Background: Increased phosphorylation of the translation initiation factor eIF2α (eIF2α-P) promotes apoptosis during hyperosmotic stress.
Results: Induction of the PP1 phosphatase subunit GADD34 (which dephosphorylates eIF2α-P) promotes adaptation.
Conclusion: GADD34 promotes survival by increasing the uptake of small neutral amino acids via the amino acid transporter SNAT2.
Significance: Rapid adaptation to changes in extracellular osmolarity is inhibited by eIF2α-P signaling.
Keywords: amino acid transport, apoptosis, eIF2, protein synthesis, stress response
Abstract
Cells respond to shrinkage induced by increased extracellular osmolarity via programmed changes in gene transcription and mRNA translation. The immediate response to this stress includes the induction of expression of the neutral amino acid transporter SNAT2. Increased SNAT2-mediated uptake of neutral amino acids is an essential adaptive mechanism for restoring cell volume. In contrast, stress-induced phosphorylation of the α subunit of the translation initiation factor eIF2 (eIF2α) can promote apoptosis. Here we show that the response to mild hyperosmotic stress involves regulation of the phosphorylation of eIF2α by increased levels of GADD34, a regulatory subunit of protein phosphatase 1 (PP1). The induction of GADD34 was dependent on transcriptional control by the c-Jun-binding cAMP response element in the GADD34 gene promoter and posttranscriptional stabilization of its mRNA. This mechanism differs from the regulation of GADD34 expression by other stresses that involve activating transcription factor 4 (ATF4). ATF4 was not translated during hyperosmotic stress despite an increase in eIF2α phosphorylation. The SNAT2-mediated increase in amino acid uptake was enhanced by increased GADD34 levels in a manner involving decreased eIF2α phosphorylation. It is proposed that the induction of the SNAT2/GADD34 axis enhances cell survival by promoting the immediate adaptive response to stress.
Introduction
The regulation of cell volume in response to changes in extracellular osmolarity is essential for cell survival. Cells respond to hyperosmotic stress by accumulating organic molecules to counteract elevated external ionic strength. This adaptation involves the synthesis of various molecular chaperones, regulatory proteins, enzymes, and transporters that promote survival (1, 2). TonEBP (NFAT5) is a major transcription factor that translocates to the nucleus during hyperosmotic stress, binds to osmotic response elements in the promoters of genes important for adaptation, and activates their transcription (1, 2). Among the targets of TonEBP is the sodium-coupled neutral amino acid transporter 2 (SNAT2)3 (3). SNAT2 is a member of the system A transporters that mediates the uptake of neutral amino acids (compatible solutes) and, therefore, plays a crucial role in survival during hyperosmotic stress (4–7).
An important component of the cellular response to hyperosmotic stress is the regulation of mRNA translation. Similar to other stresses (ER stress, UV irradiation, starvation, etc.), hyperosmotic stress leads to global inhibition of translation initiation (8, 9). This inhibition is mediated in part by phosphorylation of the α subunit of the translation initiation factor eIF2 (10, 11). eIF2 delivers the initiator tRNA methionine complex to the translation machinery. However, phosphorylation of its α subunit on Ser51 (eIF2α-P) during stress renders it inactive and causes inhibition of translation initiation (10). Although translation of the majority of mRNAs is inhibited, it is still necessary to synthesize proteins (e.g. transcription factors, chaperones, and transporters) that play an adaptive role in the stress response (10, 12, 13). This is accomplished by alternative modes of translation initiation (14) that do not require the mRNA 5′ cap. These mRNAs have internal ribosome entry sites and upstream ORFs. A prime example of the latter is the upstream ORF-mediated translation of the ATF4 mRNA, an important transcription factor of the integrated stress response (15, 16).
If stress is mild and short-lived, the adaptive response promotes survival. However, cells undergo apoptosis when stress is severe or prolonged. The necessary step in the recovery of protein synthesis is dephosphorylation of eIF2α-P and the reactivation of translation (17–19). In mammalian cells, this is mediated by protein phosphatase 1 (PP1) in complex with regulatory subunits: CReP, which is constitutively expressed and controls basal levels of eIF2α phosphorylation (20), and GADD34, which is induced late in stress responses (17).
Expression of GADD34 is regulated at multiple levels: 1) transcription, 2) translation, and 3) protein stability. 1) During ER stress, ATF4 induces GADD34 transcription (21), whereas C-Jun is the transcription factor during genotoxic stress (22). Transcriptional control in other stresses has not been evaluated. The GADD34 promoter contains sequence motifs similar to the palindromic octanucleotide cAMP response element (CRE), the binding site for different members of the ATF/cAMP response element-binding protein family (21, 22). This suggests other members of this family as potential transcription factors that induce GADD34 gene expression. 2) Phosphorylation of eIF2α during stress promotes the upstream ORF-mediated translation of GADD34 (23). 3) Phosphorylation of GADD34 has been shown to enhance its degradation (24).
The functions of GADD34 have been well studied under most stress conditions that involve its ATF4-mediated transcriptional induction (17, 21, 25). However, little is known regarding its involvement in the hyperosmotic stress response. Here we show that mild hyperosmotic stress induces GADD34 protein following transient protein synthesis inhibition. Increased levels of GADD34 are due to transcriptional induction and efficient translation. Interestingly, despite eIF2α phosphorylation, mild hyperosmotic stress does not promote ATF4 mRNA translation. Therefore, unlike other stresses, the transcriptional component of GADD34 induction is ATF4-independent.
In this study, we investigated the regulation of GADD34 and the role of this protein in the hyperosmotic stress response. We provide evidence that stress-induced GADD34 mRNA levels were partially due to transcriptional activation by JNK and posttranscriptional stabilization of the GADD34 mRNA. We have shown previously that severe hyperosmotic stress leads to eIF2α phosphorylation, which has proapoptotic functions (8). We show here that GADD34, which is also induced during severe hyperosmotic stress, antagonizes eIF2α-P. GADD34 depletion or pharmacologic inhibition of its activity led to decreased levels of the SNAT2 protein on plasma membranes, decreased transporter activity, and decreased adaptation to increased extracellular osmolarity. We conclude that GADD34 promotes survival in part via the stimulation of transport of neutral amino acids by the system A transporter SNAT2.
Materials and Methods
Cells and Biological Materials
Mouse embryonic fibroblasts (MEFs) were grown in high-glucose DMEM supplemented with 10% FBS, 2 mm l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin in 5% CO2 at 37 °C. Lentiviral particles expressing shRNA against ATF4, ATF2, GADD34, and control pLKO1 (Sigma-Aldrich) were propagated in HEK293T cells as described previously according to a standard protocol (26). After infection, cells were selected in the presence of puromycin for 5 days. For hyperosmolar treatments, cells were treated with media that were 500 or 600 mosmol/liter, prepared by adding sorbitol to growth medium (200 or 300 mm, respectively). The salubrinal derivative Sal003 (Sal) was from Tocris, and MG132 was from Enzo Life Sciences. Thapsigargin (Tg, Sigma-Aldrich) was used at 400 nm as described previously (27), and actinomycin D was from Sigma-Aldrich.
Pro Uptake
Cells were grown in 24-well plates. Transport assays were performed in Earle's balanced salt solution containing Pro (100 μm, 4 μCi/ml) at 37 °C for 2 min (27). For assays in Na+-free medium, the NaCl in Earle's balanced salt solution was replaced with choline chloride.
Caspase 3 Activity
Activity was determined with the caspase 3 substrate DEVD-7-amido-4-trifluoromethylcoumarin (Calbiochem/Millipore). Cells were lysed in 10 mm potassium HEPES (pH 7.2), 1% CHAPS, 150 mm NaCl, 0.1% sodium dodecyl sulfate, 1 mm dithiothreitol, and 1 mm EDTA. The substrate (100 μm) was added to the cell lysate (100 μg of protein) and incubated in the dark for 1 h at 37 °C. The product was measured using a fluorescence reader with excitation at 360 nm and emission at 460 nm (ThermoMax microplate reader, Molecular Devices).
Luciferase Activity
The GADD34 promoter region (152 nucleotides, containing 118 nucleotides of the promoter region and 34 nucleotides of the GADD34 5′ UTR mRNA) was cloned upstream of the luciferase ATG initiation codon into the NheI and XhoI sites of the pGL3-basic plasmid. The GADD34 promoter region was amplified from mouse genomic DNA using the primers ACG TGC TAG CTG AGA AGC TGC GGT GAC CTC AC and ACG TCT CGA GTC ACA GGC TGT AGC AAA GGC TGT C. The mutants were created by changing the CRE sequence (GTGACGTCAG) to ACTTATGTCA and the GGG into CTC in the putative NF-κB binding site (GGGACTTTCC). Site-directed mutagenesis was performed by Mutagenex. For the luciferase assays, MEFs in 10-cm plates were transfected with 8 μg of GADD34-firefly luciferase plasmid and 2 μg of Renilla luciferase plasmid using X-tremeGENE DNA transfection reagent (Roche) as described by the manufacturer. Cells were split 24 h after transfection. Cells were treated with 500 mosmol hyperosmotic medium for the indicated times. Cells were then washed with cold PBS and assayed for luciferase activity using the Dual-Luciferase reporter assay system (Promega) as described by the manufacturer. Data were analyzed as firefly luciferase expression over Renilla luciferase expression and normalized to expression in the wild-type MEFs.
Metabolic Labeling
Cells were seeded in 24-well plates (5 × 104 cells/well), grown for 24 h, and subjected to hypertonic treatment. [35S]Met/Cys was added (30 μCi/ml EXPRE35S35S protein labeling mix, PerkinElmer Life Sciences) for an additional 20 min. Cells were washed twice with PBS, followed by three changes of 5% trichloroacetic acid, 1 mm Met for 10 min on ice. Precipitates were dissolved in 200 μl of 1 m NaOH, 0.5% sodium deoxycholate, and radioactivity was determined by liquid scintillation counting. Protein was assayed in aliquots as described previously (8).
Polysome Profile Analysis and mRNA Distribution
Polysome profiles were prepared as described previously (28). Briefly, cells were seeded in 150-mm culture dishes and grown to 70% confluence. Following the indicated treatments, 100 μg/ml cycloheximide was added to cells for 3 min at 37 °C. Cells were washed twice with cold PBS containing 100 μg/ml cycloheximide, scraped, and pelleted at 4000 rpm for 10 min. The cell pellets were lysed in buffer (10 mm HEPES-KOH (pH 7.4), 2.5 mm MgCl2, 100 mm KCl, 0.25% Nonidet P-40, 200 unit/ml RNase inhibitor (RNaseOUT, Invitrogen)) and EDTA-free protease inhibitor (Roche Applied Science), kept on ice for 20 min, and then passed 15 times through a 23-gauge needle. Lysates were spun at 14,000 rpm for 15 min, and supernatants (cytosolic cell extracts) were collected. Approximately 10–15 absorbance units (260 nm) of lysates were layered over 10–50% cold sucrose gradients in buffer (10 mm potassium HEPES (pH 7.4), 2.5 mm MgCl2, and 100 mm KCl). Gradients were centrifuged at 17,000 rpm in a Beckman SW28 rotor for 15 h at 4 °C. After centrifugation, 12 fractions (1.2 ml/fraction) were collected. Cell fractionation to obtain cytoplasmic and ER-associated ribosomes was performed as described previously (29). Polysome profiles of ER-associated ribosomes were obtained by the methods described above.
RNA and mRNA Analyses
RNA from MEFs and from fractions of the polysome analysis was extracted with TRIzol or TRIzol LS (Invitrogen), respectively, according to the protocol of the manufacturer. For cDNA synthesis, SuperScript III first-strand synthesis super mix (Invitrogen) was used, and the abundance was quantified by qPCR using VeriQuest SYBR Green qPCR master mix (Affymetrix) with the StepOnePlus real-time PCR system (Applied Biosystems) as described before (30). The following primers were used: GAPDH, CGC CTG GAG AAA CCT GCC AAG TAT G (forward) and GGT GGA AGA ATG GGA GTT GCT GTT G (reverse); GADD34, TAC CCC TGT CTC TGG TAA CCT (forward) and TGG CTT TGC ATT GTA CTC ATC A (reverse); SNAT2(SLC38A2), TAA TCT GAG CAA TGC GAT TGT GG (forward) and AGA TGG ACG GAG TAT AGC GAA AA (reverse); and ATF4, GTT TGA CTT CGA TGC TCT GTT TC (forward) and GGG CTC CTT ATT AGT CTC TTG G (reverse). mRNA half-lives were determined as described previously (30).
Other Methods
Cell extracts were prepared as described previously (8). Membrane fractions were obtained as described previously (31). Biotinylated plasma membrane proteins were isolated with a cell surface protein isolation kit (Pierce) according to the instructions of the manufacturer. Protein concentration was measured with a DC protein assay kit (Bio-Rad) with bovine serum albumin as a standard. Antibodies used in this study were as follows: anti-phospho-ATF2 (catalog no. sc-8398), ATF2 (catalog no. sc-187), ATF3 (catalog no. sc-188), ATF4 (catalog no. sc-200), Calnexin (catalog no. sc-6465), C/EBP homologous protein (CHOP) (catalog no. sc-7351), GADD34 (catalog no. sc-825), eIF2 (catalog no. sc-13327), JNK1/2 (catalog no. sc-137018), and S6 (catalog no. sc-13007) were from Santa Cruz Biotechnology. Anti-caspase 3 (catalog no. 9662), phospho-ERK1/2 (catalog no. 9101), ERK1/2 (catalog no. 4695), histone H3 (catalog no. 9715), Ire1α (catalog no. 3294), phospho-JNK1/2 (catalog no. 4671), phospho-c-Jun (catalog no. 3270), c-Jun (catalog no. 9165), phospho-p38 MAPK (catalog no. 9211), p38 MAPK (catalog no. 9212), phospho-NF-κB-p65 (catalog no. 3033), phospho-S6 (catalog no. 5364), phospho-S6K Thr-389 (catalog no. 9205), and S6K (catalog no. 9202) were from Cell Signaling Technology. Anti-phospho-eIF2α (catalog no. NB110-56949) was from Novus Biologicals, anti-Snat2 (catalog no. BMP081) was from Medical and Biological Laboratories, anti-tubulin (catalog no. T9026) was from Sigma-Aldrich, and anti-ubiquitin was from Dako (catalog no. Z0458). Anti-ATPase (Na+/K+) α-1 subunit (a6F) was obtained from the Developmental Studies Hybridoma Bank, hybridoma (deposited by D. M. Fambrough). Statistical significance between groups was evaluated using Student's t test. p < 0.05 was considered significant.
Results
GADD34 Is Induced during Hyperosmotic Stress in an ATF4-independent Manner
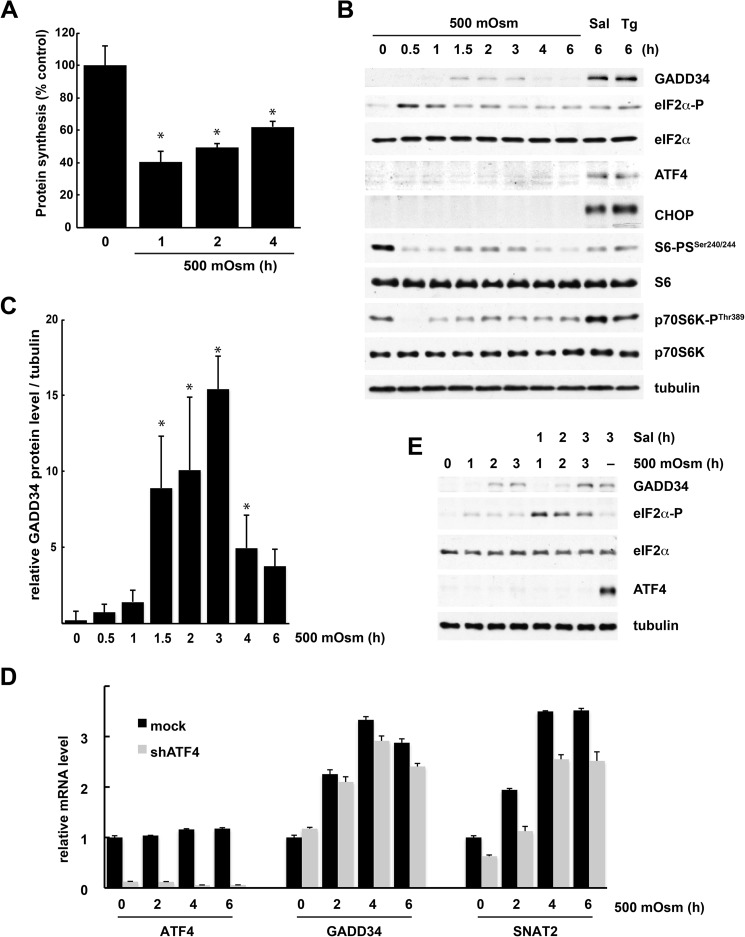
Exposure of MEFs to mild hyperosmotic stress (500 mosmol) led to an initial (1-h) decrease in the rates of protein synthesis followed by gradual recovery (Fig. 1A). After 4 h of stress, the protein synthesis rates recovered to 60% of the level in control cells. Signaling pathways known to regulate mRNA translation were modified by stress. eIF2α was phosphorylated rapidly, followed by dephosphorylation after 1.5 h of treatment (Fig. 1B). p70S6K, the protein kinase activated by mTOR, and the downstream target, the ribosomal protein S6, were transiently dephosphorylated (Fig. 1B). GADD34 mRNA and protein were increased after 1.5 h of treatment, followed by a decline at later times (Fig. 1, B–D). The induction of GADD34 was in good correlation with the kinetics of eIF2α dephosphorylation during the course of the stress. However, eIF2α phosphorylation elicited by hyperosmotic stress did not result in increased ATF4 protein (Fig. 1B). A similar phenomenon has been observed in MEFs exposed to ultraviolet light (32) or upon induction of ER stress via the proteasome inhibitor MG132 (33). As expected, when induction of eIF2α phosphorylation was promoted by the inhibitor of eIF2α dephosphorylation Sal or the ER stressor Tg, cells responded with induction of ATF4 protein (Fig. 1B). Hyperosmotic stress also did not induce the transcription factor CHOP, a proapoptotic target downstream of ATF4 (Fig. 1B). Because ATF4 activates GADD34 transcription during other stresses (21), the absence of the ATF4 program during mild hyperosmotic stress prompted us to investigate the mechanism of GADD34 induction.
FIGURE 1.
ATF4-independent induction of the PP1 phosphatase subunit protein GADD34 during mild hyperosmotic stress. A, [35S]Met/Cys incorporation into proteins in MEFs treated with 500 mosmol medium for the indicated times. Results are the means of triplicate determinations. B, Western blot analysis of extracts from cells treated with 500 mosmol medium, Sal, or Tg for the indicated times. C, quantification of the intensities of GADD34 protein levels was evaluated from three independent experiments (B) using National Institutes of Health ImageJ software. Data are expressed as the percentage of the intensities obtained from Tg-treated samples. D, qRT-PCR analysis of mRNAs from cells either left untreated or treated with 500 mosmol medium after adenovirus infection for the corresponding shRNAs. Triplicate determinations normalized to GAPDH mRNA levels are shown. E, Western blot analysis of extracts from cells treated with 500 mosmol medium alone or in the presence of Sal (15 μm) for the indicated times. *, p < 0.05 relative to untreated cells. Error bars represent mean ± S.E.
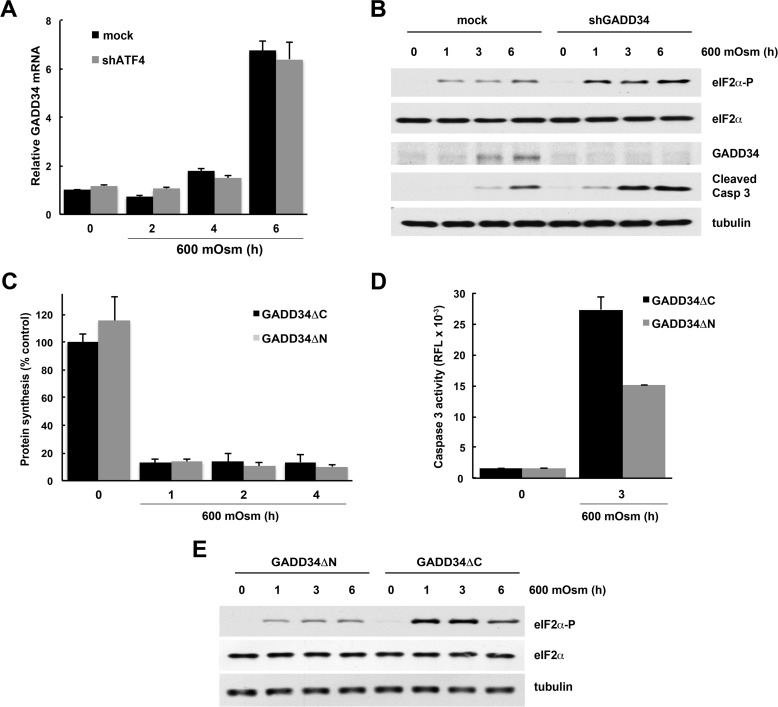
Basal levels of ATF4 protein are present in dividing cells (34). This ATF4 is required for the expression of amino acid transporter genes (35, 36). To exclude the possible contribution of these low levels of ATF4 to the induction of GADD34 by hyperosmotic stress, we knocked down ATF4 expression using shRNA and measured gene expression during osmotic stress (Fig. 1D). Although basal SNAT2 mRNA levels were decreased in shRNA-treated cells (Fig. 1D), the degree of induction of SNAT2 and GADD34 expression was not affected.
The lack of induction of ATF4 by mild hyperosmotic stress was puzzling because many stresses that trigger eIF2α phosphorylation also induce ATF4 mRNA translation. To investigate this issue, the level of eIF2α-P in stressed cells was increased with Sal treatment (Fig. 1E). Sal did not cause accumulation of ATF4 protein under these conditions although high levels of eIF2α-P were present. In contrast, ATF4 was readily detected in Sal-treated cells under isotonic conditions. These data suggest that hyperosmotic stress inhibits ATF4 protein accumulation.
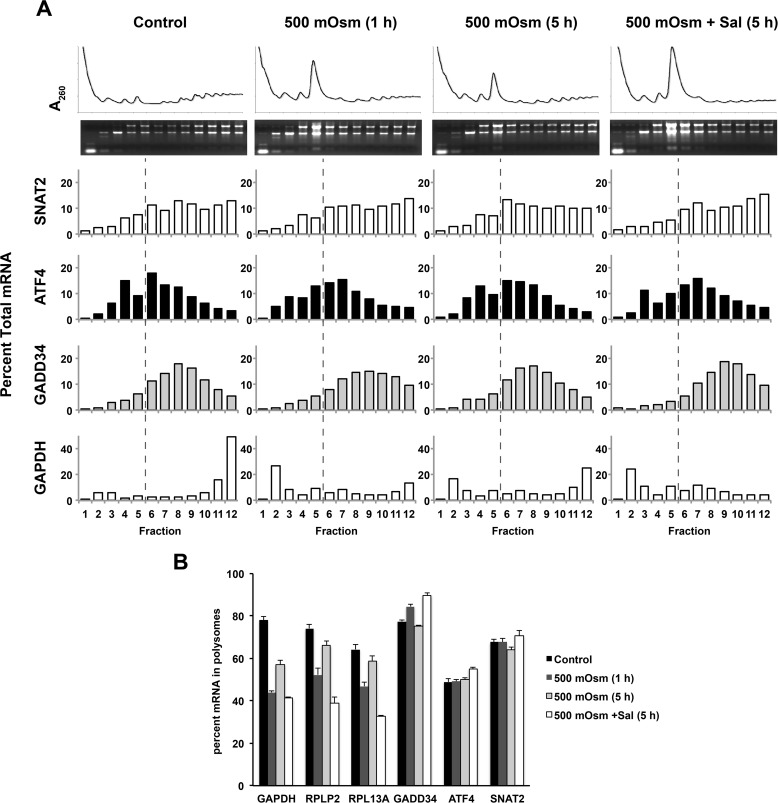
Further evidence that ATF4 mRNA is not translated more efficiently during mild hyperosmotic stress was provided by examining the distribution of mRNAs on sucrose gradients. Traces of the absorbance at 260 nm (A260) showed an initial (1 h of stress) redistribution of polyribosomes to monosomes followed by partial recovery of polyribosomes during prolonged stress (5 h of treatment). This pattern was consistent with the effects of hyperosmotic stress on protein synthesis rates (compare Figs. 1A and 2A). The sedimentation pattern of GAPDH and mTOR-regulated mRNAs that contain 5′-terminal oligopyrimidine tracts (encoding ribosomal proteins P2 and L13a) was in agreement with the A260 traces (Fig. 2B). Exposure of cells to Sal in addition to hyperosmotic stress for 5 h decreased the translation of these three mRNAs (Fig. 2, A and B), suggesting that the level of eIF2α phosphorylation has an impact on protein synthesis recovery during mild hyperosmotic conditions. The GADD34 mRNA was actively translated during hyperosmotic stress, and Sal made this process more efficient, as expected for an mRNA that uses an upstream ORF-mediated regulatory mechanism (23).
FIGURE 2.
Changes in the association of specific mRNAs with polyribosomes during mild hyperosmotic stress. A, MEFs were treated with 500 mosmol medium alone or in the presence of Sal (15 μm) for the indicated times, and the distribution of mRNAs in polysomes was analyzed using qRT-PCR. mRNA in each fraction is expressed as the percentage of total mRNA. The dashed lines indicate that fractions 6–12 contain polyribosomes. B, quantification of three independent experiments from A. Error bars represent mean ± S.E.
Finally, increased eIF2α phosphorylation in Sal-treated cells did not affect the translation of ATF4 mRNA (Fig. 2, A and B). We conclude that ATF4 protein levels do not change during mild hyperosmotic stress because mRNA levels and translational efficiency are not affected.
These results demonstrate that the modulation of eIF2α phosphorylation during mild hyperosmotic stress contributes to translational control in a manner independent of ATF4. This is different from other stresses in which the ATF4-mediated induction of GADD34 controls the phosphorylation status of eIF2α and, therefore, translational reprogramming in the stressed cells (18, 19, 21).
Signaling Pathways Induced by Hyperosmotic Stress Regulate GADD34 Expression
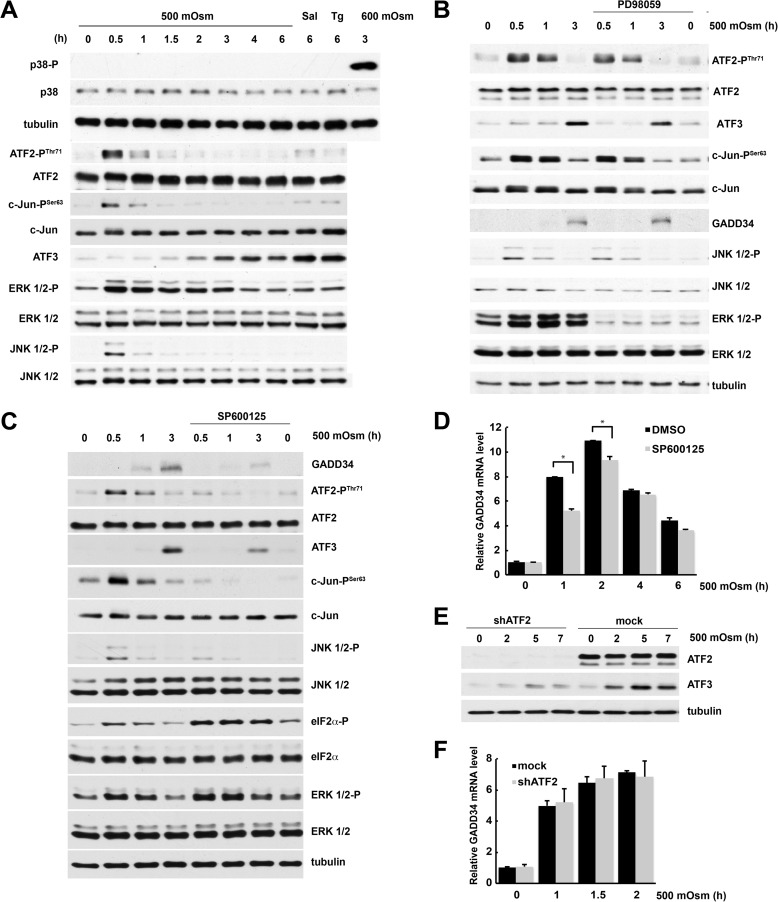
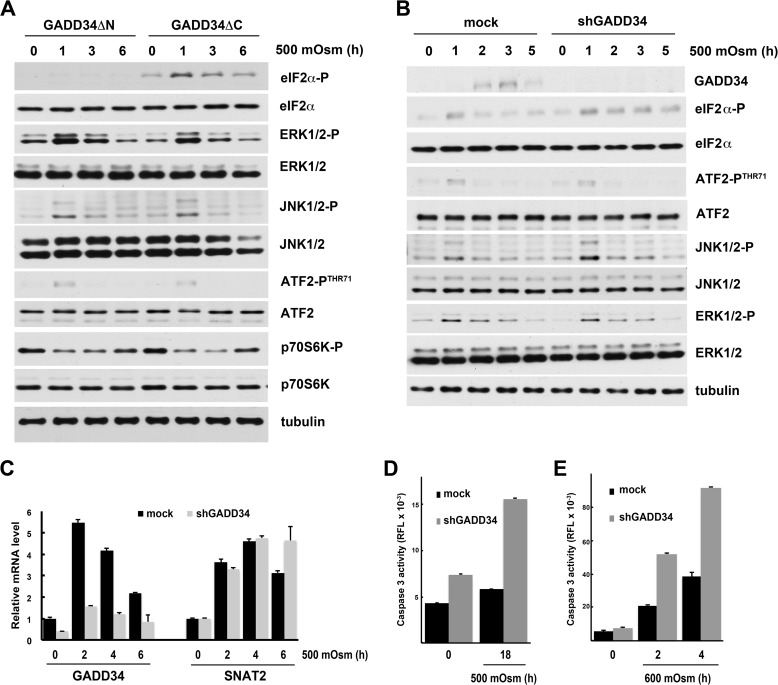
To dissect the mechanism of GADD34 induction, we studied the signaling pathways that regulate this process. First, we tested MAP kinase signaling cascades that are activated by hyperosmotic stress. In contrast to severe stress (600 mosmol), mild hyperosmotic stress (500 mosmol) did not activate p38 kinase (Fig. 3A). However, ERK1/2 and JNK1/2 were transiently phosphorylated on Thr-202/Tyr-204 and Thr-183/Tyr-185, respectively, during early stress (Fig. 3A). Activation of these kinases correlated with the early transient phosphorylation of c-Jun and ATF2 transcription factors and was followed by the delayed induction of ATF3 (Fig. 3A). We next showed that the ERK signaling pathway is not involved in GADD34 induction (Fig. 3B). Inhibition of ERK1/2 with PD98059 had a negligible effect on the phosphorylation of JNK1/2, ATF2, and c-Jun and on ATF3 expression (Fig. 3B). We therefore hypothesized that JNK1/2 are required for GADD34 induction. Treatment with SP600125, an inhibitor of JNK1/2, decreased the phosphorylation of ATF2 and c-Jun and the levels of GADD34 and ATF3 proteins (Fig. 3C). SP600125 reduced the stress-induced accumulation of GADD34 mRNA by one-third at early times. The effect of the inhibitor decreased after longer treatment (Fig. 3D). These data suggest that phosphorylation of ATF2 and c-Jun by JNK1/2 may regulate GADD34 gene expression (Fig. 3, A and C).
FIGURE 3.
Induction of GADD34 levels during mild hyperosmotic stress involves activation of the JNK kinase. A–D, MEFs were treated with 500 mosmol medium alone or in the presence of Sal or Tg (A), the ERK kinase inhibitor PD98059 (50 μm, B), or the JNK kinase inhibitor SP600125 (25 μm, C and D). A–C, Western blot analysis of cell extracts. D, qRT-PCR analysis of mRNA. Triplicate determinations were normalized to GAPDH mRNA signals. DMSO, dimethyl sulfoxide. E, Western blot analysis of extracts from cells treated with 500 mosmol medium after infection with lentivirus for the indicated shRNAs. F, qRT-PCR analysis of mRNAs isolated from cells treated with 500 mosmol medium after infection with lentivirus for the indicated shRNAs. *, p < 0.05 relative to cells without inhibitor. Error bars represent mean ± S.E.
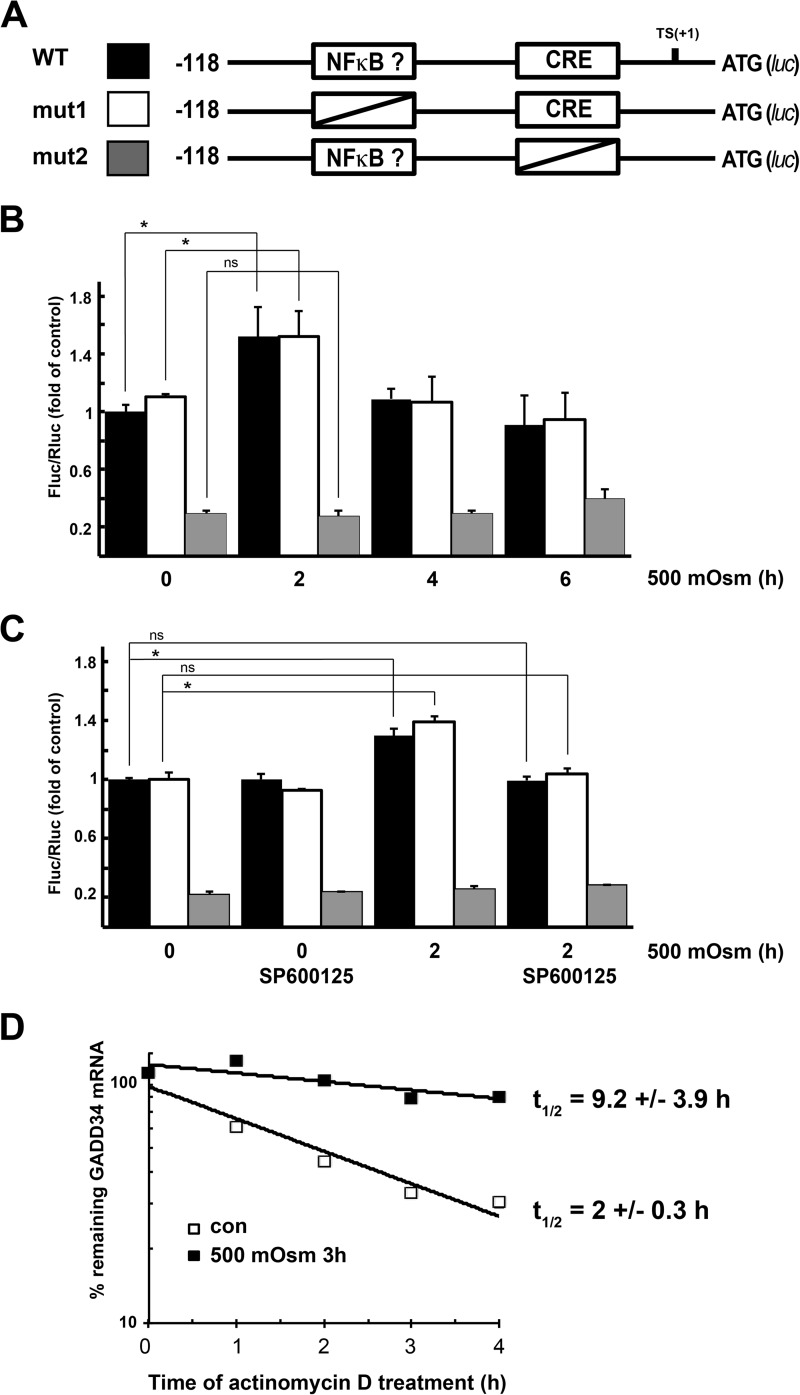
This hypothesis is supported by earlier reports that ATF2 and c-Jun can bind a conserved element in the GADD34 promoter close to the TATA box (22). To explore the potential role of ATF2 in GADD34 gene regulation during hyperosmotic stress, we knocked down ATF2 (Fig. 3E) and observed that the induction of GADD34 mRNA by hyperosmotic stress was not affected (Fig. 3F) even though the levels of its bona fide target ATF3 decreased (Fig. 3E). To further examine the involvement of JNK signaling in the regulation of GADD34 transcription, we placed the firefly luciferase gene under the control of the minimal promoter region of the GADD34 gene (Fig. 4A), shown previously to regulate its expression under genotoxic stress (22). We also generated a mutant in the CRE element known to bind c-Jun (22). Because NF-κB is known to be induced by hyperosmotic stress (37, 38), we also mutagenized a putative NF-κB binding site (−39 GGGACTTCCG −30). This putative site was predicted with TFSEARCH and was similar to the NF-κB consensus, GGGACTTTCC. WT promoter activity increased early (2 h) during hyperosmolar stress, and mutation of the putative NF-κB binding site had no effect on induction. Longer exposure to hyperosmolar medium returned the GADD34 promoter activity to its basal level (Fig. 4B). However, mutation of the CRE element decreased basal promoter activity and abolished stress-dependent induction (Fig. 4B). To further explore the involvement of the JNK signaling pathway in GADD34 gene regulation, we tested the promoter activity in the presence of the JNK inhibitor SP600125. Inhibition of JNK kinase did not change the basal activity of the promoter but abolished the stress-related induction. The inhibitor had no effect on the CRE mutant (Fig. 4C). These data explain why inhibition of JNK kinase only decreased GADD34 mRNA levels early in the response to hyperosmolar medium.
FIGURE 4.
Osmotic stress induces GADD34 promoter activity and increases GADD34 mRNA stability. A, schematic of the GADD34 promoter elements used to control firefly luciferase expression. B, induction of firefly luciferase expression normalized to Renilla luciferase expression during treatment with 500 mosmol medium. C, induction of firefly luciferase by hyperosmolar medium in the presence of the JNK inhibitor SP00125 (20 μm). Cells were pretreated with inhibitor for 30 min before treatment with hyperosmolar medium. B and C, results are the means of triplicate determinations. Error bars represent mean ± S.D. D, levels of remaining GADD34 mRNA after inhibition of transcription with Actinomycin D. Cells treated with 500 mosmol medium for 3 h or control cells were treated with Actinomycin D for the indicated times, and GADD34 mRNA was quantified by qRT-PCR. Values were normalized to GAPDH and expressed as percentages of the levels before the addition of Actinomycin D. *, p < 0.05 relative to the untreated cells; ns, not significant. Error bars represent mean ± S.D. Con, control.
To evaluate whether other mechanisms contribute to the accumulation of GADD34 mRNA during late stress, we tested its half-life in control cells and cells treated for 3 h in hyperosmolar medium. Inhibition of transcription by Actinomycin D revealed that the half-life of GADD34 mRNA increased dramatically during osmotic stress (Fig. 4D). These data suggest that the JNK activity is critical for the initial induction of GADD34 gene transcription but that the late accumulation depends on increased mRNA stability. Although it is likely that the transcription factor c-Jun is responsible for the induction of the GADD34 gene expression during early stress, other JNK-dependent transcription factors cannot be excluded.
GADD34 Contributes to eIF2α Dephosphorylation and Promotes Survival during Hyperosmotic Stress
First, we studied the contribution of GADD34 to cell survival via the modulation of eIF2α phosphorylation. We have shown previously that severe stress leads to eIF2α phosphorylation (8) but that protein synthesis inhibition under severe stress is independent of eIF2α phosphorylation (8, 9). More importantly, we have shown previously that eIF2α phosphorylation is necessary for the induction of apoptosis under these conditions (8). Severe hyperosmotic stress also induced GADD34 mRNA in an ATF4-independent manner, but the kinetics of induction were slower than for mild stress (compare Figs. 1D and 5A). GADD34 protein also accumulated with delayed kinetics (Fig. 5B). Knocking down GADD34 with shRNA increased eIF2α phosphorylation with a parallel increase in caspase 3 cleavage and, therefore, increased apoptosis (Fig. 5B). To determine GADD34 involvement, we overexpressed two forms of GADD34: active, with a truncated N-terminal region (GADD34ΔN), and inactive, without the C-terminal region (GADD34ΔC), which is unable to interact with PP1 phosphatase (17). As expected, overexpression of GADD34ΔN (active) had no impact on the protein synthesis rates in cells exposed to 600 mosmol medium (Fig. 5C) but resulted in lower caspase 3 activity (Fig. 5D). As expected, the phosphorylation of eIF2α was lower in cells expressing the active GADD34ΔN (Fig. 5E). These data suggest that attenuation of eIF2α phosphorylation by GADD34 during severe hyperosmotic stress protects cells from apoptosis in a manner independent of changes in global protein synthesis rates.
FIGURE 5.
GADD34 expression during hyperosmotic stress promotes survival. A, qRT-PCR analysis of mRNAs from cells treated with 600 mosmol medium after infection with adenovirus expressing shRNA against ATF4. GADD34 levels were normalized to the GAPDH mRNA signals. B, Western blot analysis of extracts from cells treated with 600 mosmol medium after infection with lentivirus expressing shRNA against GADD34. C, [35S]Met/Cys incorporation into proteins in cells treated with 600 mosmol medium after infection with retroviral particles expressing truncated forms of GADD34 (ΔN active and ΔC inactive). D, caspase 3 activity in extracts from cells treated with 600 mosmol medium after infection with truncated forms of GADD34 (ΔN (active) and ΔC (inactive)). E, Western blots of cells expressing GADD34ΔN and GADD34ΔC. The results in A, C, and D are the means of triplicate determinations.
We next determined the contribution of GADD34 to the regulation of eIF2α phosphorylation and apoptosis under mild hyperosmotic stress. GADD34ΔN (active) abolished eIF2α phosphorylation in response to hyperosmotic stress, whereas GADD34ΔC (inactive) had no effect (Fig. 6A). At the same time, active GADD34 did not significantly alter the ability of cells to sense the stress because the stress-induced phosphorylation of ERK1/2 and JNK1/2 was still observed (Fig. 6A). Also, the pattern of regulation of p70S6K was not affected (Fig. 6A). These data suggest that GADD34 can modulate the status of eIF2α phosphorylation independently of the activation of stress kinases in response to mild hyperosmotic stress.
FIGURE 6.
GADD34 expression during hyperosmotic stress controls the phosphorylation of eIF2α and downstream signaling but not MAP kinase signaling. MEFs were treated for the indicated times with 500 mosmol (A–D) or 600 mosmol (E) medium after infection with lentivirus-expressing shRNAs. A, Western blot of extracts from cells treated with 500 mosmol medium after infection with retroviral particles expressing truncated forms of GADD34 (ΔN active and ΔC inactive). B, Western blot analysis of extracts of cells expressing shRNA against GADD34. C, qRT-PCR analysis of the indicated mRNAs normalized to GAPDH mRNA and shown as -fold change over untreated cells. D and E, caspase 3 activity in cell extracts. Results are the means of triplicate determinations.
We confirmed these observations by decreasing GADD34 expression with an shRNA. This treatment reduced GADD34 mRNA expression by ∼70% (Fig. 6C). Induction of SNAT2 gene expression, the major facilitator of the adaptive response to mild hyperosmotic stress, was not affected by shGADD34 treatment (Fig. 6C). Attenuation of GADD34 expression also did not affect MAP kinase signaling (compare Fig. 6, A and B). In agreement with a protective role of GADD34 was the finding that MEFs lacking GADD34 were less able to adapt to prolonged mild hyperosmotic stress, as demonstrated by increased caspase 3 activation (Fig. 6D). A similar protection by GADD34 was observed in cells subjected to severe stress for short times (Fig. 6E). In conclusion, induction of GADD34 during hyperosmotic stress protects cells from apoptosis.
GADD34 Expression Increases SNAT2 Activity during Hyperosmotic Stress
We have shown previously that severe hyperosmotic stress induces apoptosis via mechanisms that involve eIF2α phosphorylation (8). One such mechanism was the inhibition of translation of antiapoptotic mRNAs (8). Conversely, the induction of the system A amino acid transporter SNAT2 is considered the major adaptive response to increasing extracellular osmolarity, especially during mild stress (6). SNAT2 drives the intracellular accumulation of amino acid osmolytes that restore cell volume (4, 7). The translation of the internal ribosome entry site-containing SNAT2 mRNA is not affected by the global inhibition of protein synthesis during stress, and its expression is mainly regulated at the level of transcription (39).
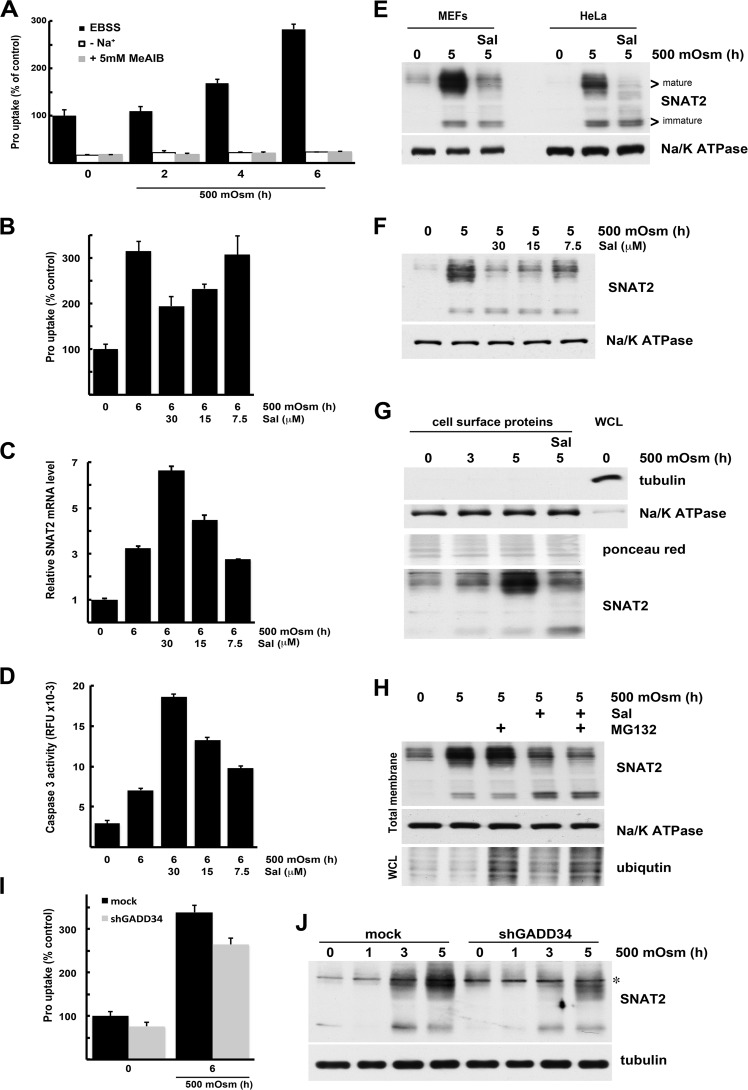
In this study, we determined whether proapoptotic eIF2α phosphorylation signaling attenuates adaptive mechanisms to mild hyperosmotic stress. We confirmed that mild stress stimulates SNAT2 activity by measuring the Na+-dependent uptake of its preferred substrate, Pro. As expected, Pro uptake gradually increased during 6 h of mild hyperosmotic stress (Fig. 7A). This Pro uptake reflects system A activity because it is Na+-dependent (Fig. 7A) and can be inhibited by high concentrations of the specific system A substrate methyl-aminoisobutyrate (Fig. 7A).
FIGURE 7.
Signaling downstream of eIF2α phosphorylation inhibits the adaptive induction of system A activity during mild hyperosmotic stress. A, Pro uptake by MEFs exposed to 500 mosmol medium for the indicated times. Uptake was measured in the presence (Earle's balanced salt solution, EBSS) or absence of Na+ ions and in the presence of 5 mm methyl-aminoisobutyrate (MeAIB). Results are the means of triplicate experiments. B, Pro uptake by cells exposed to 500 mosmol medium alone or in the presence of Sal at the indicated concentrations. Results are the means of triplicate experiments. C, cells treated as in B. qRT-PCR analysis of SNAT2 mRNA normalized to GAPDH mRNA is shown as -fold change over untreated cells. D, caspase 3 activity in extracts from cells treated as in B. Results are the means of triplicate determinations of relative fluorescence units (RFU). E, immunodetection of SNAT2 in total membrane fractions of MEFs and HeLa cell line treated with hyperosmotic medium and 30 μm Sal. F, immunodetection of SNAT2 in the total membrane fraction from MEFs treated with hyperosmolar medium and indicated the concentrations of Sal. G, immunodetection of SNAT2 in plasma membrane fractions. Biotinylated surface proteins and whole cell lysate (WCL, 10 μg) were analyzed by Western blotting. H, immunodetection of SNAT2 in total membrane fractions of cells treated with hyperosmolar medium and Sal (30 μm). MG132 (10 μm) was added during the last 2 h of hyperosmolar treatment. I, Pro uptake by stressed MEFs expressing shRNA against GADD34. J, immunodetection of SNAT2 in whole cell lysates from cells expressing shRNA against GADD34 and treated with hyperosmolar medium for indicated times. The asterisk indicates a nonspecific band recognized by antibodies.
We then examined whether GADD34 is a positive regulator of the SNAT2/System A adaptation mechanism via decreased eIF2α phosphorylation. First, we tested whether inhibiting GADD34 function with Sal caused a decrease in system A activity. Sal decreased stress-induced Pro uptake in a dose-dependent manner (Fig. 7B) without affecting SNAT2 mRNA translation measured as its association with translating ribosomes (Fig. 2). In addition, SNAT2 mRNA levels did not decrease in the presence of Sal during hyperosmotic stress (Fig. 7C). Inhibition of stress-stimulated SNAT2 activity by Sal was accompanied by increased caspase 3 activity (Fig. 7D).
To better understand how the presence of GADD34 during stress modulates the activity of SNAT2/System A, we evaluated SNAT2 protein levels. First we tested the induction of SNAT2 protein levels in MEFs and human HeLa cells to show that antibody against SNAT2 recognizes both the human and mouse proteins. In both cell lines, mild osmotic stress induced SNAT2 protein levels. Both fully glycosylated (slower migration in SDS-PAGE) and partially processed SNAT2 levels increased in the total membrane fraction of both cell lines (Fig. 7E). The increase in levels of mature SNAT2 was inhibited by Sal (Fig. 7E) in a dose-dependent manner (Fig. 7F). Because SNAT2 must be in the plasma membrane for its transport function, we isolated cell surface proteins and probed for SNAT2. We observed a gradual increase of cell surface SNAT2 protein during exposure to mild hyperosmolar stress (Fig. 7G). Only traces of immature SNAT2 were present in plasma membranes. The cytoplasmic protein tubulin was absent, and the α1 subunit of the plasma membrane protein Na+/K+ ATPase was enriched in the biotinylated fraction, demonstrating the quality of our protein isolation (Fig. 7G). Treatment with Sal during hyperosmolar treatment decreased the cell surface levels of fully processed SNAT2 transporter. However, the partially processed form was also detected (Fig. 7G).
It has been reported previously that the role of SNAT2 in the adaptation to amino acid starvation can be decreased by the presence of unsaturated fatty acids (31). This mechanism involved proteasomal degradation of the partially processed SNAT2 protein. We tested whether a similar mechanism can explain the action of Sal during hyperosmotic stress. We used the proteasome inhibitor MG132 during the last 2 h of hyperosmotic treatment with or without Sal. The accumulation of ubiquitinated proteins in total cell lysates confirmed the inhibition of proteasomal activity. However, the levels of mature and immature SNAT2 did not increase when MG132 was added to cells in hyperosmotic medium with or without Sal (Fig. 7H). These data suggest that sustained eIF2α phosphorylation during mild hyperosmolar stress did not increase proteasomal degradation of the SNAT2 protein.
Similar to the pharmacological inhibition of GADD34, GADD34 knockdown decreased stress-induced system A activity (Fig. 7I) without affecting SNAT2 mRNA induction (Fig. 6C). We confirmed this observation by testing SNAT2 protein levels in total cell lysates from osmotically stressed cells and found that induction of the mature transporter was decreased in the absence of GADD34 (Fig. 7J). Taken together, these data suggest that the GADD34/SNAT2 axis is an important part of the adaptive response to hypertonic stress. This axis promotes cell survival by GADD34-mediated dephosphorylation of eIF2α-P and increased SNAT2-mediated uptake of neutral amino acids. Sustained phosphorylation of eIF2α by inhibition of GADD34 or shRNA-mediated depletion of GADD34 decreased levels of mature SNAT2 protein. Taken together, the data suggest that GADD34 during hyperosmotic stress facilitates expression of the SNAT2 transporter to plasma membranes.
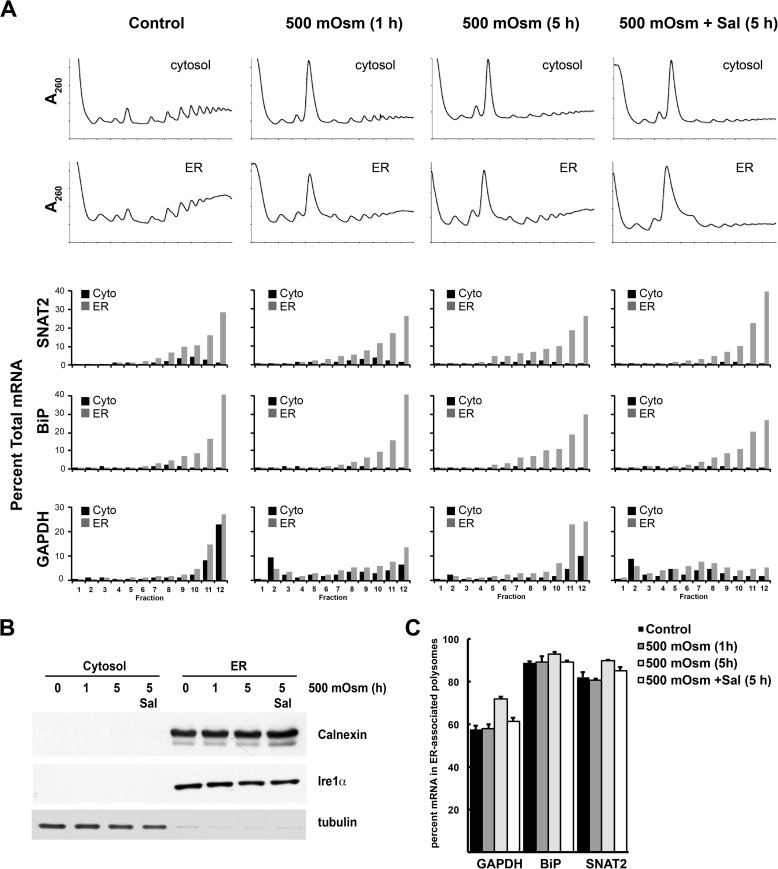
Reid et al. (29) showed recently that increased eIF2α-P during the unfolded protein response causes the transient release of translating ribosomes from the ER membrane to the cytosol. SNAT2 was one of the mRNAs that shifted from ER-associated to cytosolic polyribosomes during the UPR (29). On the basis of this finding, we tested whether mild hyperosmotic stress changes the distribution of SNAT2 mRNA on cytoplasmic and ER-associated ribosomes by analyzing these two fractions separately. The quality of subcellular fractionation was established by the detection of the ER-associated proteins calnexin and Ire1α and the cytoplasmic protein tubulin (Fig. 8B). Nearly all SNAT2 mRNA was present in ER-associated ribosomes, similar to mRNA encoding the ER chaperone BiP (Fig. 8, A and C). The association of SNAT2 mRNA with the ER ribosomes slightly increased during prolonged hyperosmotic treatment, and Sal had almost no effect on distribution of these mRNAs. The distribution of the SNAT2 mRNA in cytoplasmic and ER-associated heavy polysomes was similar, supporting efficient translation during stress, in agreement with whole-cell polysome profiles (Figs. 2 and 8). We also evaluated the distribution of GAPDH mRNA and found an increased association with the ER ribosomes that was sensitive to Sal (Fig. 8). This finding suggests that this mechanism is not important in regulating the expression of SNAT2 protein during mild hyperosmotic stress.
FIGURE 8.
Association of specific mRNAs with cytoplasmic or ER-associated polyribosomes during mild hyperosmotic stress. A, MEFs were treated with 500 mosmol medium alone or in the presence of Sal (15 μm) for the indicated times, and the distribution of mRNAs in polysomes was analyzed using qRT-PCR. Equal volumes of cytoplasmic (Cyto) or ER-associated ribosomes were loaded on gradients. The mRNA in each fraction is expressed as the percentage of total mRNA (sum of all cytoplasmic + ER-associated fractions). B, quality control of cell fractionations by immunodetection of ER and cytoplasmic marker proteins. C, quantification of three independent experiments of ER-associated polysomes for the indicated mRNAs. Error bars represent mean ± S.E.
Discussion
We described the mechanism of induction and the significance of the PP1 phosphatase regulatory subunit protein GADD34 during hyperosmotic stress. This protein is transcriptionally induced by almost all stresses that increase eIF2α phosphorylation via a mechanism that involves the transcription factor ATF4 (21). Phosphorylation of eIF2α causes global inhibition of protein synthesis, and induction of GADD34 is involved in translational recovery during stress by dephosphorylating eIF2α-P (18, 19). Under almost all stress conditions, the eIF2α-P/ATF4/GADD34 axis downstream of the stress-activated eIF2α kinases is considered the master transcriptional and translational reprogramming mechanism (40). In contrast, the cellular response to increased extracellular osmolarity has the following unique features. Despite increased eIF2α phosphorylation, ATF4 mRNA was not translated during hyperosmotic stress (shown above); the stress-sensing mechanism involves activation of MAP kinases, which cause transcriptional reprogramming via nuclear translocation of the transcription factor TonEBP (1, 2); and translational reprogramming involves mechanisms both dependent on and independent of eIF2α-P (8). We have shown earlier that eIF2α-P signaling during severe hyperosmotic stress correlates with the initiation of apoptosis (8). Because nothing was known about the significance of the eIF2α-P/ATF4/GADD34 axis during hyperosmotic stress, we investigated this network in this study.
We found that GADD34 is induced during hyperosmotic stress in an ATF4-independent manner and promotes cell survival in part by activating amino acid uptake by SNAT2. Induction of this transporter is a crucial part of the TonEBP transcription program and mediates a volume increase in response to mild hyperosmotic stress (3). The induction of GADD34 has been associated with both prosurvival and proapoptotic outcomes in response to diverse stress conditions (27, 41, 42). Several lines of evidence support prosurvival functions of GADD34 during hyperosmotic stress, especially under mild stress conditions where partial dephosphorylation of eIF2α-P could foster synthesis of proteins (e.g. chaperones) that serve to alleviate stress. The latter is true for the unfolded protein response, in which increased eIF2α-P decreases global rates of protein synthesis (18) and dephosphorylation of eIF2α permits translation of stress-induced mRNAs. In the case of hyperosmotic stress, expression of GADD34 is not involved in controlling global protein synthesis rates during severe stress. Instead, it dephosphorylates eIF2α-P and, therefore, inhibits proapoptotic eIF2α-P signaling (8).
The adaptive response to hyperosmolarity includes the increased expression of SNAT2 protein and SNAT2-mediated transport via a mechanism that is inhibited by high levels of eIF2α-P (Figs. 7 and 8). Although the mechanism by which eIF2α-P has this effect is unknown, our results eliminate several possibilities. This regulation does not involve changes in SNAT2 mRNA levels because Sal increased mRNA expression during stress. It also does not involve translation because the association of SNAT2 mRNA with membrane-bound ribosomes was only decreased slightly by Sal.
Consequently, the effect of high levels of eIF2α-P on SNAT is posttranslational. Maturation of SNAT2 protein requires its glycosylation (7), and stress-induced expression of functional SNAT2 protein can be suppressed by the glycosylation inhibitor tunicamycin (43). However, Sal caused similar decreases in total and cell surface SNAT2 protein. Moreover, the apparent size of mature SNAT2 was not affected by Sal, which is not consistent with an effect of eIF2α-P on glycosylation. It is possible that eIF2α-P induces turnover of SNAT2 protein. It has been reported that turnover of the SNAT2 protein depends on the ubiquitin proteasome system via the involvement of the ubiquitin E3 ligase NEDD4-2 (44). A recent study has suggested that proteasomal degradation explains decreased SNAT2 induction by unsaturated fatty acids under stress conditions (31). Inhibition of the proteasome caused the accumulation of immature SNAT2 but not the fully glycosylated transporter (31). In our experiments, decreased SNAT2 protein caused by Sal was not due to proteasomal degradation. Experiments are underway to determine whether the decreased turnover is due to defects in glycosylation or membrane traffic and whether autophagy or another proteolytic system is induced. Because mTOR signaling is inhibited during hyperosmotic stress (Fig. 1B and Ref. 8), autophagy may play an important role in SNAT turnover.
It has been reported that down-regulation of the constitutively expressed CReP subunit of the PP1 phosphatase complex, which dephosphorylates eIF2α, increased the phosphorylation of eIF2 and caused changes in membrane trafficking for both endo- and exocytosis. eIF2α-P was directly involved in those changes, but not via regulation of protein synthesis (45). We find these observations similar to our results, in which the posttranslational fate of SNAT2 protein is regulated by the level of eIF2α-P. We believe that the eIF2α-P status drives the induction of SNAT2 activity during hyperosmotic stress, with GADD34 controlling the phosphorylation status of eIF2α.
The mechanism of GADD34 induction during hyperosmotic stress was shown to involve JNK signaling, but neither JNK-mediated activation of ATF2 nor ATF2-mediated induction of ATF3 were involved in this process. The GADD34 promoter contains sequences immediately upstream of the transcription start site that have been shown to be essential for c-Jun-mediated transcriptional activation by DNA damage (22). In agreement with this report, induction of GADD34 promoter-driven luciferase activity and endogenous GADD34 mRNA levels during early stress were blocked by a JNK inhibitor, supporting the idea that c-Jun induces GADD34 during the early response to hyperosmotic stress. c-Jun phosphorylation was transient and occurred only during early stress. Accumulation of GADD34 mRNA during late stress can be explained by the dramatic stabilization of the GADD34 mRNA during hyperosmotic treatment. The mechanism that regulates GADD34 mRNA turnover remains unknown. The involvement of at least two mechanisms in the regulation of GADD34 expression underlines the importance of this protein phosphatase subunit in the modulation of the cellular response to hyperosmotic stress.
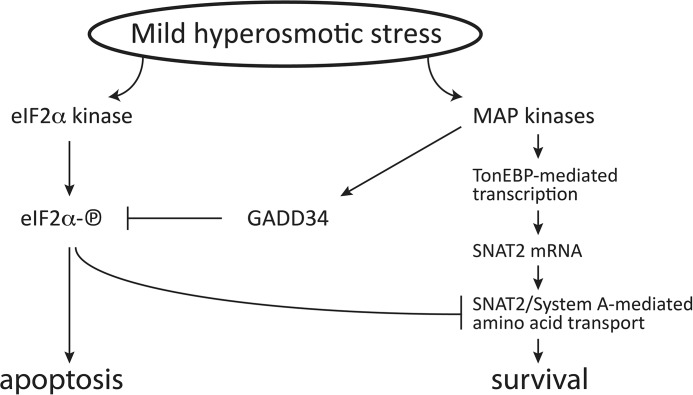
Adaptation to environmental stress is an important survival mechanism. We have shown here that rapid adaptation to hyperosmotic stress does not involve the eIF2α-P/ATF4/GADD34 axis, which is considered to be the master regulator of most stress responses (17, 19, 21, 46). Instead, it involves a novel TonEBP/MAP kinase/GADD34 pathway that leads to rapid sensing and recovery from hyperosmotic stress, involving the increase in plasma membrane uptake of compatible osmolytes (Fig. 9). All of these programmed cellular responses are meant to be adaptive. However, loss of adaptation leads to the development of disease and, in the case of hyperosmotic stress, induction of the inflammatory response. Is GADD34 involved in both adaptation (this study) and inflammation during hyperosmotic stress? Further studies are necessary to solve this interesting puzzle.
FIGURE 9.
GADD34-mediated dephosphorylation of eIF2α-P during mild hyperosmotic stress promotes adaptation. The adaptation program during hyperosmotic stress involves TonEBP-mediated transcriptional control of genes coding for transporters of compatible osmolytes, including the neutral amino acid transporter SNAT2. In contrast, induction of eIF2α phosphorylation promotes apoptosis. The model suggests that the MAP kinases signaling promote survival via induction of GADD34, which dephosphorylates eIF2α-P and, therefore, inhibits apoptosis. The proapoptotic signaling of eIF2α-P also contributes to the inhibition of the prosurvival SNAT2-mediated increased amino acid uptake.
Author Contributions
D. K. and M. H. designed the study. D. K., R. J., B. J. G., K. F., J. W., M. M., and M. G. B. performed experiments. D. K., R. J., M. D. S., and M. H. wrote the article. All authors analyzed the results and approved the final version of the manuscript.
Acknowledgments
We thank Dr. Randal Kaufman, Jasoek Han (Sanford-Burnham Medical Research Institute), and David Ron (University of Cambridge) for GADD34 retroviruses.
This work was supported, in whole or in part, by National Institutes of Health Grants R37-DK60596 and R01-DK53307 (to M. H.). The authors declare that they have no conflicts of interest with the contents of this article.
- SNAT
- sodium-coupled neutral amino acid transporter
- ATF4
- activating transcription factor 4
- ER
- endoplasmic reticulum
- CRE
- cAMP response element
- eIF2α-P
- phosphorylated eIF2α at Ser-51
- mTOR
- mammalian target of rapamycin
- MEF
- mouse embryonic fibroblast
- qRT-PCR
- real-time quantitative PCR
- Sal
- Sal003
- Tg
- thapsigargin.
References
- 1. Jeon U. S., Kim J. A., Sheen M. R., Kwon H. M. (2006) How tonicity regulates genes: story of TonEBP transcriptional activator. Acta Physiol. 187, 241–247 [DOI] [PubMed] [Google Scholar]
- 2. Burg M. B., Ferraris J. D., Dmitrieva N. I. (2007) Cellular response to hyperosmotic stresses. Physiol. Rev. 87, 1441–1474 [DOI] [PubMed] [Google Scholar]
- 3. Maallem S., Wierinckx A., Lachuer J., Kwon M. H., Tappaz M. L. (2008) Gene expression profiling in brain following acute systemic hypertonicity: novel genes possibly involved in osmoadaptation. J. Neurochem. 105, 1198–1211 [DOI] [PubMed] [Google Scholar]
- 4. Franchi-Gazzola R., Dall'Asta V., Sala R., Visigalli R., Bevilacqua E., Gaccioli F., Gazzola G. C., Bussolati O. (2006) The role of the neutral amino acid transporter SNAT2 in cell volume regulation. Acta Physiol. 187, 273–283 [DOI] [PubMed] [Google Scholar]
- 5. Franchi-Gazzola R., Gaccioli F., Bevilacqua E., Visigalli R., Dall'Asta V., Sala R., Varoqui H., Erickson J. D., Gazzola G. C., Bussolati O. (2004) The synthesis of SNAT2 transporters is required for the hypertonic stimulation of system A transport activity. Biochim. Biophys. Acta 1667, 157–166 [DOI] [PubMed] [Google Scholar]
- 6. Bevilacqua E., Bussolati O., Dall'Asta V., Gaccioli F., Sala R., Gazzola G. C., Franchi-Gazzola R. (2005) SNAT2 silencing prevents the osmotic induction of transport system A and hinders cell recovery from hypertonic stress. FEBS Lett. 579, 3376–3380 [DOI] [PubMed] [Google Scholar]
- 7. Bröer S. (2014) The SLC38 family of sodium-amino acid co-transporters. Pflugers Arch. 466, 155–172 [DOI] [PubMed] [Google Scholar]
- 8. Bevilacqua E., Wang X., Majumder M., Gaccioli F., Yuan C. L., Wang C., Zhu X., Jordan L. E., Scheuner D., Kaufman R. J., Koromilas A. E., Snider M. D., Holcik M., Hatzoglou M. (2010) eIF2α phosphorylation tips the balance to apoptosis during osmotic stress. J. Biol. Chem. 285, 17098–17111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Saikia M., Krokowski D., Guan B. J., Ivanov P., Parisien M., Hu G. F., Anderson P., Pan T., Hatzoglou M. (2012) Genome-wide identification and quantitative analysis of cleaved tRNA fragments induced by cellular stress. J. Biol. Chem. 287, 42708–42725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Malhotra J. D., Kaufman R. J. (2007) The endoplasmic reticulum and the unfolded protein response. Semin. Cell Dev. Biol. 18, 716–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang K., Kaufman R. J. (2006) The unfolded protein response: a stress signaling pathway critical for health and disease. Neurology 66, S102–S109 [DOI] [PubMed] [Google Scholar]
- 12. Wek R. C., Jiang H. Y., Anthony T. G. (2006) Coping with stress: eIF2 kinases and translational control. Biochem. Soc. Trans. 34, 7–11 [DOI] [PubMed] [Google Scholar]
- 13. Rutkowski D. T., Kaufman R. J. (2007) That which does not kill me makes me stronger: adapting to chronic ER stress. Trends Biochem. Sci. 32, 469–476 [DOI] [PubMed] [Google Scholar]
- 14. Spriggs K. A., Stoneley M., Bushell M., Willis A. E. (2008) Re-programming of translation following cell stress allows IRES-mediated translation to predominate. Biol. Cell 100, 27–38 [DOI] [PubMed] [Google Scholar]
- 15. Vattem K. M., Wek R. C. (2004) Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 101, 11269–11274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lu P. D., Harding H. P., Ron D. (2004) Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J. Cell Biol. 167, 27–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Novoa I., Zeng H., Harding H. P., Ron D. (2001) Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2α. J. Cell Biol. 153, 1011–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Novoa I., Zhang Y., Zeng H., Jungreis R., Harding H. P., Ron D. (2003) Stress-induced gene expression requires programmed recovery from translational repression. EMBO J. 22, 1180–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kojima E., Takeuchi A., Haneda M., Yagi A., Hasegawa T., Yamaki K., Takeda K., Akira S., Shimokata K., Isobe K. (2003) The function of GADD34 is a recovery from a shutoff of protein synthesis induced by ER stress: elucidation by GADD34-deficient mice. FASEB J. 17, 1573–1575 [DOI] [PubMed] [Google Scholar]
- 20. Jousse C., Oyadomari S., Novoa I., Lu P., Zhang Y., Harding H. P., Ron D. (2003) Inhibition of a constitutive translation initiation factor 2α phosphatase, CReP, promotes survival of stressed cells. J. Cell Biol. 163, 767–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ma Y., Hendershot L. M. (2003) Delineation of a negative feedback regulatory loop that controls protein translation during endoplasmic reticulum stress. J. Biol. Chem. 278, 34864–34873 [DOI] [PubMed] [Google Scholar]
- 22. Haneda M., Xiao H., Hasegawa T., Kimura Y., Nakashima I., Isobe K. (2004) Regulation of mouse GADD34 gene transcription after DNA damaging agent methylmethane sulfonate. Gene 336, 139–146 [DOI] [PubMed] [Google Scholar]
- 23. Lee Y. Y., Cevallos R. C., Jan E. (2009) An upstream open reading frame regulates translation of GADD34 during cellular stresses that induce eIF2α phosphorylation. J. Biol. Chem. 284, 6661–6673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhou W., Jeyaraman K., Yusoff P., Shenolikar S. (2013) Phosphorylation at tyrosine 262 promotes GADD34 protein turnover. J. Biol. Chem. 288, 33146–33155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Han J., Back S. H., Hur J., Lin Y. H., Gildersleeve R., Shan J., Yuan C. L., Krokowski D., Wang S., Hatzoglou M., Kilberg M. S., Sartor M. A., Kaufman R. J. (2013) ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat. Cell Biol. 15, 481–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Moffat J., Grueneberg D. A., Yang X., Kim S. Y., Kloepfer A. M., Hinkle G., Piqani B., Eisenhaure T. M., Luo B., Grenier J. K., Carpenter A. E., Foo S. Y., Stewart S. A., Stockwell B. R., Hacohen N., Hahn W. C., Lander E. S., Sabatini D. M., Root D. E. (2006) A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124, 1283–1298 [DOI] [PubMed] [Google Scholar]
- 27. Krokowski D., Han J., Saikia M., Majumder M., Yuan C. L., Guan B. J., Bevilacqua E., Bussolati O., Bröer S., Arvan P., Tchórzewski M., Snider M. D., Puchowicz M., Croniger C. M., Kimball S. R., Pan T., Koromilas A. E., Kaufman R. J., Hatzoglou M. (2013) A self-defeating anabolic program leads to β cell apoptosis in ER stress-induced diabetes via regulation of amino acid flux. J. Biol. Chem. 288, 17202–17213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Krokowski D., Gaccioli F., Majumder M., Mullins M. R., Yuan C. L., Papadopoulou B., Merrick W. C., Komar A. A., Taylor D., Hatzoglou M. (2011) Characterization of hibernating ribosomes in mammalian cells. Cell Cycle 10, 2691–2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Reid D. W., Chen Q., Tay A. S., Shenolikar S., Nicchitta C. V. (2014) The unfolded protein response triggers selective mRNA release from the endoplasmic reticulum. Cell 158, 1362–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Majumder M., Huang C., Snider M. D., Komar A. A., Tanaka J., Kaufman R. J., Krokowski D., Hatzoglou M. (2012) A novel feedback loop regulates the response to endoplasmic reticulum stress via the cooperation of cytoplasmic splicing and mRNA translation. Mol. Cell. Biol. 32, 992–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nardi F., Hoffmann T. M., Stretton C., Cwiklinski E., Taylor P. M., Hundal H. S. (2015) Proteasomal modulation of cellular SNAT2 (SLC38A2) abundance and function by unsaturated fatty acid availability. J. Biol. Chem. 290, 8173–8184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dey S., Baird T. D., Zhou D., Palam L. R., Spandau D. F., Wek R. C. (2010) Both transcriptional regulation and translational control of ATF4 are central to the integrated stress response. J. Biol. Chem. 285, 33165–33174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jiang H. Y., Wek R. C. (2005) Phosphorylation of the α-subunit of the eukaryotic initiation factor-2 (eIF2α) reduces protein synthesis and enhances apoptosis in response to proteasome inhibition. J. Biol. Chem. 280, 14189–14202 [DOI] [PubMed] [Google Scholar]
- 34. Lewerenz J., Maher P. (2009) Basal levels of eIF2α phosphorylation determine cellular antioxidant status by regulating ATF4 and xCT expression. J. Biol. Chem. 284, 1106–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Seo J., Fortuno E. S., 3rd, Suh J. M., Stenesen D., Tang W., Parks E. J., Adams C. M., Townes T., Graff J. M. (2009) Atf4 regulates obesity, glucose homeostasis, and energy expenditure. Diabetes 58, 2565–2573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Harding H. P., Zhang Y., Zeng H., Novoa I., Lu P. D., Calfon M., Sadri N., Yun C., Popko B., Paules R., Stojdl D. F., Bell J. C., Hettmann T., Leiden J. M., Ron D. (2003) An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11, 619–633 [DOI] [PubMed] [Google Scholar]
- 37. Lee J. H., Kim M., Im Y. S., Choi W., Byeon S. H., Lee H. K. (2008) NFAT5 induction and its role in hyperosmolar stressed human limbal epithelial cells. Invest. Ophthalmol. Vis. Sci. 49, 1827–1835 [DOI] [PubMed] [Google Scholar]
- 38. Eisner V., Quiroga C., Criollo A., Eltit J. M., Chiong M., Parra V., Hidalgo K., Toro B., Díaz-Araya G., Lavandero S. (2006) Hyperosmotic stress activates p65/RelB NFκB in cultured cardiomyocytes with dichotomic actions on caspase activation and cell death. FEBS Lett. 580, 3469–3476 [DOI] [PubMed] [Google Scholar]
- 39. Gaccioli F., Huang C. C., Wang C., Bevilacqua E., Franchi-Gazzola R., Gazzola G. C., Bussolati O., Snider M. D., Hatzoglou M. (2006) Amino acid starvation induces the SNAT2 neutral amino acid transporter by a mechanism that involves eukaryotic initiation factor 2α phosphorylation and cap-independent translation. J. Biol. Chem. 281, 17929–17940 [DOI] [PubMed] [Google Scholar]
- 40. Majumder M., Mitchell D., Merkulov S., Wu J., Guan B. J., Snider M. D., Krokowski D., Yee V. C., Hatzoglou M. (2015) Residues required for phosphorylation of translation initiation factor eIF2α under diverse stress conditions are divergent between yeast and human. Int. J. Biochem. Cell Biol. 59, 135–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Farook J. M., Shields J., Tawfik A., Markand S., Sen T., Smith S. B., Brann D., Dhandapani K. M., Sen N. (2013) GADD34 induces cell death through inactivation of Akt following traumatic brain injury. Cell Death Dis. 4, e754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fransson L., Sjöholm A., Ortsäter H. (2014) Inhibition of palmitate-induced GADD34 expression augments apoptosis in mouse insulinoma cells (MIN6). Cell Biochem. Funct. 32, 445–452 [DOI] [PubMed] [Google Scholar]
- 43. Guan B. J., Krokowski D., Majumder M., Schmotzer C. L., Kimball S. R., Merrick W. C., Koromilas A. E., Hatzoglou M. (2014) Translational control during endoplasmic reticulum stress beyond phosphorylation of the translation initiation factor eIF2α. J. Biol. Chem. 289, 12593–12611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hatanaka T., Hatanaka Y., Setou M. (2006) Regulation of amino acid transporter ATA2 by ubiquitin ligase Nedd4–2. J. Biol. Chem. 281, 35922–35930 [DOI] [PubMed] [Google Scholar]
- 45. Kloft N., Neukirch C., von Hoven G., Bobkiewicz W., Weis S., Boller K., Husmann M. (2012) A subunit of eukaryotic translation initiation factor 2α-phosphatase (CreP/PPP1R15B) regulates membrane traffic. J. Biol. Chem. 287, 35299–35317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Brush M. H., Weiser D. C., Shenolikar S. (2003) Growth arrest and DNA damage-inducible protein GADD34 targets protein phosphatase 1 α to the endoplasmic reticulum and promotes dephosphorylation of the α subunit of eukaryotic translation initiation factor 2. Mol. Cell. Biol. 23, 1292–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]