Abstract
We report the isolation of a Gongronella butleri species and describe it based on the analysis of the internal transcribed spacer region of rDNA and morphological characteristics. G. butleri has been reported as a high chitosan producer in the literature. This is the first record of G. butleri isolated from crop field soil in Korea.
Keywords: Chitosan, Fungi, Gongronella butleri, Molecular identification, Morphology
The Gongronella spp. are among the most commonly occurring and economically important members of the Zygomycetes class. Chitosan is an important component of the cell wall of certain Zygomycetes fungi [1]. Among the various Zygomycetes fungi, Gongronella butleri (Lendner) Peyronel & Dal Vesco (a pin mould) is the most important fungus used in chitosan production. It has been reported that G. butleri produces the highest yield of chitosan [2]. Yokoi et al. [3] extracted 730 mg/L of chitosan from the G. butleri IFO 8081 strain grown for five days in sweet potato-shochu medium.
Chitosan with a molecular weight of 5~50 kDa has received enormous worldwide attention as one of the most promising renewable polymeric materials for extensive applications in industrial and agricultural fields such as cholesterol absorption [4] and semipermeable membrane production [5], and as an antifungal agent, a plant growth elicitor [6], a protocorm-like body formation enhancer in orchid tissue culture, and as a heavy metal chelator [7]. Fungal chitosan can also be employed in medical applications, where the absence of allergenic shellfish protein is dictated [8]. Therefore, G. butleri has been considered as an alternative source for the production of chitosan with a more consistent quality.
In a recent study of fungal strains isolated from crop field soils in Korea, the authors identified one isolate as a member of the Gongronella family that has not previously been reported in Korea. Based on morphological and molecular characteristics, this species was identified as G. butleri. In this report, we describe the isolation and identification of G. butleri from crop field soil in Korea.
Soil samples (0~15 cm) were collected from crop field soil in the Samcheok province in South Korea. The fungus was isolated by conventional dilution and grown for 7 days at 25℃ on potato dextrose agar (PDA; Difco Laboratories, Detroit, MI, USA) supplemented with chloramphenicol (bacteriostat; 100 µg/mL). The pure culture was maintained on PDA slants at 4℃. Genomic DNA of strain KNU14-13-1 was extracted using the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) following the manufacturer's instructions. The internal transcribed spacer (ITS) regions of rDNA were amplified using the primers ITS1 and ITS4 [9] and the amplified PCR product was purified using a QIAquick PCR purification Kit (Qiagen, Valencia, CA, USA) following the manufacturer's instructions. The PCR product was sequenced with an ABI Prism 3730 DNA analyzer (Applied Biosystems, Foster City, CA, USA). The sequence was compared with reference ITS1-ITS4 rDNA sequences in GenBank using BLAST analysis (http://www.ncbi.nlm.nih.gov/Blast). The nucleotide sequence has been deposited in NCBI-GenBank (accession No. KP055605). The sequences of closely related strains were aligned using the MultAlin program. The phylogenetic analysis was carried out by the neighbor-joining method using MEGA software [10] with the Kimura 2-parameter model. The robustness of the tree was evaluated by 1,000 bootstrap replications. Gongronella lacrispora (strain, ATCC 24412; accession No. GU244498) was used as the outgroup.
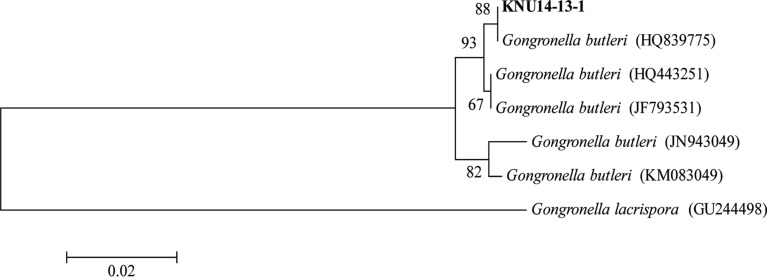
The acquired ITS sequence showed 100% similarity with a G. butleri (accession No. HQ839775) sequence available in the GenBank database, suggesting the isolate is related to G. butleri. Further, a phylogenetic tree constructed for the identification of fungus was pruned of distantly related taxa for clarity. The results revealed that the isolate was grouped in G. butleri with 88% bootstrap value support (Fig. 1). These results provide evidence that the isolate KNU14-13-1 is G. butleri.
Fig. 1. Neighbor-joining phylogenetic analysis of the partial 18S-ITS1-5.8S-ITS2-28S rDNA sequence of Gongronella butleri KNU14-13-1 obtained from crop field soil in Korea. The sequence obtained in the study is shown in boldface. Numerical values (> 50) on branches are the percentage of 1,000 bootstrap replicates that support the branch. Gongronella lacrispora was used as the outgroup. The scale represents the number of substitutions per site.
In order to confirm the molecular result, the morphology of the isolate KNU14-13-1 was compared with previous descriptions of G. butleri [11]. Only two Gongronella species exist, G. butleri and G. lacrispora Hesseltine & Ellis [12]. G. butleri was generally identified based on the following distinct characteristics: presence of rhizoids, swollen, globose apophyses growing beneath sporangia and featuring reduced columella, oval sporangiospores, and erect sporangiophores. The presence of oval sporangiospores and erect sporangiophores can distinguish G. butleri from the closest related species G. lacrispora. For morphological analysis, the strain KNU14-13-1 was grown on PDA at 25℃ for 10 days. Photomicrographs were taken with a Kodak14n digital camera (Tokyo, Japan) attached to the compound microscope or a scanning electron microscope. Slide material was mounted in water and sometimes stained with aniline blue.
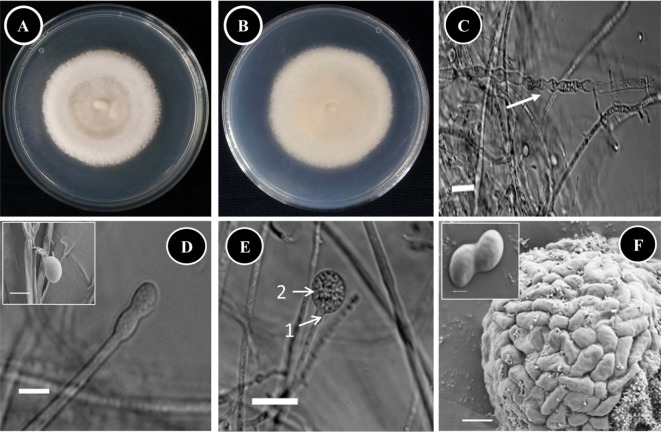
Taxonomic descriptions and microphotographs of morphological structures of the isolate are shown in Table 1 and Fig. 2. The colonies were about 3~4 mm high with a white to greyish or smokey-brown surface, and a pale brownish reverse (Fig. 2A and 2B). The colonies grew moderately rapidly, and matured within a week. The texture was woolly to cottony, and the hyphae were hyaline and septate (Fig. 2C). Intercalary and terminal chlamydospores were observed in substrate mycelium; they were smooth, thick walled, globose to ovoid-shaped, and 4.5~7 × 4.5~10 µm in size (Fig. 2C). Sporangiophores were simple or irregularly branched, hyaline, smooth to very faintly roughened, always with a septum under the apophysis beneath the sporangium and 3.0~5.5 µm in diameter (Fig. 2D and 2E). A hemispherical or cup-shaped apophysis was observed beneath the sporangium (Fig. 2E). The sporangia were globose or spherical, smooth and soluble wall and about 11.5~15.5 × 20.0~24.4 µm in diameter (Fig. 2E). Sporangiospores were smooth, oval to flattened on one side to reniform and 3.5~7.2 × 6.7~8.5 µm in size (Fig. 2F). Zygospores were not observed.
Table 1. Comparison of morphological characteristics of the study isolate with respect to reported Gongronella butleri characteristics.
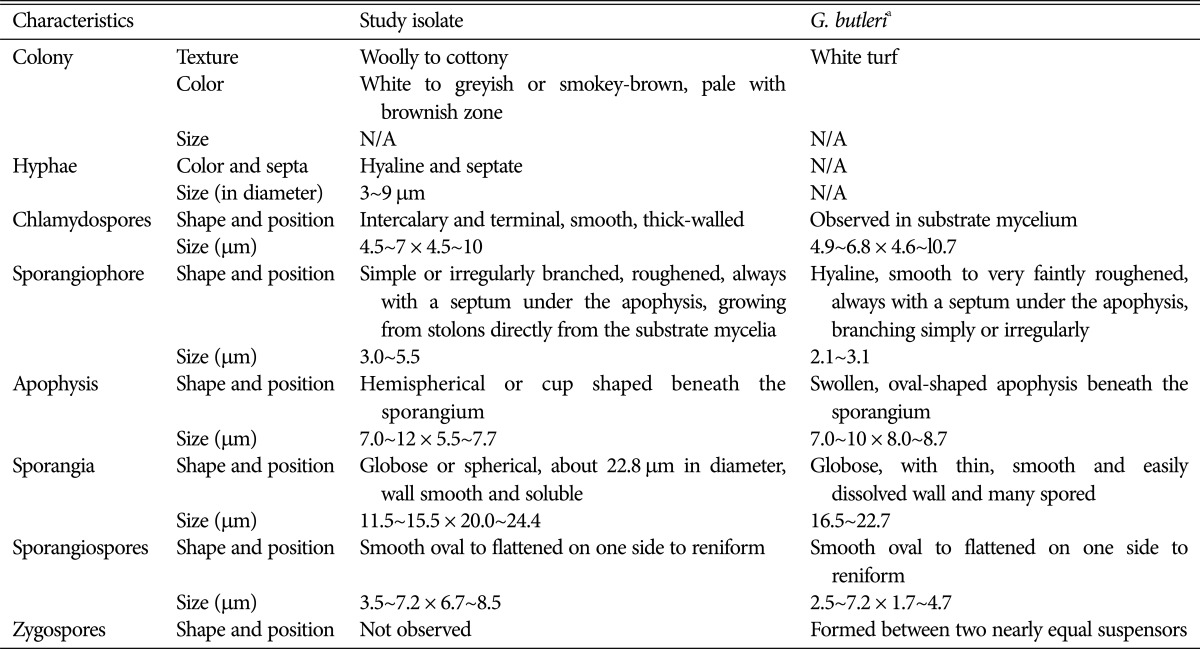
N/A, not available in the previous descriptions.
aSource of description [11].
Fig. 2. Morphology of Gongronella butleri KNU14-13-1 observed using a compound microscope and scanning electron microscope (SEM). A, Front side of the colony; B, Reverse side of the colony; C, Apical part of hyphae and chlamydospore formation during hyphal growth (white arrow) (compound microscope images); D, Immature sporangia (insert, sporangia; SEM micrograph) (compound microscope images); E, Maturing sporangia: 1, Apophysate; 2, Globose sporangia (compound microscope images); F, Aggregated sporangiospores in sporangia (SEM micrograph) (insert, sporangiospores; SEM micrograph) (scale bars: C, E = 30 µm, D = 20 µm, D insert, F = 5 µm, F insert = 1 µm).
The occurrence of G. butleri is taxonomically and ecologically remarkable, because G. butleri is the most well-known fungus to produce chitosan, which is currently being commercialized and applied in various industrial and agricultural areas. It is considered that the fungus isolated here from crop field soil may play an important role in the production of chitosan for industrial and agriculture applications; however, further studies on the production of chitosan by this fungus are needed.
ACKNOWLEDGEMENTS
This work was supported by the National Institute of Biological Resources under the Ministry of Environment, Republic of Korea, as part of a project to survey and excavate Korean indigenous fungal species.
References
- 1.Bartnicki-Garcia S. Cell wall chemistry, morphogenesis, and taxonomy of fungi. Annu Rev Microbiol. 1968;22:87–108. doi: 10.1146/annurev.mi.22.100168.000511. [DOI] [PubMed] [Google Scholar]
- 2.Tan SC, Tan TK, Wong SM, Khor E. The chitosan yield of Zygomycetes at their optimum harvesting time. Carbohydr Polym. 1996;30:239–242. [Google Scholar]
- 3.Yokoi H, Aratake T, Nishio S, Hirose J, Hayashi S, Takasaki Y. Chitosan production from shochu distillery wastewater by funguses. J Ferment Bioeng. 1998;85:246–249. [Google Scholar]
- 4.Ikeda I, Sugano M, Yoshida K, Sasaki E, Iwamoto Y, Hatano K. Effects of chitosan hydrolysates on lipid absorption and on serum and liver lipid concentration in rats. J Agric Food Chem. 1993;41:431–435. [Google Scholar]
- 5.Hirano S, Tobetto K, Hasegawa M, Matsuda N. Permeability properties of gels and membranes derived from chitosan. J Biomed Mater Res. 1980;14:477–485. doi: 10.1002/jbm.820140414. [DOI] [PubMed] [Google Scholar]
- 6.Pospieszny H, Chirkov S, Atabekov J. Induction of antiviral resistance in plants by chitosan. Plant Sci. 1991;79:63–68. [Google Scholar]
- 7.Nge KL, Nwe N, Chandrkrachang S, Stevens WF. Chitosan as a growth stimulator in orchid tissue culture. Plant Sci. 2006;170:1185–1190. [Google Scholar]
- 8.Badawy ME, Rabea EI. A biopolymer chitosan and its derivatives as promising antimicrobial agents against plant pathogens and their applications in crop protection. Int J Carbohydr Chem. 2011;2011:460381. [Google Scholar]
- 9.Govinda Rajulu MB, Thirunavukkarasu N, Babu AG, Aggarwal A, Suryanarayanan TS, Reddy MS. Endophytic Xylariaceae from the forests of Western Ghats, southern India: distribution and biological activities. Mycology. 2013;4:29–37. [Google Scholar]
- 10.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 11.Ho HM, Chen ZC. Morphological study of Gongronella butleri (Mucorales) from Taiwan. Taiwania. 1990;35:259–263. [Google Scholar]
- 12.Hesseltine CW, Ellis JJ. The genus Ahsidia: Gongronella and cylindrical-spored species of Absidio. Mycologia. 1964;56:568–601. [Google Scholar]