s stages of colorectal cancer. When monitoring the changes in urinary nucleoside concentrations of patients with colorectal cancer associated with surgery, it was found that the overall correlations with clinical assessment were 84% (27/32) and 91% (10/11) in response group and progressive group, respectively.
CONCLUSION: These findings indicate that urinary nucleosides determined by column switching high performance liquid chromatography method may be useful as biological markers for colorectal cancer.
Keywords: Nucleosides, Biological markers, Colorectal cancer, High performance liquid chromatography
INTRODUCTION
Modified nucleosides, derived predominantly from transfer ribonucleic acid (tRNA)[1-3], have been shown to be excreted in abnormal amounts in the urine of cancer patients[4-7].
Interest in these materials as potential biological markers was stimulated following evidence that tRNA methyltransferase from cancer tissue had both increased activity and capacity when compared to the enzyme derived from the corresponding normal tissue of origin[8]. Studies by Borek et al[9] also showed that tRNA from neoplastic tissue had a much more rapid turnover rate than the tRNA from the corresponding normal tissue. Evidence indicates that methylation of tRNA occurs only after synthesis of the intact macromolecule. Because there are no specific enzyme systems to incorporate the modified nucleosides into the macromolecular nucleic acid, these nucleosides once released in the process of tRNA turnover cannot be reutilized, nor are they further degraded, but are excreted in urine[10]. Studies have also shown that urinary nucleosides excretion in human beings is little affected by diet, and when normalized to urinary creatinine the daily excretion rate is remarkably constant in a healthy individual[11].
Methodically in most of the studies urinary nucleosides are isolated by phenylboronate affinity gel chromatography and separated by reverse-phase high performance liquid chromatography (HPLC)[1,5,6,12,13]. But these methods still involve elaborate and manually performed sample-processing steps due to the complexity of the sample matrix. Among the various types of cancer, colorectal cancer is known to be one of the most prevalent and its early detection is thus desirable. However, attempts were rarely made to measure the levels of urinary nucleosides from patients with colorectal cancer to date. In this study, we developed the automated column switching HPLC method to investigate the excretion pattern of urinary nucleosides associated with colorectal cancer and intestinal villous adenoma. Differences in 14 urinary nucleoside levels were quantified in 52 patients with colorectal cancer, 10 patients with intestinal villous adenoma and 60 healthy adults. The relationship of urinary excretion of these compounds from patients with colorectal cancer with the tumor size, Duke’s stages and differentiation were studied. Changes in the levels of urinary nucleosides were examined preoperatively and postoperatively in patients with colorectal cancer. These data were collected to test the utility of urinary nucleosides as biological markers for colorectal cancer.
MATERIALS AND METHODS
Chemicals and equipment
The following 14 nucleoside standards including the internal standard 8-bromoguanosine hydrate (Br8G) were obtained from Sigma (St. Louis, MO, USA): pseudouridine (Pseu), cytidine (C), uridine (U), 1-methyladenosine (m1A), inosine (I), 5-methyluridine (m5U), guanosine (G), 1-methylinosine (m1I), 1-methylguanosine (m1G), N4-acetylcytidine (ac4C), N2-methylguanosine (m2G), adenosine (A), N2, N2-methylguanosine (m22G), N6-methyladenosine (m6A). Methanol (MeOH) was HPLC-grade purchased from Tedia (Fairfield, OH, USA). Ammonium acetate (NH4AC), ammonia (NH3·H2O) and potassium dihydrogenphosphate (KH2PO4) were all analytical reagents obtained from China. Water was deionized and purified by a Milli-Q system (Millipore, Bedford, MA, USA).
The HPLC system (Figure 1) consisted of three Shimadzu LC-10ATVP pumps (Kyoto, Japan), an autoinjector model SIL 10ADVP, an SPD-10AVP UV-Vis detector and an SCL 10AVP interface. An electric six-port valve (Rheodyne, USA) was used for the automated column switching. Valve switching and data acquisition were done on Shimadzu Class-VP version 6.10 software. The column 1 (40 mm×4.0 mm ID) was packed with a laboratory prepared boronic acid-substituted silica material. It can tolerate pH values of the buffers from 2 to 12 as well as the usual organic solvents. The column 2 (250 mm×4.6 mm ID) was packed with 5 µm Hypersil ODS2 (Elite, Dalian, China).
Figure 1.
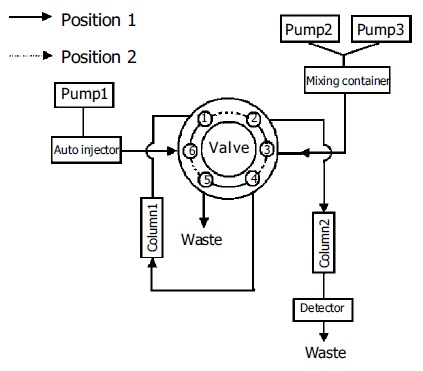
Schematic diagram of the column-switching HPLC system.
Urine samples
Sixty healthy adults (31 males, 29 females, from 21 to 71 years, median age 52 years), who have the normal physical indices including hepatic function, renal function, chest X-ray and colonoscopy during a regular physical examination period in our institute, have been chosen as control material. Fifty-two patients with colorectal cancer and 10 patients with intestinal villous adenoma were from Ruijin Affiliated Hospital of Shanghai Second Medical University, the First and Second Affiliated Hospitals of Dalian Medical University of China. No patient had received chemotherapy or radiation therapy before surgery. Diagnoses of colorectal cancer and intestinal villous adenoma were made on the basis of usual clinical and laboratory findings and were confirmed by histopathology. Table 1 shows the clinical pathological parameters of patients with colorectal cancer.
Table 1.
Clinicopathological parameters of 52 patients with colorectal carcinoma
| Clinicopathological parameters | Numbers(%) | |
| Gender | Male | 29 (55.8) |
| Female | 23 (44.2) | |
| Age (yr) | Range | 26-87 |
| Mean | 56.4 | |
| Median | 60.0 | |
| Primary site | Rectum | 15 (28.8) |
| Sigmoid colon | 6 (11.5) | |
| Colon | 31 (59.6) | |
| Duke’s stage | Duke’s A | 7 (13.5) |
| Duke’s B | 23 (44.2) | |
| Duke’s C | 15 (28.8) | |
| Duke’s D | 7 (13.5) | |
| Tumor size | ≥ 5 cm | 23 (44.2) |
| < 5 cm | 29 (55.8) | |
| Histological grade | Well differentiated tumor | 9 (17.3) |
| Moderately differentiated tumor | 32 (61.5) | |
| Poorly differentiated tumor | 11 (21.2) | |
| CEA | ≥ 5 mg/L | 15 (28.8) |
| < 5 mg/L | 37 (71.2) | |
Spontaneous urine samples were collected from healthy adults, patients with colorectal cancer and intestinal villous adenoma. In 43 patients with colorectal cancer, urine samples were also obtained 2 wk after surgery. All persons had normal renal function and were free of bacterial infection at the time when the urine was collected. After collection the samples free of preservatives were frozen immediately and stored at -20°C. Prior to analysis, the samples were thawed at room temperature and adjusted to pH 8.0 with 50 mL/L NH3·H2O and vortex for 3 min at 5 000 r/min. Aliquots of 1 mL centrifuged urine containing 30 µL of Br8G (0.30 mmol/L) were transferred to autosampler vials and samples of 150 µL were injected to a column-switching HPLC system.
Column switching HPLC method
Column 1 (Figure 1) was equilibrated for 5 min with the mobile phase delivered by pump 1. After sample injection (150 µL urine), column 1 was washed for 7 min with the same buffer. During that time, nucleosides were selectively retarded on the column 1 and the sample matrix was discharged. At the same time, column 2 was conditioned with the mobile phase delivered by pump 2 (position 1; Figure 1). After this clean-up step, column 1 was series-connected in front of the column 2. The group-specifically bound nucleosides on the column 1 were then eluted and concentrated on top of the column 2 over a period of 3 min (position 2; Figure 1). Separation of nucleosides on the column 2 was carried out with a linear gradient elution program over 40 min, while the column 1 was regenerated for a new extraction cycle (position 1; Figure 1). The nucleosides were detected at 260 nm and quantified using the internal standard method. Table 2 shows the time events used for the analytical procedure.
Table 2.
Time events for the switching of column and of mobile phase1
| Time (min) | Pump | Event | Valve position |
| 0.00 – 7.00 | Pump 1 (eluent A) | Sample matrix are discharged by column 1 | 1 |
| Pump 2 (eluent B) | Conditioning of column 2 | ||
| 7.00 – 10.00 | Pump 2 (eluent B) | Analytes are transferred from column 1 to column 2 | 2 |
| 10.00 – 50.00 | Pump 2 and Pump 3 (eluents B and C) | Analysis of nucleosides on column 2 by using a linear | 1 |
| gradient elution program | |||
| Pump 1 (eluent A) | Conditioning of column | 1 |
1Eluent A: 0.25 mol/L NH4AC (pH 8.5); eluent B: 25 mmol/L KH2PO4 (pH 4.5); eluent C: methanol:water (3:2, v/v). Flow rate: pump 1, 0.2 mL/min; pump 2, 1.2 mL/min. Detection wavelength: 260 nm.
Peak identification was performed on the basis of retention times. Standard solutions were run daily before and after the samples to monitor reproducibility of retention times. The standard addition method was also used to confirm peak identification. The levels of the urinary nucleosides were calculated by the calibration curves, and then were transformed into nmoL/µmoL creatinine. Urinary creatinine levels were determined as described by Zheng et al[7].
Data analysis
The mean excreted amounts of urinary nucleosides have been calculated using MS Excel software. Differences of urinary nucleoside concentrations of healthy adults, patients with intestinal villous adenoma and colorectal cancer were compared with SPSS 10.0 software. Spearman Correlation Analysis was used to examine the relationship of urinary nucleoside concentrations from patients of colorectal cancer with the tumor size, clinical stage and differentiation. Principal component analysis (PCA) software, a home-made pattern recognition software, was used to classify healthy controls, patients with intestinal villous adenoma and colorectal cancer. It was also used to monitor changes in the urinary excretion of nucleosides before and after surgery. Principal components plots were drawn on the basis of the first principal component analysis function (PC1) against the second principal component analysis function (PC2) of the nucleosides for each urine specimen. The oblique rotational method used in the software is the Promax method[6].
RESULTS
Analytical characteristics of the method
Figure 2A is a standard chromatogram showing the resolution of 14 nucleosides as well as the internal standard Br8G under the condition newly developed. The calibration curves for 14 nucleosides, linear responses in peak area ratios of nucleosides to the internal standard vs the nucleoside concentrations were obtained with the correlation coefficients varying from 0.995 to 0.9995. The intra- and inter-day precisions of the method were determined by five repetitive analyses of an aqueous solution of standard nucleosides on three nonconsecutive days. The relative standard deviation (RSD) of retention times was less than 1.70% (intraday) and 3.51% (interday), and that of peak areas was less than 3.84% (intraday) and 7.85% (interday). The limits of detection ranged from 0.05 to 0.56 µmol/L, which are better than the previous reports[13,15,16], thus being suitable for the quantitative analysis.
Figure 2.
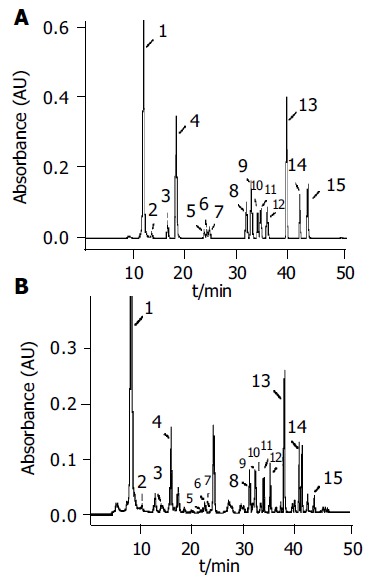
Typical chromatograms of (A) 14 standard nucleoside mixtures (B) urinary nucleosides of a normal urine obtained under the established analysis conditions. Column-switching HPLC conditions as Table 2. Peak identification: 1 Pseu; 2 C; 3 U; 4 m1A; 5 I; 6 m5U; 7 G; 8 m1I; 9 m1G; 10 ac4C; 11 m2G; 12 A; 13 m22G; 14 Br8G; 15 m6A.
When applied to the urine specimens from 60 healthy adults, 10 patients with intestinal villous adenoma and 52 patients with colorectal cancer, a total of 14 nucleosides were positively identified. A typical chromatogram of urinary nucleosides from a normal person with peak identification is given in Figure 2B.
Comparison of urinary excretion of nucleosides in healthy adults, patients with intestinal villous adenoma and colorectal cancer
Fourteen nucleoside concentrations of healthy adults (group 1), patients with intestinal villous adenoma (group 2) and patients with colorectal cancer (group 3) are listed in Table 3. In the mean values of three groups, the most abundant nucleoside was Pseu, followed by m1A and m1I. Pseu level was elevated above the normal values plus two standard deviation (s) in the 58% (30/52) of patients with colorectal cancer, while 20% (2/10) of patients with intestinal villous adenoma was elevated. Clearly, the concentrations of 12 nucleosides (except U, G) were significantly elevated in patients with colorectal cancer (P < 0.05). Only four kinds of nucleoside concentrations of patients with intestinal villous adenoma were higher in comparing with those of healthy adults. Using 14 nucleoside concentrations as the data vectors for PCA technique, 71% (37/52) patients with colorectal cancer was distinguishable from healthy adults, while healthy adults are correctly classified at 96% (58/60) specificity. Serum CEA is the tumor marker being used in clinic to diagnose colorectal cancer (cut-off level: 5 mg/L). But the sensitivity of CEA method is 29% (15/52) based on the data provided by the hospitals. Based on the equation of classification of patients with colorectal cancer and healthy adults, 14 urinary nucleoside concentrations of patients with intestinal villous adenoma were calculated and the results was marked into Figure 3A, it was found that 10% (1/10) of patients with intestinal villous adenoma were in the area of patients with colorectal cancer.
Table 3.
Comparison of levels of urinary nucleosides from healthy adults, patients with intestinal villous adenoma and colorectal can-cer (mean±SD, nmoL/µmoL creatinine)
| Nucleoside | Healthy adults | Patients with | Patients with |
| intestinal villous | colorectal cancer | ||
| adenoma | |||
| Pseu | 22.08 ± 5.11 | 23.99 ± 5.61 | 42.19 ± 22.25a |
| C | 0.15 ± 0.12 | 0.40 ± 0.21c | 0.43 ± 0.49a |
| U | 0.30 ± 0.15 | 0.31 ± 0.12 | 0.31 ± 0.23 |
| m1A | 2.04 ± 0.53 | 2.30 ± 0.62 | 2.74 ± 0.80a |
| I | 0.28 ± 0.11 | 0.27 ± 0.11 | 0.50 ± 0.35a |
| m5U | 0.04 ± 0.06 | 0.12 ± 0.06c | 0.13 ± 0.08a |
| G | 0.09 ± 0.03 | 0.08 ± 0.02 | 0.10 ± 0.04 |
| m1I | 1.25 ± 0.28 | 1.97 ± 0.50c | 2.76 ± 1.94a |
| m1G | 0.82 ± 0.24 | 1.21 ± 0.26c | 1.44 ± 0.51a |
| ac4C | 0.69 ± 0.20 | 0.70 ± 0.19 | 0.84 ± 0.30a |
| m2G | 0.55 ± 0.14 | 0.52 ± 0.28 | 0.63 ± 0.26a |
| A | 0.52 ± 0.16 | 0.59 ± 0.25 | 0.66 ± 0.30a |
| m22G | 1.25 ± 0.23 | 1.43 ± 0.27 | 1.81 ± 0.55a |
| m6A | 0.04 ± 0.02 | 0.06 ± 0.05 | 0.07 ± 0.05a |
P < 0.05 patients with colorectal cancer vs healthy adults.
P < 0.05 patients with intestinal villous adenoma vs healthy adults.
Figure 3.
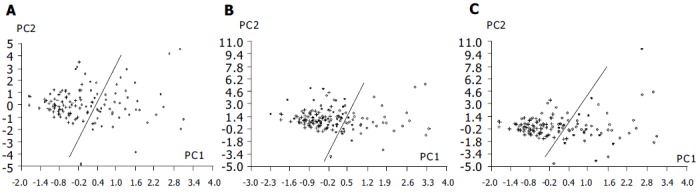
PCA based on 14 nucleoside concentrations from healthy controls (+) and patients with colorectal cancer (o). A: Positions of patients with intestinal villous adenoma (*) were marked into the figure based on classification equation from healthy controls and patients with colorectal cancer; B: Positions of 32 of responsive cases after surgery (*) were marked into the figure based on classification equation from healthy controls and patients with colorectal cancer; C: Positions of 11 of progressive cases after surgery (*) were marked into the figure based on classification equation from healthy controls and patients with colorectal cancer.
Urinary excretion of nucleosides and clinical pathological characteristics of colorectal cancer
We examined the relationship of urinary nucleoside concentrations from patients of colorectal cancer with the tumor size, Duke’s stages and differentiation. These results are listed in Table 4. The level of m1G, Pseu and m1A were positively correlated with the tumor size and Duke’s stages of colorectal cancer, respectively (P < 0.05). No significant correlation was noted in observed values with regard to tumor differentiation.
Table 4.
Relationship of the levels of urinary nucleosides from patients of colorectal cancer with the tumor size, Duke’s stage and differentiation
| Nucleoside | Tumor size | Duke’s stage | Differentiation |
| R | R | R | |
| Pseu | 0.192 | 0.325a | -0.051 |
| C | 0.199 | 0.103 | 0.109 |
| U | 0.159 | 0.19 | 0.142 |
| m1A | 0.188 | 0.303a | -0.014 |
| I | 0.241 | 0.198 | -0.009 |
| m5U | 0.056 | 0.149 | 0.033 |
| G | 0.262 | 0.135 | 0.113 |
| m1I | 0.287 | 0.202 | 0.045 |
| m1G | 0.376a | 0.256 | 0.041 |
| ac4C | 0.053 | 0.189 | 0.008 |
| m2G | 0.275 | 0.261 | 0.091 |
| A | 0.068 | 0.224 | 0.023 |
| m22G | 0.287 | 0.27 | 0.051 |
| m6A | 0.097 | 0.157 | 0.201 |
R: relationship coefficient;
P < 0.05 nucleosides vs Duke’s stage.
Preoperative and postoperative urinary excretion of nucleosides
The changes in six urinary modified nucleoside concentrations (Pseu, m1A, m1I, m1G, ac4C, m22G) before and after surgery in 43 patients with colorectal cancer were studied. The patients were classified into two groups: response group (32 persons) and progressive disease group (11 persons). Using the paired t-test, these urinary modified nucleoside concentrations of response group before surgery were significantly higher than those of patients after surgery (P < 0.05). However, the pre- and post-surgery difference in levels of these nucleosides for progressive disease group had no significant changes.
PCA technique based on 14 nucleoside concentrations as the data vectors was used to monitor changes in the urinary excretion of nucleosides for patients with colorectal cancer before and after surgery. When urinary nucleoside concentrations of patients with colorectal cancer after surgery were fed to the principal component regression and marked to the space produced by the first two principal components (PC1, PC2) of healthy adults and patients with colorectal cancer before surgery (the process was similar to the position prediction of the patients with intestinal villous adenoma), it can be seen that points of 84% (27/32) patients with effective treatment have come back to the normal person area (Figure 3B). In the mean time, points of 91% (10/11) patients with ineffective treatment have entered into the area of colorectal cancer (Figure 3C).
DISCUSSION
Urinary nucleosides as biological markers of malignancy have previously been reported for a wide variety of cancers[1-7,17,18]. However, only few reports existed regarding colorectal cancer. Our preliminary examination revealed that patients with colorectal cancer excreted in their urine significantly elevated amounts of nucleosides by HPLC method[13]. But the method analysis of urinary nucleosides involves manual sample extraction steps, resulting in artificial error and time-consumption. In the current study, we developed the automated column switching HPLC method to examine the relationship between urinary nucleosides and pathological characteristics of colorectal cancer and the potential value of these compounds in monitoring progress of the disease during surgery. The method is simple and rapid, requiring a total analysis time of 50 min per sample, with no time involved for sample processing. Its application to routine urine samples suggests utility for mass patient screening.
The present study also confirmed that significantly elevated levels of urinary nucleosides were detected in patients with colorectal cancer by the column-switching HPLC method developed. Our data revealed that the mean nucleoside concentrations from patients with intestinal villous adenoma were significantly lower than those from patients with colorectal cancer. Only 10% (1/10) of patients with intestinal villous adenoma were in the area of patients with colorectal cancer. This is in accordance with the previous studies showing that patients with noncancerous diseases and acute infections[19,20] do not excrete significantly elevated levels of urinary nucleosides. A current study on urinary nucleosides in colorectal cancer patients demonstrate that in the malignant disease not just one but several of the nucleosides is elevated. In such a multi-component alteration of the nucleoside levels, a pattern recognition method could reveal more information on difference among healthy adults, patients with intestinal villous adenoma and colorectal cancer than the evaluation of single components. The PCA method was applied for evaluation of the nucleoside levels in the three groups. The sensitivity of urinary nucleosides in patients with colorectal cancer was 71%, whereas the sensitivity of currently used tumor marker CEA is 29%. Urinary nucleosides may be a satisfactory biological marker for this disease.
It was also found that the level of m1G, Pseu and m1A were positively correlated with the tumor size and Duke’s stages of colorectal cancer, respectively. These nucleosides may be useful as prognostic factors. Modified nucleosides have a much shorter half-life in the body than protein. A faster response to therapy and recurrence of disease is therefore feasible. Our results show that the changes in urinary nucleoside concentrations of patients with colorectal cancer almost paralleled with the change of disease status of patients after surgery (Figures 3B and C). The results are in agreement with the previous studies about urinary nucleosides being useful for monitoring progress of lymphoma and small cell carcinoma of the lung[21,22]. These facts indicated that urinary nucleosides may be of value for a rapid assessment of disease course in monitoring colorectal cancer.
These studies have indicated the potential utility of urinary nucleosides as biological marker for colorectal cancer, especially when the column switching HPLC method developed is combined with the PCA data processing method. Because the whole-body RNA turnover correlates quite well with the protein turnover[23], attention has to be paid to the alterations of urinary nucleosides under various catabolic conditions other than those occurring in malignancies, e.g. malnutrition, endocrine abnormalities, alcoholism and stress. More works are currently in progress to evaluate the usefulness of urinary nucleosides in differentiating cancer from other disease status.
ACKNOWLEDGMENTS
We are very grateful to Professor K.-S. Boos of Institute of Clinical Chemistry, University Hospital Grosshadern, Munich, Germany for the denotation of boronic acid-substituted silica column.
Footnotes
Supported by the High-tech R and D Plan, No. 2003AA223061 and the Sociality Commonweal Project of State Ministry of Science and Technology of China, the Knowledge Innovation Program of the Chinese Academy of Sciences, No. K2003A16 and Liaoning Province Foundation of Science and Technology
Science Editor Guo SY Language Editor Elsevier HK
References
- 1.Xu G, Di Stefano C, Liebich HM, Zhang Y, Lu P. Reversed-phase high-performance liquid chromatographic investigation of urinary normal and modified nucleosides of cancer patients. J Chromatogr B Biomed Sci Appl. 1999;732:307–313. doi: 10.1016/s0378-4347(99)00296-0. [DOI] [PubMed] [Google Scholar]
- 2.Liebich HM, Xu G, Di Stefano C, Lehmann R. Capillary electrophoresis of urinary normal and modified nucleosides of cancer patients. J Chromatogr A. 1998;793:341–347. doi: 10.1016/s0021-9673(97)00915-1. [DOI] [PubMed] [Google Scholar]
- 3.Xu G, Lu X, Zhang Y, Lu P, Di Stefano C, Lehmann R, Liebich H. Two approaches for determining the urinary excretion patterns of nucleosides--HPLC and CE. Se Pu. 1999;17:97–101. [PubMed] [Google Scholar]
- 4.Zhao R, Xu G, Yue B, Liebich HM, Zhang Y. Artificial neural network classification based on capillary electrophoresis of urinary nucleosides for the clinical diagnosis of tumors. J Chromatogr A. 1998;828:489–496. doi: 10.1016/s0021-9673(98)00589-5. [DOI] [PubMed] [Google Scholar]
- 5.Xu G, Liebich HM, Lehmann R, Müller-Hagedorn S. Capillary electrophoresis of urinary normal and modified nucleosides of cancer patients. Methods Mol Biol. 2001;162:459–474. doi: 10.1385/1-59259-055-1:459. [DOI] [PubMed] [Google Scholar]
- 6.Xu G, Schmid HR, Lu X, Liebich HM, Lu P. Excretion pattern investigation of urinary normal and modified nucleosides of breast cancer patients by RP-HPLC and factor analysis method. Biomed Chromatogr. 2000;14:459–463. doi: 10.1002/1099-0801(200011)14:7<459::AID-BMC7>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 7.Zheng YF, Xu GW, Liu DY, Xiong JH, Zhang PD, Zhang C, Yang Q, Lv S. Study of urinary nucleosides as biological marker in cancer patients analyzed by micellar electrokinetic capillary chromatography. Electrophoresis. 2002;23:4104–4109. doi: 10.1002/elps.200290027. [DOI] [PubMed] [Google Scholar]
- 8.Borek E, Kerr SJ. Atypical transfer RNA's and their origin in neoplastic cells. Adv Cancer Res. 1972;15:163–190. doi: 10.1016/s0065-230x(08)60374-7. [DOI] [PubMed] [Google Scholar]
- 9.Borek E, Baliga BS, Gehrke CW, Kuo CW, Belman S, Troll W, Waalkes TP. High turnover rate of transfer RNA in tumor tissue. Cancer Res. 1977;37:3362–3366. [PubMed] [Google Scholar]
- 10.Mandel LR, Srinivasan PR, Borek E. Origin of urinary methylated purines. Nature. 1966;209:586–588. doi: 10.1038/209586a0. [DOI] [PubMed] [Google Scholar]
- 11.Sander G, Topp H, Heller-Schöch G, Wieland J, Schöch G. Ribonucleic acid turnover in man: RNA catabolites in urine as measure for the metabolism of each of the three major species of RNA. Clin Sci (Lond) 1986;71:367–374. doi: 10.1042/cs0710367. [DOI] [PubMed] [Google Scholar]
- 12.Liebich HM, Di Stefano C, Wixforth A, Schmid HR. Quantitation of urinary nucleosides by high-performance liquid chromatography. J Chromatogr A. 1997;763:193–197. doi: 10.1016/s0021-9673(96)00757-1. [DOI] [PubMed] [Google Scholar]
- 13.Zheng YF, Chen YJ, Pang T, Shi XZ, Kong HW, Lü S, Yang Q, Xu GW. [Investigation of urinary nucleosides excretion of intestinal cancer patients by reversed-phase high performance liquid chromatography] Se Pu. 2002;20:498–501. [PubMed] [Google Scholar]
- 14.Gehrke CW, Kuo KC, Waalkes TP, Borek E. Patterns of urinary excretion of modified nucleosides. Cancer Res. 1979;39:1150–1153. [PubMed] [Google Scholar]
- 15.Liebich HM, Xu G, Di Stefanoc C, Lehmann R, Haring HU, Lu P, Zhang Y. Analysis of normal and modified nucleosides in Urine by Capillary Electrophoresis. Chromatographia. 1997;45:396–401. [Google Scholar]
- 16.Zheng YF, Zhang Y, Liu DY, Guo XL, Mei SR, Xiong JH, Kong HW, Zhang C, Xu G. Analysis of Urinary Nucleosides by Micellar Electrokinetic Chromatography. Chemical J Chinese Universities. 2001;22:912–915. [Google Scholar]
- 17.Kim KR, La S, Kim A, Kim JH, Liebich HM. Capillary electrophoretic profiling and pattern recognition analysis of urinary nucleosides from uterine myoma and cervical cancer patients. J Chromatogr B Biomed Sci Appl. 2001;754:97–106. doi: 10.1016/s0378-4347(00)00585-5. [DOI] [PubMed] [Google Scholar]
- 18.La S, Cho JH, Kim JH, Kim KR. Capillary electrophoretic profiling and pattern recognition analysis of urinary nucleo-side from throid cancer patients. Anal Chim Acta. 2003;486:171–182. [Google Scholar]
- 19.Borek E, Sharma OK, Buschman FL, Cohn DL, Penley KA, Judson FN, Dobozin BS, Horsburgh CR, Kirkpatrick CH. Altered excretion of modified nucleosides and beta-aminoisobutyric acid in subjects with acquired immunodeficiency syndrome or at risk for acquired immunodeficiency syndrome. Cancer Res. 1986;46:2557–2561. [PubMed] [Google Scholar]
- 20.Fischbein A, Sharma OK, Selikoff IJ, Borek E. Urinary excretion of modified nucleosides in patients with malignant mesothelioma. Cancer Res. 1983;43:2971–2974. [PubMed] [Google Scholar]
- 21.Rasmuson T, Björk GR. Urinary excretion of pseudouridine and prognosis of patients with malignant lymphoma. Acta Oncol. 1995;34:61–67. doi: 10.3109/02841869509093640. [DOI] [PubMed] [Google Scholar]
- 22.Waalkes TP, Abeloff MD, Ettinger DS, Woo KB, Gehrke CW, Kuo KC, Borek E. Biological markers and small cell carcinoma of the lung: a clinical evaluation of urinary ribonucleosides. Cancer. 1982;50:2457–2464. doi: 10.1002/1097-0142(19821201)50:11<2457::aid-cncr2820501134>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 23.Sander G, Hülsemann J, Topp H, Heller-Schöch G, Schöch G. Protein and RNA turnover in preterm infants and adults: a comparison based on urinary excretion of 3-methylhistidine and of modified one-way RNA catabolites. Ann Nutr Metab. 1986;30:137–142. doi: 10.1159/000177186. [DOI] [PubMed] [Google Scholar]


