Abstract
Lichen-forming fungal proteins have been seldom searched due to many difficulties in their extraction. Phenols, quinones, proteases, and other components released during cell disruption have been known to be the greatest challenges related to protein extraction from lichens. To overcome these problems and maintain good electrophoretic resolution and high protein concentration, an extraction buffer containing polyvinylpolypyrrolidone, ascorbic acid, Triton X-100, polyethylene glycol, proteinase, and oxidase inhibitors in sodium phosphate buffer was developed. This extraction buffer showed high efficiency for all lichen species tested in the study.
Keywords: Electrophoresis, Optimization method, Phenols, Proteins, Quinones
Lichens are a symbiotic organism found in association with fungi, algae, and cyanobacteria, or possibly all three. Lichens have been known to produce many unique compounds and thus considered to be valuable source for commercially interesting compounds. Lichens and their natural products have been used in a wide variety of applications throughout human history. With aid of advanced analytical technology, discovering and utilizing useful metabolites of lichens became feasible nowadays s, and, accordingly, attention to lichen proteins has increased in recent years [1,2].
Although lichen proteins are no longer used for chemotaxonomy, they are currently considered as potential biologically active substances with unique biochemical properties. Recently, new candidate compounds for drugs (antibiotics and proteins with anti-prion activity), UVB protection, oxidative enzymes for the pulp industry, and antifreeze proteins for frozen foods, as well as proteins from symbiotic cyanobacteria have been discovered [3,4]. Nevertheless, the proteome of lichens has rarely been studied and is still not completely elucidated.
Lichen proteome studies are hampered by several factors. Specifically, lichens have a small volume of cytoplasm, low protein content, and strong cell walls. However, the main problem associated with such studies is the presence of large amounts of phenolic compounds and the high activity of their proteolytic and oxidative enzymes such as phenoloxidases. Phenoloxidases catalyze the aerobic oxidation of certain phenolic compounds to quinones, which subsequently leads to auto-oxidation resulting in dark brown pigments or to the formation of insoluble dark-colored melano-protein complexes. In addition to covalent interactions involving oxidative enzymes, phenolic compounds can react with proteins by forming hydrogen or ionic bonds and via hydrophobic interactions. In all cases, these interactions result in changes to the properties and activities of proteins [5,6,7]. However, lichen thalli are promising sources of biologically active proteins that have been virtually unstudied to date. Currently, there is no universal procedure for the extraction of proteins from lichens. Thus, this study was conducted to develop an effective procedure for protein extraction from lichens.
MATERIALS AND METHODS
Lichen material
The thalli of five lichen species from different taxonomic groups were used in this study. All samples were macrolichens (fruticose or foliose lichens), which have sufficient biomass to extract high quantities of proteins for further analysis compared to microlichens (crustose lichens). The five lichens were collected in different continents and contain many different lichen acids. The samples of Usnea antarctica Du Rietz were collected from King Sejong Antarctic Research Station, Antarctica; Pseudocyphellaria faveolata (Delise) Malme samples were collected from Lago Balmaceda & Lago Pinto, Patagonia, South America; and Cetraria ericetorum Opiz, Evernia divaricata (L.) Ach., and Flavocetraria cucullata (Bellardi) Kärnefelt & A. Thell samples were collected from Lacul Bolboci, Mt. Bucegi, Romania, Europe. All samples were stored at 4℃ until further analysis.
Protein extraction
To remove extracellular acetonesoluble phenolics, the lichen thalli were washed several times with acetone (approximately 25 mL/g) and then air-dried. The samples were subsequently ground in a cold mortar with liquid nitrogen until a very fine powder was obtained. Next, 100 mg of this powder was amended with ascorbic acid, polyvinylpolypyrrolidone (PVPP), and 1 mL of extracting solution (1 : 10 weight : volume) containing 5 mM ethylenediaminetetraacetic acid (EDTA), 2 mM phenylmethylsulfonyl fluoride (PMSF), and a protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN, USA) at concentrations recommended by the manufacturer. Extraction was then conducted by stirring the samples at 180 rpm and 15℃ for 2 hr. The samples were subsequently centrifuged at 10,000×g for 30 min, after which the pellet was discarded and the supernatant was centrifuged again if necessary. Finally, the absorbance of the supernatant was measured at 280 nm and 320 nm using an Optizen 3220UV spectrophotometer (Mecasys Co., Daejeon, Korea).
Sample preparation
Proteins in the obtained extracts were precipitated with trichloroacetic acid (TCA) at a final concentration of 12.5%. The protein precipitates were then washed three times with acetone and dissolved in a minimum amount of 0.1M phosphate-buffered saline (PBS), pH 7.4, or in buffer for electrophoretic samples.
Electrophoresis
Sodium dodecyl sulfate (SDS)-polyacrylamide gel (PAAG) electrophoresis was conducted on 12% acrylamide gels containing 0.2% bisacrylamide using the buffer system described by Laemmli [8]. The electrophoresis conditions were 20 mA for 10 min followed by 35 mA for 30~40 min. The gels were then treated with 50% ethanol and 10% acetic acid for at least 1 hr to fix the proteins. Proteins were detected by silver staining using the method described by Heukeshoven and Dernick [9], with minor modifications. Qualitative and quantitative analysis of electropherograms was performed using TotalLab 2.01 (MH Biotech Solutions, http://biosol2000.tripod.com).
Statistical analysis
The experimental results presented in the graphs are expressed as the mean ± SD for all studied lichens species. Data were analyzed using SPSS ver. 18.0 (SPSS Inc., Chicago, IL, USA) and a p < 0.05 was considered significant for all analyses. Differences between treatments were analyzed by one-way ANOVA followed by Tukey's honest significant difference. All experiments were performed in triplicate. Representative data are shown for the corresponding experiments.
RESULTS AND DISCUSSION
Extracting solution
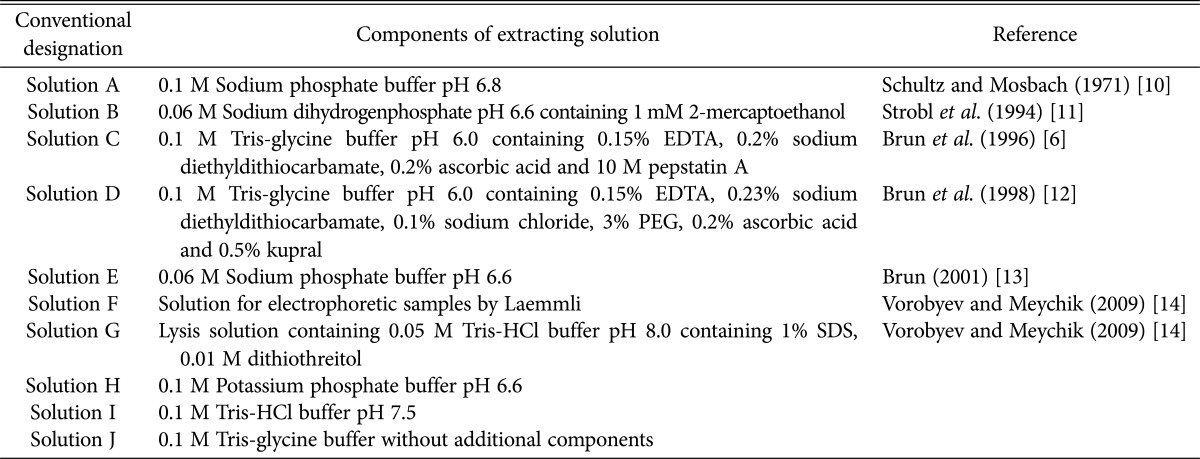
There are few descriptions for obtaining protein extracts from lichen thalli, and each recommends the use of different solutions. A list of these solutions and conventional designation letter codes, which are referred to hereafter, is provided in Table 1 [6,10,11,12,13,14].
Table 1. Solutions for protein extraction from lichen.
SDS, sodium dodecyl sulfate.
The solutions shown in Table 1 (with slight modifications) were used to prepare protein extracts from the lichen thalli of U. antarctica, P. faveolata, C. ericetorum, E. divaricata, and F. cucullata. In addition, 0.1M potassium phosphate buffer at pH 6.6 (solution H), 0.1M Tris-HCl buffer at pH 7.5 (solution I), and 0.1M Tris-glycine buffer without additional components (solution J) were used for comparison of extraction properties.
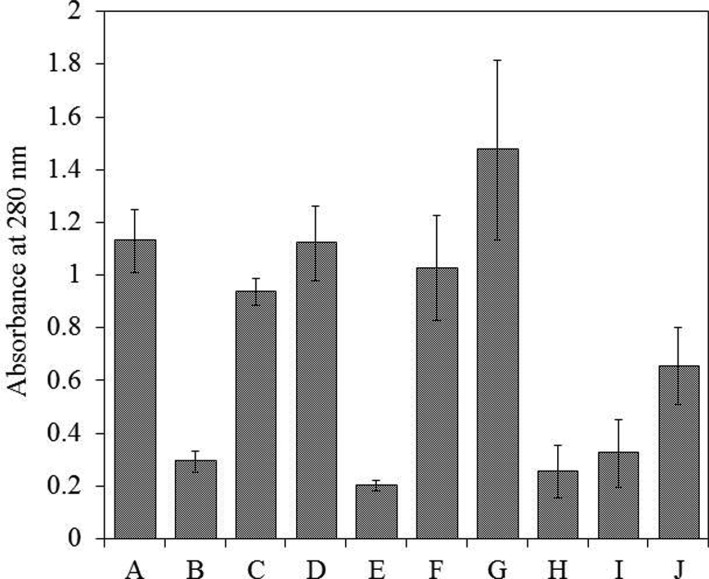
Effectiveness of extraction was assessed based on the protein concentration in the extracts, which was determined by the absorbance of the solution and by the density of protein bands on the PAAG. The absorbance was relatively high in extracts prepared using solutions A, F, and G, whereas solutions B, E, H, I, and J showed low extraction properties. The multicomponent solutions C and D showed increased absorbance of extracts when compared with solution J (Fig. 1). These findings indicate that the presence of additional components in the extracting solution improved the extraction. Despite the high absorbance, we excluded the possibility of using a lysis solution (G) and electrophoretic buffer (F) for samples because they contain SDS, and its removal can result in a portion of the protein being lost or irreversibly denatured. Moreover, the presence of excess SDS led to a strong background signal on the gel resulting in poor resolution of the protein bands. Electrophoresis of the samples prepared using solutions without inhibitors (A, B, E, H, I, and J) resulted in an almost complete absence of protein band resolution. The addition of inhibitors to these solutions improved the resolution; however, there was a strong background signal, which made it impossible to identify the individual protein bands clearly. This phenomenon may have been due to the presence of phenolic compounds and their derivatives, which prevented the resolution of proteins on the PAAG [5,7]. The best results were obtained using solutions C and D, but the protein bands were still unclear and only present in low amounts. Overall, sodium phosphate buffer (solution A) provided the highest concentration of protein in the extract. Therefore, we concluded that the optimal extracting solution should be based on a phosphate buffer system and contain components that can effectively reduce the influence of phenolics and derivatives in the extract, as well as promote the resolution of high-quality protein bands on the PAAG.
Fig. 1. Effect of extraction buffer on the amount of extracted proteins. The amount of extracted proteins was measured by the absorbance at 280 nm. The buffer compositions are summarized in Table 1. Values with the same letter are not significantly different according to Tukey's honest significant difference test. Error bars represent the standard deviations.
Effects of phenolic compounds and quinones on protein extraction
To avoid interaction of phenols and quinones with proteins, we used the protein extraction protocols described for plant tissues that are rich in phenolic compounds [7,15,16,17]. The main approach used in the isolation of plant proteins is the removal of phenolic compounds and other secondary metabolites as rapidly as possible while concurrently preventing the formation of covalent complexes formed between quinones and proteins (e.g., phenoloxidases inhibition).
Various polymers can be used for the removal of phenolic compounds, including polyvinylpyrrolidone (PVP) and water-insoluble PVPP [5,7,15,17]. The use of PVP-40 in the process of protein extraction from lichens was not successful, because TCA precipitation resulted in an extremely thick, insoluble precipitate. However, when PVPP was used, the properties of this polymer were not changed in the presence of TCA, and it could be removed from the protein extract by centrifugation.
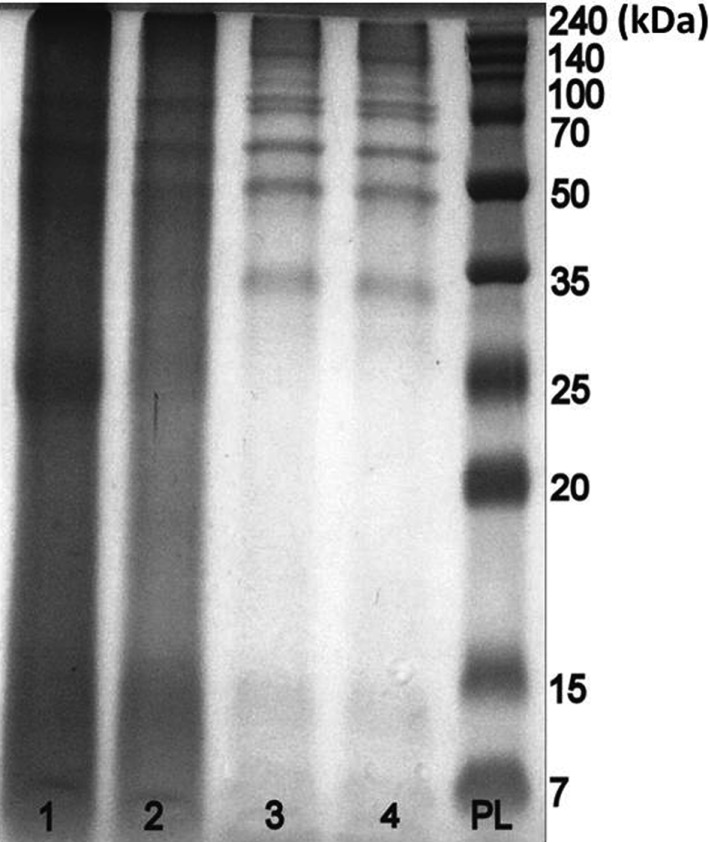
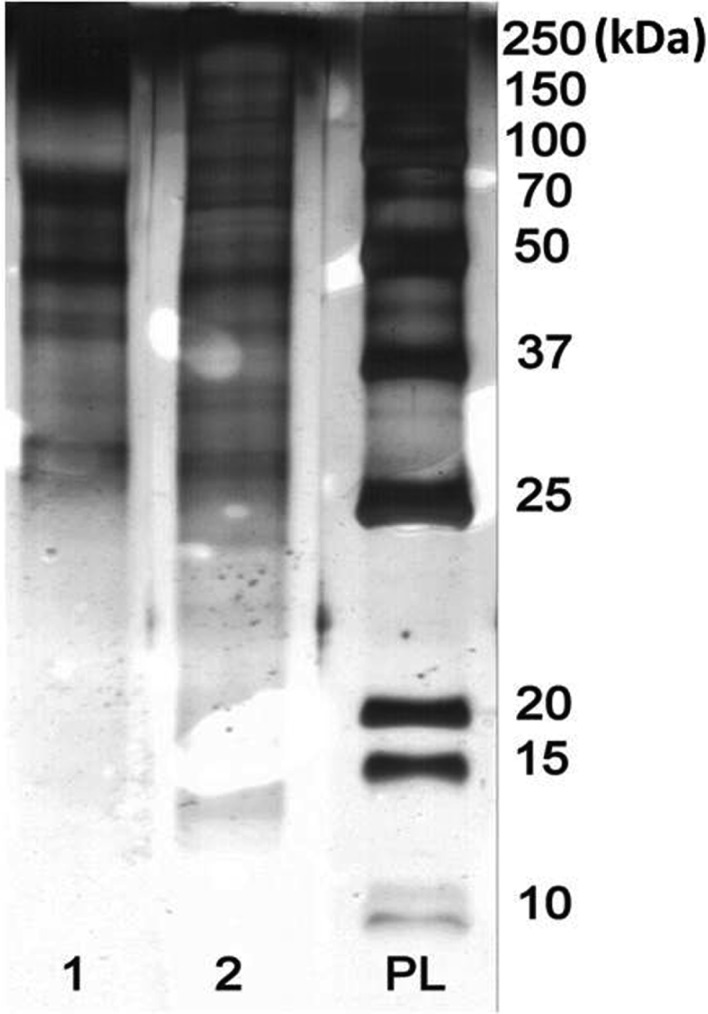
Ascorbic acid reduces quinones, leading to their regeneration to phenols. To obtain a colorless extract with satisfactory resolution on the PAAG, the concentration of ascorbate must be different for each lichen species when ascorbic acid is used in the process of protein extraction from lichens. In the present study, 5 mg/mL was used for extracts from E. divaricata, 70 mg/mL was used for P. faveolata, and 50 mg/mL was used for the other species. Ascorbic acid was added to the ground material in dry form immediately before addition of the buffer solution. Colorless extracts with better resolution on the PAAG were obtained using 1% PVPP in combination with an effective amount of ascorbic acid than when these components were used separately (Fig. 2).
Fig. 2. Effect of different additives on the extraction of Pseudocyphellaria faveolata proteins. Lane 1, 0.1M phosphate-buffered saline at pH 7.4 containing 0.2% Triton X-100, 1 mM polyethylene glycol 8000, and inhibitors; lane 2, in the presence of 1% polyvinylpolypyrrolidone (PVPP); lane 3, both 1% PVPP and 70 mg/mL ascorbic acid; lane 4, or ascorbic acid alone; PL, protein ladder (7~240 kDa).
Effect of buffer concentration and pH on protein extraction
Subsequent optimization of the extracting solution was conducted to achieve a higher quality of the extract and protein band resolution on the PAAG.
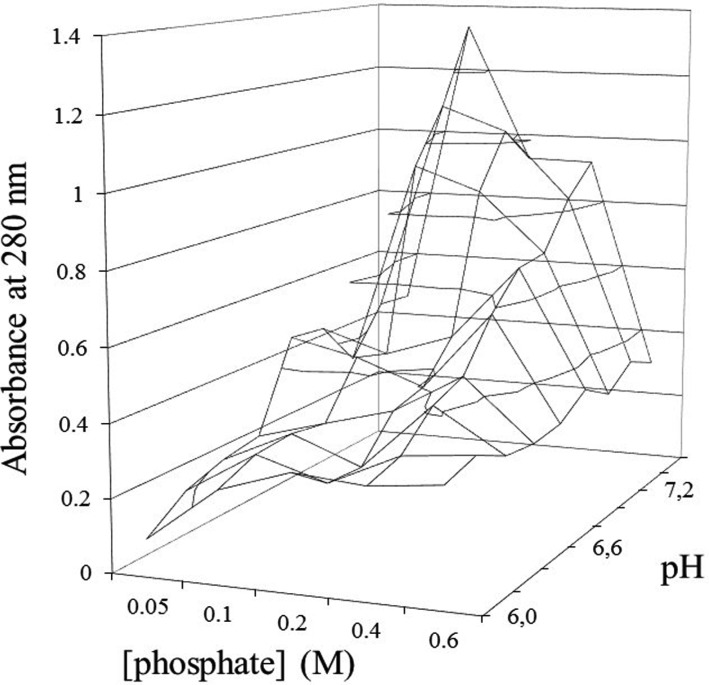
Initially, the optimum pH of the buffer solution was determined. Phosphate buffer solutions cover a range of pH values from 6.0 to 7.7. Comparison of the protein extraction efficiency using sodium phosphate buffer solutions with different molar concentrations and pH values indicated that the optimum parameters did not differ greatly among the tested lichens. Specifically, the maximum protein concentration in the extract from E. divaricata was observed using the extracting solution at 0.1M and pH 7.6 (Fig. 3). The maximum protein concentrations in extracts from U. antarctica and F. cucullata were observed at 0.2M and pH 7.4, and at 0.1M and pH 7.4 for P. faveolata, and at 0.1M and pH 7.2 for C. ericetorum. Such differences are important in obtaining a large amount of protein for any targeted study. However, these differences were ignored in the present study since we were attempting to optimize the overall system. Accordingly, a 0.1M sodium phosphate buffer at pH 7.4 containing inhibitors and other components was used as the universal extracting solution for lichens.
Fig. 3. Dependence of the protein extraction on pH and the concentration of phosphate buffer.
Effects of additives on protein extraction
The glycine and sodium chloride contents in the extracting solution (as in those contained in solutions C and D) did not influence the results of the extraction.
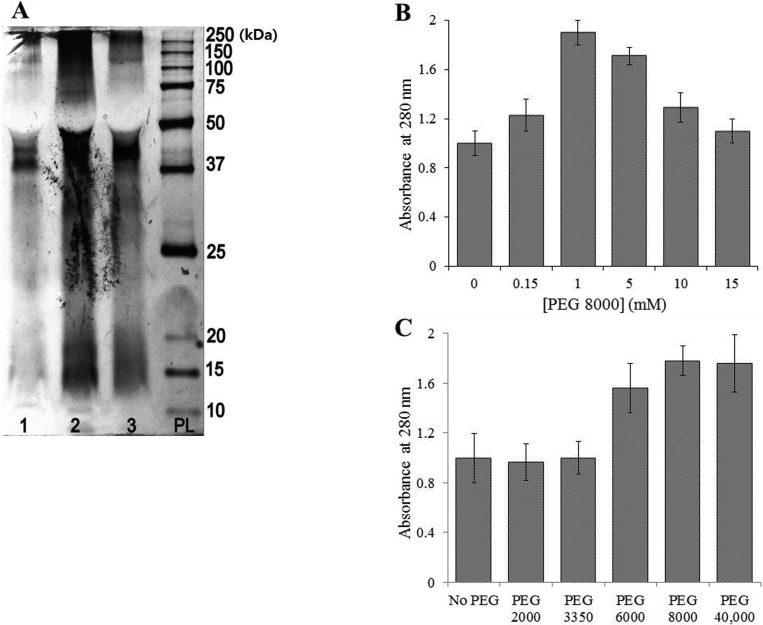
The presence of polyethylene glycol (PEG), which has exclusion properties, in the extraction solution increased the efficiency of the extraction process. Comparison of the efficacy of PEG 2000, PEG 3350, PEG 6000, PEG 8000, and PEG 40,000 revealed that the greatest efficiency occurred when PEG 6000 and PEG 8000 were used. PEG 2000 and PEG 3350 had almost no effect on the extraction parameters. The use of PEG 40,000 significantly increased the concentration of the protein, but also created an artifact on the electropherograms, thereby preventing effective separation of proteins on the PAAG (Fig. 4). In addition, PEG 8000 at 1 mM was found to be the most effective concentration among the tested range from 0.15 mM to 15.0 mM (Fig. 5).
Fig. 4. Effect of polyethylene glycol (PEG) on the extraction of lichen proteins. A, Sodium dodecyl sulfate-polyacrylamide gel electrophoresis image of the protein extracts obtained using a solution containing PEG 40,000: 1, Cetraria ericetorum; 2, Evernia divaricata; 3, Flavocetraria cucullata; PL, protein ladder (10~250 kDa); B, The dependence of the extract absorbance on concentrations of PEG; C, Effect of the molecular weight of PEG on protein concentration in the extracts. Values with the same letter are not significantly different according to Tukey's honest significant difference test. Error bars represent the standard deviations.
Fig. 5. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis image of the protein extracts obtained using 0.1M sodium phosphate buffer, pH 7.4: 1, containing 1% polyvinylpolypyrrolidone, 5% ascorbic acid, 0.2% Triton X100, and 1 mM polyethylene glycerol 8000; 2, P-PER (commercial protein extraction kit; Thermo); PL, protein ladder (7~240 kDa) (silver staining).
The presence of detergents in the extracting solution improved the results of extraction. However, extracts containing Tween 20 caused the saturated color to change from yellow to brown. No such effect occurred when Triton X-100 was used. The presence of detergent had no effect on the protein concentration, but improved separation of the precipitate from the extract upon centrifugation. The protein resolution on the PAAG was better when Triton X-100 was used than when Tween 20 was used.
Overall, the results of this study indicate that it is necessary to carefully select the conditions for achieving the maximum effective extraction of proteins for each individual lichen, especially with respect to the amount of ascorbic acid and buffer solution parameters such as the pH and molarity. However, we suggest the use of 0.1M sodium phosphate buffer at pH 7.4 containing 0.2% Triton X-100, 1 mM PEG 8000, an oxidase, protease inhibitors (EDTA, PMSF, inhibitor cocktail), 1% PVPP, and 5% ascorbic acid, which should be added immediately before extraction, as a universal solution for the extraction of proteins from lichen thalli.
Hearst et al. [18] described the use of a plant protein extraction kit (P-PER; Thermo, Rockford, IL, USA) to extract proteins from fungi. Comparison of the extraction properties using P-PER with the results obtained using our recommended solution revealed that the protein concentration in the P-PER extract was lower than that obtained in the PBS extract, but the protein resolution on the PAAG showed superior quality than that of the PBS extract. It should be noted that the use of commercial kits is ideal for characterization of the protein composition and investigation of proteomes by electrophoresis and isoelectric focusing. However, since manufacturers often do not indicate the exact composition of their proposed solutions, in our opinion, it is better to use solutions for which all components and concentrations are known for investigations of the biological activity and properties of proteins.
The aim of this study was to optimize the protein extraction from lichens. The results presented herein serve as a first step towards identification of new biologically active proteins from lichens.
ACKNOWLEDGEMENTS
This work was supported by a grant from the National Research Foundation of Korea (2013K2A4A1043370) and the Korea National Research Resource Center Program (NRF, 2012M3A9B8021726). We are grateful to Dr. J.A. Kim (Sunchon, South Korea) for kind support during this work, and to Dr. S.-O. Oh (Sunchon, South Korea) for providing fresh material.
References
- 1.Rodriguez CM, Bennett JP, Johnson CJ. Lichens: unexpected anti-prion agents? Prion. 2012;6:11–16. doi: 10.4161/pri.6.1.17414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zambare VP, Christopher LP. Biopharmaceutical potential of lichens. Pharm Biol. 2012;50:778–798. doi: 10.3109/13880209.2011.633089. [DOI] [PubMed] [Google Scholar]
- 3.Oksanen I. Ecological and biotechnological aspects of lichens. Appl Microbiol Biotechnol. 2006;73:723–734. doi: 10.1007/s00253-006-0611-3. [DOI] [PubMed] [Google Scholar]
- 4.Anshakova VV, Karataeva EV, Kershengoltc BM. Bakery products quality improvement by means of the mechanically activated bioadditives from lichens. Fund Res. 2011;8:593–596. [Google Scholar]
- 5.Gan YY. Preparation of extracts from tropical plants for electrophoresis: an analysis of methodology. Pertanika. 1985;8:9–20. [Google Scholar]
- 6.Brun GO, Navrotskaya IL, Gizbullina VK. Protein complexes of some species of lichens of genus Cladina Nyl. Ukr Bot J. 1996;53:68–73. [Google Scholar]
- 7.Pierpoint WS. The extraction of enzymes from plant tissues rich in phenolic compounds. Methods Mol Biol. 1996;59:69–80. doi: 10.1385/0-89603-336-8:69. [DOI] [PubMed] [Google Scholar]
- 8.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 9.Heukeshoven J, Dernick R. Simplified method for silver staining of proteins in polyacrylamide gels and the mechanism of silver staining. Electrophoresis. 1985;6:103–112. [Google Scholar]
- 10.Schultz J, Mosbach K. Studies on lichen enzymes. Purification and properties of an orsellinate depside hydrolase obtained from Lasallia pustlata. Eur J Biochem. 1971;22:153–157. doi: 10.1111/j.1432-1033.1971.tb01526.x. [DOI] [PubMed] [Google Scholar]
- 11.Strobl A, Türk R, Thalhamer J. Investigations on the protein composition of the lichen Pseudevernia furfuracea (L.) Zopf var. ceratea (Ach.) Hawksw. from different altitudes. Phyton. 1994;34:67–83. [Google Scholar]
- 12.Brun GO, Navrotskaya IL, Gizbullina VK. Chemotaxonomical study of the lichen genera Cladonia Hill ex Brown and Cladina (Nyl.) Harm. Kyiv: M.H. Kholodny Institute of Botany; 1998. [Google Scholar]
- 13.Brun GO. Comparative investigation on proteins of Cladonia arbuscula (Wallr.) Flot. subsp. mitis (Sandst.) Ruoss. Ukr Bot J. 2001;58:210–215. [Google Scholar]
- 14.Vorobyov DV, Meychik NR. The composition of ion-exchange groups in the cell walls of mycobiont and photobiont comprising ternary lichen Peltigera aphthosa (L.) Willd. Immunopathol Allergol Infect. 2009;1:18–19. [Google Scholar]
- 15.King BJ, Lee LS, Rackemann RG, Scott PT. Preparation of extracts for electrophoresis from citrus leaves. J Biochem Biophys Methods. 1994;29:295–305. doi: 10.1016/0165-022x(94)90040-x. [DOI] [PubMed] [Google Scholar]
- 16.Giavalisco P, Nordhoff E, Lehrach H, Gobom J, Klose J. Extraction of proteins from plant tissues for two-dimensional electrophoresis analysis. Electrophoresis. 2003;24:207–216. doi: 10.1002/elps.200390016. [DOI] [PubMed] [Google Scholar]
- 17.Charmont S, Jamet E, Pont-Lezica R, Canut H. Proteomic analysis of secreted proteins from Arabidopsis thaliana seedlings: improved recovery following removal of phenolic compounds. Phytochemistry. 2005;66:453–461. doi: 10.1016/j.phytochem.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 18.Hearst M, Nelson D, McCollum G, Ballard LM, Millar BC, Moore S, McClean S, Moore JE, Rao JR. Antimicrobial properties of protein extracts from wild mushroom fungi and native plant species against hospital pathogens. J Pharmacogn Phytother. 2010;2:103–107. [Google Scholar]