Abstract
Recently, a model was proposed to assess hepatic insulin sensitivity during a meal, i.e., the ability of insulin to suppress glucose production (EGP), SIP. The model was developed on EGP data obtained from a triple-tracer meal and the tracer-to-tracee clamp technique and validated against the euglycemic hyperinsulinemic clamp. The aim of this study was to assess whether SIP can be obtained from plasma concentrations measured after a single-tracer meal by incorporating the above EGP model into the oral glucose minimal model by describing both glucose production and disposal (OMMPD). Triple-tracer meal data of two databases (20 healthy and 60 healthy and prediabetic subjects) were used. Virtually model-independent EGP estimates were calculated. OMMPD was identified on exogenous and endogenous glucose concentrations, providing indices of SIP, disposal insulin sensitivity (SID), and EGP. The model fitted the data well, and SIP and SID were estimated with precision in both databases (SIP = 5.48 ± 0.54 10−4 dl·kg−1·min−1 per μU/ml and SID = 9.93 ± 2.18 10−4 dl·kg−1·min−1 per μU/ml in healthy; SIP = 5.41 ± 3.55 10−4 dl·kg−1·min−1 per μU/ml and SID = 5.34 ± 6.17 10−4 dl·kg−1·min−1 per μU/ml, in healthy and prediabetic subjects). Estimated SIP and that derived from the triple-tracer EGP model were very similar on average. Moreover, the time course of EGP normalized to basal EGP (EGPb), and EGP/EGPb agreed with the results obtained using the triple-tracer method. In this study, we have demonstrated that SIP, SID, and EGP/EGPb time course can be estimated reliably from a single-tracer meal protocol in both healthy and prediabetic subjects.
Keywords: glucose kinetics, tracer, meal, insulin resistance
the primary source of endogenous glucose production (EGP) in the body is the liver, which modulates glucose release into the circulation influenced by prevailing glucose, insulin, glucagon, and other hormone concentrations. In a healthy subject, when plasma glucose and insulin concentrations rise, e.g., after a meal, EGP is suppressed, whereas if plasma glucose and insulin concentrations decrease below a given threshold (such as during prolonged fasting), more glucose is delivered to avoid potentially dangerous hypoglycemia. In pathological situations, such as type 2 diabetes, insulin regulation of EGP is altered (17). Thus, quantifying the effect of insulin in suppressing EGP [hepatic insulin sensitivity (SIP)] could be a useful tool for studying the pathophysiology of metabolic disorders, including various stages of diabetes and efficacy of therapeutic approaches.
Among the methods proposed in the literature to estimate SIP in vivo, the labeled euglycemic hyperinsulinemic clamp (16), and surrogate indices of hepatic insulin resistance (1) have been used. However, both methods suffer from important limitations. Briefly, the clamp method calculates SIP as the difference between net (derived from steady-state glucose infusion rate, plasma glucose, and insulin concentrations) and disposal insulin sensitivity (derived from steady-state tracer infusion rate, plasma glucose, tracer glucose, and insulin concentrations). Its main drawbacks are that it is labor intensive and nonphysiological. The index proposed by Abdul-Ghani et al. (1) is the product of the area under glucose and insulin concentrations measured in the first 30 min of an oral glucose tolerance test (OGTT). It utilizes a simpler method and is more physiological. However, it is based on the erroneous assumptions that glucose utilization minimally increases during the initial 30 min after food ingestion and that glucose rate of appearance in the first 30 min is zero.
An important step forward was a model-based method to estimate SIP during a meal (14). The model was fitted against model-independent estimates of EGP from the triple-tracer meal protocol (4) and has recently been validated against the labeled euglycemic hyperinsulinemic clamp (13). Of note, only two of the three tracers available in Basu et al. (4) were used to reconstruct the EGP time course, i.e., one oral to segregate the endogenous from the exogenous source of glucose and the other intravenous to mimic the expected EGP pattern, keeping the tracer-to-tracee (TTR) ratio constant (5). Thus, the state-of-the-art method to estimate SIP during a meal uses a dual-tracer approach.
However, a less labor-intensive method that is able to provide an estimation of SIP would be particularly appealing for large-scale studies. One possibility could be to derive SIP as the difference between net (SI) and disposal insulin sensitivity (SID) derived from the labeled oral glucose minimal model (12), but, as already pointed out in Dalla Man et al. (12), this would lead to a nonphysiological result in a not insignificant percentage of the subjects (19% of them showed net insulin sensitivity lower than disposal insulin sensitivity), as shown in Fig. 7 in Ref. 9. This was proven to be due largely to a suboptimal description of EGP in the minimal model (14).
The aim of this study was to combine the EGP model proposed by Dalla Man et al. (14) with the oral minimal model (12) to enable an accurate estimation of the SIP index from a single-tracer meal protocol. First, we identified and validated the model on healthy subjects, and then we evaluated the performance of the method in prediabetes.
MATERIALS AND METHODS
Subjects and Protocols
In this analysis, two different databases were used. The first database (healthy) was employed to build and validate the new oral minimal model. It consisted of 20 healthy subjects (age = 32 ± 4 yr, BMI = 25 ± 1 kg/m2), already described in Dalla Man et al. (14), who received a triple-tracer mixed meal (10 kcal/kg: 45% carbohydrate, 15% protein, 40% fat) containing 1.00 ± 0.02 g/kg glucose (4). [1-13C]glucose was administered orally together with the meal, and [6,6-2H2]glucose and [6-3H]glucose were infused intravenously, as described below. Blood samples were drawn frequently to measure plasma glucose (G), insulin (I), and tracer concentrations, as reported previously (4). In particular, labeling the meal with [1-13C]glucose (G*) allowed for deriving the exogenous, i.e., coming from the meal, glucose (Gexo) concentration as
| (1) |
with zmeal denoting the TTR ratio in the meal. Endogenous glucose (Gend) was then calculated as difference between G, Gexo, and the intravenously infused tracer [6,6-2H2]glucose (GD2):
| (2) |
It is worth noting that, in the case of a single-tracer protocol, GD2 in Eq. 2 is zero.
The [6-3H]glucose and [6,6-D2]glucose tracers were infused intravenously at variable rates, to clamp, respectively, the specific activity, i.e., the ratio between [6-3H]glucose and [1-13C]glucose, and the TTR, i.e., the ratio between [6.6-2H2]glucose and Gend. More details on the experimental protocol can be found in Basu et al. (4). Thanks to this protocol, it was possible to simultaneously estimate both EGP and meal glucose rate of appearance (Ra meal) using Radziuk's model (18) by minimizing the non-steady-state model error.
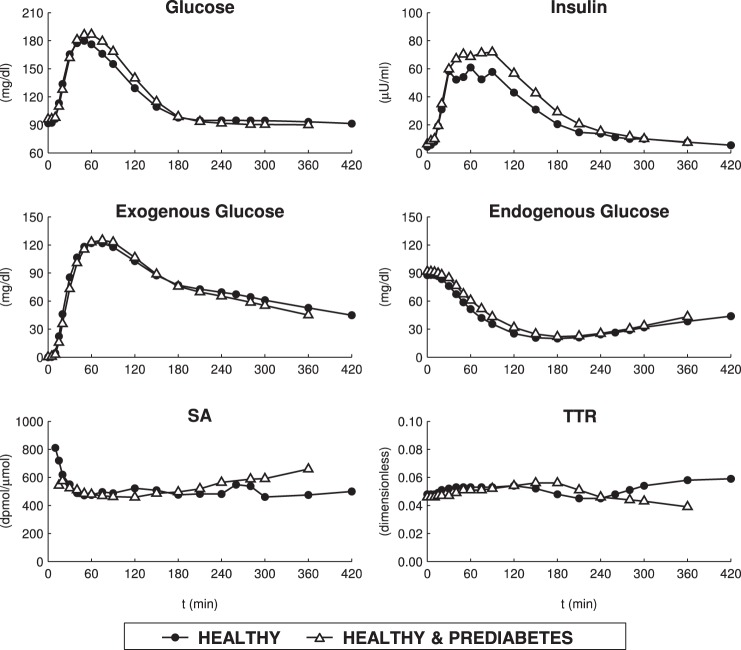
The second database (healthy and prediabetic subjects) was used to test the new method in prediabetes. In particular, this database consisted of 32 subjects (age = 54 ± 1 yr, BMI = 31 ± 1 kg/m2) with impaired fasting glucose (IFG) and 28 subjects (age = 51 ± 2 yr; BMI = 28 ± 1 kg/m2) with normal fasting glucose (NFG) (6) classified in subgroups on the basis of their glucose tolerance, i.e., 16 NFG subjects (age = 50 ± 2 yr, BMI = 27 ± 1 kg/m2) with normal glucose tolerance (NFG/NGT), 12 NFG subjects (age = 53 ± 3 yr, BMI = 30 ± 1 kg/m2) with impaired glucose tolerance (NFG/IGT), seven IFG subjects (age = 53 ± 3 yr, BMI = 31 ± 2 kg/m2) with normal glucose tolerance (IFG/NGT), 17 IFG subjects (age = 54 ± 2 yr, BMI = 31 ± 1 kg/m2) with impaired glucose tolerance (IFG/IGT), and eight IFG subjects (age = 54 ± 2 yr, BMI = 32 ± 1 kg/m2) with diabetes (IFG/DM). These subjects underwent the same triple-tracer mixed meal protocol described above, except for the glucose amount, fixed here at 75 g. Average time courses of G, I, Gexo, Gend, SA and TTR are shown in Fig. 1 for both databases. For more details on protocol we refer to Bock et al. (6).
Fig. 1.
Mean concentrations of plasma glucose and insulin (top), exogenous and endogenous glucose (middle), and specific activity (SA) and clamped tracer-to-tracee ratio (TTR) (bottom) in healthy database (n = 20) and healthy and prediabetes database (n = 60).
A Production and Disposal Minimal Model
A production and disposal oral minimal model (OMMPD) is presented, which permits description of G, Gexo, and Gend using a single, orally administered tracer. The model, shown in Fig. 2, is built on the mass balance principle
| (3) |
Fig. 2.
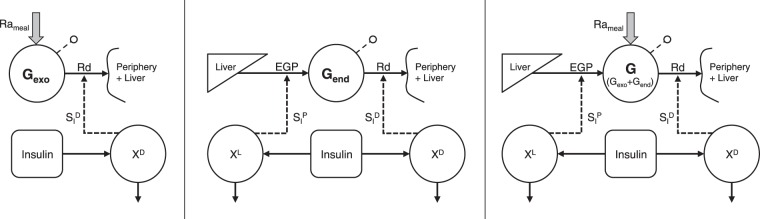
Scheme of the production and disposal oral minimal model (OMMPD). Left: submodel of exogenous glucose (Gexo). Middle: submodel of endogenous glucose (Gend). Right: submodel of total glucose (G = Gexo + Gend).
where Gb is basal glucose, V is the distribution volume, EGP is the endogenous glucose production, Ra D2 is the [6,6-2H2]glucose rate of appearance, Ra meal is the meal glucose rate of appearance, and Rd is the rate of glucose disposal. It is of note that, in the case of single-tracer approach, Ra D2 is zero and Eq. 3 becomes
| (4) |
Since Eq. 2 holds, Eq. 3 can be split to distinguish the exogenous and endogenous components
| (5) |
with Gendb the basal endogenous glucose and Rd exo and Rd end the disposal rates of Gexo and Gend, respectively. Like in Cobelli et al. (9) and Dalla Man et al. (12), OMMPD assumes that Ra meal is described by a piecewise-linear function with known break points ti and unknown amplitudes αi
| (6) |
and Rd is linearly dependent on G and controlled by insulin in a remote compartment:
| (7) |
where SGD is the fractional (i.e., per unit distribution volume) disposal glucose effectiveness (GED), I is the plasma insulin concentration (with Ib its basal value), and XD is the insulin action on glucose disposal, and k1D and k2D are rate constants describing its dynamics and magnitude. Similarly, Rd exo and Rd end are defined as in Eq. 7, substituting G with Gexo and Gend, respectively.
In addition, OMMPD incorporates the EGP description proposed in Dalla Man et al. (14), which is based on the assumption that EGP is suppressed by plasma glucose and a delayed insulin action, and it is also promptly inhibited by portal insulin. In particular, portal insulin is substituted with insulin secretion rate, which can be modeled as the sum of two components, i.e., one proportional to the glucose rate of change (which takes into account of the newly secreted insulin) and one to the above-basal glucose concentration. Therefore, EGP suppression is described by three components: one proportional to glucose rate of change (through kGR), one proportional to glucose concentration (through kG), and a delayed insulin action (XP):
| (8) |
with
| (9) |
and EGPb related to the other model parameters through the steady-state constraint
| (10) |
Hence, OMMPD is described by Eqs. 3 and 5-10. The production and disposal components of the insulin sensitivity indices (SIP, SID) and glucose effectiveness (GEP, GED) are derived from steady-state model equations as follows:
| (11) |
| (12) |
| (13) |
| (14) |
where AUCG is the area under plasma glucose curve.
Model Identification
Identifiability.
OMMPD is a priori nonidentifiable; in particular, parameter V is not identifiable and parameter SGD is not uniquely identifiable. The a priori knowledge required for model identification was obtained paralleling that done in Dalla Man et al. (12). The model of Eqs. 3, 5, and 7-10 was identified using Ra meal as known input (available from the triple-tracer protocol), and parameter V and SGD were estimated in each subject. Then, to identify the OMMPD, V and SGD were fixed to their respective mean values in the population (V = 1.61 ± 0.06 dl/kg, SGD = 0.0115 ± 0.0007 min), and the constraint
| (15) |
was imposed to the area under estimated Ra meal, where D is the dose of ingested glucose, f is the fraction of absorbed glucose, and BW is the body weight of the subject. However, due to Eq. 10, fixing both V and SGD would make it impossible to individualize EGPb in a given subject. Hence, GED (and thus SGD) was decomposed into its insulin-independent [i.e., glucose effectiveness on glucose disposal at zero insulin (GEZID)] and insulin-dependent components (3):
| (16) |
| (17) |
By fixing GEZID to its mean value (GEZID = 0.021 ± 0.001 dl·kg−1·min−1 in healthy, GEZID = 0.017 ± 0.001 dl·kg−1·min−1 in healthy and prediabetic subjects), estimated with an approach similar to that presented in Dalla Man et al. (12), we were able to ensure a certain degree of freedom for SGD (SID is derived from k2D in Eq. 12), therefore providing an estimation of EGPb for each subject. It is worth noting that the steady-state plasma clearance rate results were underestimated using a single compartment (7); as a consequence, the estimated EGPb, and thus also the estimation of absolute EGP, is partially incorrect. However, to overcome this drawback, one can consider the EGP/EGPb ratio, which is more robust (see results).
Parameter estimation.
The model was numerically identified on Gexo and Gend by nonlinear least squares (8, 10), as implemented in SAAM II (2). Measurement errors on exogenous and endogenous glucose were assumed to be independent, Gaussian, with zero mean and known standard deviations calculated by error propagation from primary measurements. Plasma insulin concentration was the model-forcing function and was assumed to be known without error.
Model Validation
As already pointed out in Subjects and Protocols, the healthy database used for model identification is the same as that employed in Dalla Man et al. (14). Therefore, in addition to assessing model performance in terms of ability to fit the data and provide precise parameter estimates, it has been possible to validate OMMPD in normal subjects by comparing the estimated SIP with that provided in Dalla Man et al. (14). Moreover, the comparison between the estimated EGP time course and that obtained with the triple-tracer method provided a further validation of OMMPD.
To evaluate the performance OMMPD in healthy and prediabetic subjects, SIP and SID were compared with those presented in Bock et al. (6). The EGP was compared in terms of prediction of time course and by calculating a suppression index S%, defined as
| (18) |
Statistical Analysis
Data are presented as means ± SE. Two sample comparisons were done by Wilcoxon signed-rank test, and Shapiro-Wilk test was used to verify whether parameters are normally distributed (significance level set to 5%). Pearson's correlation was used to evaluate univariate correlation. Two-way ANOVA, including both the main effects and a term for interaction, was used to assess difference in prediabetes between type of data (triple-tracer estimates vs. OMMPD predictions) among the subgroups (NFG/NGT, NFG/IGT, IFG/NGT, IFG/IGT, and IFG/DM). In particular, we focused on the significance of the interaction term, indicating whether the differences among the subgroups were affected or not by the factor “type of data” (significance level set to 5%).
RESULTS
Model Fit and Indices in Healthy Subjects
The OMMPD provided a good fit of the data, as demonstrated by the weighted residual time courses shown in Fig. 3, left. Parameters were estimated with precision: SGD = 0.0151 ± 0.0003 min [coefficient of variation (CV) = 1.1 ± 0.4%], k1D = 0.0248 ± 0.0062 min (CV = 40.5 ± 9.4%), k2D = 0.0006 ± 0.0001 min·μU−1·ml−1 (CV = 8.4 ± 2.7%), kG = 0.0174 ± 0.0033 dl·kg−1·min−1 (CV = 9.5 ± 1.1%), kGR = 0.1210 ± 0.0426 dl/kg (CV = 28.1 ± 6.5%), k1P = 0.0199 ± 0. 0017 min (CV = 5.1 ± 0.6%), k2P = 0.0469 ± 0.0045 mg·kg−1·min−1 per μU/ml (CV = 3.4 ± 0.8%), EGPb = 2.01 ± 0.05 mg·kg−1·min−1 (CV = 1.1 ± 0.5%), GEP = 0.019 ± 0.003 dl·kg−1·min−1 (CV = 10.6 ± 2.6%), GED = 0.024 ± 0.001 dl·kg−1·min−1 (CV = 1.1 ± 0.4%), SIP = 5.48 ± 0.54 10−4 dl·kg−1·min−1 per μU/ml (CV = 3.4 ± 0.8%), and SID = 9.93 ± 2.18 10−4 dl·kg−1·min−1 per μU/ml (CV = 8.4 ± 2.7%).
Fig. 3.
Weighted residuals (vertical bars represent SD) of OMMPD fit on Gexo and Gend. Left: results in healthy database (n = 20). Right: results in healthy and prediabetes database (n = 60).
It is worth noting that GEP and GED account for 44 and 56% of total glucose effectiveness, respectively, whereas SIP and SID account for 36 and 64% of total insulin sensitivity, respectively.
Model Validation
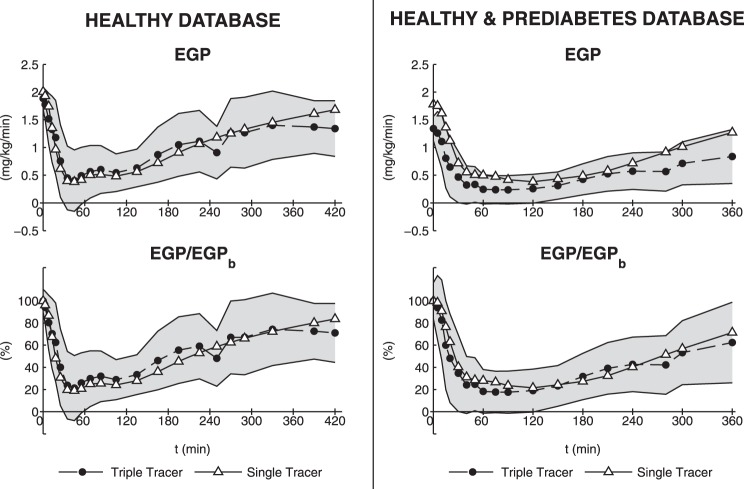
The comparison between SIP estimated with OMMPD and that estimated in Dalla Man et al. (14) using triple-tracer-derived EGP is reported in Fig. 4; mean values are very similar (5.34 ± 0.47 vs. 5.48 ± 0.54 10−4 dl·kg−1·min−1 per μU/ml), and correlation is very good (r = 0.87, P < 0.0001). Furthermore, percentages of production and disposal SI are in agreement with those presented in Dalla Man et al. (14). Moreover, the time course of EGP derived with OMMPD is reproduced reliably with that obtained by the triple-tracer method (Fig. 5, top left), except for EGPb (2.01 ± 0.05 vs. 1.88 ± 0.04 mg·kg−1·min−1). On the other hand, if EGP/EGPb (%) is considered, the comparison between single-tracer model-based predictions and triple-tracer estimates is satisfactory (Fig. 5, bottom left).
Fig. 4.
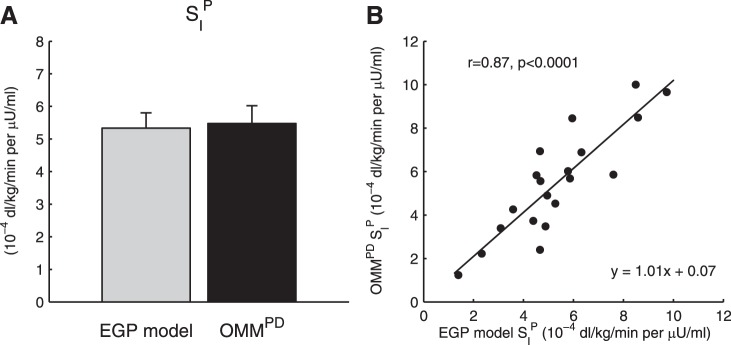
A: comparison between hepatic insulin sensitivity (SIP) estimated from OMMPD (black bar) and endogenous glocose production (EGP) model of Dalla Man et al. (14) (gray bar) in the healthy database (n = 20). B: correlation plot.
Fig. 5.
OMMPD predictions (solid line and △) vs. triple-tracer estimates (dashed line and ●) with SD variability range (gray region). Left: absolute EGP and EGP normalized to basal EGP (EGPb) in healthy database (n = 20). Right: absolute EGP and EGP normalized to EGPb in healthy and prediabetes database (n = 60).
Model Fit and Indices in Prediabetes
The results of OMMPD identification were satisfactory also in prediabetes, providing good fit of the data (Fig. 3, right). Parameters were estimated with precision: SGD = 0.0117 ± 0.0021 min (CV = 1.0 ± 1.9%), k1D = 0.0283 ± 0.0365 min (CV = 51.5 ± 40.7%), k2D = 0.0003 ± 0.0004 min·μU−1·ml−1 (CV = 3.3 ± 2.7%), kG = 0.0116 ± 0.0081 dl·kg−1·min−1 (CV = 13.3 ± 15.2%), kGR = 0.1046 ± 0.1327 dl/kg (CV = 29.7 ± 29.6%), k1P = 0.0177 ± 0.0090 min (CV = 5.3 ± 3.3%), k2P = 0.0515 ± 0.0337 mg·kg−1·min−1 per μU/ml (CV = 3.3 ± 2.7%), EGPb = 1.75 ± 0.34 mg·kg−1·min−1 (CV = 1.1 ± 2.0%), GEP = 0.013 ± 0.008 dl·kg−1·min−1 (CV = 11.3 ± 12.6%), GED = 0.019 ± 0.003 dl·kg−1·min−1 (CV = 1.0 ± 1.9%), SIP = 5.41 ± 3.55 10−4 dl·kg−1·min−1 per μU/ml (CV = 3.3 ± 2.7%), and SID = 5.34 ± 6.17 10−4 dl·kg−1·min−1 per μU/ml (CV = 12.2 ± 20.6%). Figure 6 shows the average SIP and SID obtained for each subgroup. In particular, the distributions of SIP and SID reflected those presented in Bock et al. (6), with both SIP and SID significantly lower (P < 0.05) in the IFG group in IFG/IGT and IFG/DM compared with NFG/NGT. On the other hand, SIP and SID in NFG/IGT and IFG/NGT were lower than NFG/NGT, but not significantly. In terms of model fit, exogenous and endogenous glucose profiles were predicted well, but the EGP time course was overestimated if compared with the triple-tracer estimate (Fig. 5, top right), similar to what was found in the 20 healthy subjects. Conversely, also in this case the EGP/EGPb time course was similar to that obtained from the triple-tracer protocol (Fig. 5, bottom right). S% obtained with OMMPD was 54.4 ± 2.7 (NFG/NGT), 56.1 ± 2.8 (NFG/IGT), 55.7 ± 5.2 (IFG/NGT), 62.7 ± 2.7 (IFG/IGT), and 62.8 ± 2.4% (IFG/DM), and S% derived from the triple-tracer data was 56.7 ± 5.7 (NFG/NGT), 61.0 ± 2.7 (NFG/IGT), 61.0 ± 4.4 (IFG/NGT), 65.2 ± 2.2 (IFG/IGT), and 60.6 ± 5.9% (IFG/DM). S% from OMMPD was identical to that derived from the triple-tracer data in all of the subgroups.
Fig. 6.
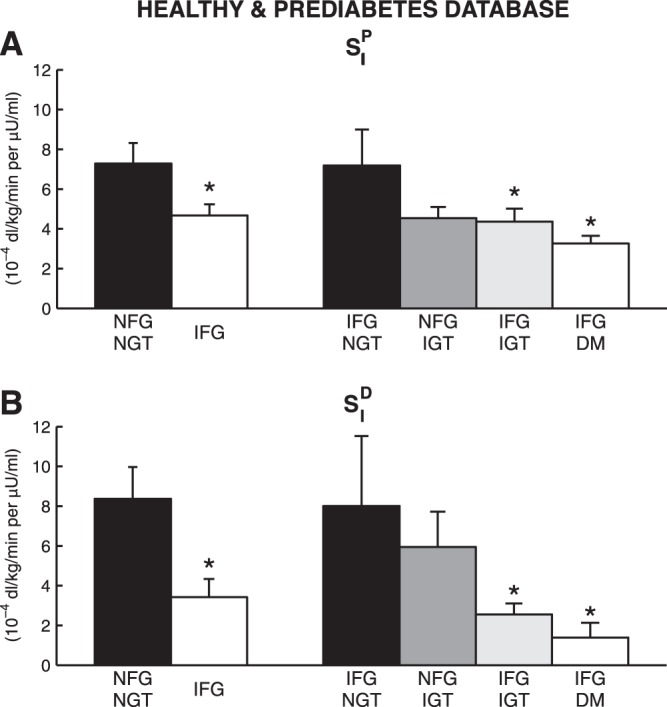
SIP (A) and disposal insulin sensitivity (SID) (B) estimated with OMMPD in healthy and prediabetes database, i.e., subjects with normal fasting glucose/normal glucose tolerance (NFG/NGT; n = 16), impaired fasting glucose (IFG; n = 32), IFG/NGT (n = 7), NFG/impaired glucose tolerance (IGT) (n = 12), IFG/IGT (n = 17), and IFG/DM (n = 8). *P < 0.05 vs. NFG/NGT.
DISCUSSION
The labeled euglycemic hyperinsulinemic clamp is the gold standard method to measure SIP in humans. However, this procedure is laborious, thus precluding its use in large-scale studies. In addition, during a clamp the liver is exposed to a nonphysiological glucose and insulin milieu. Conversely, the oral tests (OGTT and meal), besides being easier to perform, better reproduce physiological conditions and are thus potentially usable to assess SIP index in vivo. Currently, the state-of-the-art method to estimate SIP during a meal employs a dual-tracer approach. The first tracer, ingested with the meal, allows the segregation of plasma glucose into its endogenous and exogenous components; the second tracer, infused intravenously, mimics EGP time course and allows EGP estimation in a model-independent way (5). The EGP is then used to identify a mathematical model [recently validated against clamp (13)], providing an estimate of SIP. This technique requires the use of two isotopes. Moreover, achieving a quasi-constant TTR, thus obtaining a model-independent estimate of EGP, is not an easy task. Hence, an alternative, less labor-intensive method to assess insulin action on EGP is highly desirable.
Thus the purpose of this study was to develop a method to estimate SIP from a single-tracer oral test (OMMPD). To do so, we incorporated the EGP model of Dalla Man et al. (14) into the oral minimal model (12) and identified it on the data of 20 healthy subjects studied with a triple-tracer mixed meal (4). The model fitted the data well, and its parameters were estimated with good precision. Furthermore, SIP obtained with OMMPD from a single-tracer meal performed well against that estimated with a dual-tracer method and the EGP model (Fig. 4).
The performance of OMMPD was also judged in terms of EGP prediction; the comparison of model-derived EGP with that derived from triple-tracer was satisfactory despite EGPb being slightly overestimated from the model (Fig. 5, top left), as expected. This is likely due to the inability of a single-compartment model to correctly measure the steady-state plasma clearance rate, and certainly it represents a limitation of the OMMPD. Nevertheless, the comparison with the model-independent profiles definitely improved if one looks at the EGP/EGPb time course (Fig. 5, bottom left).
To test the performance of OMMPD also in subjects with quite varying stages of prediabetes into type 2 diabetes, we applied OMMPD on a population of IFG and NFG subjects, classified on the basis of their glucose tolerance, i.e., normal (NGT), impaired (IGT), or diabetes (DM) (6). For all of these subjects, GEZID was available from the analysis performed by Bock et al. (6). Hence, it was considered as appropriate to apply OMMPD on this population, setting GEZID at the respective average value (GEZID = 0.017 ± 0.001 dl·kg−1·min−1). It is of note that, even if GEZID distributes among the subjects with a certain degree of variability, the use of the parameter mean value is sufficient to have a satisfactory model performance. In fact, SIP and SID estimates in the subgroups reflected the pattern observed by Bock et al. (6) (Fig. 6). In particular, whole body SI obtained with OMMPD, derived as SI = SIP + SID, was higher than the triple-tracer-related values, as expected, due to the significant contribution of SIP.
Moreover, similarly to what was observed in the healthy subjects, OMMPD predicted the EGP/EGPb time course well (Fig. 5, bottom right) and provided values of EGP suppression (S%), which were identical to those derived from the triple-tracer data. Therefore, using the more appropriate GEZID value, we were able to successfully apply OMMPD on different populations.
Despite the satisfactory results, the proposed method has limitations. First of all, the prediction of EGP suffers from the EGPb overestimation. As a consequence, the Rd also is partially incorrect, since it consists of its endogenous (Rd end) and exogenous (Rd exo) components. To have good estimation of EGP, an intravenous tracer needs to be infused to extract the individual EGPb, or regression models could be used to derive EGPb on the basis of patient's characteristics (basal concentrations, anthropometric measurements, etc.). However, most of the clinical studies are focused on quantifying the EGP suppression, and thus the provided EGP/EGPb could be sufficient for this purpose. However, this limitation does not affect the estimation of either SIP, which was proven to be in agreement with that of Dalla Man et al. (14), or SID, which was independent of EGPb, as derived from Rd exo.
It is important to point out that the proposed model assumes that EGP depends on plasma glucose and a delayed insulin action but does not account for a direct effect of plasma insulin. As discussed in Dalla Man et al. (14), given the similarity between patterns of portal insulin and plasma glucose, a model that includes direct effect of both plasma insulin and glucose is not identifiable; i.e., it is unable to distinguish the two components of glucose effectiveness on the liver. On the other hand, considering plasma glucose also as a surrogate of portal insulin allows a better description of EGP suppression, particularly in the last portion of the meal. Moreover, the EGP model assumes that both the direct control of plasma glucose and the indirect control of portal insulin on EGP suppression are regulated by the same parameter, i.e., kG. However, this problem can be overcome by interpreting the parameter GEP as an overall measure of the ability of glucose to inhibit EGP both directly and indirectly. It is also important to underline that, in calling SIP as index of hepatic insulin sensitivity, a strong correlation between portal insulin action and SIP itself is implied. To support this assumption, we compared SIP with portal insulin action (i.e., parameter k3 of Eq. 12 reported in Ref. 14), and indeed, a good correlation between the two was found (r = 0.85, P < 0.0001).
Another limitation concerning the minimal model is that the distribution volume (V) needs to be fixed. Therefore, error in the assumed V might introduce error in estimation of Ra meal and Rd, as already stated in Basu et al. (5). However, such methods represent the state of the art for the estimation of postprandial glucose fluxes, and a previous work revealed that V can be fixed to the population average without introducing appreciable bias (15).
Finally, the model requires an observation interval of ≥240 min. To apply OMMPD on shorter protocols, such as the reduced OGTT (120 min), we would need to modify the description of Ra meal, as described previously (11). Moreover, prior studies have shown that fixing first-pass hepatic extraction of glucose absorption may affect whole body insulin sensitivity. However, as shown in Fig. 4, SIP estimated with the model compares well with that estimated from EGP model presented in Dalla Man et al. (14), which is independent from the first-pass hepatic extraction.
In conclusion, we have presented a model capable of reliably estimating both production and disposal contribution of insulin sensitivity index from a single-tracer meal (or OGTT) protocol in both normal individuals and individuals with prediabetes. The single-tracer approach, being less complex than the dual-tracer meal and labeled euglycemic hyperinsulinemic clamp methods, is a candidate as the method of choice to measure the index SIP in clinical studies.
GRANTS
This study was supported by National Institute of Diabetes and Digestive and Kidney Diseases Grant DK-29953 and Italian Ministero dell'Università e della Ricerca, FIRB 2008.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
R.V., C.D.M., and C.C. conception and design of research; R.V. analyzed data; R.V. prepared figures; R.V. and C.D.M. drafted manuscript; R.V., C.D.M., R.B., A.B., R.A.R., and C.C. edited and revised manuscript; R.V., C.D.M., R.B., A.B., R.A.R., and C.C. approved final version of manuscript; R.B. and A.B. performed experiments.
REFERENCES
- 1.Abdul-Ghani MA, Matsuda M, Balas B, DeFronzo RA. Muscle and liver insulin resistance indexes derived from the oral glucose tolerance test. Diabetes Care 30: 89–94, 2007. [DOI] [PubMed] [Google Scholar]
- 2.Barrett P, Bell B, Cobelli C, Golde H, Schumitzky A, Vicini P, Foster D. SAAM II: simulation, analysis, and modeling software for tracer and pharmacokinetic studies. Metabolism 47: 484–492, 1998. [DOI] [PubMed] [Google Scholar]
- 3.Basu A, Dalla Man C, Basu R, Toffolo G, Cobelli C, Rizza RA. Effects of type 2 diabetes on insulin secretion, insulin action, glucose effectiveness, and postprandial glucose metabolism. Diabetes Care 32: 866–872, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basu R, Dalla Man C, Campioni M, Basu A, Klee G, Jenkins G, Toffolo G, Rizza RA, Cobelli C. Effect of age and sex on postprandial glucose metabolism: difference in glucose turnover, insulin secretion, insulin action, and hepatic insulin extraction. Diabetes 55: 2001–2014, 2006. [DOI] [PubMed] [Google Scholar]
- 5.Basu R, Di Camillo B, Toffolo G, Basu A, Shah P, Vella A, Rizza R, Cobelli C. Use of a novel triple-tracer approach to assess postprandial glucose metabolism. Am J Physiol Endocrinol Metab 284: E55–E69, 2003. [DOI] [PubMed] [Google Scholar]
- 6.Bock G, Dalla Man C, Campioni M, Chittilapilly E, Basu R, Toffolo G, Cobelli C, Rizza R. Pathogenesis of pre-diabetes: mechanisms of fasting and postprandial hyperglycemia in people with impaired fasting glucose and/or impaired glucose tolerance. Diabetes 55: 3536–3549, 2006. [DOI] [PubMed] [Google Scholar]
- 7.Caumo A, Vicini P, Zachwieja JJ, Avogaro A, Yarasheski K, Bier DM, Cobelli C. Undermodeling affects minimal model indexes: insights from a two-compartment model. Am J Physiol Endocrinol Metab 276: E1171–E1193, 1999. [DOI] [PubMed] [Google Scholar]
- 8.Cobelli C, Carson ER. Introduction to Modeling in Physiology and Medicine. San Diego, CA: Academic, 2008. [Google Scholar]
- 9.Cobelli C, Dalla Man C, Toffolo G, Basu R, Vella A, Rizza RA. The oral minimal model method. Diabetes 63: 1203–1213, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cobelli C, Foster D, Toffolo G. Tracer Kinetics in Biomedical Research: From Data to Model. New York: Kluwer Academic/Plenum Publishers, 2000. [Google Scholar]
- 11.Dalla Man C, Campioni M, Polonsky KS, Basu R, Rizza RA, Toffolo G, Cobelli C. Two-hour seven-sample oral glucose tolerance test and meal protocol: minimal model assessment of beta-cell responsivity and insulin sensitivity in nondiabetic individuals. Diabetes 54: 3265–3273, 2005. [DOI] [PubMed] [Google Scholar]
- 12.Dalla Man C, Caumo A, Basu R, Rizza R, Toffolo G, Cobelli C. Measurement of selective effect of insulin on glucose disposal from labeled glucose oral test minimal model. Am J Physiol Endocrinol Metab 289: E909–E914, 2005. [DOI] [PubMed] [Google Scholar]
- 13.Dalla Man C, Piccinini F, Basu R, Basu A, Rizza RA, Cobelli C. Modeling hepatic insulin sensitivity during a meal: validation against the euglycemic hyperinsulinemic clamp. Am J Physiol Endocrinol Metab 304: E819–E825, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dalla Man C, Toffolo G, Basu R, Rizza RA, Cobelli C. Use of labeled oral minimal model to measure hepatic insulin sensitivity. Am J Physiol Endocrinol Metab 295: E1152–E1159, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dalla Man C, Yarasheski KE, Caumo A, Robertson H, Toffolo G, Polonsky KS, Cobelli C. Insulin sensitivity by oral glucose minimal models: validation against clamp. Am J Physiol Endocrinol Metab 289: E954–E959, 2005. [DOI] [PubMed] [Google Scholar]
- 16.DeFronzo RA, Tobin JD, Andres R. Glucose clamp technique: a method for quantifying insulin secretion and resistance. Am J Physiol Endocrinol Metab Gastrointest Physiol 237: E214–E223, 1979. [DOI] [PubMed] [Google Scholar]
- 17.Firth RG, Bell PM, Marsh HM, Hansen I, Rizza RA. Postprandial hyperglycemia in patients with noninsulin-dependent diabetes mellitus. Role of hepatic and extrahepatic tissues. J Clin Invest 77: 1525–1532, 1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Radziuk J, Norwich KH, Vranic M. Experimental validation of measurements of glucose turnover in nonsteady state. Am J Physiol Endocrinol Metab Gastrointest Physiol 234: E84–E93, 1978. [DOI] [PubMed] [Google Scholar]