Background: Lin28 is an evolutionary conserved RNA-binding protein that regulates miRNAs and mRNAs; Lin28 overexpression is linked to poor prognosis in cancer.
Results: Lin28 binds to guanine-rich miRNAs and mRNAs that contain G-quartet structural features and remodels these structures.
Conclusion: Lin28 recognition of G-quartets is a unifying feature of Lin28-regulated RNAs.
Significance: Small molecules that bind to G-quartets might be useful inhibitors of Lin28 function.
Keywords: cancer, embryonic stem cell, fluorescence, microRNA (miRNA), nuclear magnetic resonance (NMR), G-quartet, Lin28, N-methyl mesoporphyrin IX (NMM), let-7
Abstract
Lin28 is an evolutionarily conserved RNA-binding protein that inhibits processing of pre-let-7 microRNAs (miRNAs) and regulates translation of mRNAs that control developmental timing, pluripotency, metabolism, and tumorigenesis. The RNA features that mediate Lin28 binding to the terminal loops of let-7 pre-miRNAs and to Lin28-responsive elements (LREs) in mRNAs are not well defined. Here we show that Lin28 target datasets are enriched for RNA sequences predicted to contain stable planar structures of 4 guanines known as G-quartets (G4s). The imino NMR spectra of pre-let-7 loops and LREs contain resonances characteristic of G4 hydrogen bonds. These sequences bind to a G4-binding fluorescent dye, N-methyl-mesoporphyrin IX (NMM). Mutations and truncations in the RNA sequence that prevent G4 formation also prevent Lin28 binding. The addition of Lin28 to a pre-let-7 loop or an LRE reduces G4 resonance intensity and NMM binding, suggesting that Lin28 may function to remodel G4s. Further, we show that NMM inhibits Lin28 binding. Incubation of a human embryonal carcinoma cell line with NMM reduces its stem cell traits. In particular it increases mature let-7 levels, decreases OCT4, HMGA1, CCNB1, CDK4, and Lin28A protein, decreases sphere formation, and inhibits colony formation. Our results suggest a previously unknown structural feature of Lin28 targets and a new strategy for manipulating Lin28 function.
Introduction
Lin28 regulates networks of RNAs that control developmental timing (1–3), pluripotency (4, 5), metabolism (6, 7), and tumorigenesis (8–10). Lin28 was first identified as a heterochronic gene coordinating the “larva-to-adult” switch in Caenorhabditis elegans (1, 2, 11). All bilaterian animals possess a Lin28 homologue. It is abundantly expressed in embryos and stem cells, but absent in most terminally differentiated cells with the exception of skeletal and cardiac muscle (12, 13). In embryonic stem cells, Lin28 contributes to pluripotency by promoting self-renewal and blocking differentiation (14–17). It is part of a small unique set of proteins that include OCT4, SOX2, and Nanog that can reprogram terminally differentiated fibroblasts to induced pluripotent stem cells that resemble embryonic stem cells (18). Lin28 is the only protein that is not a transcription factor identified thus far to assist in reprogramming, suggesting that Lin28-targeted RNAs regulate stem cell properties.
Lin28 overexpression is associated with many human cancers and is linked to poor prognosis (16). Approximately 15% of primary tumors and human cancer cell lines have increased Lin28 expression (8, 9, 16, 19). Elevated Lin28 is associated with aggressiveness of germ cell tumors, hepatocellular carcinoma, and cancers of the breast, colon, thyroid, esophagus, ovaries, prostate, and head and neck (8, 9, 20–22). Ectopic Lin28 expression facilitates cellular transformation, and Lin28-overexpressing cells can form tumors in mice (9). Expression of the Lin28 target let-7 inhibits tumor formation and metastasis (23–25). Uncovering the mechanism by which Lin28 selects RNA targets and regulates them is critical to understanding the function of Lin28.
The Lin28 protein contains three nucleic acid-binding domains, a cold-shock domain (CSD)3 and two zinc finger (ZnF) motifs. Lin28 is the only known animal protein that contains both a CSD and ZnFs. Lin28 binds to the loops of pre-let-7 miRNAs to block processing by Dicer (17, 26, 27). The loop sequences are diverse in size and sequence. Upon binding to let-7 pre-miRNAs, Lin28 recruits ZCCHC11, also known as TUTase4 (TUT4), a uridylyl transferase, which adds a 3′-uridine tail to pre-let-7s (27–30). Terminal uridylation promotes pre-let-7 degradation.
Although much of Lin28 research has focused on its role in regulating let-7 biogenesis, mRNAs constitute the overwhelming majority of RNAs bound to Lin28 in cells (31). There is no agreement in the literature about common features of Lin28-binding sites. In vitro binding assays suggested that specific guanine (26, 32, 33), adenosine (26), uracil (34), and cytosine (35) nucleotides affect Lin28 binding. The crystal structures of Lin28 bound to pre-let-7 loop oligonucleotides suggested that the CSD and ZnF domains form critical contacts with guanine and adenine bases in distinct G-rich regions in the loops of pre-let-7s (33, 36, 37). In particular the ZnF domain was proposed to bind to a GGAG motif in the loop sequence of pre-let-7 miRNAs. However, the NMR structure of isolated ZnFs complexed with AGGAGAU suggested that an exact GGAG sequence is not required (36). The Lin28 CSD appears to have little sequence specificity because it forms crystals with d(T)6 and r(U)6 (37). Sequencing of Lin28-bound mRNAs suggested that Lin28 might bind to sequences containing GGAGA, AAGNNG, AAGNG(N), or (N)UGUG(N) where N is any nucleotide (31, 38). Molecular modeling instead suggested that Lin28 recognizes a critical “A bulge” flanked by two G:C bonds in an extended double-stranded region of the LREs of mRNAs (39). A single A to U mutation in OCT4, HMGA1, and RPS19 mRNAs inhibited the ability of Lin28 to stimulate their translation (39). However, the “A bulge” is not present in pre-let-7 miRNAs.
These structural, sequencing, and modeling data have suggested short Lin28 recognition elements. Such sequences are abundant in mRNAs throughout the transcriptome and hence are unlikely to uniquely define Lin28 binding specificity. None of these motifs are useful for predicting LREs of Lin28-regulated mRNAs. Thus, the key RNA features that determine Lin28 binding remain unclear.
We hypothesized that Lin28-regulated miRNAs and mRNAs share common structural features. We observed that Lin28 targets have an abundance of guanine and guanine repeats. Because guanine rich nucleic acids can form stable planar structures called G-quartets (G4s) by G:G:G:G hydrogen bonding, we hypothesized that Lin28 might recognize structured RNAs that contain G4s. To address this hypothesis, we used bioinformatics, NMR spectroscopy, a fluorescent dye that specifically binds to G4s, and gel shift assays to analyze the structural features of the pre-let-7s and their loops, which contain the 3′-GGAG motif; the LREs of DSG2, HSPA5, and SUN1, which contain the putative Lin28 consensus sequences AAGNNG, AAGNG(N), and (N)UGUG(N), respectively; and the LRE of OCT4, which contains the proposed “A bulge.” We provide evidence that a G-quartet structure is a unifying feature of Lin28 RNA targets. Further, we demonstrate that Lin28 may function to unwind G4s and that G4-stabilizing agents represent a novel class of Lin28 inhibitors.
Experimental Procedures
RNA Sequences
Sequences of RNA used in this study are as follows: pre-let-7d, 5′-CCUAGGAAGAGGUAGUAGGUUGCAUAGUUUUAGGGCAGGGAUUUUGCCCACAAGGAGGUAACUAUACGACCUGCUGCCUUUCUUAGG-3′; let-7d-loop, 5′-UUAGGGCAGGGAUUUUGCCCACAAGGAGGU-3′;pre-let-7g, 5′-AGGCUGAGGUAGUAGUUUGUACAGUUUGAGGGUCUAUGAUACCACCCGGUACAGGAGAUAACUGUACAGGCCACUGCCUUGCCA-3′; let-7g-loop, 5′-GAGGGUCUAUGAUACCACCCGGUACAGGAG-3′; DSG2 LRE, 5′-GGUCUGCUAUGUGGGCAGAGG-3′; HSPA5 LRE, 5′-GGCGUGAGGUAGAAAAGGCUAAGAGAGCCUUGUCUU-3′; SUN1 LRE, 5′-UAUCCAGAAGGAGCUGGAAG-3′; OCT4 LRE, 5′-GCAGAAGAGGAUCACCCUGGGAUAUACACAGGCCGAUGUGGGGCUCACCCUGGGGGUUCUAUUUGGGAAGGUAUUCAGCCAAACGACCAUCUGCC-3′; miR-21, 5′-GAUGUUGACUGUUGAAUCUCAUGGCAACAC-3′.
RNA Synthesis and Purification
For NMR studies, template DNA containing the T7 promoter, sequence of interest, and restriction enzyme cleavage site for linearization (SmaI or DraI) were generated by PCR and inserted into the pUC19 plasmid after EcoRI and BamHI digestions. After linearizing the plasmid with SmaI or DraI, the templates were amplified by PCR, extracted with phenol-chloroform, and precipitated with ethanol.
RNAs were synthesized from this template by in vitro transcription using T7 RNA polymerase, in 10–20-ml reactions. After denaturation at 95 °C for 2 min, the RNAs were precipitated with ethanol and purified by electrophoresis on urea-containing polyacrylamide denaturing gels. Note that these RNAs have 1–3 additional guanosines at their 5′ termini for efficient transcription, and one additional uracil (HSPA5) or 1–3 additional cytosines (the others) at their 3′ termini for linearization of the template by DraI or SmaI, respectively.
NMR Spectroscopy
NMR spectra were acquired on Bruker 500- or 600-MHz spectrometers equipped with cryoprobes. One-dimensional 1H NMR spectra were acquired after incubating 100–500 μm RNA in 10 mm sodium phosphate buffer, pH 6.4, 100 mm KCl, 4 mm MgCl2 in 90% H2O/10% D2O. Samples were equilibrated at 5 °C. Data were accumulated with 1,024 transients over a frequency width of 10.33 kHz with 32,768 data points. Spectra were processed using MestReNova.
N-Methyl Mesoporphyrin IX (NMM) Fluorescence Assay
NMM was obtained from Frontier Scientific (Logan, UT). Fluorescence assays were performed in 30 μl of 10 mm sodium phosphate buffer, pH 6.4, 100 mm KCl, 4 mm MgCl2, and 5 μm NMM. The RNA concentration ranged from 0 to 100 μm. All fluorescence experiments were performed using a FlexStation III plate reader with excitation and emission wavelengths of 399 nm and 614 nm, respectively. Fluorescence measurements were repeated three times for each sample, and the intensities were averaged and corrected by running a buffer control without RNA before each series of experiments. Fluorescence intensities were normalized to the maximum intensity of UG4U. Results shown are the average of 3–5 independent replicates. Error bars represent the standard deviation between experiments.
Lin28 Expression and Purification
The cDNA for mouse Lin28A were obtained from Addgene (Cambridge, MA). The region containing the CSD and ZnFs (residues 36–186) was PCR amplified and cloned into the bacterial expression vector pET15b, adding a C-terminal His6 tag. Lin28-His6 was overexpressed in Escherichia coli Rosetta (DE3) cells (Millipore). Cells were grown in Luria Broth (LB) at 25 °C to an A600 of 0.9 and induced with 1.0 mm isopropyl d-1-thiogalactopyranoside. After 3 h, cells were harvested by centrifugation. Cells were lysed by sonication in 50 mm Tris-Cl, pH 8.0, 1 m NaCl, 5 mm β-mercaptoethanol, 0.1 mm ZnCl2, and 10 units of Benzonase nuclease (Sigma). Lin28-His6 was purified by nickel affinity chromatography followed by size-exclusion chromatography on a Superdex 75 prep-grade column in 50 mm Tris-Cl, pH 6.4, 200 mm NaCl, 5 mm β-mercaptoethanol. All purification steps were carried out at 4 °C.
Electrophoretic Mobility Shift Analysis
EMSAs were conducted using 2 × 105 cpm 32P-5′-end-labeled RNA probes. Binding buffer contained 50 mm Tris (pH 7.6), 100 mm KCl, 5% glycerol, 40 units of RNaseOUT (Invitrogen), and 10 mm β-mercaptoethanol. Binding reactions were conducted in 20 μl with increasing amounts of recombinant Lin28-His6 (0, 0.15, 0.3, 0.6, 1.25, 2.5, 5.0, 10 μm). Bound complexes were resolved on native 5% polyacrylamide gels and visualized by autoradiography.
Unwinding Assay
Unwinding assays were carried out with constant amounts of let-7g loop or HSPA5 LRE (50 μm) in 10 mm sodium phosphate buffer, pH 6.4, 100 mm KCl, 4 mm MgCl2, and 5 μm NMM to which recombinant Lin28 or BSA protein was added. The protein concentration ranged from 0 to 6.25 μm. After the addition of protein, samples were allowed to equilibrate at least 4 h at 4 °C. Fluorescence intensities were recorded using a FlexStation III plate reader, as in the NMM fluorescence assay. Protein only samples were recorded in 10 mm sodium phosphate buffer, pH 6.4, 100 mm KCl, 4 mm MgCl2, and 5 μm NMM without RNA. Fluorescence intensities were normalized to the maximum intensity without protein. Results shown are the mean value and standard deviation of 3 replicates.
Competition EMSA
Competition EMSAs were conducted using 2 × 105 cpm 32P-5′-end-labeled let-7g loop or HSPA5 LRE probes and a constant rLin28A (10 μm). Competition reactions were conducted in the same EMSA binding buffer with increasing amounts of NMM (0, 1.5, 3, 6, 12.5, 25, 50, 100 μm). Bound complexes were resolved on native 5% polyacrylamide gels and visualized using autoradiography.
NMM Addition to Human NCCIT Embryonal Carcinoma Cells
Human NCCIT cells were cultured alone or in the presence of 100 μm NMM. RNA and protein were harvested after 3 and 48 h, respectively. miRNA levels were assessed using TaqManTM miRNA probes. The protein levels of Lin28 mRNA targets were measured by Western blot (OCT4, GeneTex GTX100622; HMGA1, Abcam ab129153; c-Myc, Cell Signaling 9402; CCNB1, Santa Cruz Biotechnology sc-718; CDK4, Cell Signaling 2906; Lin28, Abcam ab46020). Mature, precursor, and primary miRNA levels were measured after 3 h of NMM incubation by quantitative RT-PCR using the TaqManTM miRNA and pri-miRNA assays (Life Technologies) and miScript precursor assay (Qiagen). Cell viability was assessed by CellTiter-Glo (Promega) after 48 h. For the sphere-forming assay, 1,000/ml viable cells were cultured in suspension in serum-free DMEM/F12 1:1 (Invitrogen), supplemented with EGF (20 ng/ml, BD Biosciences), B27 (1:50, Invitrogen), 0.4% bovine serum albumin (Sigma), and 4 μg/ml insulin (Sigma). Spheres were counted after 1 or 2 weeks. For the colony-forming assay, 1,000 viable cells were plated on 10-cm plates in serum-containing medium. Medium was replaced every 3 days. After 8–14 days, cells were fixed in methanol (−20 °C) and stained with crystal violet.
Luciferase Reporter Assay
Human NCCIT cells were transfected alone, with 5 nm wild-type pre-let-7g (5′-AGGCUGAGGUAGUAGUUUGUACAGUUUGAGGGUCUAUGAUACCACCCGGUACAGGAGAUAACUGUACAGGCCACUGCCUUGCCA-3′), or 5 nm mutant pre-let-7g (5′-AGGCUGAGGUAGUAGUUUGUACAGUUUGAGAAUCUAUGAUACCACCCGGUACAAAAGAUAACUGUACAGGCCACUGCCUUGCCA-3′) and 25 ng of psiCHECK3 (Promega) vector containing two copies of the let-7-binding site cloned into the multiple cloning site (NotI and XhoI) of Renilla luciferase. After 48 h, cells were incubated with 100 μm NMM for 3 h and luciferase activities were measured using the Dual-Luciferase assay system (Promega). Data were normalized to firefly luciferase.
Results
Lin28-regulated Gene Sequences Are Enriched for G-quartet Motifs
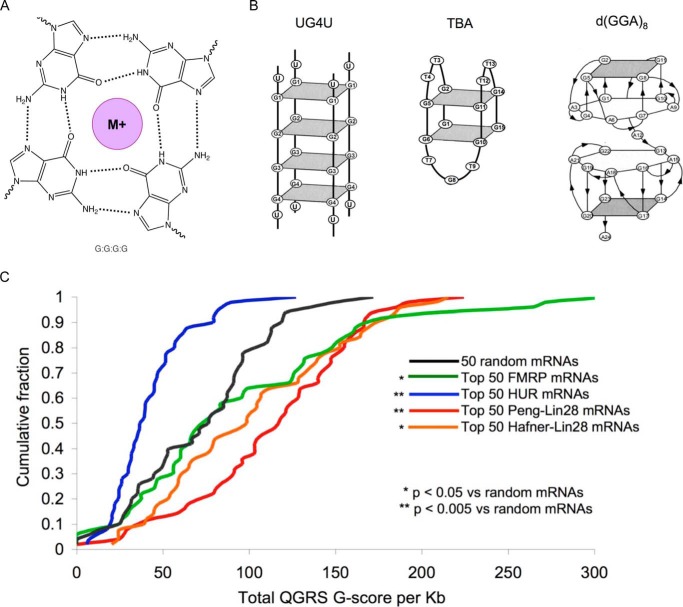
We examined the sequences of the 12 let-7 loops and of all 15 Lin28 targets with an experimentally defined LRE (Table 1). Many of the LREs are enriched for Gs and have multiple stretches of 2–6 consecutive guanines. Those LREs that do not have an excess of Gs are mostly longer (200–3270 nucleotides) and may not correspond to the minimal LRE sequence. G-rich nucleic acids can form structures containing G4s (Fig. 1A). Stacked G4s adopt unique structures known as G-quadruplexes that form diverse structures (Fig. 1B) depending on the number of stacked G4s, strand orientation, loop structures, and whether G4s are formed via inter- or intramolecular interactions (40–42). To determine whether Lin28 RNA targets might contain G4s, we first utilized the Quadruplex G-Rich Sequence (QGRS) Mapper, which predicts G4s in nucleic acids by identifying potential G-quartet sequences that match the following motif: GxNy1GxNy2GxNy3Gx, where x is the number of G-repeats and y1,y2,and y3 are the lengths of the gaps (43). The program generates a G-score such that sequences with higher scores are more likely to form G4s. QGRS was applied to the 50 most enriched mRNAs immunoprecipitated with Lin28 antibody in human embryonic stem cells (5) and the 50 most enriched Lin28-bound mRNAs identified by photoactivatable ribonucleoside-enhanced cross-linking and immunoprecipitation (PAR-CLIP) in FLAG/HA-Lin28-overexpressing HEK293 cells (34) (Fig. 1C). (We did not apply this analysis to putative Lin28 mRNA targets identified by two other studies that gave large numbers of Lin28-interacting mRNAs (6,000 and 13,000) without any ranking (31, 38).) The G scores of Lin28-bound RNAs were compared with the scores of the 50 mRNA most enriched in binding to FMRP (fragile X mental retardation protein-1), which is known to contain G4s (44–46), as positive control, or HuR, which binds AU-rich mRNAs (47), as negative control. These datasets were compared with the G-scores of 50 randomly chosen mRNAs. As expected, FMRP-binding mRNAs had significantly higher G-scores than random mRNAs (p = 0.017), and the HuR mRNAs had significantly lower G-scores than random mRNAs (p < 0.0001) (Fig. 1C). The top 50 Lin28-bound mRNAs in the immunoprecipitation and PAR-CLIP datasets had significantly higher G-scores than the random mRNAs (p < 0.0001 and p = 0.017, respectively). The increased G-scores of Lin28-bound mRNAs suggests that a common feature might be the ability to form G4s.
TABLE 1.
Lin28 recognition elements are enriched for guanine
The frequency of each base was calculated. The most enriched nucleotide is shown in red. The number and length of G-repeats were also tabulated.
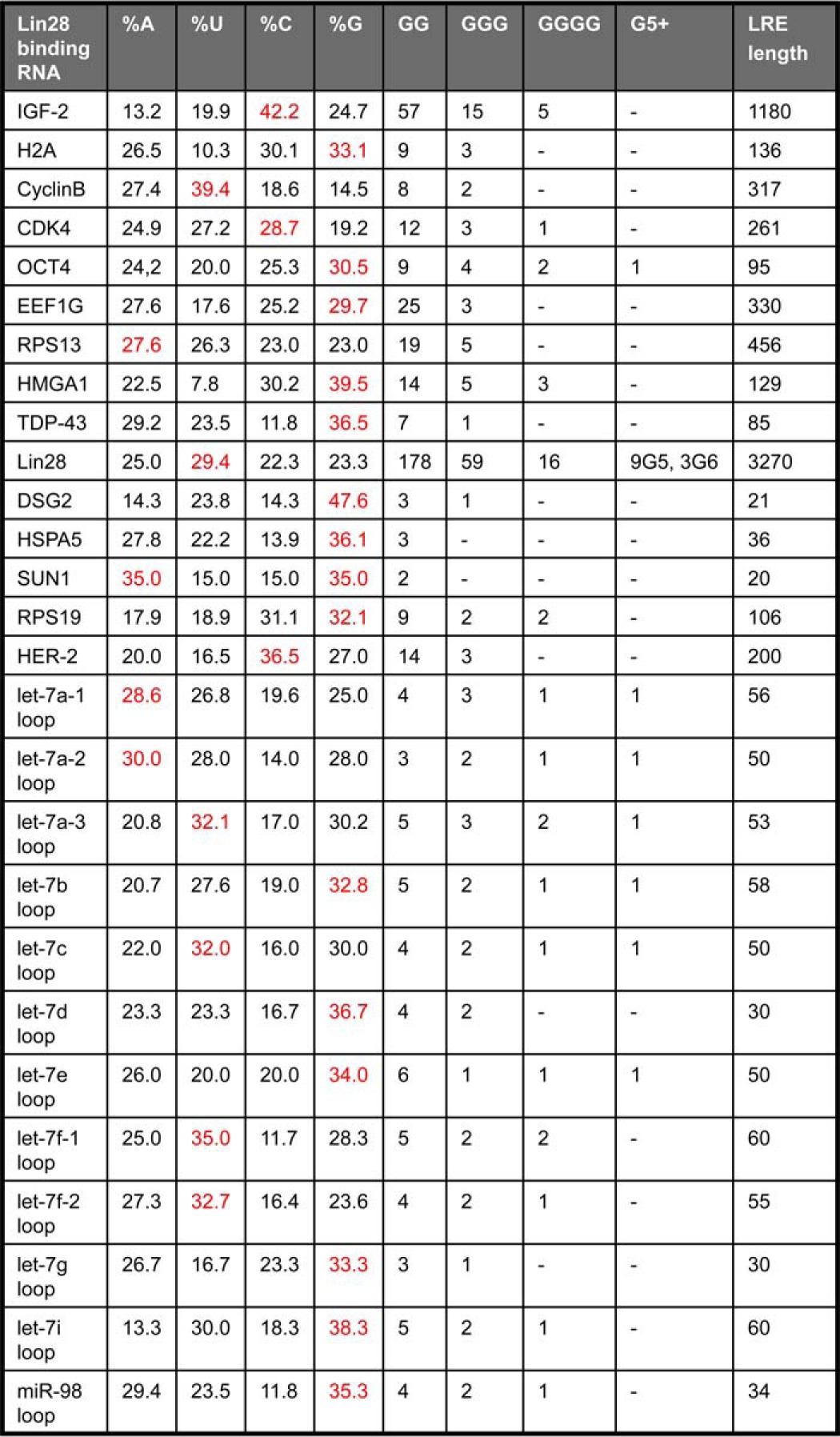
FIGURE 1.
Lin28 target RNAs are predicted to form G4s. A, G4s are composed of at least four guanines interconnected through Hoogsteen hydrogen bonds. A monovalent ion (M+) binds within the central cavity of G4s. G-quadruplexes form distinct conformations based on the number of stacked G4s, the loop sequences, and whether they are formed by inter- or intramolecular G4s. B, shown are the structures of known quadruplexes UG4U with sequence UGGGGU, the thrombin-binding aptamer (TBA) with sequence d(GGTTGGTGTGGTTGG), and d(GGA)8. C, cumulative distribution plot of G-scores for the top 50 FMRP target mRNAs (green), HuR mRNA targets (blue), Lin28-bound mRNAs in the Peng dataset (red) (5) and Lin28 mRNA targets in the Hafner dataset (orange) (34). p values were calculated using a Kolmogorov-Smirnov test.
Lin28-regulated miRNAs and mRNAs Contain G-quartet Features
By monitoring imino protons, NMR can detect the hydrogen-bonding network that governs nucleic acid structure. Labile imino proton resonances are only observed when they are protected from solvent exchange by hydrogen bonding. In Watson-Crick base pairing, A:U hydrogen bonds have a chemical shift at ∼13 ppm, whereas G:C hydrogen bonds are closer to 12.7 ppm (48). G:U wobbles and other non-canonical hydrogen bonds occur upfield of 12.7 ppm (48, 49). The unusual hydrogen-bonding pattern of G4s (Fig. 1A) gives rise to characteristic imino resonances at ∼10–12.2 ppm (48, 49). Peaks within this region are highly suggestive of G4 formation. We recorded one-dimensional proton NMR spectra for Lin28-binding RNAs (pre-let-7d, the let-7d loop, pre-let-7g, the let-7g loop, and the LREs of DSG2, SUN1, HSPA5 and OCT4). All of the RNAs had imino resonances indicating the presence of non-canonical hydrogen bonds (Fig. 2A). In addition to putative A:U and G:C bonds, each Lin28 RNA target had resonances in the non-canonical hydrogen bond region from ∼10.0 to 12.2 ppm. Imino resonances in this region suggest that pre-let-7d, the let-7d loop, pre-let-7g, the let-7g loop, and LREs of DSG2, SUN1, HSPA5, and OCT4 might contain G4 features. We attempted to use multidimensional NMR methods including NOESY experiments to unambiguously map the hydrogen bond connectivity of these Lin28-binding RNAs. However, from PAGE we observed that these RNAs oligomerize and/or dynamically change structure in solution, making additional NMR analysis difficult to interpret. Thus, although the imino resonance pattern suggests the presence of G4s, we were unable to obtain definitive structural evidence.
FIGURE 2.
Lin28 RNA targets contain G4 features. A, 1H imino proton spectra for Lin28 targets contain resonances indicative of non-canonical hydrogen bonds similar to G4s (boxed region). B, Lin28 targets bind to NMM, a G4-specific fluorescent dye, but poly(U), yeast tRNA, and the loop of miR-21 do not. Error bars indicate ± S.D.
To gain further insight as to whether Lin28 RNAs contain G4s, we made use of NMM, a dye that binds selectively to G4s (52–55). When it binds to G4s, NMM is excited at 399 nm and fluoresces at 614 nm (55). We therefore compared NMM fluorescence of these Lin28-binding RNAs (pre-let-d, the let-7d loop, pre-let-7g, the let-7g loop, and the LREs of DSG2, HSPA5, SUN1, and OCT4) with that of RNAs that lack a G4 structure. Strong NMM fluorescence was measured for the known G4 RNA UG4U (UGGGGU) and for all of the Lin28-binding RNAs measured, but little to no fluorescence was detected when NMM was mixed with yeast-tRNA, poly(U), or the loop of miR-21 (Fig. 2B). Taken together with the one-dimensional NMR results, NMM fluorescence suggests that Lin28-binding miRNAs and mRNAs form G4s.
RNA Mutations That Disrupt Lin28 Binding Disrupt NMM Fluorescence
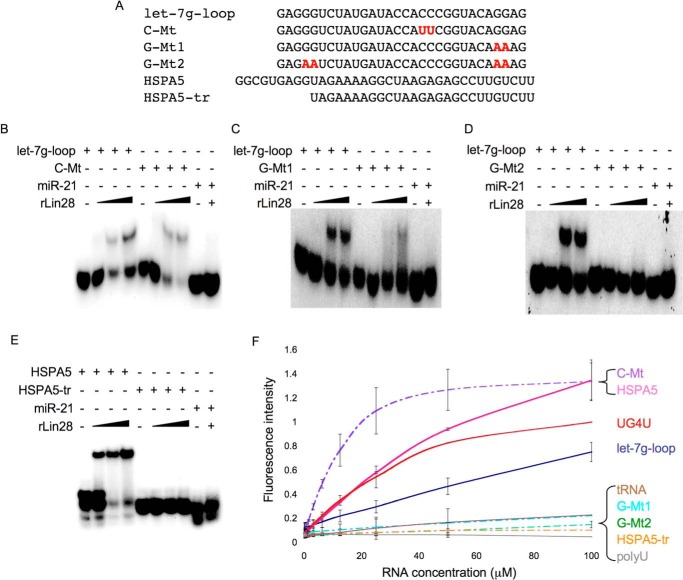
To probe whether Lin28 binding to RNA correlates with G4 formation, we generated RNAs that were truncated or contained mutations that might affect G4 formation. For the let-7g loop, we mutated C18C19 to U18U19 (C-Mt), G27G28 to A27A28 (G-Mt1), and G4G5 and G27G28 to A4A5 and A27A28 (G-Mt2). We also synthesized an HSPA5 LRE truncation by removing 9 nucleotides from the 5′-end containing the first two GG repeats (HSPA5-tr). We tested the ability of each mutant to bind Lin28 by EMSA and to bind to NMM by fluorescence assay (Fig. 3). When compared with the wild-type let-7g-loop, the C-Mts had increased affinity for Lin28 as suggested by the relative intensity of the free let-7g loop with the gel-shifted band and increased NMM fluorescence. G-Mt1 had reduced Lin28 binding and weaker NMM fluorescence. G-Mt2 did not bind to Lin28 and showed no NMM fluorescence. HSPA5-tr did not cause a Lin28 gel shift or NMM fluorescence, suggesting that G-repeats are critical for Lin28 binding and that Lin28 binding correlates with NMM fluorescence. These findings suggest that Lin28 and NMM recognize a shared structural feature of RNA. Because NMM is only known to bind G4s, Lin28 likely recognizes a G4 structure in its targets.
FIGURE 3.
Lin28 binding correlates with NMM fluorescence. A, let-7g loop mutations and HSPA5 truncation evaluated for gel shift and NMM binding. B–E, EMSAs comparing the Lin28 interaction with wild-type and mutant let-7g loop (B–D) and full-length and truncated HSAP5 LRE (E). F, NMM fluorescence assay for wild-type and mutant let-7g loop and full-length and truncated HSPA5 LRE. Error bars indicate ± S.D.
Lin28 Binding Disrupts the Ability of Lin28 RNA Targets to Interact with N-Methyl Mesoporphyrin IX
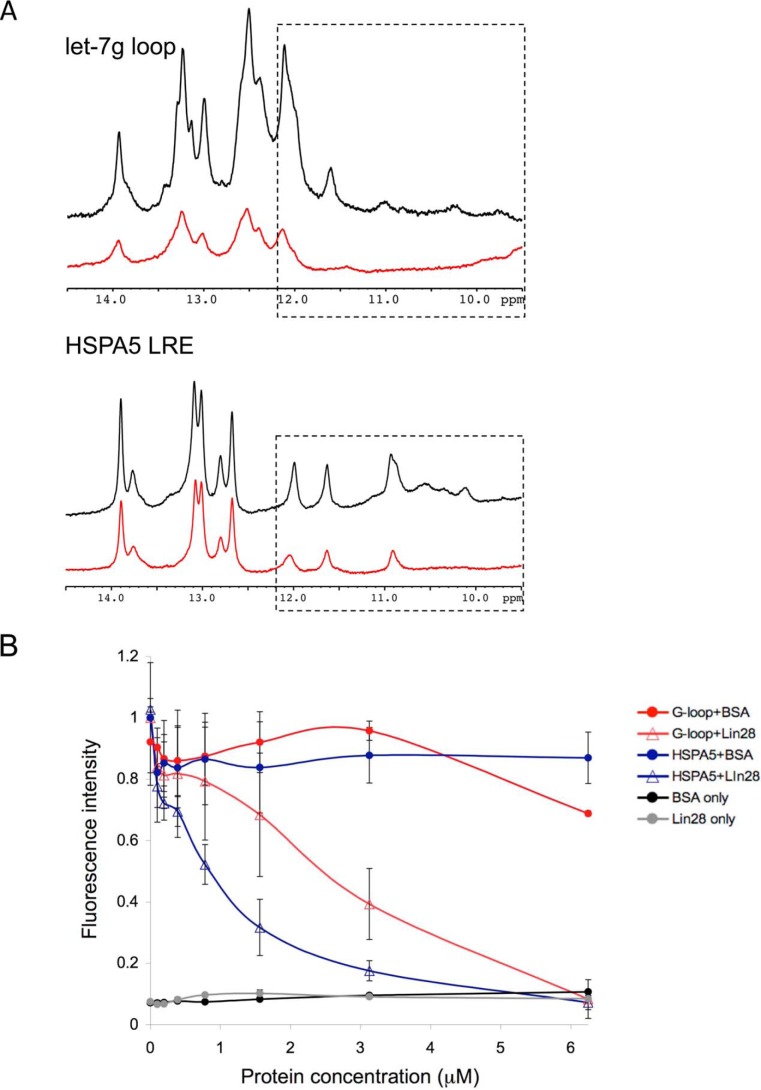
Upon binding, Lin28 induces a conformational change in its RNA targets (32, 36, 37, 56). We hypothesized that the Lin28-induced RNA rearrangement might remodel the RNA G4 structure. To test this idea, we monitored changes to the imino protons of the let-7g loop and the HSPA5 LRE upon the addition of Lin28 (Fig. 4A). When saturating amounts of Lin28 were incubated with these RNAs, the intensity of the iminos decreased and the G4-like imino signals were greatly reduced. This suggests that Lin28 directly binds to these target RNAs and that binding disrupts the hydrogen-bonding network. To further examine the ability of Lin28 to unwind G4s, we performed a modified NMM assay in which the let-7g-loop or HSPA5 LRE was incubated with a constant concentration of NMM and increasing amounts of rLin28A or BSA (Fig. 4B). The let-7g-loop and HSPA5 LRE exhibited strong NMM fluorescence on their own, but when Lin28 was added, NMM fluorescence was lost. The addition of BSA had no effect. Thus, in the presence of Lin28, the RNA targets are no longer able to interact with NMM. These results together with the NMR data suggest that Lin28 binding remodels the G4s.
FIGURE 4.
Lin28 binding unwinds G4s. A, 1H imino proton spectra of the let-7g-loop and HSPA5 LRE (50 μm) (black) and in the presence of rLin28A (75 μm) (red). The intensities of imino resonances decrease and the predicted G4 iminos (boxed region) broaden in the presence of Lin28. B, 50 μm of the let-7g-loop or HSPA5 LRE was incubated with a constant amount of NMM (5 μm). When increasing amounts of rLin28A were added, NMM fluorescence diminished. BSA had no effect. Lin28 or BSA on their own did not fluoresce. Error bars indicate ± S.D.
NMM Inhibits Lin28
If remodeling G4s is crucial to Lin28 function, the G4 feature of Lin28-binding RNAs could represent a novel site to manipulate Lin28 activity. By binding to G4s within gene promoters and within regulatory sites of mRNAs, G4-intercalating agents, like NMM, inhibit transcription and translation (57–59). We performed a competition EMSA to examine whether NMM inhibits Lin28 binding (Fig. 5A). Both 32P-labeled let-7g-loop and HSPA5 LRE bound rLin28A as measured by the presence of a gel shift. However, adding increasing amounts of NMM led to a decrease in the gel shift and an increase in the signal of free RNA. Thus, NMM inhibits the ability of Lin28 to bind RNA. To our knowledge, small-molecule inhibitors of Lin28 binding have not been described before. The miR-21 loop and HSPA5 LRE truncation (HSPA5-tr) did not bind Lin28 or NMM at the highest concentration.
FIGURE 5.
NMM inhibits Lin28 binding and function. A, 32P-labeled let-7g loop (left) and HSPA5 LRE (right) were incubated with a constant amount of rLin28A (10 μm) known to form a Lin28·RNA gel shift. Increasing amounts of NMM (0–100 μm) diminished the amount of rLin28A-shifted product and increased signal of the free [32P]RNA. NMM and rLin28A had no effect on miR-21 or HSPA5-tr. B and C, effect of NMM on cell viability after 48 h (B) and on levels of pri-miRNAs, pre-miRNAs, and mature miRNAs after 3 h in NCCIT cells (C). D, luciferase reporter assay using a Dual-Luciferase reporter in which the Renilla luciferase 3′-UTR contains let-7-binding elements. NCCIT cells were cotransfected with the luciferase reporter plasmid and wild-type (WT) or mutant (MT) pre-let-7g and were then treated or not with NMM. NMM enhanced the activity of WT, but not MT, pre-let-7g. Renilla luciferase activity was normalized to firefly luciferase activity. E and F, NMM suppresses the protein levels (E), but not the mRNA levels (F), of Lin28 mRNA targets. G and H, incubation of NCCIT cells with NMM (100 μm for 48 h) decreases the number of sphere-forming cells (G) and blocks colony formation (H). *, p value < 0.005, **, <0.001 using a two-tailed Student's t test. Error bars indicate ± S.D.
We next sought to determine whether NMM could inhibit Lin28 function in cells. Human NCCIT embryonal carcinoma cells, which, like embryonic stem cells, express high levels of Lin28, were incubated with 100 μm NMM. NMM had no significant effect on cell viability (Fig. 5B). However, exposure to NMM for 3 h increased levels of mature let-7a and let-7g miRNAs, but did not affect miR-21 or miR-34a control miRNAs (Fig. 5C). This increase was specific to mature let-7 miRNAs as neither pri-let-7 nor pre-let-7 levels changed with NMM treatment. Thus, NMM alleviates the Lin28-induced block in let-7 processing.
Next we used a cell reporter assay to determine whether adding NMM increases let-7 function (Fig. 5D). NCCIT cells were transfected with a luciferase reporter containing let-7-binding sites in the 3′-UTR and either wild-type pre-let-7g or a mutant pre-let-7g, in which 4 guanines were replaced with adenosines. These mutations do not affect the mature let-7 sequence (Fig. 5D, underlined) but prevent G4 formation (Fig. 3F). The addition of both wild-type and mutant pre-let-7g decreased luciferase activity. The mutant pre-let-7g showed greater luciferase inhibition, which could result from diminished binding of Lin28 to the mutant pre-let-7g sequence (Fig. 3D). Both wild-type and mutant pre-let-7g decreased luciferase activity, but mutant pre-let-7g inhibited activity more. NMM had no effect on mutant pre-let-7, but wild-type pre-let-7 now inhibited luciferase assay as well as mutant pre-let-7. Thus, the increase of NMM in let-7 processing led to better gene knockdown.
Lin28 also promotes translation of stem cell target genes. With NMM, we observed a decrease in the protein levels of Lin28 mRNA targets OCT4, HMGA1, CCNB1, CDK4, and Lin28A (Fig. 5E), but there was no effect on actin or tubulin protein levels. The mRNAs of the Lin28 target genes were unchanged, with the exception of MYC and Lin28 mRNA, which were significantly decreased (Fig. 5F). MYC transcription is known to be affected by G4 agents. The decrease in Lin28 mRNA suggests that NMM may also interfere with Lin28 transcription.
Because of increases in let-7 miRNAs and decreased levels of proteins associated with pluripotency, we next examined whether NMM affects stem-like properties using sphere-forming and colony-forming assays. The ability to form spheres in vitro depends on the presence of self-renewing stem cells within a population. We counted the number of spheres embryonal carcinoma cells formed in the presence and absence of NMM. Incubation with NMM significantly reduced the number of sphere-forming cells by ∼75% (Fig. 5G). This suggests that NMM inhibits self-renewal. We also tested NMM in a colony formation assay, where the ability of single cells to undergo unlimited division is assessed. NMM nearly abolished the ability to form colonies (Fig. 5H). This further supports our hypothesis that NMM inhibits self-renewal and stem-like properties. Because NMM binds broadly to G4-containing RNAs and DNAs, it is unlikely that these effects are due solely to the ability of NMM to inhibit Lin28. Nonetheless, these results suggest that G4-intercalating agents can inhibit Lin28 binding to RNA and inhibit stem-like properties of cells.
Discussion
Lin28 is an evolutionarily conserved RNA-binding protein that regulates cell differentiation (12, 16). It has been unclear how Lin28 identifies its target RNAs and distinguishes them from other RNAs. Although multiple RNA sequence motifs have been suggested to influence Lin28 binding to RNA, the motifs are small and abundant in the transcriptome. Our data strongly suggest that Lin28 recognizes a common structural feature in its RNA targets; Lin28-regulated miRNAs and mRNAs share G-quartet features.
Lin28-bound mRNAs are enriched for sequences with increased potential to form G4s. The Peng dataset of Lin28 targets contains 1,200 mRNAs that were enriched at least 2.5-fold in a Lin28 immunoprecipitation when compared with a pre-immune control immunoprecipitation in human embryonic stem cells (5). No significantly enriched linear sequence was identified within this list of Lin28 targets. However, we found that the top 50 most enriched Lin28 mRNAs from this dataset are highly enriched for sequences likely to form G4s. In addition, the top 50 Lin28 mRNA targets from the Hafner dataset of Lin28-bound mRNAs identified by PAR-CLIP in FLAG/HA-Lin28-overexpressing HEK 293 cells were also predicted to have an increased likelihood to form G4s (34). The Hafner dataset contains 1,800 Lin28A mRNA targets. Although the authors proposed a AYYHY consensus sequence where Y is a pyrimidine and H is any nucleotide except guanine, they did observe that ∼15% of the Lin28 targets were enriched for guanines (34). The increased potential to form G4s within multiple Lin28 target datasets suggests that the G4 structure might be common to all Lin28-binding RNAs.
By one-dimensional NMR spectroscopy, we found that Lin28-binding RNAs contain imino resonances from 10 to 12.2 ppm, indicative of non-canonical hydrogen bonds (48, 51, 60). The imino resonance pattern is characteristic of the G:G:G:G hydrogen-bonding networks found in G4s (48, 51, 61). We were unable to obtain high-resolution two-dimensional NMR spectra that could provide further structural support. G4-containing RNAs are prone to aggregate (51, 62–64), which may have foiled our efforts.
Lin28-binding RNAs interact with the highly specific G4 fluorescent dye, NMM (54, 55). Lin28 RNA targets, but not control RNAs such as the loop of miR-21, induced NMM fluorescence. Mutations and truncations of Lin28 RNA targets that hindered Lin28 binding also led to decreased NMM fluorescence. These results suggest that Lin28 and NMM recognize a similar structure. Because NMM selectively binds only to G4s, Lin28 RNA targets likely contain G4 features that are important for Lin28 recognition.
Recent atomic structures of Lin28 in complex with fragments of pre-let-7 loop sequences did not detect G4s (33, 36). However, the structure of the free RNA may not be the same as that bound to Lin28 because our data suggest that Lin28 unwinds G4 quartets. This is consistent with footprinting assays of pre-let-7g, which previously suggested that Lin28 binding induces a conformational change within the RNA (32). Isolated studies with the ZnF domains also suggest that they partially unfold pre-let-7 targets (56). Additionally, a FRET study revealed that the CSD of Lin28 remodels the terminal loop of pre-let-7 by unwinding the upper stem region (37). We propose that the Lin28-mediated remodeling of RNA disrupts the G4 structure because in the presence of Lin28, the Lin28-binding RNAs no longer induce NMM fluorescence.
A recent study found that another CSD protein, YB-1, binds to G4s in tRNA fragments (65). Truncation analysis showed that the CSD is responsible for G4 binding of YB-1. That study and our data suggest that the CSD, a nucleic acid-binding domain, which first arose in bacteria and forms an antiparallel β barrel, functions by binding and remodeling G4s. Further studies are needed to explore this hypothesis.
Remodeling G4s could be essential for Lin28 function (Fig. 6). During let-7 biogenesis, Lin28 recruits TUT4 to pre-let-7 transcripts and then TUT4 uridylates pre-let-7s to promote their degradation (28, 29). TUT4 does not bind to pre-let-7 in the absence of Lin28 (29). Remodeling of the pre-let-7 G4 by Lin28 could be required for the TUT4 interaction and subsequent uridylation. Because the G4 structure might interfere with ribosome scanning, remodeling G4s in mRNAs could also explain the ability of Lin28 to stimulate mRNA translation.
FIGURE 6.
Model of Lin28 RNA regulation. A and B, we propose that Lin28 binds and remodels G4s in pre-let-7 miRNAs (A) and in LREs of mRNAs (B). Remodeling pre-let-7s might be required for TUT4 binding, subsequent uridylation, and degradation of pre-let-7s. Remodeling G4s in LREs may increase ribosome scanning and stimulate mRNA translation. The Lin28 CSD is in blue, and the ZnFs are in green.
G-quadruplexes are found within DNAs in eukaryotic telomeres (66); the promoter regions of some cancer-related genes, including BCL2 (67), KIT (68), MYB (69), MYC (70), HIF1A (71), hTERT (72), KRAS (73), RB (74), and VEGF (75); immunoglobulin gene switch regions (76); and ribosomal genes (77). G4-intercalating agents have been assessed for anticancer therapy (57, 50, 51). TMPyP4, a porphyrin analogue of NMM that binds G4s and double-stranded DNA, blocks MYC transcription and decreases tumor growth and increases survival in tumor xenograft studies (57, 58). By blocking transcription and translation of oncogenes, G4-intercalating agents have demonstrated anticancer activity (57, 50, 51). Similar agents could be designed to target the G4 feature of Lin28-binding RNAs, preventing Lin28 remodeling G4s and inhibiting Lin28 activity.
Here, we have shown that NMM inhibits Lin28 binding to RNAs in a gel shift. This is the first example of a small-molecule inhibitor of Lin28. NMM may also inhibit Lin28 in cells. The addition of NMM to embryonal carcinoma cells increased let-7 miRNAs levels and decreased protein levels of Lin28 mRNA targets OCT4, HMGA1, CCNB1, CDK4, and Lin28. NMM also decreased the number of sphere-forming cells and inhibited colony formation. These findings suggest that NMM inhibits self-renewal. Because many human cancers, especially those with the worst prognosis, are often undifferentiated and have increased Lin28, NMM or other G4-intercalating agents may possess anticancer activity in these tumors. Our findings add new insight to understanding how Lin28 selects and regulates its miRNA and mRNA targets and suggest a new strategy for therapeutic intervention.
Author Contributions
E. O. conceived the study, conducted the experiments in Table 1, Fig. 2, Fig. 3, Fig. 4, Fig. 5, A, B, E, F, G, and H, and Fig. 6, and wrote the paper. M. T. N. L. performed experiments in Fig. 5, C and D. S. I. synthesized all the RNA constructs. S. M. T. performed experiments in Fig. 1C. R. K. and O. H. provided technical advice. H. A. helped to design and to execute the NMR and NMM experiments. G. W. and J. L. analyzed the data, coordinated the studies, and edited the paper. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Victoria D'Souza and members of her laboratory for technical assistance including the in vitro transcription of RNA constructs and NMR acquisition and data analysis. We also thank Gary Ruvkun and Jack Szostak for their helpful insights and suggestions.
This work was supported by the Department of Defense Breast Cancer Research Program Grant W81 XWH-09-1-0058 (to J. L.) and National Institutes of Health Grant GM047467 (to G. W.). The authors declare that they have no conflicts of interest with the contents of this article.
- CSD
- cold-shock domain
- ZnF
- zinc finger
- TUT4
- TUTase4
- G4
- G-quartet
- NMM
- N-methyl mesoporphyrin IX
- miRNA
- microRNA
- miR
- microRNA
- LRE
- Lin28-responsive element
- PAR-CLIP
- photoactivatable ribonucleoside-enhanced cross-linking and immunoprecipitation
- FMRP
- fragile X mental retardation protein-1.
References
- 1. Ambros V., Horvitz H. R. (1984) Heterochronic mutants of the nematode Caenorhabditis elegans. Science 226, 409–416 [DOI] [PubMed] [Google Scholar]
- 2. Liu Z. C., Ambros V. (1989) Heterochronic genes control the stage-specific initiation and expression of the dauer larva developmental program in Caenorhabditis elegans. Genes Dev. 3, 2039–2049 [DOI] [PubMed] [Google Scholar]
- 3. Moss E. G., Lee R. C., Ambros V. (1997) The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell 88, 637–646 [DOI] [PubMed] [Google Scholar]
- 4. Zheng K., Wu X., Kaestner K. H., Wang P. J. (2009) The pluripotency factor LIN28 marks undifferentiated spermatogonia in mouse. BMC Dev. Biol. 9, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Peng S., Chen L.-L., Lei X.-X., Yang L., Lin H., Carmichael G. G., Huang Y. (2011) Genome-wide studies reveal that Lin28 enhances the translation of genes important for growth and survival of human embryonic stem cells. Stem Cells 29, 496–504 [DOI] [PubMed] [Google Scholar]
- 6. Zhu H., Shyh-Chang N., Segrè A. V., Shinoda G., Shah S. P., Einhorn W. S., Takeuchi A., Engreitz J. M., Hagan J. P., Kharas M. G., Urbach A., Thornton J. E., Triboulet R., Gregory R. I., DIAGRAM Consortium, MAGIC Investigators, Altshuler D., Daley G. Q. (2011) The Lin28/let-7 axis regulates glucose metabolism. Cell 147, 81–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhu H., Shah S., Shyh-Chang N., Shinoda G., Einhorn W. S., Viswanathan S. R., Takeuchi A., Grasemann C., Rinn J. L., Lopez M. F., Hirschhorn J. N., Palmert M. R., Daley G. Q. (2010) Lin28a transgenic mice manifest size and puberty phenotypes identified in human genetic association studies. Nat. Genet. 42, 626–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. West J. A., Viswanathan S. R., Yabuuchi A., Cunniff K., Takeuchi A., Park I.-H., Sero J. E., Zhu H., Perez-Atayde A., Frazier A. L., Surani M. A., Daley G. Q. (2009) A role for Lin28 in primordial germ-cell development and germ-cell malignancy. Nature 460, 909–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Viswanathan S. R., Powers J. T., Einhorn W., Hoshida Y., Ng T. L., Toffanin S., O'Sullivan M., Lu J., Phillips L. A., Lockhart V. L., Shah S. P., Tanwar P. S., Mermel C. H., Beroukhim R., Azam M., Teixeira J., Meyerson M., Hughes T. P., Llovet J. M., Radich J., Mullighan C. G., Golub T. R., Sorensen P. H., Daley G. Q. (2009) Lin28 promotes transformation and is associated with advanced human malignancies. Nat. Genet. 41, 843–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Meirelles K., Benedict L. A., Dombkowski D., Pepin D., Preffer F. I., Teixeira J., Tanwar P. S., Young R. H., MacLaughlin D. T., Donahoe P. K., Wei X. (2012) Human ovarian cancer stem/progenitor cells are stimulated by doxorubicin but inhibited by Mullerian inhibiting substance. Proc. Natl. Acad. Sci. U.S.A. 109, 2358–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ambros V. (1989) A hierarchy of regulatory genes controls a larva-to-adult developmental switch in C. elegans. Cell 57, 49–57 [DOI] [PubMed] [Google Scholar]
- 12. Moss E. G., Tang L. (2003) Conservation of the heterochronic regulator Lin-28, its developmental expression and microRNA complementary sites. Dev. Biol. 258, 432–442 [DOI] [PubMed] [Google Scholar]
- 13. Polesskaya A., Cuvellier S., Naguibneva I., Duquet A., Moss E. G., Harel-Bellan A. (2007) Lin-28 binds IGF-2 mRNA and participates in skeletal myogenesis by increasing translation efficiency. Genes Dev. 21, 1125–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xu B., Zhang K., Huang Y. (2009) Lin28 modulates cell growth and associates with a subset of cell cycle regulator mRNAs in mouse embryonic stem cells. RNA 15, 357–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Qiu C., Ma Y., Wang J., Peng S., Huang Y. (2010) Lin28-mediated post-transcriptional regulation of Oct4 expression in human embryonic stem cells. Nucleic Acids Res. 38, 1240–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Viswanathan S. R., Daley G. Q. (2010) Lin28: A microRNA regulator with a macro role. Cell 140, 445–449 [DOI] [PubMed] [Google Scholar]
- 17. Viswanathan S. R., Daley G. Q., Gregory R. I. (2008) Selective blockade of microRNA processing by Lin28. Science 320, 97–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yu J., Vodyanik M. A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J. L., Tian S., Nie J., Jonsdottir G. A., Ruotti V., Stewart R., Slukvin I. I., Thomson J. A. (2007) Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920 [DOI] [PubMed] [Google Scholar]
- 19. Iliopoulos D., Hirsch H. A., Struhl K. (2009) An epigenetic switch involving NF-κB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell 139, 693–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. King C. E., Cuatrecasas M., Castells A., Sepulveda A. R., Lee J.-S., Rustgi A. K. (2011) LIN28B promotes colon cancer progression and metastasis. Cancer Res. 71, 4260–4268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xue D., Peng Y., Wang F., Allan R. W., Cao D. (2011) RNA-binding protein LIN28 is a sensitive marker of ovarian primitive germ cell tumours. Histopathology 59, 452–459 [DOI] [PubMed] [Google Scholar]
- 22. Dangi-Garimella S., Yun J., Eves E. M., Newman M., Erkeland S. J., Hammond S. M., Minn A. J., Rosner M. R. (2009) Raf kinase inhibitory protein suppresses a metastasis signalling cascade involving LIN28 and let-7. EMBO J. 28, 347–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yu F., Yao H., Zhu P., Zhang X., Pan Q., Gong C., Huang Y., Hu X., Su F., Lieberman J., Song E. (2007) let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 131, 1109–1123 [DOI] [PubMed] [Google Scholar]
- 24. Chang T.-C., Zeitels L. R., Hwang H.-W., Chivukula R. R., Wentzel E. A., Dews M., Jung J., Gao P., Dang C. V., Beer M. A., Thomas-Tikhonenko A., Mendell J. T. (2009) Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation. Proc. Natl. Acad. Sci. U.S.A. 106, 3384–3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Johnson S. M., Grosshans H., Shingara J., Byrom M., Jarvis R., Cheng A., Labourier E., Reinert K. L., Brown D., Slack F. J. (2005) RAS is regulated by the let-7 microRNA family. Cell 120, 635–647 [DOI] [PubMed] [Google Scholar]
- 26. Newman M. A., Thomson J. M., Hammond S. M. (2008) Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA 14, 1539–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Heo I., Joo C., Cho J., Ha M., Han J., Kim V. N. (2008) Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol. Cell 32, 276–284 [DOI] [PubMed] [Google Scholar]
- 28. Hagan J. P., Piskounova E., Gregory R. I. (2009) Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 16, 1021–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Heo I., Joo C., Kim Y.-K., Ha M., Yoon M.-J., Cho J., Yeom K.-H., Han J., Kim V. N. (2009) TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell 138, 696–708 [DOI] [PubMed] [Google Scholar]
- 30. Lehrbach N. J., Armisen J., Lightfoot H. L., Murfitt K. J., Bugaut A., Balasubramanian S., Miska E. A. (2009) LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 16, 1016–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cho J., Chang H., Kwon S. C., Kim B., Kim Y., Choe J., Ha M., Kim Y. K., Kim V. N. (2012) LIN28A is a suppressor of ER-associated translation in embryonic stem cells. Cell 151, 765–777 [DOI] [PubMed] [Google Scholar]
- 32. Lightfoot H. L., Bugaut A., Armisen J., Lehrbach N. J., Miska E. A., Balasubramanian S. (2011) A LIN28-dependent structural change of pre-let-7g directly inhibits Dicer processing. Biochemistry 50, 7514–7521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nam Y., Chen C., Gregory R. I., Chou J. J., Sliz P. (2011) Molecular basis for interaction of let-7 microRNAs with Lin28. Cell 147, 1080–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hafner M., Max K. E. A., Bandaru P., Morozov P., Gerstberger S., Brown M., Molina H., Tuschl T. (2013) Identification of mRNAs bound and regulated by human LIN28 proteins and molecular requirements for RNA recognition. RNA 19, 613–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Piskounova E., Viswanathan S. R., Janas M., LaPierre R. J., Daley G. Q., Sliz P., Gregory R. I. (2008) Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J. Biol. Chem. 283, 21310–21314 [DOI] [PubMed] [Google Scholar]
- 36. Loughlin F. E., Gebert L. F. R., Towbin H., Brunschweiger A., Hall J., Allain F. H.-T. (2012) Structural basis of pre-let-7 miRNA recognition by the zinc knuckles of pluripotency factor Lin28. Nat. Struct. Mol. Biol. 19, 84–89 [DOI] [PubMed] [Google Scholar]
- 37. Mayr F., Schütz A., Döge N., Heinemann U. (2012) The Lin28 cold-shock domain remodels pre-let-7 microRNA. Nucleic Acids Res. 40, 7492–7506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wilbert M. L., Huelga S. C., Kapeli K., Stark T. J., Liang T. Y., Chen S. X., Yan B. Y., Nathanson J. L., Hutt K. R., Lovci M. T., Kazan H., Vu A. Q., Massirer K. B., Morris Q., Hoon S., Yeo G. W. (2012) LIN28 binds messenger RNAs at GGAGA motifs and regulates splicing factor abundance. Mol. Cell 48, 195–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jin J., Jing W., Lei X.-X., Feng C., Peng S., Boris-Lawrie K., Huang Y. (2011) Evidence that Lin28 stimulates translation by recruiting RNA helicase A to polysomes. Nucleic Acids Res. 39, 3724–3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sen D., Gilbert W. (1988) Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature 334, 364–366 [DOI] [PubMed] [Google Scholar]
- 41. Burge S., Parkinson G. N., Hazel P., Todd A. K., Neidle S. (2006) Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 34, 5402–5415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Patel D. J., Phan A. T., Kuryavyi V. (2007) Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 35, 7429–7455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kikin O., D'Antonio L., Bagga P. S. (2006) QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 34, W676–W682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bole M., Menon L., Mihailescu M.-R. (2008) Fragile X mental retardation protein recognition of G quadruplex structure per se is sufficient for high affinity binding to RNA. Mol. Biosyst. 4, 1212–1219 [DOI] [PubMed] [Google Scholar]
- 45. Brown V., Jin P., Ceman S., Darnell J. C., O'Donnell W. T., Tenenbaum S. A., Jin X., Feng Y., Wilkinson K. D., Keene J. D., Darnell R. B., Warren S. T. (2001) Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell 107, 477–487 [DOI] [PubMed] [Google Scholar]
- 46. Darnell J. C., Jensen K. B., Jin P., Brown V., Warren S. T., Darnell R. B. (2001) Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell 107, 489–499 [DOI] [PubMed] [Google Scholar]
- 47. Mukherjee N., Corcoran D. L., Nusbaum J. D., Reid D. W., Georgiev S., Hafner M., Ascano M., Jr., Tuschl T., Ohler U., Keene J. D. (2011) Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol. Cell 43, 327–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Webba da Silva M. (2007) NMR methods for studying quadruplex nucleic acids. Methods 43, 264–277 [DOI] [PubMed] [Google Scholar]
- 49. Adrian M., Heddi B., Phan A. T. (2012) NMR spectroscopy of G-quadruplexes. Methods 57, 11–24 [DOI] [PubMed] [Google Scholar]
- 50. Mikami-Terao Y., Akiyama M., Yuza Y., Yanagisawa T., Yamada O., Yamada H. (2008) Antitumor activity of G-quadruplex-interactive agent TMPyP4 in K562 leukemic cells. Cancer Lett. 261, 226–234 [DOI] [PubMed] [Google Scholar]
- 51. Murat P., Singh Y., Defrancq E. (2011) Methods for investigating G-quadruplex DNA/ligand interactions. Chem Soc Rev 40, 5293–5307 [DOI] [PubMed] [Google Scholar]
- 52. Nicoludis J. M., Barrett S. P., Mergny J.-L., Yatsunyk L. A. (2012) Interaction of human telomeric DNA with N-methyl mesoporphyrin IX. Nucleic Acids Res. 40, 5432–5447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nicoludis J. M., Miller S. T., Jeffrey P. D., Barrett S. P., Rablen P. R., Lawton T. J., Yatsunyk L. A. (2012) Optimized end-stacking provides specificity of N-methyl mesoporphyrin IX for human telomeric G-quadruplex DNA. J. Am. Chem. Soc. 134, 20446–20456 [DOI] [PubMed] [Google Scholar]
- 54. Ren J., Chaires J. B. (1999) Sequence and structural selectivity of nucleic acid binding ligands. Biochemistry 38, 16067–16075 [DOI] [PubMed] [Google Scholar]
- 55. Arthanari H., Basu S., Kawano T. L., Bolton P. H. (1998) Fluorescent dyes specific for quadruplex DNA. Nucleic Acids Res. 26, 3724–3728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Desjardins A., Yang A., Bouvette J., Omichinski J. G., Legault P. (2012) Importance of the NCp7-like domain in the recognition of pre-let-7g by the pluripotency factor Lin28. Nucleic Acids Res. 40, 1767–1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Grand C. L., Han H., Muñoz R. M., Weitman S., Von Hoff D. D., Hurley L. H., Bearss D. J. (2002) The cationic porphyrin TMPyP4 down-regulates c-MYC and human telomerase reverse transcriptase expression and inhibits tumor growth in vivo. Mol. Cancer Ther. 1, 565–573 [PubMed] [Google Scholar]
- 58. Phan A. T., Kuryavyi V., Gaw H. Y., Patel D. J. (2005) Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter. Nat. Chem. Biol. 1, 167–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bugaut A., Rodriguez R., Kumari S., Hsu S.-T. D., Balasubramanian S. (2010) Small molecule-mediated inhibition of translation by targeting a native RNA G-quadruplex. Org. Biomol. Chem. 8, 2771–2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fürtig B., Richter C., Wöhnert J., Schwalbe H. (2003) NMR spectroscopy of RNA. Chem. Biol. Chem. 4, 936–962 [DOI] [PubMed] [Google Scholar]
- 61. Lane A. N., Chaires J. B., Gray R. D., Trent J. O. (2008) Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 36, 5482–5515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Smargiasso N., Rosu F., Hsia W., Colson P., Baker E. S., Bowers M. T., De Pauw E., Gabelica V. (2008) G-quadruplex DNA assemblies: loop length, cation identity, and multimer formation. J. Am. Chem. Soc. 130, 10208–10216 [DOI] [PubMed] [Google Scholar]
- 63. Marsh T. C., Henderson E. (1994) G-wires: self-assembly of a telomeric oligonucleotide, d(GGGGTTGGGG), into large superstructures. Biochemistry 33, 10718–10724 [DOI] [PubMed] [Google Scholar]
- 64. Krishnan-Ghosh Y., Liu D., Balasubramanian S. (2004) Formation of an interlocked quadruplex dimer by d(GGGT). J Am Chem Soc 126, 11009–11016 [DOI] [PubMed] [Google Scholar]
- 65. Ivanov P., O'Day E., Emara M. M., Wagner G., Lieberman J., Anderson P. (2014) G-quadruplex structures contribute to the neuroprotective effects of angiogenin-induced tRNA fragments. Proc. Natl. Acad. Sci. U.S.A. 111, 18201–18206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Henderson E., Hardin C. C., Walk S. K., Tinoco I., Jr., Blackburn E. H. (1987) Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine-guanine base pairs. Cell 51, 899–908 [DOI] [PubMed] [Google Scholar]
- 67. Dai J., Dexheimer T. S., Chen D., Carver M., Ambrus A., Jones R. A., Yang D. (2006) An intramolecular G-quadruplex structure with mixed parallel/antiparallel G-strands formed in the human BCL-2 promoter region in solution. J. Am. Chem. Soc. 128, 1096–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Fernando H., Reszka A. P., Huppert J., Ladame S., Rankin S., Venkitaraman A. R., Neidle S., Balasubramanian S. (2006) A conserved quadruplex motif located in a transcription activation site of the human c-kit oncogene. Biochemistry 45, 7854–7860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Palumbo S. L., Memmott R. M., Uribe D. J., Krotova-Khan Y., Hurley L. H., Ebbinghaus S. W. (2008) A novel G-quadruplex-forming GGA repeat region in the c-myb promoter is a critical regulator of promoter activity. Nucleic Acids Res. 36, 1755–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Simonsson T., Pecinka P., Kubista M. (1998) DNA tetraplex formation in the control region of c-myc. Nucleic Acids Res. 26, 1167–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. De Armond R., Wood S., Sun D., Hurley L. H., Ebbinghaus S. W. (2005) Evidence for the presence of a guanine quadruplex forming region within a polypurine tract of the hypoxia inducible factor 1α promoter. Biochemistry 44, 16341–16350 [DOI] [PubMed] [Google Scholar]
- 72. Palumbo S. L., Ebbinghaus S. W., Hurley L. H. (2009) Formation of a unique end-to-end stacked pair of G-quadruplexes in the hTERT core promoter with implications for inhibition of telomerase by G-quadruplex-interactive ligands. J Am Chem Soc 131, 10878–10891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Cogoi S., Xodo L. E. (2006) G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 34, 2536–2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Xu Y., Sugiyama H. (2006) Formation of the G-quadruplex and i-motif structures in retinoblastoma susceptibility genes (Rb). Nucleic Acids Res. 34, 949–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sun D., Guo K., Rusche J. J., Hurley L. H. (2005) Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucleic Acids Res. 33, 6070–6080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Maizels N. (2006) Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat. Struct. Mol. Biol. 13, 1055–1059 [DOI] [PubMed] [Google Scholar]
- 77. Drygin D., Siddiqui-Jain A., O'Brien S., Schwaebe M., Lin A., Bliesath J., Ho C. B., Proffitt C., Trent K., Whitten J. P., Lim J. K. C., Von Hoff D., Anderes K., Rice W. G. (2009) Anticancer activity of CX-3543: a direct inhibitor of rRNA biogenesis. Cancer Res. 69, 7653–7661 [DOI] [PubMed] [Google Scholar]