Background: Different methods are established for identifying prostate stem cells (P-SCs). However, the relationship of these P-SCs is not fully clear.
Results: Sphere-forming cells were from the basal compartment and also formed organoids. However, organoid-derived P-SCs cannot form prostaspheres.
Conclusion: The basal P-SCs represent more primitive P-SCs than luminal P-SCs.
Significance: The finding helps define the hierarchy of P-SCs.
Keywords: cell signaling, fibroblast growth factor (FGF), fibroblast growth factor receptor (FGFR), prostate, stem cells
Abstract
Prostate stem cells (P-SCs) are capable of giving rise to all three lineages of prostate epithelial cells, including basal, luminal, and neuroendocrine cells. Multiple methods have been used to identify P-SCs in adult prostates. These include in vivo renal capsule implantation of a single epithelial cell with urogenital mesenchymal cells, in vitro prostasphere and organoid cultures, and lineage tracing with castration-resistant Nkx3.1 expression (CARN), in conjunction with expression of cell type-specific markers. Both organoid culture and CARN tracing show the existence of P-SCs in the luminal compartment. Although prostasphere cells predominantly express basal cell-specific cytokeratin and P63, the lineage of prostasphere-forming cells in the P-SC hierarchy remains to be determined. Using lineage tracing with P63CreERT2, we show here that the sphere-forming P-SCs are P63-expressing cells and reside in the basal compartment. Therefore we designate them as basal P-SCs (P-bSCs). P-bSCs are capable of differentiating into AR+ and CK18+ organoid cells, but organoid cells cannot form spheres. We also report that prostaspheres contain quiescent stem cells. Therefore, the results show that P-bSCs represent stem cells that are early in the hierarchy of overall prostate tissue stem cells. Understanding the contribution of the two types of P-SCs to prostate development and prostate cancer stem cells and how to manipulate them may open new avenues for control of prostate cancer progression and relapse.
Introduction
The prostate is a male reproductive organ. It is comprised of epithelial and stromal compartments. The epithelial compartment contains basal, luminal, and neuroendocrine cells. The stromal compartment is mainly composed of fibroblasts and smooth muscle cell-like cells. The development, maintenance, and function of the prostate are androgen dependent. Androgen deprivation leads to prostatic atrophy. The majority of luminal epithelial cells undergo apoptosis within a few days. In mice, when androgens are replenished, a limited number of surviving cells undergo rapid proliferation within a few hours and the prostate recovers to its original size in 2 weeks. This regression/regeneration cycle can be repeated multiple times, suggesting the existence of prostate stem cells (P-SCs)2 that survive under androgen deprivation and contribute to prostate regeneration (1–3).
Multiple methods have been used to identify and propagate P-SCs, and to pinpoint their location in prostate tissue. These include cell lineage tracing in vivo, expression of either basal or luminal epithelial cell-specific proteins, and prostaspheres or organoid cultures. However, the compartment in which prostate stem cells reside has remained debatable.
During prostate postnatal development, CK14-positive basal cells have been shown to give rise to basal, luminal, and neuroendocrine cells (4). P63, a basal cell-specific protein, is required for prostate development and support the notion that the basal cell compartment contains prostate stem cells (5). A luminal epithelial cell population known as CARNs (castration-resistant Nkx3.1-expressing cells) has demonstrated bipotential stem cell functions during prostate regeneration (6). In addition, both luminal and basal compartments contain single-lineage P-SCs that only give rise to daughter cells in their respective compartments (7, 8). Interestingly, the basal cells display both symmetrical and asymmetrical divisions, whereas luminal cells only exhibit symmetrical divisions (9). Asymmetrical divisions of stem cells reflect their capacity to self-renew and produce daughter cells for differentiated progeny. Symmetrical division is an indicator of late stage lineages in which both daughter cells terminally differentiate. Both lineages of cells have been shown capable of development into prostate cancers with distinct aggressiveness and molecular signatures (8, 10–13).
Prostasphere cell cultures have been extensively used for propagation of P-SCs. Prostasphere cells express basal cell cytokeratins and P63, and are capable of self-renewal and multilineage differentiation. However, the lineages of cells in prostaspheres is not clear. Moreover, whether the prostasphere forming cells represent P-SCs is still controversial. Methods for growth of luminal-resident P-SCs in vitro remained problematic until two groups recently developed distinct culture systems (14, 15). One group shows that both CARNs and normal prostate luminal epithelial cells can form prostate organoids and exhibit functional androgen receptors (AR) in culture. The other group shows that a three-dimensional culture system supports expansion of primary mouse and human prostate organoids, which are comprised of fully differentiated CK5+ basal and CK8+ luminal cells and also exhibit responses to androgen. Although both basal and luminal cells give rise to organoids, organoids derived from luminal cells more closely resemble prostate glands (15).
Herein we report that prostaspheres are derived from P63-expressing basal cells, designated as basal prostate stem cells (P-bSCs) to be distinguished from luminal P-SCs (P-lSCs). Prostaspheres were capable of forming organoids with differentiated marker CK18 and functional AR. However, organoid-derived cells could not form prostaspheres. The results suggest that P-bSCs represent more primitive P-SCs than P-lSCs. We also showed that both the prostate and prostaspheres had a reservoir of quiescent stem cells, which possessed a high self-renewal capacity.
Materials and Methods
Animals
Mice were housed in the Program of Animal Resources of the Institute of Biosciences and Technology, Texas A&M Health Science Center, in accordance with the principles and procedure of the Guide for the Care and Use of Laboratory Animals. All animal procedures were approved by the Institutional Animal Care and Use Committee. Mice carrying the Nkx3.1cre, P63CreERT2, K5H2B/GFP, Lgr5EGFP-CreERT2, and ROSA26LacZ reporter alleles were bred and genotyped as previously described (16–20). Prostates were harvested immediately after the animals were euthanized by CO2 asphyxiation.
For inducible gene activation, mice bearing P63CreERT2 and the reporter alleles, as well as their wild type littermates were injected intraperitoneally with 20 mg/ml of tamoxifen (Sigma; diluted in corn oil) at 100 mg/kg. For gene ablations in vitro, cells were treated with 5 mm 4-hydroxytamoxifen in ethanol (Sigma) at a final concentration of 500 nm.
Prostaspheres and Organoid Cultures
The conditions for preparing, culturing, and passaging prostate spheres were developed based on published protocols (21). Prostates were dissected from 6- to 8-week-old male mice immediately after euthanasia and minced with a pair of steel scissors. The minced tissues were then incubated with 10 ml of DMEM with 10% FBS containing 1 mg/ml of collagenase (Sigma) at 37 °C for 90 min. The cells were then washed with PBS, and incubated with 0.25% trypsin/EDTA for 10 min at 37 °C, followed by passing several times through a 25-gauge syringe. After adding FBS to the cell suspension to inactivate trypsin, the cells were passed through a 40-μm cell strainer, washed with Dulbecco's PBS (Sigma), and counted. Prostate cells were suspended with 50 μl of the prostate epithelial growth medium (Lonza, Walersville) at a density of 6 × 105/ml and then mixed with Matrigel (BD Biosciences) at a ratio of 1:1. The mixtures (100 μl) were plated around the rim of wells in a 12-well plate. After incubation at 37 °C for 30 min or until the Matrigel solidified, 1 ml of prostate epithelial growth medium was added to each well, and the plates were transferred to a 5% CO2 tissue culture incubator. The cells were then cultured at 37 °C for 8–10 days with the medium replenished every other day until spheres reached over 100 μm in diameter. The Matrigel was then digested by incubation in 1 ml of 1 mg/ml of dispase solution (Invitrogen) at 37 °C for 30 min. The spheres were harvested for analyses or subculture by centrifugation. For subculture, the pellets were digested with 1 ml of 0.05% trypsin/EDTA (Invitrogen) for 5 min at 37 °C, followed by inactivation of trypsin by FBS. The cells were passed through a 40-μm cell strainer, counted, and replated.
For organoid culture in hepatocyte medium as described (14), the cells were resuspended in hepatocyte culture medium (designated as Medium H) that consisted of hepatocyte medium supplemented with 10 ng/ml of EGF (PeproTech), 10 μm fasudil (Tocris), 1× Glutamax (Gibco), 5% Matrigel (Corning), and 5% charcoal-stripped FBS (Gibco) that had been heat inactivated at 55 °C for 1 h. Dissociated cells (10,000) were resuspended in Medium H and plated in ultralow attachment 96-well plates (Corning) in the presence of 100 nm dihydroxytesterone. Fresh Medium H (100 μl) was added to the wells every other day. The cells were incubated at 37 °C for 8–10 days. The organoids were dissociated for subculture by incubation in trypsin/EDTA with the addition of 10 μm fasudil for 5 min at 37 °C.
For organoid culture in the DMEM/F-12 medium (15), prostate cells (3 × 104) isolated as above were suspended in 50 μl of the DMEM/F-12 medium (designated as Medium DF), which was supplemented with 1× B27 (Invitrogen), 10 mm HEPES, Glutamax (Invitrogen), penicillin/streptomycin, 10 ng/ml of EGF, 500 ng/ml of recombinant R-spondin1, 100 ng/ml of recombinant Noggin (PeproTech), the TGF-β/Alk inhibitor A83-01 (Tocris), and 1 nm dihydrotestosterone (Sigma). The cell suspension was mixed with Matrigel (1:1) and plated around the rim of the wells in a 12-well plate. After gels solidified by incubation at 37 °C for 30 min, 1 ml of Medium DF was added to each well. The cells were incubated at 37 °C for 8–10 days with half of the medium replenished every other day. The organoids were dissociated for passaging by trypsinization with trypsin/EDTA with the addition of 10 μm fasudil for 5 min at 37 °C.
BrdU Labeling
BrdU (Sigma) was added to the culture medium at a final concentration of 2.5 μm. After incubation at 37 °C for 40 h, the spheres were harvested as described and BrdU positive cells were identified by immunostaining. For long-term retention of BrdU, the medium was aspirated and the culture were washed 3 times with fresh medium to remove free BrdU after 24 h exposure. The cells were cultured in fresh medium for another 7 days prior to being harvested for analyses. Immunostaining with anti-BrdU antibody (1:500 dilution; Sigma) was used to detect BrdU-labeled cells.
Flow Cytometry and Cell Sorting
Dissociated single prostate cells were incubated with FITC-conjugated anti-CD31, -CD45, -Ter119 antibodies, and phycoerythrin-conjugated anti-Sca-1 antibody (eBioscience), or Alexa 647-conjugated anti-CD49f antibody (Biolegend) diluted in the cell sorting buffer (2% FBS in PBS). Primary antibody labeling was conducted by incubation for 20 min in ice-cold conditions in a volume of 100 μl/105-108 cells with antibody dilutions according to the manufacturer's suggestions. The cells were then washed in 1 ml of ice-cold cell sorting buffer to remove unbound antibodies, followed by resuspension in the cell sorting buffer for fluorescence-activated cell sorting on a BD FACSAria I SORP. The sorted cells were collected in DMEM containing 20% FBS.
Histology
Prostaspheres or cells were fixed with 4% paraformaldehyde/PBS solution for 30 min at room temperature. The spheres were pelleted and suspended in Histogel. After solidifying at room temperature, the pellets were ethanol dehydrated, paraffin embedded in paraffin, and sectioned at 5-μm thickness as described (21). For histological analyses, sections were re-hydrated for H&E staining. For immunostaining, the slides were incubated in boiling citrate buffer (pH 8.0) for 20 min for antigen retrieval. The source and concentration of primary antibodies are: anti-cytokeratin 8 (1:15 dilution) and anti-cytokeratin 5 (1:500 dilution) from Fitzgerald Industries International (Concord, MA); anti-cytokeratin 18 (1:200 dilution) and anti-AR (1:200 dilution) from Abcam; mouse anti-BrdU (1:500 dilution) from Sigma; mouse anti-cytokeratin 14 (1:100 dilution) and anti-p63 (1:150 dilution) from Santa Cruz (Santa Cruz, CA), rabbit anti-β-catenin (1:200 dilution), and anti-cleaved caspase 3 (1:200 dilution) from Cell Signaling Technology. After washing, the specifically bound antibodies were detected with fluorescence-conjugated secondary antibodies (Invitrogen) or the ExtraAvidin Peroxidase System (Sigma). For immunofluorescence staining, the nuclei were counterstained with To-Pro 3 before being observed under a confocal microscope (Zeiss LSM 510). For LacZ staining, the intestine, prostates, or prostaspheres were lightly fixed with 0.2% glutaraldehyde for 1 h at room temperature, and then incubated overnight with 1 mg/ml of X-Gal at room temperature. After washing with PBS, the tissues or prostaspheres were post-fixed with 4% PFA for 1 h, ethanol dehydrated, paraffin-embedded, and sectioned for subsequent analyses.
Western Blotting
The prostate spheres were pelleted by centrifugation, resuspended in RIPA buffer (50 mm Tris-HCl buffer, pH 7.4, 1% Nonidet P-40, 150 mm NaCl, 0.25% sodium deoxycholate, 1 mm EGTA, 1 mm PMSF), and homogenized with a tissue homogenizer. The protein extracts were harvested by centrifugation. Samples containing 30 μg of proteins were separated by SDS-PAGE and electroblotted onto PVDF membranes. The membranes were then incubated with the indicated antibodies at 4 °C overnight. The dilutions and the sources of the antibodies were: anti-phosphorylated ERK1/2, 1:1000; anti-phosphorylated AKT, 1:1000; anti-ERK1/2, 1:1000; and anti-AKT, 1:1000 (Cell Signaling Technology). After being washed with the TBST buffer three times to remove nonspecific antibodies, the membranes were then incubated with the horseradish peroxidase-conjugated secondary antibody at room temperature for 1 h, followed by washing with the TBST buffer three times to remove unbound antibodies. Specifically bound antibody was visualized by using the ECL-Plus chemoluminescent reagents. The films were scanned with a densitometer for quantitation.
RNA Expression
Total RNA was isolated from prostaspheres using TRIzol RNA isolation reagents (Life Technologies). The SuperScript III reverse transcriptase (Invitrogen) and random primers were used for first-strand cDNAs synthesis according to the manufacturer's protocols. Real-time PCR analyses were carried out using the Fast SYBR Green Master Mix (Life Technologies). After normalization to the β-actin internal control, relative abundance of mRNA was calculated using the comparative threshold (CT) cycle method. The mean ± S.D. from at least three individual experiments are shown.
Statistical Analysis
Statistical analysis was performed using the two tailed t test, with significance set to p < 0.05. Error bars indicate standard deviation.
Results
Prostaspheres Are Comprised of Basal Epithelial Cells Derived from the Nkx3.1-expressing Lineage
To establish prostasphere cultures, single cell suspensions were prepared from adult prostate tissues and plated in Matrigel as described (21). Small, solid spheres were visible at day 5 and two-layer spheres around days 7 to 10, which reached an average size of 100 μm or larger in diameter. The outermost layer of the spheres and the layers closely underneath were aligned concentrically around a central cavity filled with acellular substances (Fig. 1A). The NKX3.1 homeobox protein is the earliest marker for prostate epithelial precursor cells (22). Cell-lineage tracing with the Nkx3.1cre-activated ROSA26LacZ reporter demonstrated that prostasphere cells were of epithelial origin as illustrated by LacZ staining (Fig. 1B). Most cells of the spheres expressed basal cell-specific cytokeratin (CK) 5; only a few cells in the inner layers expressed luminal cell-specific CK8 (Fig. 1C). However, when the cells were transferred to regular two-dimensional culture conditions, an increasing number of cells displayed luminal CK8 staining (Fig. 1D). We then carried out FACS analyses to determine cell compositions in prostate and prostaspheres based on the profile of Lin (CD45, Ter119, and CD31), Sca-1, and CD49f. Basal cells were Lin−/Sca-1+/CD49f+; luminal cells were Lin−/Sca-1−/CD49f+; and stromal cells were Lin−/Sca-1+/CD49f−. FACS analysis revealed that 74% of the cells in the spheres were Sca-1+/CD49f+. In contrast, only 28% of primary prostate cells were Sca-1+/CD49f+, indicating that the sphere culture enriched cells with basal cell properties (Fig. 1E). Real-time RT-PCR analysis also showed that CK14 expression in the 1st and 8th generation of spheres was increased up to 80-fold and CK18 expression was reduced in the 8th generation of spheres (Fig. 1F).
FIGURE 1.
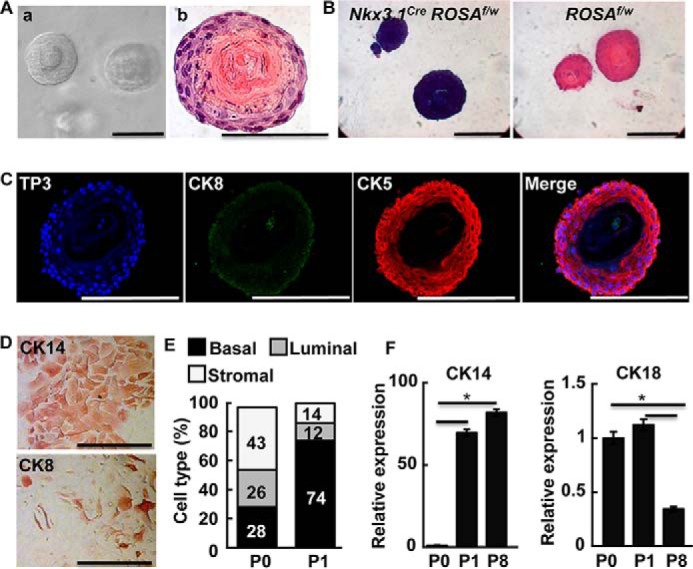
Prostaspheres are derived from basal cells and maintain basal cell properties. A, phase-contrast image of representative prostaspheres in Matrigel (a) and H&E staining of a prostasphere section (b). B, X-Gal staining of prostaspheres derived from mice with the indicated genotype. C, immunostaining of prostaspheres with luminal cell-specific CK8 and basal cell-specific CK5. D, disassociated cells from prostaspheres were cultured in tissue culture dishes for 3 days and stained with anit-CK14 or -CK8 antibodies as indicated. E and F, FACS analysis of the primary and 1st generation prostaspheres. Numbers indicate percentages of the indicated cell type (E). Real-time RT-PCR analyses of CK14 and CK18 expression in primary prostate cell suspensions (P0) and P1 or P8 prostaspheres (F). f, floxed; w, wild type; CK5, cytokeratin 5; CK8, cytokeratin 8; CK14, cytokeratin 14; TP3, To-Pro3; *, p ≤ 0.05; scale bars, 100 μm.
Prostaspheres Are Derived from P63-expressing Basal Stem Cells
Even though prostaspheres were derived from the Nkx3.1-expressing progenitors, expression of Nkx3.1 in prostaspheres was below the detection limit of RT-PCR (Fig. 2A). Because Nkx3.1 is expressed in luminal epithelial cells of adult prostates (23), the data indicate that prostaspheres do not contain or only contain scanty terminally differentiated luminal epithelial cells. P63 is expressed in prostate basal cells and is required for prostate development (5). Lineage tracing with the ROSA26LacZ reporter activated by P63CreERT2 at 2 weeks after birth showed that LacZ positive progeny were distributed in both basal and luminal compartments (Fig. 2B, a). This indicated that P63-expressing cells gave rise to both luminal and basal cells as reported (24).
FIGURE 2.
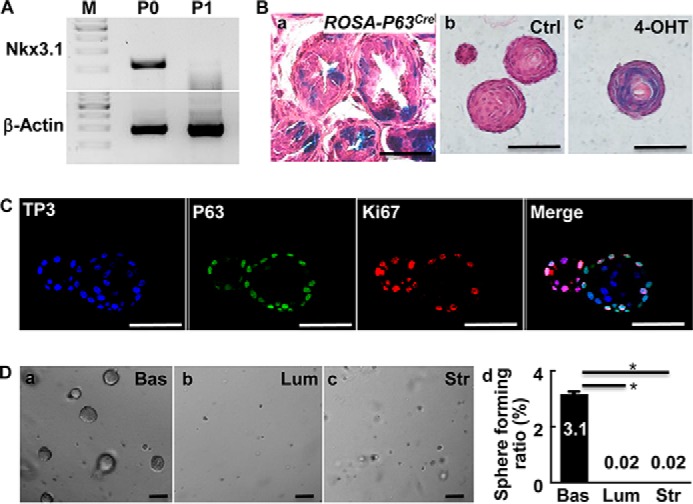
Prostaspheres are derived from P63-expressing basal cells. A, RT-PCR analyses of Nkx3.1 in primary (P0) and 1st generation prostaspheres (P1). B, X-Gal staining of prostate (a) or prostasphere (b and c) sections from the mice bearing ROSA26LacZ and P63CreERT2 reporter alleles. Panel a, the mouse was injected intraperitoneal with tamoxifen 2 weeks after birth and the prostates were harvested at 6 weeks. Panels b and c, prostaspheres were treated with alcohol or 4-OHT at day 1 after the inoculation. C, prostasphere sections were immunostained with anti-P63 and Ki67 antibodies. D, prostasphere forming assays for FACS-fractionated cells. P0, primary prostate cells; P1, 1st generation of prostaspheres; TP3, To-Pro 3 staining for nuclei; Ctrl, control; Bas, basal cells; Lum, luminal cells; Str, stromal cells; *, p ≤ 0.05; scale bars, 100 μm.
To determine whether prostaspheres were derived from P63 expressing basal cells, the prostaspheres derived from P63CreERT2-ROSA26LacZ mice were treated with 4-hydroxytamoxifen (4-OHT) at day 1 to activate the ROSA26LacZ reporter. X-Gal staining revealed that almost all prostaspheres were homogeneously LacZ+ (Fig. 2B, b and c). Thus, the spheres were derived from P63-expressing basal stem cells. Immunostaining revealed that P63 was expressed in the outer layer but not inner layer cells. About 80% of the cells in the outermost layer were P63+ (Fig. 2C). Together with the data that inner cells expressed luminal CK8, the results suggested that inner cells were more differentiated and lost P63 expression. Moreover, Ki67 was located at the outermost layer of the spheres and co-localized with P63. This indicated that most proliferating cells were P63-expressing cells (Fig. 2C). To determine which population of cells had sphere-forming capacity, primary prostate cells were fractionated by FACS based on the aforementioned strategy. Sphere cultures of FACS-fractionated cells demonstrated that ∼3.17% of basal cells formed spheres, whereas only 0.02% luminal and stromal cells formed spheres (Fig. 2D, d). Together, the data indicated that prostaspheres were derived from basal cells but not luminal cells. The cells are hereafter designated as P-bSCs.
The Label-retaining Cells in Prostaspheres Have High Capacity to Form Prostaspheres
To determine whether prostaspheres had label-retaining slow cycling cells, BrdU was used to label and trace the cells engaged in DNA synthesis. Approximately 90% of cells at the outer layer displayed BrdU 40 h after the labeling (Fig. 3A, a). As expected, a few BrdU+ cells at the outer layer were still visible even at 7 days after labeling. This confirmed that prostaspheres contained rare long-term label retaining cells (Fig. 3A, b).
FIGURE 3.
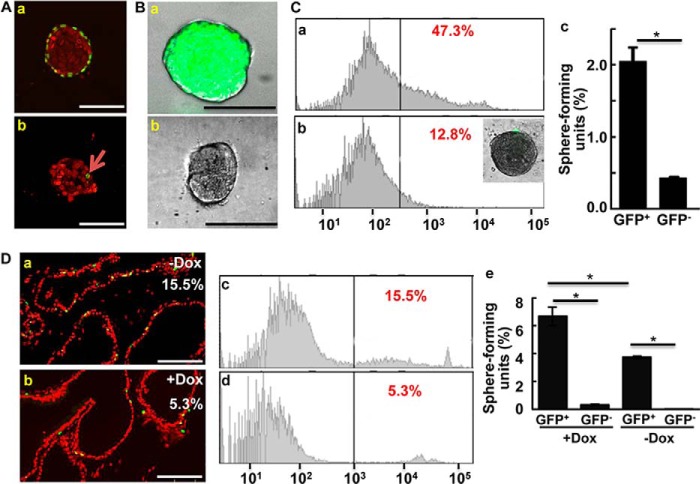
Prostaspheres contain slow-cycling P-bSCs. A, prostaspheres were labeled with BrdU for 40 h (a), or a 24-h pulse and 7-day chase (b). The labeled cells were detected by anti-BrdU immunostaining. The arrows indicate label-retaining cells. B, merged images of phase-contrast and fluorescent microscopy of prostaspheres bearing the K5H2B/GFP reporter showing GFP+ prostaspheres (a) and GFP− prostaspheres (b). C, FACS analyses of prostaspheres carrying the K5H2B/GFP reporter in the absence or presence of doxycycline (a and b). Numbers indicate percent of GFP+ cells. Inset, doxycycline-treated prostasphere derived from cells carrying the K5H2B/GFP allele. The FACS-fractionated cells from doxycycline-treated prostaspheres were cultured in Matrigel and the average sphere numbers derived from 100 cells were showed as mean ± S.D. (c). D, the prostates bearing the K5H2B/GFP reporter allele with or without doxycycline were sectioned and GFP was detected by confocal microscopy (panels a and b), or subjected to FACS analyses (panels c and d). The sphere forming activity of the 4 groups of cells was assessed and presented as numbers of spheres per 100 cells (e). Dox, doxycycline; *, p ≤ 0.05; scale bars, 100 μm.
To further determine whether these slow cycling cells had high self-renewal ability, the mice that carried the tetracycline-regulated K5H2B/GFP reporter were used as described (25). Over 80% of prostaspheres derived from the reporter-bearing prostate were GFP+ in the absence of doxycycline (Fig. 3B). The data further indicated that prostaspheres were derived from basal cells that expressed CK5. In contrast to the majority of GFP+ prostasphere cells in its absence, the presence of doxycycline resulted in only 1 or 2 GFP+ cells at the outer layer, which represented long-term label-retaining cells (Fig. 3C, a and b). These long-term label-retaining GFP+ cells showed 4-fold higher capacity to form spheres than GFP− cells (Fig. 3C, c).
Consistently, in the absence of doxycycline, the majority of prostate basal cells in vivo were GFP+ (Fig. 3D, a and c), which accounted for 15.5% of total prostate cells. However, 1 month after feeding with doxycycline to repress GFP expression, only 5.3% of the cells in the prostate were still GFP+ (Fig. 3D, b and d). Prostates with or without doxycycline treatment were then dissociated into single cells. The GFP+ and GFP− cells were separated by cell sorting and cultured in Matrigel to assess their sphere-forming capacity. The results showed that the GFP+ cells from the doxycycline-treated group, which represented the long-term label-retaining cells in the prostates, had a higher capacity in forming prostaspheres than the general population of basal cells (Fig. 3D, e). None of the GFP− cells formed spheres with a diameter larger than 50 μm. The results further demonstrate that label-retaining basal cells have a high capacity to form prostaspheres.
P-bSCs Have the Capacity to Form Organoids, but Organoid Cells Cannot Form Spheres
Two recent reports show that the luminal compartment contains P-SCs that form prostate-like organoids in two different in vitro conditions. To determine whether the organoid-forming P-SCs and P-bSCs were interchangeable and shared the same properties, P-bSCs were transferred to the two organoid culture conditions, namely Medium H and Medium DF as described under “Materials and Methods.” Interestingly, P-bSCs formed organoid-like structures with a morphology similar to reported earlier in both media (Fig. 4A, a and b). Immunostaining showed that the resulting organoids in Medium H contained cells expressing AR and luminal cell-specific CK18, but no longer P63 (Fig. 4A, c–e). The organoids generated in Medium DF contained cells expressing AR and CK18. Only a few cells located at the outer layer expressed P63 (Fig. 4A, f-h). Thus, the results indicate that prostaspheres contain organoid-forming cells, which can differentiate into luminal cells under the organoid culture conditions.
FIGURE 4.
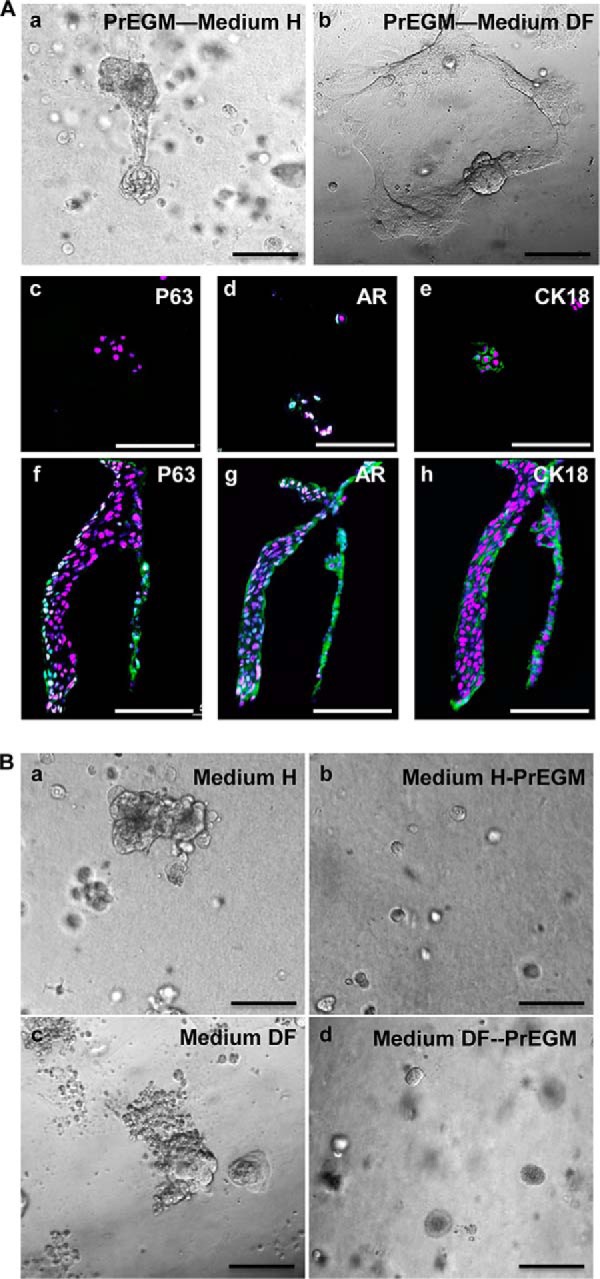
Prostasphere cells have the capacity to form prostate organoids, but organoid cells cannot form prostaspheres. A, prostaspheres cultured in Matrigel with prostasphere culture medium (PrEGM) were dissociated and inoculated in organoid culture with hepatocyte (Medium H) or DMEM/F-12 (Medium DF) as described. Panels a and b are phase-contrast microscopy images. Panels c–e and f–h are immunostaining of organoid sections from Medium H or Medium DF with the indicated antibodies. B, organoids cultured in the indicated medium were dissociated and inoculated in Matrigel/PrGEM. Scale bars, 100 μm.
To determine whether the organoids contained prostasphere-forming cells, primary prostate cells were first cultured in organoid conditions as reported. The organoids cells were then harvested and grew either in the organoid or sphere culture conditions. Although the cells continued to form organoids in the organoid culture conditions, none of them formed prostaspheres in the prostasphere culture condition (Fig. 4B). Therefore, it was unlikely that the organoids contained prostasphere-forming P-bSCs. Because prostasphere-forming P-bSCs were capable of forming both prostaspheres and organoids, but organoid forming cells cannot form prostaspheres, the results indicate that P-bSCs sit in a higher position in the prostate stem cell hierarchy than luminal resident organoid-forming P-SCs.
Neither the Prostate Epithelium nor the Prostasphere Contains Lgr5-expressing Cells
Emerging evidence has indicated the coexistence of quiescent and active adult stem cells in mammals (26). The leucine-rich repeat containing G-protein-coupled receptor 5 (Lgr5) has been reported to be expressed in many active, but not quiescent, tissue stem cells (27, 28). To determine whether prostaspheres contained Lgr5+ cells, RT-PCR was employed to detect Lgr5 expression at the mRNA level in primary prostate cells and first generation prostaspheres. The results showed that prostaspheres had no detectable Lgr5 expression although primary prostate cells exhibited a very low level of Lgr5 expression (Fig. 5A). In contrast, β-catenin expression was comparable between primary prostate cells and prostaspheres. Consistently, no cells in prostaspheres derived from Lgr5EGFP-CreERT2/ROSA26LacZ mice were LacZ positive with or without administration of 4-OHT to activate CreERT2 (Fig. 5B). This further confirmed there were no Lgr5 expressing cells in prostaspheres. Lineage tracing with Lgr5EGFP-CreERT2/ROSA26LacZ reporter alleles in vivo failed to demonstrate progeny derived from Lgr5-expressing cells in the prostate (Fig. 5C). Combined with FACS analyses, RT-PCR data also showed that Lgr5 was only expressed in stromal cells, but not epithelial cells (Fig. 5D). This indicates that unlike other active tissue SCs, active P-bSCs do not express Lgr5.
FIGURE 5.
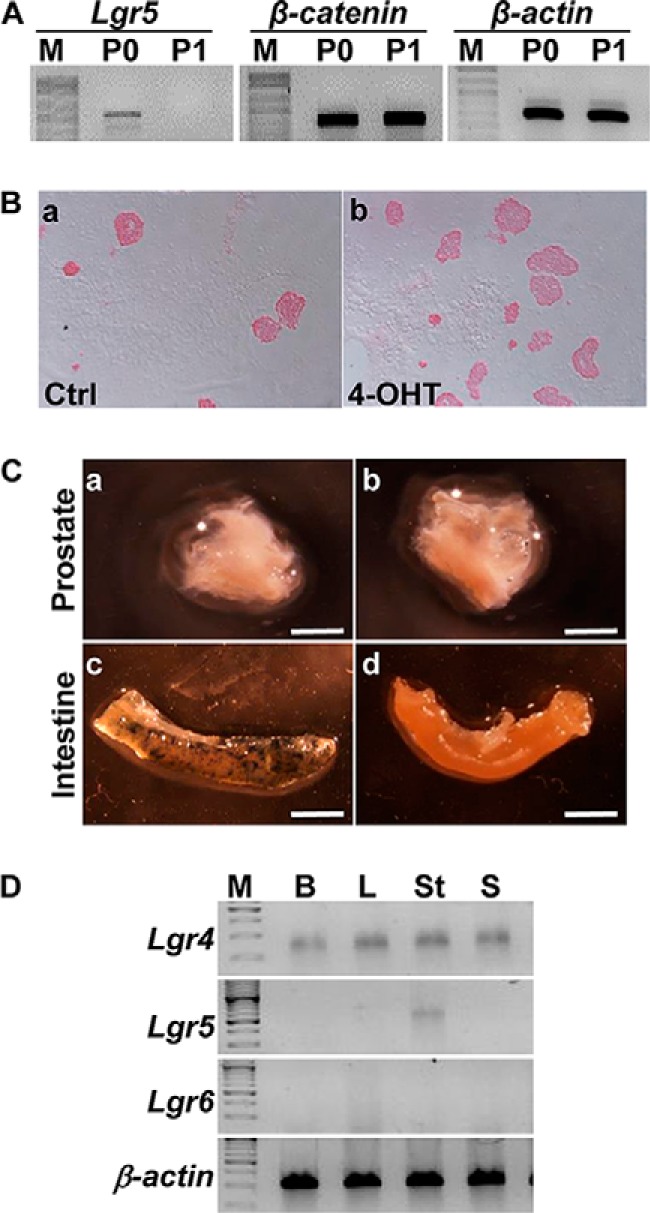
Lgr5 is not expressed in prostate epithelial cells and prostaspheres. A, RT-PCR analyses of the indicated genes in primary prostate cells (P0) and 1st generation prostaspheres (P1). B, prostaspheres derived from Lgr5EGFP/CreERT2;RosalacZ prostate were treated with 4-OHT treatment, whole mount stained with X-Gal, and then sectioned. C, whole mount X-Gal staining of prostates and intestines from Lgr5EGFP/CreERT2;RosalacZ mice with or without tamoxifen administration. Note that the blue LacZ staining is visible in intestine, but not in prostate. D, RT-PCR analyses of Lgr4, Lgr5, and Lgr6 expression in primary prostate cells fractioned by FACS and in prostaspheres. β-Actin was used as a loading control. P0, primary prostate cells; P1, 1st generation prostaspheres; M, molecular weight markers; B, basal cells; L, luminal cells; St, stromal cells; S, spheres derived from basal cells; scale bars, 2 mm.
Discussion
Prostasphere culture has been widely used for study and quantitation of self-renewal activity of both human and mouse P-SCs (21, 29, 30). Although it has been reported that prostaspheres expressed basal cell cytokeratins and P63, the precise placement of prostasphere cells in the overall prostate stem cell lineages and hierarchy has not been reported. Recent reports indicate that prostate organoid cultures contain P-SCs derived from luminal cells (14, 15). Together with urogenital mesenchymal cells, both the sphere- and organoid-forming cells can give rise to a glandular structure resembling the prostate in vivo (29). In this report, we showed that prostaspheres are derived from P63-expressing basal stem cells, designated as P-bSCs. The P63-expressing P-bSCs were able to give rise to cells expressing either luminal epithelial-specific CK8, or basal epithelial-specific cells CK14. We also showed that P-bSCs were capable of forming organoids. In contrast, the organoid-forming cells were not able to form prostaspheres. Therefore, the results indicate that P-bSCs represent high hierarchy tissue stem cells in the prostate, which can give rise to both luminal and basal stem cells in the prostate.
The prostate consists of the stromal and epithelial compartments, which are separated by basement membranes. The epithelial compartment of mature prostates consists of basal epithelial cells that express cytokeratin 5/14 and well differentiated luminal epithelial cells that express cytokeratin 8/18, and neuroendocrine cells that express synaptophysin. Basal cells are small and localized between the basement membrane and luminal cells, which account for ∼10% of cells in the mature prostate epithelium. The origin of basal and luminal cell populations has been controversial. However, it has been reported that P63, a basal cell marker, is required for prostate epithelial lineage commitment and prostate development (5, 31). Recent reports with P63CreERT2 lineage tracing reveal that both luminal and basal cells are derived from P63-expressing cells (20, 24). Cell lineage tracing with luminal epithelial-specific Nkx3.1cre or K8CreERT2 and basal epithelial-specific K5CreERT2 or K14CreERT2-mediated activation of reporter genes demonstrates that both basal and luminal compartments contain P-SCs. Both luminal and basal P-SCs give rise to multiple lineages of cells either in a tumor microenvironment or co-implanted with urogenital mesenchymal cells. Our results indicate that the prostasphere-forming P-SCs were from P63-expressing basal cells we designate as P-bSCs. The organoid-forming P-SCs are located in the luminal compartment, and therefore, represent P-lSCs.
Expression of Nkx3.1 in prostate progenitor cells starts at embryonic day 17.0, when the cells are actively engaged in proliferation and branching morphogenesis. Although it is expressed in all prostate epithelial cells during early prostate branching morphogenesis, Nkx3.1 is only expressed in luminal epithelial cells in adult prostate and is not expressed in prostaspheres. Data in Fig. 1B showed that prostaspheres are derived from the Nkx3.1 lineage. Lineage tracing also demonstrates that both basal and luminal cells in mouse prostate are derived from the cells that express P63 (20). Therefore, both basal and luminal cells are derived from the lineage. However, in adult prostate, we did not observe cells that expressed both Nkx3.1 and P63. Whether there are cells in developing prostate that express both characteristic proteins is interesting and remains to be determined. Because no cells in prostaspheres expressed Nkx3.1, we also did not observe cells that expressed both Nkx3.1 and P63. Because both P-bSCs and P-lSCs are from the Nkx3.1-expressing cell lineage, the data that P-bSCs were able to form organoids and prostaspheres and that P-lSCs could not form prostaspheres indicates that in addition to self-renewal, P-bSCs can also give rise to P-lSCs. Therefore, P-bSCs sit at a higher position in the prostate stem cell hierarchy than P-lSCs, although both populations of P-SCs can give rise to multiple cell lineages in the prostate epithelium.
Cancer stem cells have been suggested to underlie relapse of some malignancies. It has been reported that by loss of tumor suppressor genes, both P-bSCs and P-lSCs can give rise to prostate cancer (12, 13). Therefore, understanding how to manipulate the two populations of P-SCs to either remain silent in a dormant state or undergo terminal differentiation should provide new avenues to retrain prostate cancer progression and relapse. In the accompanying article (32), we report that FGF signaling is required for self-renewal and stemness of P-bSCs. Treatment of cells with FGF7 or FGF10 increases sphere formation. Ablation of Fgfr2 or Frs2α in P-bSCs abrogates the sphere-forming activity and induces differentiation of the cells. In conclusion, prostaspheres are derived from the P63-expressing P-bSCs, which contain both quiescent and cycling cells that can either differentiate to basal epithelial or luminal epithelial cells.
Author Contributions
Y. H., T. H., J. L., C. W., L. A., P. Y., and J. Y. F. performed experiments and analyzed the data; J. X., W. L. M., and F. W. analyzed and evaluated the results and wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Dr. Michael Shen for sharing the Nkx3.1cre knock-in mice, Dr. Li Xin for constructive suggestions and discussions, Alon Azares for FACS cell sorting and data analysis, and Dr. Stefan Siwko for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants CA96824 and DE023106 (to F. W.), and CA140388 (to W. L. M., and F. W.), and Cancer Prevention and Research Institution of Texas Grant CPRIT110555 (to F. W. and W. L. M.), Natural Science Foundation of Zhejiang Province of China Grant Y2110492, and the National Natural Science Foundation of China Grants 81101712, 31371470, and 81270761 (to C. W.). The authors declare that they have no conflicts of interest with the contents of this article.
- P-SCs
- prostate stem cells
- CARN
- castration-resistant Nkx3.1-expressing cells
- AR
- androgen receptor
- P-bSC
- basal prostate stem cell
- P-lSC
- luminal P-SC
- CK
- cytokeratin
- 4-OHT
- 4-hydroxytamoxifen.
References
- 1. English H. F., Santen R. J., Isaacs J. T. (1987) Response of glandular versus basal rat ventral prostatic epithelial cells to androgen withdrawal and replacement. Prostate 11, 229–242 [DOI] [PubMed] [Google Scholar]
- 2. Evans G. S., Chandler J. A. (1987) Cell proliferation studies in the rat prostate: II. the effects of castration and androgen-induced regeneration upon basal and secretory cell proliferation. Prostate 11, 339–351 [DOI] [PubMed] [Google Scholar]
- 3. Sugimura Y., Cunha G. R., Donjacour A. A. (1986) Morphological and histological study of castration-induced degeneration and androgen-induced regeneration in the mouse prostate. Biol. Reprod. 34, 973–983 [DOI] [PubMed] [Google Scholar]
- 4. Ousset M., Van Keymeulen A., Bouvencourt G., Sharma N., Achouri Y., Simons B. D., Blanpain C. (2012) Multipotent and unipotent progenitors contribute to prostate postnatal development. Nat. Cell Biol. 14, 1131–1138 [DOI] [PubMed] [Google Scholar]
- 5. Signoretti S., Waltregny D., Dilks J., Isaac B., Lin D., Garraway L., Yang A., Montironi R., McKeon F., Loda M. (2000) p63 is a prostate basal cell marker and is required for prostate development. Am. J. Pathol. 157, 1769–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang X., Kruithof-de Julio M., Economides K. D., Walker D., Yu H., Halili M. V., Hu Y. P., Price S. M., Abate-Shen C., Shen M. M. (2009) A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature 461, 495–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu J., Pascal L. E., Isharwal S., Metzger D., Ramos Garcia R., Pilch J., Kasper S., Williams K., Basse P. H., Nelson J. B., Chambon P., Wang Z. (2011) Regenerated luminal epithelial cells are derived from preexisting luminal epithelial cells in adult mouse prostate. Mol. Endocrinol. 25, 1849–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Choi N., Zhang B., Zhang L., Ittmann M., Xin L. (2012) Adult murine prostate basal and luminal cells are self-sustained lineages that can both serve as targets for prostate cancer initiation. Cancer Cell 21, 253–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang J., Zhu H. H., Chu M., Liu Y., Zhang C., Liu G., Yang X., Yang R., Gao W. Q. (2014) Symmetrical and asymmetrical division analysis provides evidence for a hierarchy of prostate epithelial cell lineages. Nat. Commun. 5, 4758. [DOI] [PubMed] [Google Scholar]
- 10. Wang Z. A., Mitrofanova A., Bergren S. K., Abate-Shen C., Cardiff R. D., Califano A., Shen M. M. (2013) Lineage analysis of basal epithelial cells reveals their unexpected plasticity and supports a cell-of-origin model for prostate cancer heterogeneity. Nat. Cell Biol. 15, 274–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lu T. L., Huang Y. F., You L. R., Chao N. C., Su F. Y., Chang J. L., Chen C. M. (2013) Conditionally ablated Pten in prostate basal cells promotes basal-to-luminal differentiation and causes invasive prostate cancer in mice. Am. J. Pathol. 182, 975–991 [DOI] [PubMed] [Google Scholar]
- 12. Lawson D. A., Zong Y., Memarzadeh S., Xin L., Huang J., Witte O. N. (2010) Basal epithelial stem cells are efficient targets for prostate cancer initiation. Proc. Natl. Acad. Sci. U.S.A. 107, 2610–2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang Z. A., Toivanen R., Bergren S. K., Chambon P., Shen M. M. (2014) Luminal cells are favored as the cell of origin for prostate cancer. Cell Rep. 8, 1339–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chua C. W., Shibata M., Lei M., Toivanen R., Barlow L. J., Bergren S. K., Badani K. K., McKiernan J. M., Benson M. C., Hibshoosh H., Shen M. M. (2014) Single luminal epithelial progenitors can generate prostate organoids in culture. Nat. Cell Biol. 16, 951–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Karthaus W. R., Iaquinta P. J., Drost J., Gracanin A., van Boxtel R., Wongvipat J., Dowling C. M., Gao D., Begthel H., Sachs N., Vries R. G., Cuppen E., Chen Y., Sawyers C. L., Clevers H. C. (2014) Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell 159, 163–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lin Y., Zhang J., Zhang Y., Wang F. (2007) Generation of an Frs2α conditional null allele. Genesis 45, 554–559 [DOI] [PubMed] [Google Scholar]
- 17. Trokovic R., Trokovic N., Hernesniemi S., Pirvola U., Vogt Weisenhorn D. M., Rossant J., McMahon A. P., Wurst W., Partanen J. (2003) FGFR1 is independently required in both developing mid- and hindbrain for sustained response to isthmic signals. EMBO J. 22, 1811–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yu K., Xu J., Liu Z., Sosic D., Shao J., Olson E. N., Towler D. A., Ornitz D. M. (2003) Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development 130, 3063–3074 [DOI] [PubMed] [Google Scholar]
- 19. Lin Y., Liu G., Zhang Y., Hu Y. P., Yu K., Lin C., McKeehan K., Xuan J. W., Ornitz D. M., Shen M. M., Greenberg N., McKeehan W. L., Wang F. (2007) Fibroblast growth factor receptor 2 tyrosine kinase is required for prostatic morphogenesis and the acquisition of strict androgen dependency for adult tissue homeostasis. Development 134, 723–734 [DOI] [PubMed] [Google Scholar]
- 20. Lee D. K., Liu Y., Liao L., Wang F., Xu J. (2014) The prostate basal cell (BC) heterogeneity and the p63-positive BC differentiation spectrum in mice. Int. J. Biol. Sci. 10, 1007–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xin L., Lukacs R. U., Lawson D. A., Cheng D., Witte O. N. (2007) Self-renewal and multilineage differentiation in vitro from murine prostate stem cells. Stem Cells 25, 2760–2769 [DOI] [PubMed] [Google Scholar]
- 22. Bhatia-Gaur R., Donjacour A. A., Sciavolino P. J., Kim M., Desai N., Young P., Norton C. R., Gridley T., Cardiff R. D., Cunha G. R., Abate-Shen C., Shen M. M. (1999) Roles for Nkx3.1 in prostate development and cancer. Genes Dev. 13, 966–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shen M. M., Abate-Shen C. (2003) Roles of the Nkx3.1 homeobox gene in prostate organogenesis and carcinogenesis. Dev. Dyn. 228, 767–778 [DOI] [PubMed] [Google Scholar]
- 24. Pignon J. C., Grisanzio C., Geng Y., Song J., Shivdasani R. A., Signoretti S. (2013) p63-expressing cells are the stem cells of developing prostate, bladder, and colorectal epithelia. Proc. Natl. Acad. Sci. U.S.A. 110, 8105–8110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tumbar T., Guasch G., Greco V., Blanpain C., Lowry W. E., Rendl M., Fuchs E. (2004) Defining the epithelial stem cell niche in skin. Science 303, 359–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li L., Clevers H. (2010) Coexistence of quiescent and active adult stem cells in mammals. Science 327, 542–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Barker N., van Es J. H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P. J., Clevers H. (2007) Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 [DOI] [PubMed] [Google Scholar]
- 28. Jaks V., Barker N., Kasper M., van Es J. H., Snippert H. J., Clevers H., Toftgård R. (2008) Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat. Genet. 40, 1291–1299 [DOI] [PubMed] [Google Scholar]
- 29. Garraway I. P., Sun W., Tran C. P., Perner S., Zhang B., Goldstein A. S., Hahm S. A., Haider M., Head C. S., Reiter R. E., Rubin M. A., Witte O. N. (2010) Human prostate sphere-forming cells represent a subset of basal epithelial cells capable of glandular regeneration in vivo. Prostate 70, 491–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rybak A. P., He L., Kapoor A., Cutz J. C., Tang D. (2011) Characterization of sphere-propagating cells with stem-like properties from DU145 prostate cancer cells. Biochim. Biophys. Acta 1813, 683–694 [DOI] [PubMed] [Google Scholar]
- 31. Signoretti S., Pires M. M., Lindauer M., Horner J. W., Grisanzio C., Dhar S., Majumder P., McKeon F., Kantoff P. W., Sellers W. R., Loda M. (2005) p63 regulates commitment to the prostate cell lineage. Proc. Natl. Acad. Sci. U.S.A. 102, 11355–11360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huang Y., Hamana T., Liu J., Wang C., An L., You P., Chang J. Y., Xu J., Jin C., Zhang Z., McKeehan W. L., Wang F. (2015) Type 2 fibroblast growth factor receptor signaling preserves stemness and prevents differentiation of prostate stem cells from the basal compartment. J. Biol. Chem. 290, 17753–17761 [DOI] [PMC free article] [PubMed] [Google Scholar]