Background: Polyamines are ubiquitous compounds present in most organisms.
Results: Polyamines increase the level of RpoS, the σ38-RNA-polymerase subunit, even in non-amber-strain, to induce glutamate-dependent-acid resistance (GDAR) in Escherichia coli via gadE.
Conclusion: GDAR activation by polyamines is the indirect effect of increased RpoS level.
Significance: Polyamines are not only important for acid survival, but polyamine-dependent increase in RpoS levels will have pleiotropic effects.
Keywords: gene expression, microarray, post-transcriptional regulation, RNA polymerase, spermidine
Abstract
To study the physiological roles of polyamines, we carried out a global microarray analysis on the effect of adding polyamines to an Escherichia coli mutant that lacks polyamines because of deletions in the genes in the polyamine biosynthetic pathway. Previously, we have reported that the earliest response to polyamine addition is the increased expression of the genes for the glutamate-dependent acid resistance system (GDAR). We also presented preliminary evidence for the involvement of rpoS and gadE regulators. In the current study, further confirmation of the regulatory roles of rpoS and gadE is shown by a comparison of genome-wide expression profiling data from a series of microarrays comparing the genes induced by polyamine addition to polyamine-free rpoS+/gadE+ cells with genes induced by polyamine addition to polyamine-free ΔrpoS/gadE+ and rpoS+/ΔgadE cells. The results indicate that most of the genes in the E. coli GDAR system that are induced by polyamines require rpoS and gadE. Our data also show that gadE is the main regulator of GDAR and other acid fitness island genes. Both polyamines and rpoS are necessary for the expression of gadE gene from the three promoters of gadE (P1, P2, and P3). The most important effect of polyamine addition is the very rapid increase in the level of RpoS sigma factor. Our current hypothesis is that polyamines increase the level of RpoS protein and that this increased RpoS level is responsible for the stimulation of gadE expression, which in turn induces the GDAR system in E. coli.
Introduction
Polyamines (putrescine, spermidine, or spermine) are polycationic aliphatic bases that are abundantly present in all organisms from bacteria to humans. Polyamines have been implicated in many biological processes, including nucleic acid and protein synthesis and structure, cell growth, binding to membrane phospholipids, N-methyl-d-aspartate receptors, and protection against oxygen toxicity (1–8). However, despite many studies in many laboratories, the specific in vivo targets of the polyamines and the detailed mechanism of most of their effects have been uncertain.
To answer these questions, we have studied the effects of adding polyamines to a mutant strain of Escherichia coli that contains no polyamines because of deletions in the genes responsible for polyamine biosynthesis. We have used microarray techniques to study which genes are induced by addition of polyamines to this polyamine-deficient mutant. We found that the genes most rapidly induced by polyamines are the genes involved in the glutamate-dependent acid resistance pathway (GDAR)2 and that polyamines are required for their induction (9).
The systems involved in the acid resistance of bacteria have been of particular interest over many years, particularly because resistance to the high acidity of the stomach contents is a prerequisite for host colonization in food-borne pathogens (10–12). Bacteria have developed a number of systems for protection against death from exposure to low pH. Four of these acid resistance systems depend on the decarboxylation of amino acids by pyridoxal phosphate-dependent decarboxylase such as glutamate decarboxylase (13–15), and acid-inducible arginine (adiA) (16), lysine (cadA) (17), and ornithine (speF) decarboxylase (18). Of these, the glutamate-dependent acid resistance system is the most effective in protecting cells against an extreme acid environment and has been extensively studied, especially by Foster and others (13–15). The mechanism of the glutamate-dependent acid resistance involves the decarboxylation of glutamate by two glutamic acid decarboxylase (encoded by gadA and gadB) and export of the resultant γ-aminobutyric acid by the antiporter gadC, with the resultant loss of intracellular protons (14, 15).
In our earlier studies, direct enzymatic assays were made to assess the changes in glutamate decarboxylase levels after polyamine addition to polyamine-deficient cells (9). We found that polyamines are essential for this induction, confirming other studies by Jung and Kim (19). During these studies, we presented preliminary evidence that two regulatory factors rpoS and gadE are important for this induction by polyamines of the glutamate-dependent-acid resistance system. However, it was not clear how these genes were involved in the polyamine induction of the GDAR system.
The significance of RpoS was of particular interest because of its role as a regulatory subunit of bacterial RNA polymerase. In addition to five core subunits (two α, one β, one β′, and one ω), bacterial RNA polymerase contains one of the seven regulatory subunits called sigma, whose composition varies slightly in different versions of the polymerase to form the holoenzyme. One of these versions is called sigma38 (RpoS), and it has been studied extensively because it is markedly induced in response to various stress factors and in cells reaching stationary growth phase (20–28).
How gadE is activated by polyamines in regulating the GDAR system is of interest because of the extensive literature (unrelated to polyamines) showing the regulatory action of gadE on many components of the GDAR and other acid resistance genes (29–31). The gadE gene is located between hdeD and yhiU at the center of a 12-gene acid fitness island and contains an upstream sensory region comprising three different promoters (32).
The present paper reports our current studies on the involvement of these two factors in the polyamine response in greater detail. We postulate a cascade model in which the primary action of the polyamine addition to the polyamine-deficient cells is the very rapid increase in the RpoS level with subsequent induction of gadE expression that in turn increases the GDAR system. This demonstration of the effect of polyamines in rapidly increasing the RpoS level is of importance not only for understanding the requirement of polyamines for the glutamate-dependent acid response system but also for the likely role of the polyamines in regulating the large number of systems already known to involve RpoS and to be induced by various stress conditions.
Experimental Procedures
Bacterial Strains, Medium, and Growth
The E. coli strains and plasmids and their source are listed in Table 1. An E. coli mutant lacking polyamines was developed by sequential deletion of seven genes in the polyamine biosynthetic pathway to construct HT873 strain (see footnote to Table 1). All strains were constructed in the background of BW25113 (CGSC7636), parent of the Keio collection (33, 34) (obtained from the E. coli Genetics Stock Collection at Yale University). P1 transductions from mutant strains in this collection were carried out essentially as described by Miller (36). The kanamycin marker from the transduced strains was excised by Flp recombinase as described in Ref. 34.
TABLE 1.
Bacterial strains and plasmids used in this study
All the strains obtained in this study for relevant gene deletion (Keio collection) were obtained from the Yale E. coli genetic stock center. The pQEgadE plasmid was obtained from Dr. John Foster (University of South Alabama, Mobile, AL), and the pArpoS was obtained from Dr. Udo Bläsi (University of Vienna, Vienna, Austria).
| Strain or plasmid | Relevant genotype | Source |
|---|---|---|
| Strains | ||
| HT779 | F′,Δ (araD-araB)567, ΔlacZ4787 (::rrnB-3), λ-, rph-1,Δ (rhaD-rhaB) 568, hsdR514 | Parent of Keio collection (BW25113; CGSC 7636) (33) |
| HT873 | ΔspeA ΔspeC ΔspeD ΔldcC ΔspeF ΔadiA ΔcadA | Seven sequential gene deletions of polyamine biosynthetic pathway in HT779 straina |
| HT874 | ΔspeA ΔspeC ΔspeD ΔldcC ΔspeF ΔadiA ΔcadA ΔrpoS | HT873 X ΔrpoS |
| HT875 | ΔspeA ΔspeC ΔspeD ΔldcC ΔspeF ΔadiA ΔcadA ΔgadE | HT873 X ΔgadE |
| HT894 | ΔspeA ΔspeC ΔspeD ΔldcC ΔspeF ΔadiA ΔcadA ΔrpoS/pgadE | HT874/pgadE |
| HT895 | ΔspeA ΔspeC ΔspeD ΔldcC ΔspeF ΔadiA ΔcadA ΔgadE/pgadE | HT875/pgadE |
| HT896 | ΔspeA ΔspeC ΔspeD ΔldcC ΔspeF ΔadiA ΔcadA ΔrpoS ΔgadE | HT874 X ΔgadE |
| HT898 | ΔspeA ΔspeC ΔspeD ΔldcC ΔspeF ΔadiA ΔcadA ΔclpX | HT873 X ΔclpX |
| HT901 | ΔspeA ΔspeC ΔspeD ΔldcC ΔspeF ΔadiA ΔcadA ΔrpoS/prpoS | HT874/prpoS |
| HT902 | ΔspeA ΔspeC ΔspeD ΔldcC ΔspeF ΔadiA ΔcadA ΔgadE/prpoS | HT875/prpoS |
| HT903 | ΔspeA ΔspeC ΔspeD ΔldcC ΔspeF ΔadiA ΔcadA ΔrpoS ΔgadE/prpoS | HT896/prpoS |
| HT904 | ΔspeA ΔspeC ΔspeD ΔldcC ΔspeF ΔadiA ΔcadA ΔrpoS ΔgadE/pgadE | HT896/pgadE |
| HT905 | ΔspeA ΔspeC ΔspeD ΔldcC ΔspeF ΔadiA ΔcadA ΔrpoS ΔgadE/prpoS/pgadE | HT896/prpoS/pgadE |
| Plasmids | ||
| pCP20 | FLP recombinase | Ref. 34 |
| pArpoS | rpoS overexpression | Ref. 35 |
| pQE-gadE | gadE overexpression | Ref. 29 |
a The seven genes sequentially deleted in HT873 are speC → speA → speD → cadA → ldcC → speF → adiA.
Strains were grown in amine-free M9 (36) or Vogel Bonner minimal medium (37) containing 0.4% glucose and supplemented with different concentrations of amines as indicated. Incubations were at 37 °C with shaking in air. For strains containing plasmids, the appropriate antibiotic was added: 20 μg/ml chloramphenicol for strains containing the pArpoS plasmid or 100 μg/ml of carbenicillin for the strain containing the pQEgadE plasmid.
RNA Isolation, rRNA Removal, and Microarray Analyses
For the microarray experiments, polyamine-free mutants (HT873, HT874, and HT875) were inoculated from an LB plate in triplicate into amine-free M9 minimal medium. The cultures were deprived of contaminating amines (obtained from the prior growth in LB) by growing them overnight in amine-free medium. Then the cultures were diluted to A600 of 0.2 and grown until the A600 reached 1.0. Each culture was divided into two parts. To one part, a mixture of putrescine and spermidine was added to a final concentration of 100 μm; the other part was left untreated and used as a control. Both cultures were harvested 60 min after amine addition. Total RNA from each sample was isolated as described previously (38). Briefly, for each RNA isolation, 109–1010 cells were resuspended in Tris-EDTA buffer (100 mm Tris, 10 mm EDTA, pH 8.0) containing 2 mg/ml lysozyme (Sigma), and RNA was isolated according to the protocol described in the RNeasy mini kit (Qiagen). The mRNAs were enriched from total RNA by removing the 16S and 23S ribosomal RNAs using the MICROBExpress method and kit (Ambion/Life Technologies; part AM1905) and used for microarray analysis in the NIDDK genomic core facility. The enriched mRNA (40 ng/reaction) was reverse transcribed and amplified using an Ovation WTA pico-V2 kit (part 3302; NuGEN, San Carlos, CA) according to the manufacturer's protocol. 2.5 μg of cDNA was fragmented, biotinylated and then hybridized for 18 h to E. coli GeneChip arrays (Genome 2.0 array; Affymetrix Santa Clara, CA; n = 3 each for each sample). Hybridized arrays were washed and stained with Affymetrix WHS kit (catalog no. 900720) using Fluidic Station 450. Arrays were scanned using a GeneChip scanner 3000 7G (Affymetrix) operated by Affymetrix GeneChip Command Console software. Analysis of variance was performed and p values were calculated using Partek Pro software (Partek, St. Louis, MO) and plotted as the negative log on the y axis against the Affimetrix signal ratios for each probe set on the x axis. Up- and down-regulated genes were selected for further study based on p values of <0.05 and fold change >+2 or −2.
To identify the motif of GadE and RpoS binding sites on the DNA of the induced genes, the pattern search tool in coliBASE was used. The published GAD box sequence (5′-TTAGGATTTTGTTATTTAAA-3′) (39, 40) and RpoS binding sequence (5′-TCTATAAT-3′) (41) were used to identify the promoter regions of the genes showing increased transcription after polyamine treatment and differentially regulated in rpoS+/gadE+, ΔrpoS/gadE+ and rpoS+/ΔgadE polyamine-deficient strains. The sequence logos for the frequency plot were created as described in WebLogo (42).
Quantitative Real Time PCR and Reverse Transcriptase-PCR Analysis
cDNAs from the microarray experiments were used to confirm the gene expression by quantitative real time PCR analysis. Three cDNA replicates from each of rpoS+/gadE+ and mutant groups, ΔrpoS/gadE+ and rpoS+/ΔgadE were used for quantitative real time PCR assays. Six of the highly induced genes in the acid response pathway were selected to confirm the expression. The E. coli glyceraldehyde-3-phosphate dehydrogenase-A gene (gapA) was taken for all quantitative real time PCR analysis as a control for input mRNA. Gene-specific primer sequences were designed using the Primer 3 program (all oligonucleotide primers used in this study are listed in Table 2).
TABLE 2.
List of oligonucleotides used in this study
The primers were designed using the Primer 3 program.
| Gene names | Sequences (from 5′ to 3′) |
|---|---|
| gadA-up | TTACCAGGTTGCCGCTTATC |
| gadA-down | ACGCAGACGTTCAGAGAGGT |
| gadB-up | TTACCAGGTTGCCGCTTATC |
| gadB-down | ACGCAGACGTTCAGAGAGGT |
| gadC-up | TCGAAGACCTTCTTCCCTGA |
| gadC-down | CATTACCCGGAATGACCATC |
| gadE-up | TGCCCCATAAGAATTCACAA |
| gadE-down | GTGACGATGTCGCTCATACG |
| hdeD-up | CCGGTATTATCCGGTTTCCT |
| hdeD-down | CTTTCATTGAACGCTGACGA |
| slp-up | GCGAAGCCTGATATTGAAGC |
| slp-down | GGATGCCCTGCATATTCACT |
| gapA-up | AGGTCTGATGACCACCGTTC |
| gapA-down | GGAACGCCATACCAGTCAGT |
| rpoS-up | CCTGCACAAAATTCCACCGTTGC |
| rpoS-down | GATGGGCATCGGACCTTTTATT |
| gadE-P1-up (−195)a | AGGAATCTTACTTAGGATCAATAT |
| gadE-P1-down (+49)a | GCCAAAAGCCCTGTAAAAGAAAAGAATC |
| gadE-P2-up (−360)a | TTGCCAGCTTAAGTCGAAACAAGG |
| gadE-P2down (−190)a | ATTCCTGGTTGTTATCAGCTTGTA |
| gadE-P3-up (−532)a | GCGTTGATGCTATGGGCGGTTAAAT |
| gadE-P3-down (−310)a | ATGTAATCCGATTTAAATATCGAG |
a For the primers of gadE promoters (P1, P2, and P3), the positions are denoted based on +1 as the starting position and designed according to Sayed and Foster (32) and obtained from Sigma Life Sciences.
The Bio-Rad CFX96 real time PCR system was used for the assay. For each reaction 10 μl of 2× SyBr Green master mix (Bio-Rad), 400 nmol from each 5′ and 3′ primer, and 500 pg of cDNA were mixed in a total volume of 20 μl. Each reaction was set up in triplicate in a 96-well plate. Samples were heated at 95 °C for 10 min to activate the enzymes followed by 95 °C denaturing for 15 s and 60 °C annealing and extension for 75 s, and the cycle was repeated 40 times. Melt curve analysis (65–95 °C, 0.5 °C increments) showed a single amplified product for each well. The data were analyzed using Bio-Rad CFX manager (version 3.1) software, and relative expression levels were derived using HT873, without added polyamines as the controls.
For further analysis of the level of rpoS gene induction by polyamines (see Fig. 5, A and B), cultures were grown without amines as mentioned above, and when the cultures reached an A600 of 1.0, in part of the culture, polyamines (100 μm each of putrescine and spermidine) were added, and the cultures were harvested after 10, 20, and 60 min. Another part of the culture was untreated and used as minus amine culture. RNA was isolated as above, ribosomal RNAs were removed, and cDNAs were synthesized using SuperScript Vilo (Life Technologies). 1.0 ng of cDNA was used for each quantitative real time PCR analyses using gapA as control, following the Bio-Rad protocol as mentioned above.
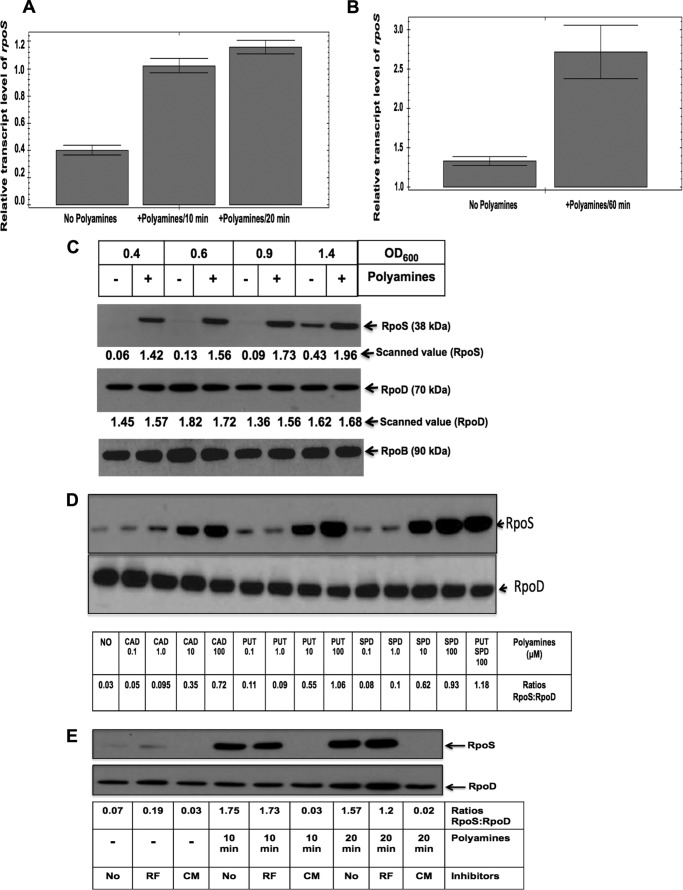
FIGURE 5.
Polyamine addition to polyamine-deficient cells induced a large and rapid increase in RpoS level. rpoS+/gadE+ cells were grown without any amines to an A600 of 1.0; in part of the culture, polyamines (100 μm each of putrescine and spermidine) were added, and the cultures were harvested after 10-, 20-, and 60-min intervals. RNA was isolated as above, ribosomal RNAs were removed, and cDNAs were synthesized using Superscript Vilo. 1.0 ng of cDNA was used for each quantitative real time PCR analyses with rpoS gene-specific primers; gapA gene expression was used as a control to calculate relative transcript level. A and B, the relative rpoS induction after 10 and 20 min (A) and 60 min (B) is shown. C, rpoS+/gadE+ polyamine mutant cells were grown without any amines, and aliquots of the cultures at different growth phases were treated with 100 μm of putrescine plus 100 μm of spermidine, and cultures were harvested after 20 min incubation at 37 °C. Protein extracts (5 μg/lane) were resolved by SDS-polyacrylamide gel electrophoresis; blotted into PVDF membranes; probed with anti-RpoS antibody, anti-RpoD antibody, or anti-RpoB antibody; and subjected to immunological detections as described in the text. The autoradiograms containing the RpoS and RpoD protein intensities were scanned in Bio-Rad GS-800 densitometer, and the scanned values are presented below each band of RpoS and RpoD. D, rpoS+/gadE+ cells were grown without any amines to an A600 of 1.0, and part of the cultures were divided into 14 parts to treat with different amines or left untreated as indicated for 20 min. Western blot and immune detections were performed as described. The relative ratio of RpoS:RpoD is shown below each band after scanning the Western blot data. E, rpoS+/gadE+ cells were grown without any amines to an A600 of 1.0, and part of the cultures were divided into nine parts to treat with or without amines and different inhibitors as indicated. Rifampicin (RF, 10 μg/ml) and chloramphenicol (CM, 200 μg/ml) were added 10 min before the addition of polyamine to the indicated cultures, and then amines were added as indicated, and samples were harvested after 10- and 20-min intervals. For minus amine cultures, the same amount of inhibitors were used, and samples were harvested after a total time of 30 min. Protein samples were extracted and assayed by Western blot as above, and the relative ratio of RpoS:RpoD is shown below each band after scanning the Western blot data.
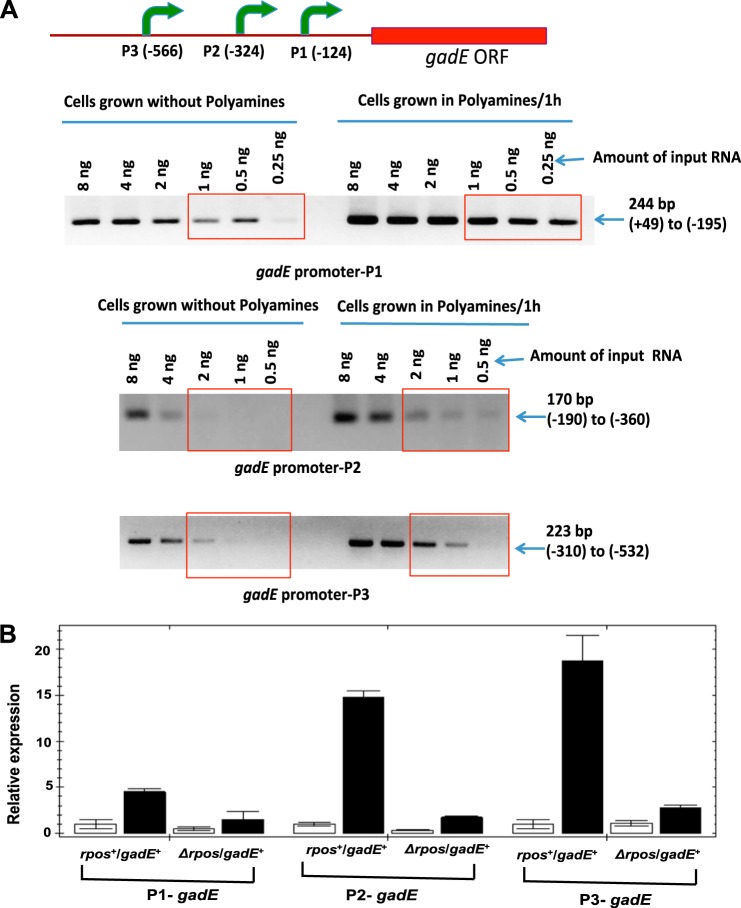
One-step reverse transcriptase-PCR method was used to amplify mRNA using Superscript III and platinum Taq high fidelity polymerase mixtures according to the manufacturer's protocol (Life Technologies). For this PCR analysis (see Fig. 4A), different amounts of enriched mRNA samples from polyamine-treated and untreated cells were used as a template to amplify gadE promoter regions using the primers described by Sayed and Foster (32).
FIGURE 4.
Polyamines and RpoS are essential for polyamine-induced activation of gadE promoters. A, single-step RT-PCR analysis of three gadE promoters (P1, P2, and P3) using enriched mRNA as a template. The different amounts of mRNAs isolated from cells grown with or without polyamines from rpoS+/gadE+ culture were taken for semiquantitative RT-PCR analysis using the gadE primers for each promoters (32) as described under “Experimental Procedures.” B, to identify the role of rpoS, polyamine mutant cultures of rpoS+/gadE+ or ΔrpoS/gadE+ cells were grown with (filled bars) or without (open bars) polyamines and after 60 min, cells were harvested, and RNA was isolated; cDNA was synthesized from enriched mRNA, and quantitative RT-PCR assays were performed using oligonucleotides specific for each gadE promoter according to the methods described under “Experimental Procedures.” The data are means of triplicate assays ± S.D.
Acid Stress and Cell Survival Assay
Polyamine mutant cultures were inoculated from LB plates into Vogel Bonner medium without amines and grown overnight to deplete intracellular amines. The cultures were further diluted in Vogel Bonner medium with and without different amine supplements as indicated and grown for 20–24 h to full growth. The cultures were harvested and resuspended in Vogel Bonner medium (pH 7.0 with 0.4% glucose) at a cell density of 1 × 1010 cells/ml and used for acid survival assays for 30 min at pH 2.5 (9).
Glutamate Decarboxylase Assay
Glutamate decarboxylase activity was measured by direct measurement of decarboxylation of l-glutamic acid as described previously (9). Briefly, for each assay 4–10 μg of protein was mixed with 100 μl of assay buffer, and the reaction was initiated by the addition of 10 μl of 10 mm l-glutamic acid containing 10 μCi/ml of l-[14C(U)]glutamic acid (260 mCi/mmol, #NEC 290; PerkinElmer Life Sciences) in a total volume of 125 μl in a 1.5-ml locked microcentrifuge tube. The reaction was incubated at 37 °C for 20 min with intermittent shaking, terminated by the addition of 125 μl of 6 n HCl to inactivate the enzyme, and incubated further for 30 min to release dissolved CO2. Radioactivity was measured in a liquid scintillation counter (LS6500; Beckman), and enzyme activity was expressed in μmol of CO2 released per mg of protein per h. Protein contents of the cell extracts were assayed by Bradford assay (43) using Bio-Rad reagent.
Western Blot Analysis
Cells were grown and deprived of amines and then incubated for 10 or 20 min with or without amines as indicated. The cultures were then harvested for protein extraction. Western blot analyses were performed with either anti-RpoS mouse monoclonal antibody (ab81737; Abcam, Cambridge, MA), anti-RpoD mouse monoclonal antibody (SP-WP004; Neoclone, Madison, WI), or anti-RpoB mouse antibody (12087; Abcam) (all 1:10,000 dilutions) using cell extracts containing 5 μg protein/lane. Anti-mouse, horseradish peroxidase-conjugated secondary antibody (GE Healthcare) was used at 1:20,000 dilution. Western blots were detected using ECL prime detection reagent (GE Healthcare) according to the manufacturer's protocol. The autoradiograms were scanned using Bio-Rad GS-800 calibrated densitometer, and the relative intensity of RpoS protein was calculated on the basis of RpoD levels.
Results
Polyamine-induced Expression of GDAR Genes and Genes in the E. coli Acid Fitness Island Is Dependent on the Presence of rpoS and gadE Genes
In the current paper, we first studied the relative importance of rpoS and gadE for the effect of polyamines in inducing the acid response genes by performing a series of microarrays. The global gene expression pattern was measured after polyamine addition to rpoS+/gadE+, ΔrpoS/gadE+, and rpoS+/ΔgadE polyamine-deficient strains. For this purpose, we first constructed a new polyamine-free mutant using deletion strains from the Keio collection and inserted an additional ΔrpoS or ΔgadE mutation (Table 1). The use of the Keio collection has the advantage that all of the mutated genes used for the construction of the different mutated strains came from the same genetic background, and thus the strains used were isogenic except for the specific deletions. In addition, we carried out these new studies in cultures grown to early stationary phase (1.0 A600) because we found that the expression of acid resistance genes and of the polyamine effects are greater in such cultures than log phase cultures (9). Because many E. coli strains contain an amber mutation in codon 33 of the rpoS gene, we confirmed by sequencing that the HT873 strain does not have this mutation (supplemental Fig. S1).
The mutant cells were grown in the absence of polyamines to an A600 of 1.0, and then the cultures were divided into two parts; to one part a mixture of 100 μm of putrescine and spermidine was added, and the other part remained untreated. The cells were harvested after 1 h, and microarrays were performed from isolated, enriched mRNA. Polyamine addition resulted in greater than 3-fold increased expression of ∼85 genes, and it also suppressed 70 genes at least 3-fold. The full microarray data are available under GEO accession number GSE67119 and in supplemental Data Sets S1 and S2.
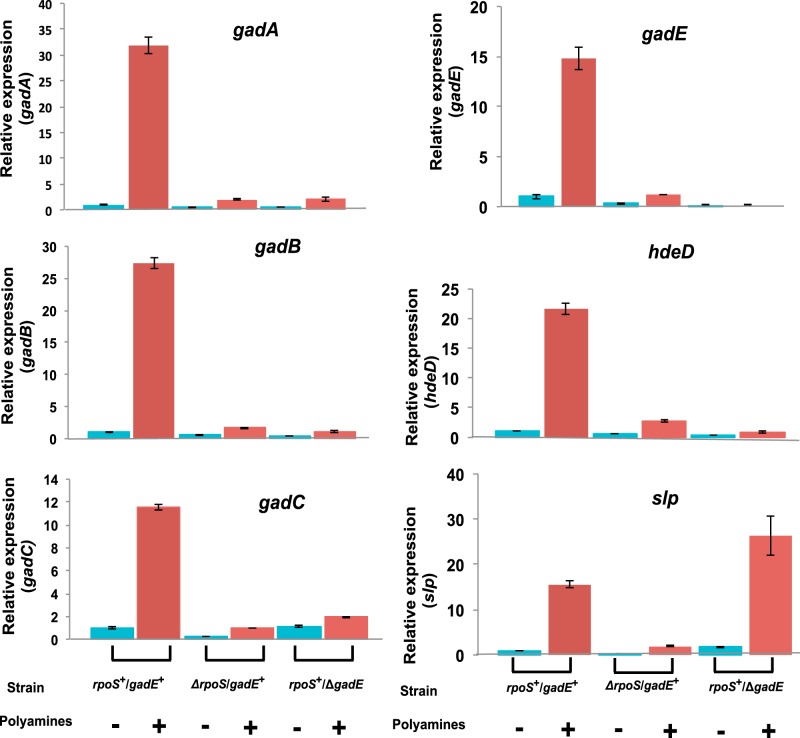
Among the pathways most affected by the addition of polyamines were the genes of the E. coli acid response pathway and genes in the acid fitness island including gadA, gadB, gadC, gadE, hdeD, hdeA, slp, dctR, and yhiD; the data for fold changes are presented in Table 3. Most striking were the very large increases in gadA and gadB expression (20- and 26-fold), confirming our previous results. Many of these inductions were markedly decreased if the cells contained a deletion in rpoS or gadE.
TABLE 3.
Effect of polyamine addition to an E. coli mutant lacking polyamines on the expression of genes in the acid fitness island and other acid response pathways and the relative roles of rpoS and gadE gene deletions
| Gene names | Functions | Fold change in gene expression (after addition of polyamines)a |
||
|---|---|---|---|---|
| rpoS+/gadE+ | ΔrpoS/gadE+ | ΔgadE/rpoS+ | ||
| gadB | Glutamate decarboxylase B | 25.9 | 8.2 | 8 |
| gadA | Glutamate decarboxylase A | 20.7 | 3 | 3 |
| gadE | Regulator of GDAR | 8.6 | 3 | |
| hdeD | Acid resistance, putative membrane transporter | 7.2 | 3 | 2.2 |
| slp | Acid resistance, outer membrane lipoprotein | 6.8 | 4 | 6 |
| dctR | Putative LuxR family repressor for C4-dicarboxylate transporter | 6 | 2 | 6.9 |
| yhiD | Predicted Mg2+ transport ATPase | 5.4 | 2 | 3.8 |
| gadC | Antiporter GABA/glutamate | 4.8 | 3 | 1.4 |
| mdtF | Anaerobic multidrug efflux transporter | 3.9 | 2 | −1.8 |
| hdeA | Periplasmic chaperone of acid denatured proteins | 3.8 | 3 | 2.3 |
| yhiU | Anaerobic multidrug efflux transporter | 2.3 | 1 | −1.7 |
| gadX | Regulator of GDAR | 1.9 | 2 | 2 |
| gadY | Small RNA regulator of GDAR | 1.9 | 1 | 2.1 |
| hdeB | Periplasmic chaperone of acid-denatured proteins | 1.7 | 2 | 1.8 |
| rpoS | Sigma factor (σ38) for RNA polymerase | 1.2 | 1.3 | |
a The data are averages of three independent microarray experiments using Affymetrix E. coli chip 2.0 (p < 0.05). The data presented for rpoS+/gadE+ strain in this table are from the microarray experiments comparing rpoS+/gadE+ and ΔgadE/rpoS+ cultures. The complete microarray data are available as supplemental Data Sets S1 and S2. Functional annotations are based on the Ecogene Database.
To supplement the microarray data, we also carried out quantitative real time PCR analyses on the effect of rpoS or gadE deletions on polyamine-induced expression of six of these mostly affected genes (Fig. 1). Polyamine addition resulted in increased expression of all of these genes in rpoS+/gadE+ strains from 12- to 32-fold, but, except for slp expression, there was no significant induction in the strains containing deletions in the rpoS or gadE genes. These results confirm the microarray data that gadE and rpoS, as well as polyamines, are critical for the induced expression of most of the genes in the E. coli acid response system.
FIGURE 1.
Quantitative real time PCR analyses showing the effects of rpoS or gadE deletion on polyamine-induced gene expression of six genes in the acid response pathway. Polyamine mutant cultures of rpoS+/gadE+ or ΔrpoS/gadE+ or rpoS+/ΔgadE strains were grown in M9 medium to deplete polyamines as described under “Experimental Procedures.” When the cultures reached 1.0 A600, the cultures were divided into two parts; to one part of each culture, 100 μm putrescine and 100 μm spermidine (+ polyamines; red bars) were added; the other part was untreated (− polyamines, blue bars). Samples were harvested after 60 min, RNA was isolated, cDNA was synthesized from enriched mRNA, and quantitative RT-PCR assays were performed using oligonucleotides specific for each gene according to the methods described under “Experimental Procedures.” The data are means of triplicate assays ± S.D.
In Fig. 2, we carried out pattern searches on putative GAD box sequences or rpoS binding sequences in the upstream sequences of these genes. In Fig. 2A, we show that the five genes induced more than 4-fold by polyamines and most of those affected by a gadE deletion all contain a conserved GAD box in the −80 to −100 bp upstream sequence region. This GAD box has previously been described as critical for the genes involved in the acid response pathway (39, 40). In Fig. 2B, we show that the eight genes induced more than 4-fold by polyamines and most of those affected by an rpoS deletion all contain a putative rpoS binding region −14 to −6 bp upstream. This RpoS binding sequence has previously been described by Weber et al. (41) and Landini et al. (27) in positions −14 to −4 in putative promoters of RpoS-controlled genes.
FIGURE 2.
Identification of GAD box and RpoS binding sites on the genes expressed differentially by polyamines. A, alignment of putative GAD box sequence from sequences upstream of five genes differentially expressed in gadE+ versus ΔgadE cells after polyamine induction. B, alignment of putative sigma38 (RpoS) binding sites from sequences upstream of acid fitness island genes and glutamate decarboxylase genes differentially expressed in rpoS+ versus ΔrpoS cells after polyamine induction. These putative binding sequences were identified by pattern search tools of coliBASE, and the frequency plots were developed by WebLogo (42).
Relative Importance of rpoS and gadE for the Polyamine-induced Glutamate Decarboxylase Activity and Acid Survival
Because the above experiments showed that both rpoS and gadE are essential for the action of the polyamines in inducing the expression of various genes in the glutamate decarboxylase acid resistance system, we then studied the role of each of these two genes for the induction of the glutamate decarboxylase protein.
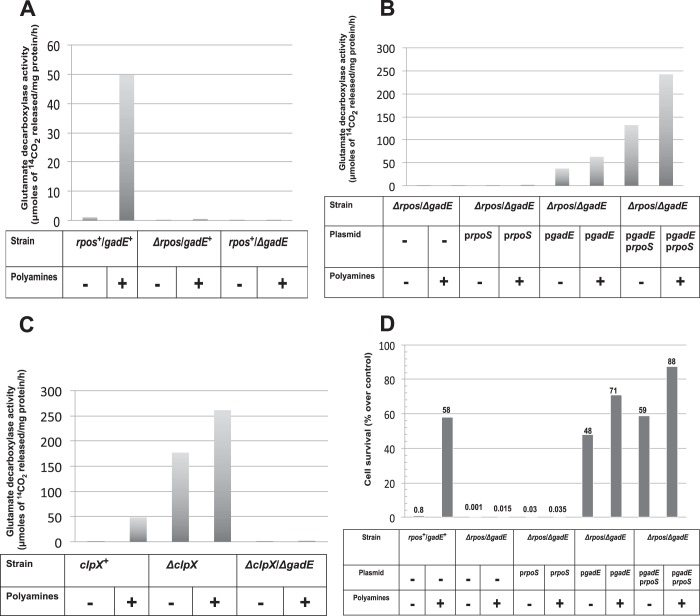
The critical importance of rpoS and gadE for the polyamine-induced expression of glutamate decarboxylase in stationary phase is shown (Fig. 3A) by comparing the amount of glutamate decarboxylase obtained upon polyamine addition to a rpoS+gadE+ and either ΔrpoS gadE+ or rpoS+ΔgadE strains. Polyamine addition increased glutamate decarboxylase activity 50–70-fold in a stationary phase culture of rpoS+/gadE+ cells; in contrast the ΔrpoS gadE+ and the rpoS+ ΔgadE strains had no activity, even if the culture contained polyamines.
FIGURE 3.
Relative role of rpoS and gadE regulators on polyamine-induced glutamate decarboxylase activities; gadE is the direct regulator of GAD activation and acid survival. A, polyamine mutants (rpoS+/gadE+) and their isogenic rpoS mutant (ΔrpoS/gadE+) and gadE mutant (rpoS+/ΔgadE) cells were grown overnight, and glutamate decarboxylase activities from the cell extracts were assayed as described in the text. B, double mutants of both ΔrpoS and ΔgadE were made in the polyamine mutant background (Table 1), and cells were transformed with either pArpoS, pQEgadE or both plasmids as indicated. Cultures were grown in Vogel Bonner medium (with necessary antibiotics) with or without polyamine supplementation (100 μm spermidine plus 100 μm putrescine) to 0.4 A600. pRpoS plasmid was constitutively expressed, whereas gadE expression was induced by adding 0.5 mm isopropyl β-d-thiogalactopyranoside, and the cultures were grown for an additional 24 h. Cell extracts were made from these cells and assayed for glutamate decarboxylase activity. The results are averages of three different assays expressed in μmol of 14CO2 released per mg of protein per h. C, deletion of either clpX or both clpX and gadE were introduced in the polyamine mutant (HT873). The cultures were deprived of amines and then grown with or without amines overnight for glutamate decarboxylase activity assays as mentioned above. D, the cultures were grown and induced by 0.5 m isopropyl β-d-thiogalactopyranoside as described above in Fig. 3B but with the inclusion of glutamate (300 μm) in the media. At the end of the 24-h incubation, the cells were tested for acid survival at pH 2.5 for 30 min.
In the above experiments, the ΔrpoS cells still contained a genomic copy of gadE, and the ΔgadE cells still had a genomic copy of rpoS. Therefore, we constructed a polyamine mutant that lacked both the rpoS and gadE genes and repeated the above experiments in the presence of prpoS, pgadE, or both. As shown in Fig. 3B, overexpression of the RpoS protein from a plasmid had no effect in the absence of the gadE gene. In contrast, overexpression of a gadE plasmid was able to partially replace an rpoS deletion. The addition of plasmids overexpressing both RpoS and GadE proteins from both plasmids plus polyamine addition resulted in a very large increase in glutamate decarboxylase activity. Hence, gadE is most directly involved in the synthesis of and activity of glutamate decarboxylase via the activation of gadA and gadB, and rpoS acts by stimulating gadE synthesis.
The importance of gadE is also shown in Fig. 3C. In this figure, we show that if the polyamine-deficient strain also contains a deletion in clpX, a mutant that is known to result in a large increase in the RpoS level by inhibition of RpoS degradation (44, 45), there is a very large increase in the glutamate decarboxylase level, even in the absence of any added polyamines. However, in the absence of gadE, there is no increase in glutamate decarboxylase activity (Fig. 3C).
The above conclusion on the direct involvement of gadE for the GDAR system is further supported by survival assays at pH 2.5 (Fig. 3D). The ΔrpoS-ΔgadE cells were highly sensitive to acid stress, and the addition of polyamines did not prevent their death. Overexpression of RpoS resulted in a trivial increase in acid survival; in contrast, GadE overexpression alone resulted in a large increase in acid survival, both in the presence or in the absence of polyamines, indicating clearly that overexpression of GadE is effective on survival even in the absence of RpoS. Overexpression of both RpoS and GadE and the addition of polyamines resulted in nearly complete protection of cells from acid stress (Fig. 3D).
Effect of rpoS and Polyamine Addition on Induction of gadE Promoters
From the above experiments, it is clear that gadE expression is directly correlated with polyamine-induced expression of glutamate decarboxylase activity. Sayed and Foster (32) have shown that the gadE gene of E. coli contains an upstream 798-bp sensory region containing three different promoters. Using a RT-PCR method, we have used the RNA from our polyamine-free mutant strain (HT873-rpoS+/gadE+) to study the effect of polyamine addition on each of the three promoters. As shown in Fig. 4A, all three promoters showed enhanced activity after polyamine treatment, particularly P2. Because we have shown (next section) that polyamines increase the RpoS level, it seemed possible that the effect of polyamines on the promoter induction was the result of the effect of polyamines on increasing the endogenous RpoS protein level. Therefore, we next tested the effect of an rpoS deletion (HT874-ΔrpoS/gadE+) on the polyamine induction of gadE promoters by quantitative real time PCR. In the absence of rpoS, there was no induction by polyamines of any of the three promoters of gadE (Fig. 4B). This is parallel to the finding that there was no induction of glutamate decarboxylase by polyamines in ΔrpoS cells (Fig. 3A).
Polyamine Addition Induces a Very Rapid Increase in the RpoS Protein Level
The data from the above experiments supported the postulation that the rpoS gene or RpoS protein could be a direct target of polyamines and that this would trigger the expression of genes in the GDAR pathway. In the next experiment, we added polyamines to cells deficient in polyamines and checked the level of rpoS gene transcription 10, 20, and 60 min after polyamine addition by quantitative real time PCR. Although there was only a 1.2-fold increase in the rpoS expression after polyamine addition in the in microarray data (Table 3), there was a modest 2.3–2.8-fold increase in rpoS expression after polyamine addition as shown by the quantitative real time PCR data (Fig. 5, A and B), when the data were normalized against the expression of control gene glyceraldehyde-3-phosphate dehydrogenase-A (gapA).
More strikingly, we found that polyamine addition markedly increased the RpoS protein level within 20 min after the addition of polyamines to the amine-deprived culture (Fig. 5C). Because RpoS protein levels differ during different stages of growth, being very low in cells during logarithmic growth and very high during stationary phase (44, 46), we tested the effect of polyamine addition at different growth phases. We found that in the early growth phase (from A600 of 0.4–1.0), there was nearly undetectable amount of RpoS protein in the absence of polyamines. However, the addition of polyamines (100 μm of putrescine plus 100 μm spermidine) for a very short time (20 min) resulted in a large increase in RpoS protein level at all optical densities (Fig. 5C). Note that polyamine addition did not change the RpoD or RpoB level (Fig. 5C). We also tested the effect of addition of different amines (putrescine, spermidine, or cadaverine) at different concentrations on the RpoS protein level and found that at least 10 μm cadaverine, putrescine, or spermidine is effective in inducing RpoS protein within 20 min (24–35-fold increase), when the assays were performed at A600 of 1.0. There was no large difference in the effect of cadaverine, putrescine, or spermidine, although spermidine showed the highest level of induction among the amines used (Fig. 5D). We also tested the effects of addition of inhibitors of RNA synthesis (10 μg/ml of rifampicin) or protein synthesis (chloramphenicol at 200 μg/ml). These inhibitors were added 10 min before polyamine addition and then after 10 or 20 min, samples were assayed for RpoS level. As shown in Fig. 5E, new protein synthesis is required for polyamine-induced expression of RpoS protein, because addition of chloramphenicol completely prevented the stimulation of RpoS synthesis. Rifampicin addition essentially showed no inhibition at 10 min and only resulted in a mild inhibition after 20 min.
Discussion
In this paper, we have extended our previous studies on the effect of polyamines in inducing the glutamate decarboxylase acid resistance pathway. We conclude that the primary effect of polyamines on this induction is the extremely rapid increase in the level of the RpoS subunit of RNA polymerase. Our results indicate that polyamines do not directly increase the synthesis of the glutamate decarboxylase enzymes; instead, the increase in the RpoS protein stimulates the synthesis of the GadE regulatory protein and other components of the glutamate decarboxylase acid resistance system.
The induction of the GDAR system is of particular importance because it is necessary for the survival of various pathogenic and nonpathogenic bacteria when exposed to the acids present in the stomach (14). Four different amino acid-dependent acid resistance systems have been reported, and each consists of two parts: a cytosolic pyridoxal phosphate-dependent decarboxylase that catalyzes decarboxylation of substrate amino acid and an inner membrane substrate/product antiporter that enables the continued operation of the system by exchanging external substrate for internal decarboxylase product (14). The GDAR system consists of the highly homologous, inducible glutamate decarboxylase GadA/GadB (pH optimum 3.7–3.8), and the glutamate/GABA antiporter GadC. The arginine-dependent acid resistance (ADAR) system consists of the acid inducible arginine decarboxylase AdiA (pH optimum 4.9–5.2) and the arginine/agmatine antiporter AdiC. The lysine-dependent acid resistance (LDAR) system is composed of the inducible lysine decarboxylase LdcI (CadA, pH optimum 5.7) and the lysine/cadaverine antiporter CadB; the ornithine-dependent acid resistance (ODAR) system consists of the inducible ornithine decarboxylase SpeF (pH optimum 7.0) and the ornithine/putrescine antiporter PotE (17, 47). Among these four, the GDAR and ADAR systems provide highest protection against extreme acid stress (11, 48, 49). The LDAR and ODAR systems provide protection during mild acid stress conditions (14, 17, 18, 47). Thus, the efficiencies of the acid resistance systems are associated with the pH optimum of the decarboxylase (GDAR > ADAR > LDAR > ODAR) (14, 47). Interestingly, three of these (acid-inducible arginine decarboxylase (adiA) (16), lysine decarboxylase (cadA) (17), and ornithine decarboxylase (speF) (18) result in the formation of amine products. However, these decarboxylase are necessarily excluded from our studies because our polyamine-deficient strain has deletions in the genes for these decarboxylase (Table 1). Thus, at this point, we have no evidence that polyamines also regulate the expression of other acid inducible decarboxylase or that the products of these three systems are helpful in inducing the acid response systems needed for colonization of E. coli cells in its human habitat. Certainly, E. coli is capable of developing a wide range of acid stress response systems, and it is possible that all four systems coordinate their response to provide protection from various levels of acid stress during colonization and survival in human host (47).
RpoS is an alternate sigma subunit of E. coli RNA polymerase (sigma38), and there have been many papers on its role in response to a variety of stresses and in adaptation to stationary phase (20–22, 24, 25, 27, 28). RpoS has been shown to control the expression of over 100 genes (50, 51). The activation and regulation of RpoS have been shown to be a tightly regulated complex process, involving transcription, mRNA turnover, small RNA regulation, translation, and proteolytic degradation (reviewed in Refs. 22, 44, 52, and 53).
GadE is a Lux-R family regulator that has been shown by various studies to be involved in the regulation of various acid stress-related genes (29, 31). A ΔgadE strain has been reported as lacking the glutamate decarboxylase acid-resistant system (30). In one virulent strain of E. coli (O157:H7), a deletion in gadE was reported to decrease the expression of more than 100 genes (54). The GadE protein binds a specific 20-bp consensus sequence located upstream to the gadA and gadB genes (called a “GAD box”) (39, 40). Sayed and Foster (32) have shown that the gadE gene contains a long 798-bp upstream sequence containing three tandem promoter sites that are bound by many regulators.
Our studies differ in several ways from most of the published studies on the function and regulation of RpoS and GadE and on the role of polyamines in various systems. Most published studies were carried out in crude media, such as LB, that contain polyamines or with strains that contain large amounts of endogenous polyamines because of the presence of many genes in the polyamine biosynthetic pathway. Our experiments were carried out in strains deleted in genes of the polyamine biosynthetic pathway and in purified media that do not contain polyamines. In addition, most of the previous studies on the effect of polyamine addition (including some of our own previous experiments) have compared cells grown for their entire growth period in the presence or absence of polyamines. Because the cells grown with polyamines grow faster than the polyamine-depleted cells, some of the effects found might have been related to these differences in growth rate rather than a specific effect of the added polyamines. Where indicated, the critical experiments in our current work were carried out after relatively short exposure of the polyamine-deprived cells to added polyamines. For example, in the experiments reported in Fig. 5, the increases in the RpoS levels were seen within 10–20 min after the addition of polyamines, and this induction is mostly at the level of translation.
A complication in interpreting some of the published papers involves the reports from several laboratories showing that there is a marked variation in different E. coli strains in the sequence of codon 33 (in 97–99 nucleotides of the rpoS gene) (53, 55–57). We have made a search of the rpoS gene sequences in the current NCBI database and have confirmed these findings. Many strains have a GAG codon (coding for glutamic acid), some have a CAG codon (coding for glutamine), and some have a TAG amber termination codon in position 33. Another complication in interpreting some of the published papers involves an important finding reported by Yoshida et al. (56), showing that polyamines facilitate the read-through of an amber mutation present in codon 33 of the rpoS gene, which was due to an increase in both the level of suppressor tRNA (supE) and the binding affinity of Gln-tRNA (supE) for ribosomes. This result is similar to the finding that polyamines are required for optimal translation of amber codons in vivo in a bacteriophage T7-E. coli system (58). Because some of the strains used in the literature, including those used to develop the concept of a polyamine modulon (MA261) (59) contain an amber mutation in codon 33, further work is needed to determine which strains have this amber mutation and to evaluate whether the effect of added polyamines is solely the result of read-through of such an amber mutation. Thus, Tkachenko et al. (60) in their studies on bacterial persister cells specifically showed that putrescine increased RpoS levels both in rpoS amber and non-amber cells.
The strain used in the current paper has the CAG codon coding for glutamine (i.e. derived from the Keio collection (BW25113) (34)). We also confirmed by sequence analysis that the strain used in our work contains the CAG codon in position 33 (supplemental Fig. S1) and thus does not contain the amber mutation.
As mentioned above, our data show that the effect of increased RpoS levels on the glutamate-dependent acid-resistant system is mediated through the expression of gadE; i.e. GadE rather than RpoS is the more direct regulator of the glutamate-dependent acid resistance system. This conclusion is based on our findings that 1) there is no expression of glutamate decarboxylase in ΔrpoS or ΔgadE cells even in the presence of added polyamines (Fig. 3); 2) deletion of gadE does not affect the level of rpoS expression (Table 3); 3) overexpression of RpoS protein either from a plasmid or as a result of deletion in the clpX/P protease system only results in a large increase in the level of glutamate decarboxylase if gadE is present (Fig. 3); and 4) most important, polyamines induce the expression of gadE promoters in rpoS+ but not in ΔrpoS cells (Fig. 4B).
In conclusion, we postulate a model in which an important function of polyamines is to increase the level of RpoS, and this increased level of RpoS leads to an increase in gadE level and subsequent expression of GDAR genes and acid resistance (Fig. 6). Thus, our study demonstrates that polyamines play a major role in the expression of RpoS protein in E. coli in addition to other factors that have been reported earlier, such as small RNAs and RNA chaperon Hfq (52, 61).
FIGURE 6.
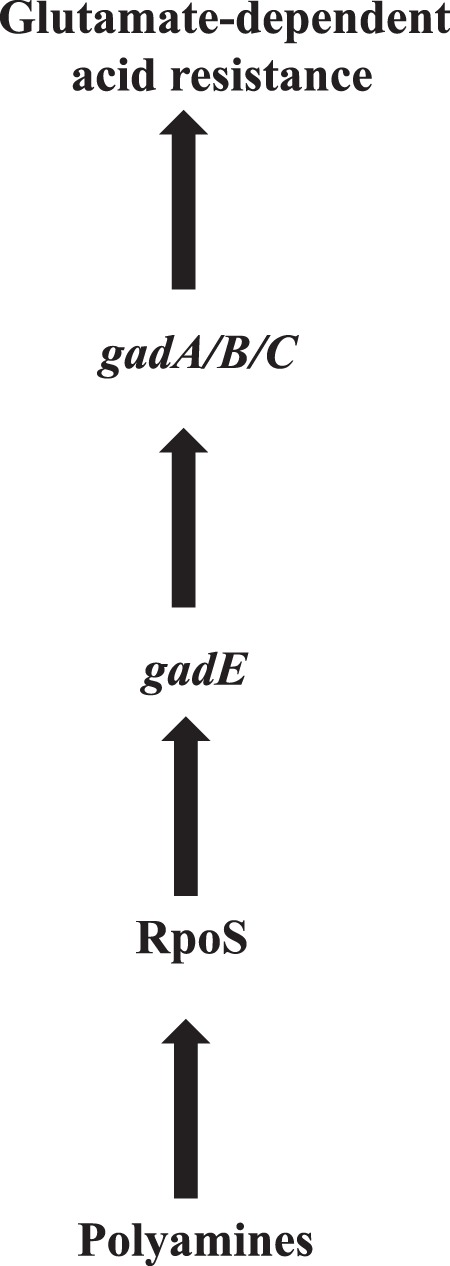
Postulated model for polyamine-induced glutamate-dependent acid response system.
Although our work has concentrated on the action of polyamines in inducing the glutamate decarboxylase-dependent acid response system and in particular on the role of RpoS and GadE, these findings will probably have a broader significance because it is well known that both RpoS and GadE have numerous effects in controlling a variety of other genes. Therefore, it is likely that our findings on the effect of polyamines on the levels of RpoS and GadE will be shown to be relevant to the control by polyamines of a variety of other systems.
Author Contributions
M. K. C. and H. T. designed the experiments; analyzed and acquired all the data, except the microarray data; and wrote and revised the manuscript. C. N. K. contributed the microarray and real time PCR analysis. W. C. helped in microarray data analysis. All of the authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Dr. John Foster (University of South Alabama, Mobile, AL) for the pQEgadE plasmid and Dr. Udo Bläsi (University of Vienna, Vienna, Austria) for the pACYCRpoS and control plasmids. We also thank Dr. Herman Edskes (Laboratory of Biochemistry and Genetics, NIDDK, NIH, Bethesda, MD) and Dr. Michael Cashel (Laboratory of Molecular Genetics, NICHD, NIH, Bethesda, MD) for critical reading of and comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health NIDDK Intramural Research Program. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Fig. S1 and Data Sets S1 and S2.
- GDAR
- glutamate-dependent acid resistance
- GAD
- glutamic acid decarboxylase
- ADAR
- arginine-dependent acid resistance
- LDAR
- lysine-dependent acid resistance
- ODAR
- ornithine-dependent acid resistance.
References
- 1. Tabor C. W., Tabor H. (1984) Polyamines. Annu. Rev. Biochem. 53, 749–790 [DOI] [PubMed] [Google Scholar]
- 2. Cohen S. S. (1998) A Guide to the Polyamines, Oxford University Press, New York [Google Scholar]
- 3. Wallace H. M., Fraser A. V., Hughes A. (2003) A perspective of polyamine metabolism. Biochem. J. 376, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Igarashi K., Kashiwagi K. (2010) Modulation of cellular function by polyamines. Int. J. Biochem. Cell Biol. 42, 39–51 [DOI] [PubMed] [Google Scholar]
- 5. Pegg A. E., Casero R. A., Jr. (2011) Current status of the polyamine research field. Methods Mol. Biol. 720, 3–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Williams K. (1997) Interactions of polyamines with ion channels. Biochem. J. 325, 289–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chattopadhyay M. K., Tabor C. W., Tabor H. (2003) Polyamines protect Escherichia coli cells from the toxic effect of oxygen. Proc. Natl. Acad. Sci. U.S.A. 100, 2261–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chattopadhyay M. K., Tabor C. W., Tabor H. (2006) Polyamine deficiency leads to accumulation of reactive oxygen species in a spe2Delta mutant of Saccharomyces cerevisiae. Yeast 23, 751–761 [DOI] [PubMed] [Google Scholar]
- 9. Chattopadhyay M. K., Tabor H. (2013) Polyamines are critical for the induction of the glutamate decarboxylase-dependent acid resistance system in Escherichia coli. J. Biol. Chem. 288, 33559–33570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gorden J., Small P. L. (1993) Acid resistance in enteric bacteria. Infect. Immun. 61, 364–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lin J., Lee I. S., Frey J., Slonczewski J. L., Foster J. W. (1995) Comparative analysis of extreme acid survival in Salmonella typhimurium, Shigella flexneri, and Escherichia coli. J. Bacteriol. 177, 4097–4104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. De Biase D., Tramonti A., Bossa F., Visca P. (1999) The response to stationary-phase stress conditions in Escherichia coli: role and regulation of the glutamic acid decarboxylase system. Mol. Microbiol. 32, 1198–1211 [DOI] [PubMed] [Google Scholar]
- 13. Richard H. T., Foster J. W. (2003) Acid resistance in Escherichia coli. Adv. Appl. Microbiol. 52, 167–186 [DOI] [PubMed] [Google Scholar]
- 14. Foster J. W. (2004) Escherichia coli acid resistance: tales of an amateur acidophile. Nat. Rev. Microbiol. 2, 898–907 [DOI] [PubMed] [Google Scholar]
- 15. De Biase D., Pennacchietti E. (2012) Glutamate decarboxylase-dependent acid resistance in orally acquired bacteria: function, distribution and biomedical implications of the gadBC operon. Mol. Microbiol. 86, 770–786 [DOI] [PubMed] [Google Scholar]
- 16. Stim-Herndon K. P., Flores T. M., Bennett G. N. (1996) Molecular characterization of adiY, a regulatory gene which affects expression of the biodegradative acid-induced arginine decarboxylase gene (adiA) of Escherichia coli. Microbiology 142, 1311–1320 [DOI] [PubMed] [Google Scholar]
- 17. Kanjee U., Gutsche I., Alexopoulos E., Zhao B., El Bakkouri M., Thibault G., Liu K., Ramachandran S., Snider J., Pai E. F., Houry W. A. (2011) Linkage between the bacterial acid stress and stringent responses: the structure of the inducible lysine decarboxylase. EMBO J. 30, 931–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kashiwagi K., Suzuki T., Suzuki F., Furuchi T., Kobayashi H., Igarashi K. (1991) Coexistence of the genes for putrescine transport protein and ornithine decarboxylase at 16 min on Escherichia coli chromosome. J. Biol. Chem. 266, 20922–20927 [PubMed] [Google Scholar]
- 19. Jung I. L., Kim I. G. (2003) Polyamines and glutamate decarboxylase-based acid resistance in Escherichia coli. J. Biol. Chem. 278, 22846–22852 [DOI] [PubMed] [Google Scholar]
- 20. Sammartano L. J., Tuveson R. W., Davenport R. (1986) Control of sensitivity to inactivation by H2O2 and broad-spectrum near-UV radiation by the Escherichia coli katF locus. J. Bacteriol. 168, 13–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lange R., Hengge-Aronis R. (1991) Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol. Microbiol. 5, 49–59 [DOI] [PubMed] [Google Scholar]
- 22. Hengge-Aronis R. (1993) Survival of hunger and stress: the role of rpoS in early stationary phase gene regulation in E. coli. Cell 72, 165–168 [DOI] [PubMed] [Google Scholar]
- 23. Gentry D. R., Hernandez V. J., Nguyen L. H., Jensen D. B., Cashel M. (1993) Synthesis of the stationary-phase sigma factor sigma s is positively regulated by ppGpp. J. Bacteriol. 175, 7982–7989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Small P., Blankenhorn D., Welty D., Zinser E., Slonczewski J. L. (1994) Acid and base resistance in Escherichia coli and Shigella flexneri: role of rpoS and growth pH. J. Bacteriol. 176, 1729–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dukan S., Nyström T. (1999) Oxidative stress defense and deterioration of growth-arrested Escherichia coli cells. J. Biol. Chem. 274, 26027–26032 [DOI] [PubMed] [Google Scholar]
- 26. Brown L., Gentry D., Elliott T., Cashel M. (2002) DksA affects ppGpp induction of RpoS at a translational level. J. Bacteriol. 184, 4455–4465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Landini P., Egli T., Wolf J., Lacour S. (2014) sigmaS, a major player in the response to environmental stresses in Escherichia coli: role, regulation and mechanisms of promoter recognition. Environ. Microbiol. Rep. 6, 1–13 [DOI] [PubMed] [Google Scholar]
- 28. Schellhorn H. E. (2014) Elucidating the function of the RpoS regulon. Future Microbiol. 9, 497–507 [DOI] [PubMed] [Google Scholar]
- 29. Masuda N., Church G. M. (2003) Regulatory network of acid resistance genes in Escherichia coli. Mol. Microbiol. 48, 699–712 [DOI] [PubMed] [Google Scholar]
- 30. Ma Z., Gong S., Richard H., Tucker D. L., Conway T., Foster J. W. (2003) GadE (YhiE) activates glutamate decarboxylase-dependent acid resistance in Escherichia coli K-12. Mol. Microbiol. 49, 1309–1320 [DOI] [PubMed] [Google Scholar]
- 31. Hommais F., Krin E., Coppée J. Y., Lacroix C., Yeramian E., Danchin A., Bertin P. (2004) GadE (YhiE): a novel activator involved in the response to acid environment in Escherichia coli. Microbiology 150, 61–72 [DOI] [PubMed] [Google Scholar]
- 32. Sayed A. K., Foster J. W. (2009) A 750 bp sensory integration region directs global control of the Escherichia coli GadE acid resistance regulator. Mol. Microbiol. 71, 1435–1450 [DOI] [PubMed] [Google Scholar]
- 33. Datsenko K. A., Wanner B. L. (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K. A., Tomita M., Wanner B. L., Mori H. (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vecerek B., Beich-Frandsen M., Resch A., Bläsi U. (2010) Translational activation of rpoS mRNA by the non-coding RNA DsrA and Hfq does not require ribosome binding. Nucleic Acids Res. 38, 1284–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Miller J. H. (1992) A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria, pp. 263–278 and 437, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 37. Vogel H. J., Bonner D. M. (1956) Acetylornithinase of Escherichia coli: partial purification and some properties. J. Biol. Chem. 218, 97–106 [PubMed] [Google Scholar]
- 38. Chattopadhyay M. K., Chen W., Tabor H. (2013) Escherichia coli glutathionylspermidine synthetase/amidase: phylogeny and effect on regulation of gene expression. FEMS Microbiol. Lett. 338, 132–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Castanie-Cornet M. P., Foster J. W. (2001) Escherichia coli acid resistance: cAMP receptor protein and a 20 bp cis-acting sequence control pH and stationary phase expression of the gadA and gadBC glutamate decarboxylase genes. Microbiology 147, 709–715 [DOI] [PubMed] [Google Scholar]
- 40. Ma Z., Masuda N., Foster J. W. (2004) Characterization of EvgAS-YdeO-GadE branched regulatory circuit governing glutamate-dependent acid resistance in Escherichia coli. J. Bacteriol. 186, 7378–7389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Weber H., Polen T., Heuveling J., Wendisch V. F., Hengge R. (2005) Genome-wide analysis of the general stress response network in Escherichia coli: sigmaS-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 187, 1591–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. (2004) WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 44. Battesti A., Majdalani N., Gottesman S. (2011) The RpoS-mediated general stress response in Escherichia coli. Annu. Rev. Microbiol. 65, 189–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bougdour A., Gottesman S. (2007) ppGpp regulation of RpoS degradation via anti-adaptor protein IraP. Proc. Natl. Acad. Sci. U.S.A. 104, 12896–12901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bougdour A., Cunning C., Baptiste P. J., Elliott T., Gottesman S. (2008) Multiple pathways for regulation of sigmaS (RpoS) stability in Escherichia coli via the action of multiple anti-adaptors. Mol. Microbiol. 68, 298–313 [DOI] [PubMed] [Google Scholar]
- 47. Kanjee U., Houry W. A. (2013) Mechanisms of acid resistance in Escherichia coli. Annu. Rev. Microbiol. 67, 65–81 [DOI] [PubMed] [Google Scholar]
- 48. Castanie-Cornet M. P., Penfound T. A., Smith D., Elliott J. F., Foster J. W. (1999) Control of acid resistance in Escherichia coli. J. Bacteriol. 181, 3525–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lin J., Smith M. P., Chapin K. C., Baik H. S., Bennett G. N., Foster J. W. (1996) Mechanisms of acid resistance in enterohemorrhagic Escherichia coli. Appl. Environ. Microbiol. 62, 3094–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Patten C. L., Kirchhof M. G., Schertzberg M. R., Morton R. A., Schellhorn H. E. (2004) Microarray analysis of RpoS-mediated gene expression in Escherichia coli K-12. Mol. Genet. Genomics 272, 580–591 [DOI] [PubMed] [Google Scholar]
- 51. Lacour S., Landini P. (2004) SigmaS-dependent gene expression at the onset of stationary phase in Escherichia coli: function of sigmaS-dependent genes and identification of their promoter sequences. J. Bacteriol. 186, 7186–7195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gottesman S. (2004) The small RNA regulators of Escherichia coli: roles and mechanisms. Annu. Rev. Microbiol. 58, 303–328 [DOI] [PubMed] [Google Scholar]
- 53. Lange R., Hengge-Aronis R. (1994) The cellular concentration of the sigma S subunit of RNA polymerase in Escherichia coli is controlled at the levels of transcription, translation, and protein stability. Genes Dev. 8, 1600–1612 [DOI] [PubMed] [Google Scholar]
- 54. Kailasan Vanaja S., Bergholz T. M., Whittam T. S. (2009) Characterization of the Escherichia coli O157:H7 Sakai GadE regulon. J. Bacteriol. 191, 1868–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Atlung T., Nielsen H. V., Hansen F. G. (2002) Characterisation of the allelic variation in the rpoS gene in thirteen K12 and six other non-pathogenic Escherichia coli strains. Mol. Genet. Genomics 266, 873–881 [DOI] [PubMed] [Google Scholar]
- 56. Yoshida M., Kashiwagi K., Kawai G., Ishihama A., Igarashi K. (2002) Polyamines enhance synthesis of the RNA polymerase sigma 38 subunit by suppression of an amber termination codon in the open reading frame. J. Biol. Chem. 277, 37139–37146 [DOI] [PubMed] [Google Scholar]
- 57. Subbarayan P. R., Sarkar M. (2004) A comparative study of variation in codon 33 of the rpoS gene in Escherichia coli K12 stocks: implications for the synthesis of sigma (s). Mol. Genet. Genomics 270, 533–538 [DOI] [PubMed] [Google Scholar]
- 58. Tabor H., Tabor C. W. (1982) Polyamine requirement for efficient translation of amber codons in vivo. Proc. Natl. Acad. Sci. U.S.A. 79, 7087–7091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yoshida M., Kashiwagi K., Shigemasa A., Taniguchi S., Yamamoto K., Makinoshima H., Ishihama A., Igarashi K. (2004) A unifying model for the role of polyamines in bacterial cell growth, the polyamine modulon. J. Biol. Chem. 279, 46008–46013 [DOI] [PubMed] [Google Scholar]
- 60. Tkachenko A. G., Kashevarova N. M., Karavaeva E. A., Shumkov M. S. (2014) Putrescine controls the formation of Escherichia coli persister cells tolerant to aminoglycoside netilmicin. FEMS Microbiol. Lett. 361, 25–33 [DOI] [PubMed] [Google Scholar]
- 61. Soper T., Mandin P., Majdalani N., Gottesman S., Woodson S. A. (2010) Positive regulation by small RNAs and the role of Hfq. Proc. Natl. Acad. Sci. U.S.A. 107, 9602–9607 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.