Background: D4 is a uracil-DNA glycosylase (UNG) and an essential component of the vaccinia virus DNA polymerase holoenzyme.
Results: The crystal structure of D4 in complex with DNA is presented.
Conclusion: The D4·DNA contacts exhibit major differences compared with the human UNG·DNA complex.
Significance: This work allows a better understanding of the structural determinants required for UNG function.
Keywords: base excision repair (BER), crystal structure, DNA viruses, viral polymerase, viral replication, uracil-DNA glycosylase, vaccinia virus
Abstract
Vaccinia virus polymerase holoenzyme is composed of the DNA polymerase catalytic subunit E9 associated with its heterodimeric co-factor A20·D4 required for processive genome synthesis. Although A20 has no known enzymatic activity, D4 is an active uracil-DNA glycosylase (UNG). The presence of a repair enzyme as a component of the viral replication machinery suggests that, for poxviruses, DNA synthesis and base excision repair is coupled. We present the 2.7 Å crystal structure of the complex formed by D4 and the first 50 amino acids of A20 (D4·A201–50) bound to a 10-mer DNA duplex containing an abasic site resulting from the cleavage of a uracil base. Comparison of the viral complex with its human counterpart revealed major divergences in the contacts between protein and DNA and in the enzyme orientation on the DNA. However, the conformation of the dsDNA within both structures is very similar, suggesting a dominant role of the DNA conformation for UNG function. In contrast to human UNG, D4 appears rigid, and we do not observe a conformational change upon DNA binding. We also studied the interaction of D4·A201–50 with different DNA oligomers by surface plasmon resonance. D4 binds weakly to nonspecific DNA and to uracil-containing substrates but binds abasic sites with a Kd of <1.4 μm. This second DNA complex structure of a family I UNG gives new insight into the role of D4 as a co-factor of vaccinia virus DNA polymerase and allows a better understanding of the structural determinants required for UNG action.
Introduction
Uracil, one of the four bases present in RNA, can also appear in DNA. Uracil arises in DNA either via misincorporation of dUTP during DNA synthesis or by spontaneous deamination of cytosine leading to U:A base pairs or U:G mismatches that could potentially give rise to C to T transition after the next round of replication (1). To prevent the deleterious effect of these lesions on genome integrity, organisms have developed DNA repair pathways to actively remove the inappropriate bases. The major pathway leading to the removal of uracil bases, called base excision repair, is initiated by uracil-DNA glycosylases (UNG)3 (2). These ubiquitous DNA repair enzymes specifically recognize and excise uracil bases by hydrolyzing the N-glycosidic bond of deoxyuridine in both single- and double-stranded DNA (3). The newly generated abasic site is a substrate for an AP endonuclease that will generate a strand break leaving a 3′OH group and a 5′-deoxyribose phosphate. The latter is then removed, and the gap is filled by a DNA polymerase and sealed by a DNA ligase (4).
Over the years, numerous structural and enzymatic studies have helped to elucidate the molecular basis by which UNGs discriminate between normal and uracil bases within DNA. UNGs share a common mechanism in which bases are flipped out from the DNA duplex into a selective active site pocket, the structure of which allows accurate detection of uracil bases (5). However, one issue that remains to be clarified is how UNGs examine each base in the genome to identify with efficiency the rare uracil bases that are present within the large excess of normal bases. Our current understanding favor a model that includes both hopping and sliding on DNA (6). According to this model, UNGs would exist in two distinct conformational states: the closed conformation would interact nonspecifically with the phosphate backbone favoring one-dimensional protein sliding along the DNA over short distances. In contrast, in the open conformation, UNGs would dissociate from DNA and hop to another region of the genome (7, 8).
Viruses belonging to the Herpesviridae and Poxviridae families are sensitive to the presence of uracil bases in their DNA genome. Thus, like cells from all living organisms, these pathogens encode UNGs to protect their genetic information. UNGs encoded by several herpesviruses were shown to be dispensable for replication in cell culture (9). Indeed, deletion mutants lacking UNG were replication-competent and viable (10–13). In contrast, the poxvirus UNG is an obligate component of the replication machinery. Studies performed on D4 (the UNG encoded by vaccinia virus (VACV)) demonstrated that the presence of the protein is crucial for DNA replication as knock-out mutants lacking D4 are not viable (14, 15), whereas its glycosylase activity is dispensable (16). Later, data obtained from the Traktman group revealed that D4 is a subunit of the VACV DNA polymerase holoenzyme (17). D4 forms together with A20 a heterodimeric complex whose function is essential for processive DNA synthesis by E9, the catalytic DNA polymerase. It is believed that the DNA binding activity of D4 tethers the holoenzyme to the DNA template and that D4 and E9 move together along the genome allowing at the same time an efficient search for uracil bases. Thus, for poxviruses, replication and base excision repair appear to be coupled (18).
VACV D4, as well as Escherichia coli, human and herpes simplex virus type 1 (HSV-1) UNGs are grouped in the family I UNGs, the most extensively studied enzymes from the UNG superfamily. D4 is the most evolutionarily distant member of family I sharing only ∼20% sequence identity with the other UNGs (5). However, despite the relatively low degree of homology, the overall fold of D4 is well conserved when compared with other family members (19, 20). Furthermore, sequence alignments of VACV D4 with related UNGs identified conserved active site residues that are predicted to form the uracil recognition pocket (i.e. Tyr-70, Phe-79, and Asn-120) in addition to Asp-68 and His-181 that are needed for glycosyl bond cleavage (16, 21). When the crystal structure of D4 is superimposed onto the human enzyme bound to DNA, these residues match perfectly their human counterparts (20).
Nonetheless, structure-based sequence alignments of D4 show differences in a number of motifs that are generally highly conserved among UNGs (5). In particular, the important leucine residue of the intercalation loop is replaced in poxviruses by an arginine residue (16). So far, only enzymes carrying a leucine residue have been studied in detail showing its central role in the catalytic mechanism. Indeed, upon binding, human UNG (hUNG) changes from an open to a closed conformation. The protein binds in the minor groove of dsDNA and inserts the hydrophobic leucine residue of the intercalation loop into the DNA base stack, replacing the extrahelical base, whereas the DNA is kinked by ∼45°, and the base is extruded from the double helix (22). If a cognate uracil base is present, it is bound in the uracil recognition site in a distorted conformation enabling hydrolysis (23), whereas the enzyme continues to bind to the newly formed abasic site (24). The replacement of Leu-272 with alanine completely abrogate enzyme activity highlighting the importance of this residue for catalysis (25).
The current study presents the crystal structure of His-D4·A201–50 (i.e. N-terminal His-tagged D4 bound to the first 50 residues of A20) in complex with a 10-mer oligonucleotide duplex containing a single abasic site resulting from the cleavage of a uracil base. The structure determined at a resolution of 2.7 Å revealed in great detail how D4 binds to DNA. We observed that the DNA binding properties of the viral enzyme exhibited major differences compared with hUNG, which drove a study of the interaction of His-D4·A201–50 with various DNA substrates by surface plasmon resonance (SPR). In light of these new data, we discuss the function of D4 in context of the poxvirus DNA replication machinery.
Experimental Procedures
Protein Expression and Purification
The complex His-D4·A201–50 (with a Thr to Ala change in A201–50 at position 2) from VACV (Copenhagen strain) was expressed in E. coli BL21(DE3) strain and purified as described by Contesto-Richefeu et al. (20). The complex was concentrated to 8 mg/ml prior to crystallization.
Crystallization of His-D4·A201–50
Crystals were obtained using the hanging drop vapor diffusion method in 0.1 m Bicine, pH 8.7, 1.5 m ammonium sulfate as described (20). The uracil complex was obtained by transferring a crystal into a 5-μl drop of cryoprotectant solutions (10% (v/v) and 20% (v/v) glycerol/reservoir solution) containing 20 mm uracil and allowing it to soak for several minutes before flash-freezing in liquid nitrogen.
Crystallization of the His-D4·A201–50·DNA Complex
Two 10-mer DNA oligomers (5′-AAG ATA ACA G-3′ and 5′-CTG TUA TCT T-3′) were synthesized and PAGE-purified by Eurogentec. Equimolar concentrations of the primers were mixed in 100 mm NaCl, 50 mm Tris-HCl, pH 7.5, and heated to 95 °C for 5 min. The mixture was allowed to cool slowly to room temperature. The DNA duplex was mixed in a 3:1 molar ratio with His-D4·A201–50 and incubated at room temperature for at least 30 min. Crystals were initially grown at the High-Throughput Crystallisation Laboratory at EMBL Grenoble (HTX Lab) using the sitting drop vapor diffusion technique as previously described (26). The complex crystallized in the PEG/ion Screen (Hampton Research) condition: 0.2 m ammonium chloride, 20% w/v PEG 3350, pH 6.3, at 20 °C. Conditions were manually optimized, and the best crystals were observed in 0.2 m ammonium chloride, 10% (w/v) PEG 3350 with a 1 μl:1 μl protein:reservoir ratio. Prior to data collection, crystals were successively transferred to cryoprotectant solutions containing 10% (v/v) and 20% (v/v) glycerol in the reservoir solution before flash-freezing in liquid nitrogen.
Data Collection
Diffraction data were collected on Beamlines ID23-1 and ID29 at the European Synchrotron Radiation Facility (Grenoble, France). The data were processed with XDS (27) and scaled with the task AIMLESS from the CCP4 suite (28).
Phase Determination, Refinement, and Structure Analysis
The structures were solved by molecular replacement using the His-D4·A201–50 structure from Protein Data Bank entry 4OD8 (20) as a search model in PHASER (29). Clear extra electron density corresponding to a uracil molecule, and the DNA duplex was visible. The DNA oligomer was built using the DNA of the hUNG·DNA complex structure from Protein Data Bank entry 1SSP (25). The models were manually modified using COOT (30) and refined using REFMAC5 (31). Data collection, refinement statistics, and model composition are shown in Table 1. Protein/DNA interactions were analyzed with PISA (32) and visually checked using PyMOL (The PyMOL Molecular Graphics System, version 1.4.1; Schrödinger), which was also used for structure superpositions and to generate all structure-related figures. Rms calculations used SUPERPOSE (28).
TABLE 1.
Data collection and refinement statistics
| His-D4/A201–50/uracil | His-D4/A201–50/DNA | |
|---|---|---|
| Data collection and processing | ||
| Beamline | ESRF ID23-1 | ESRF ID29 |
| Space group | P312 | P65 |
| Unit cell parameters | ||
| a, b, c (Å) | 92.79, 92.79, 146.7 | 136.1, 136.1, 161.1 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 |
| Wavelength (Å) | 0.9786 | 0.9763 |
| Resolution (Å) | 46.4–1.85 (1.89–1.85) | 48.86–2.70 (2.79–2.70) |
| No. of observed reflections | 459,786 (28,120) | 231,967 (23,392) |
| No. of unique reflections | 60,959 (3639) | 46,345 |
| Completeness (%) | 96.4 (94.0) | 99.9 (100) |
| Multiplicity | 7.5 (7.7) | 5.0 (5.2) |
| Mean I/σ(I) | 23.2 (3.6) | 19.1 (2.9) |
| Rmerge | 0.052 (0.597) | 0.045 (0.492) |
| Refinement and model composition | ||
| Rwork/Rfree | 0.192 (0.36)/0.222 (0.37) | 0.181 (0.31)/0.229 (0.29) |
| No. of residues | ||
| Protein | 539 | 810 |
| DNA | 60 | |
| Water | 378 | 54 |
| Uracil | 2 | 3 |
| rms deviations | ||
| Bond lengths (Å) | 0.013 | 0.014 |
| Bond angles (°) | 1.63 | 1.83 |
| Average B-factor (Å2) | ||
| Protein | 34.7 | 72.9 |
| Water | 38.4 | 55.6 |
Surface Plasmon Resonance
Different DNA duplexes were obtained after annealing of the 5′-biotinylated primer 5′-CCG AAT CAG GAA GAT AAC AGC GGT TTA GCC-3′ with site-specifically modified complementary DNA strands containing (opposite to the underlined A) either a thymine base, a uracil base, a noncleavable pseudo-uracil base (ΨU), or a chemically synthesized tetrahydrofuran (THF) abasic site. Surface plasmon resonance acquisitions were carried out on CM5 sensor chips on a BIAcore 3000 instrument (GE Healthcare). All experiments were performed in HBS-P running buffer (150 mm NaCl, 10 mm HEPES, pH 7.4, 0.005% (v/v) Surfactant P20) at a flow rate of 15 μl/min. Approximately 2500 resonance units of streptavidin (Sigma-Aldrich) were coated on the EDC-NHS-activated surfaces. DNA oligomers were then injected at 1 μg/ml into one flow cell for 10 min (see Table 2). A second flow cell (without bound DNA) was used for background subtraction. 2-fold serial dilutions (20–0.625 μm) of His-D4·A201–50 dialyzed in HBS-P running buffer were injected onto the streptavidin bound DNA for 180 s (association phase) and then a 150-s dissociation phase was allowed. Three independent flow cells were used for experiments involving DNA containing a uracil base as it is hydrolyzed irreversibly during the experiment. Background-subtracted signals were exported from the Biologic software (GE Healthcare) and imported into LibreOffice Calc for curve fitting using the Solver function and for the preparation of figures.
TABLE 2.
Kinetic constants derived from SPR data
The KD, kd, and RU-bound DNA values were obtained experimentally, whereas the other values were calculated.
| DNA oligomer | KD | ka | kd | R | RU-bound DNA |
|---|---|---|---|---|---|
| × 10−6 m | s−1 m−1 | s−1 | μm−1 | ||
| dsDNA | >50 | 20.3 | 800 | ||
| ssDNA | >50 | 18.9 | 580 | ||
| Pseudouracil dsDNA | >50 | 18.9 | 813 | ||
| Abasic tetra-hydrofuran dsDNA opposite to A | 1.4 | 4.9 × 105 | 0.69 | 780 | |
| Abasic site in dsDNA from U:A | 0.39 | 4.7 × 105 | 0.175 | 780 | |
| Abasic site in dsDNA from U:G | 1.5 | 2.5 × 105 | 0.37 | 1280 | |
| U:A in dsDNA | kobs/[E]a, 4.1 × 104 | 0.16 | 680, 680, 780 | ||
| U:G in dsDNA | kobs/[E], 2.8 × 105 | 0.36 | 1280, 1280, 1280 | ||
| Abasic site in ssDNA after U cleavage | ∼8 | 490 |
a kobs/[E] indicates the slope of a linear fit of the dependence of kobs upon the enzyme concentration [E].
Results
Crystal Structure of His-D4·A201–50·DNA Complex
The complex formed by His-D4 and the first 50 amino acids of A20 (His-D4·A201–50) bound to a 10-mer DNA duplex containing an abasic site formed upon UNG activity crystallized at pH 6.3 in space group P65 with unit cell parameters a = b = 136.1, c = 161.1 Å. The structure was solved by molecular replacement using the His-D4·A201–50 structure from Protein Data Bank entry 4OD8 (20) and refined at a resolution of 2.7 Å (Table 1). Three His-D4·A201–50·DNA complexes were present per asymmetric unit whose Cα atoms superposed with less than 0.35 Å rms. As expected from modeling (20), A201–50 binds in proximity but on a different side of D4 than the dsDNA molecule (Fig. 1A). Stacking interactions of the DNA strands extremities with symmetry-related DNA molecules form three contiguous DNA strands only interrupted by the absence of phosphodiester linkages between individual molecules. For this reason, the DNA molecules are well defined up to their extremities. The entire DNA duplex could be modeled and refined (Fig. 1A). In the structure, the deoxyribose of the abasic product site is bound in an α-anomeric conformation as expected for a glycosylase acting with inversion of configuration through a nucleophilic attack with a water molecule. The carboxylate of Asp-68, the carbonyl of Pro-69, and the hydroxyl group of Thr-130 form the binding pocket accommodating the OH1′ group of the α-deoxyribose after cleavage (Fig. 1B), whereas the cleaved uracil base remains bound next to it in the uracil recognition pocket (Fig. 1C).
FIGURE 1.
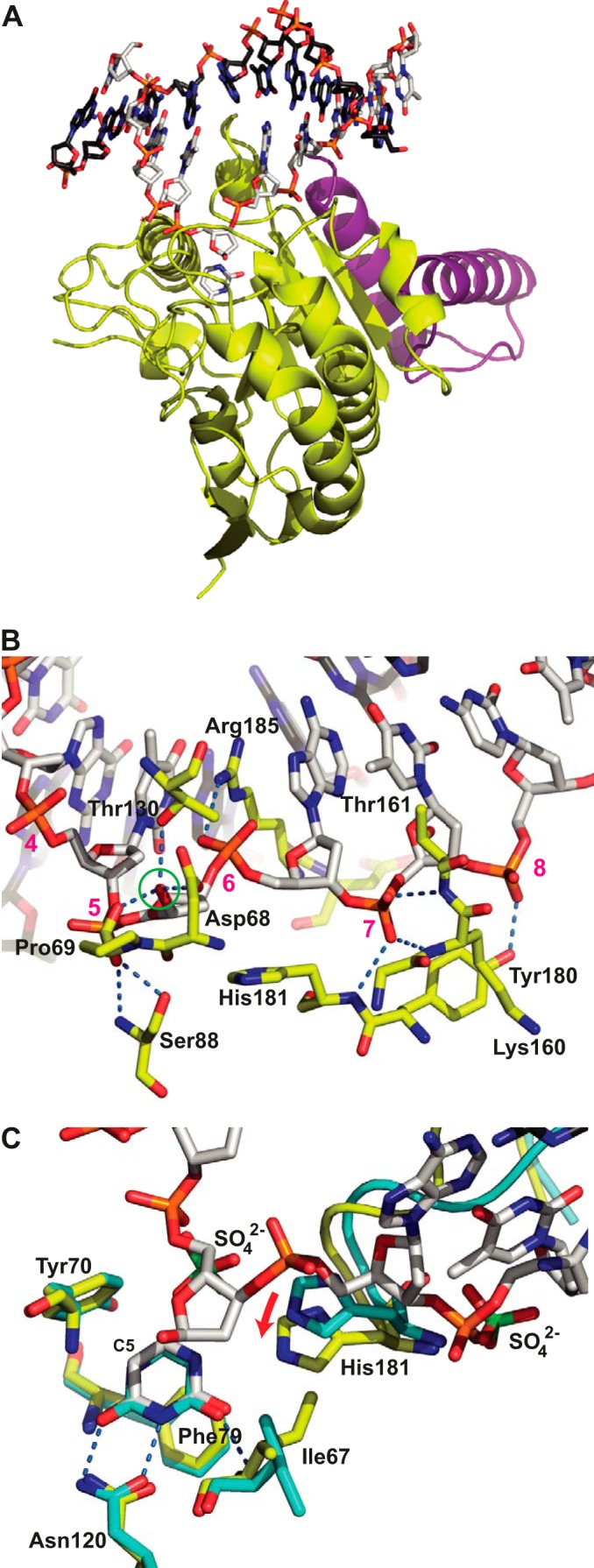
Structure of the His-D4·A201–50·DNA complex. A, D4 is shown with yellow carbon atoms, A201–50 is in violet, the uracil molecule is in stick representation with white carbon atoms, and the dsDNA oligomer is in stick representation with white carbon atoms for the cognate strand and black carbon atoms for the complementary strand. B, detail of the D4·DNA interaction. The residues from D4 involved in hydrogen bonds with the phosphate backbone or the OH1′ hydroxyl group of the extrahelical α-deoxyribose (green circle) are shown in stick representation using the coloring scheme described above for A. Their hydrogen bonds are shown as dotted blue lines. Nucleotides of the cognate strand are numbered in magenta. C, comparison of the uracil binding site of the 1.85 Å structure of His-D4·A201–50 soaked with uracil (cyan carbon atoms) to the same site of the His-D4·A201–50·DNA complex colored as before. The hydrogen bonds involving the uracil molecule are indicated. Sulfate ions (green sulfur atoms) arising from the crystallization solution in the His-D4·A201–50 structure are shown together with the DNA in the DNA-complex structure. Note the 1.6 Å movement of His-181 upon DNA binding (red arrow). The active site residues Asp-68 and Pro-69 are hidden for clarity. The complex formed by chains A, B, C, and D of the crystal structure (Protein Data Bank entry 4YIG) has been used throughout the analysis.
Comparison of the D4·DNA and hUNG·DNA Complexes
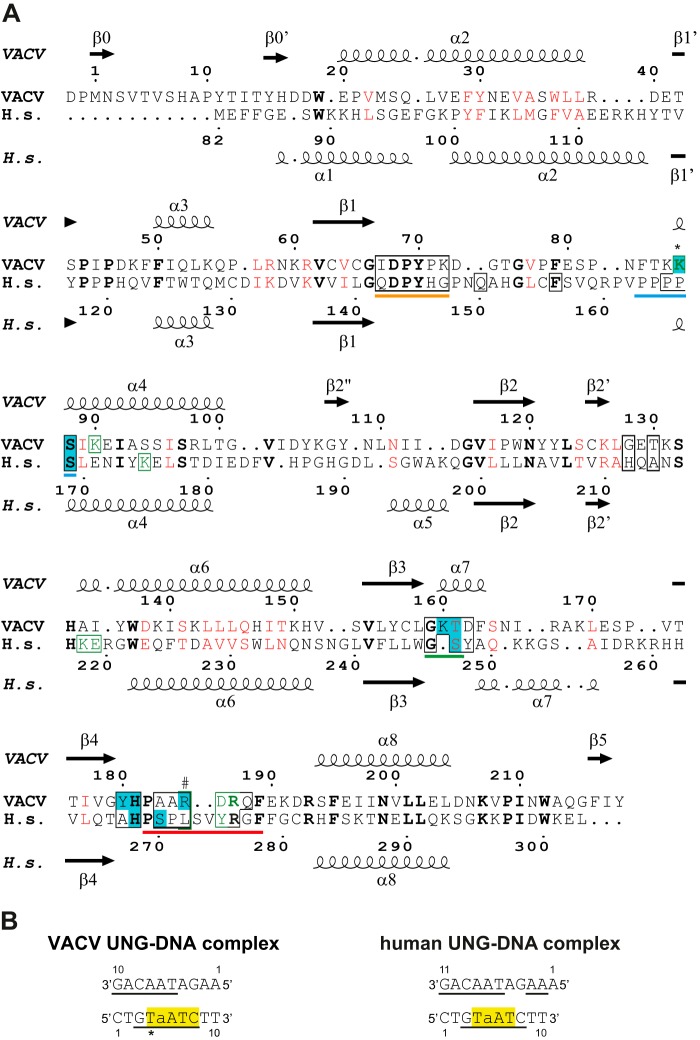
At the protein/DNA interface, the degree of sequence conservation between VACV and human UNG is low (Fig. 2A). The principal conserved contacts between the bound DNA and the UNG concern (i) the deoxyribose of the abasic site involving the conserved active site residues 68–70 (VACV numbering) of the water activation loop (Fig. 1, B and C); (ii) conserved residues Gly-159 and Tyr-70, which contact the DNA backbone outside the active site and contribute to the shape complementarity; and (iii) Ser-88 and His-181 forming hydrogen bonds with backbone phosphates (Fig. 1B). The remaining residues that contact the DNA, in particular the ones of the intercalation loop, do not show any conservation compared with the human enzyme (Fig. 2A).
FIGURE 2.
D4·DNA and hUNG·DNA contacts. A, structural alignment of the hUNG sequence with the one from VACV D4 (done with PDBeFold (46) and drawn with Espript (48)). Matched residues are printed in bold and black, and positions with similar residues are in red. Residues contacting the cognate DNA strand are boxed in black, residues contacting the complementary strand are boxed and printed in green, and residues marked with a # contact both strands. Residues involved in hydrogen bonds with DNA are printed with a cyan background. An asterisk marks a hydrogen bond to a thymidine base, which is specific to the DNA oligomer used. Residues corresponding to the water-activating loop are underlined in orange, the ones belonging to the 4-Pro loop are underlined in blue, the ones of the Gly-Ser loop are underlined in green, and residues belonging to the intercalation loop are underlined in red. B, sequences of DNA oligomers present in the complexes. Nucleotides involved in van der Waals contacts are underlined, and nucleotides involved in hydrogen bonds are highlighted in yellow. An asterisk marks a hydrogen bond, which is specific to the DNA oligomer used. a, abasic site. The analysis was done with PISA (32).
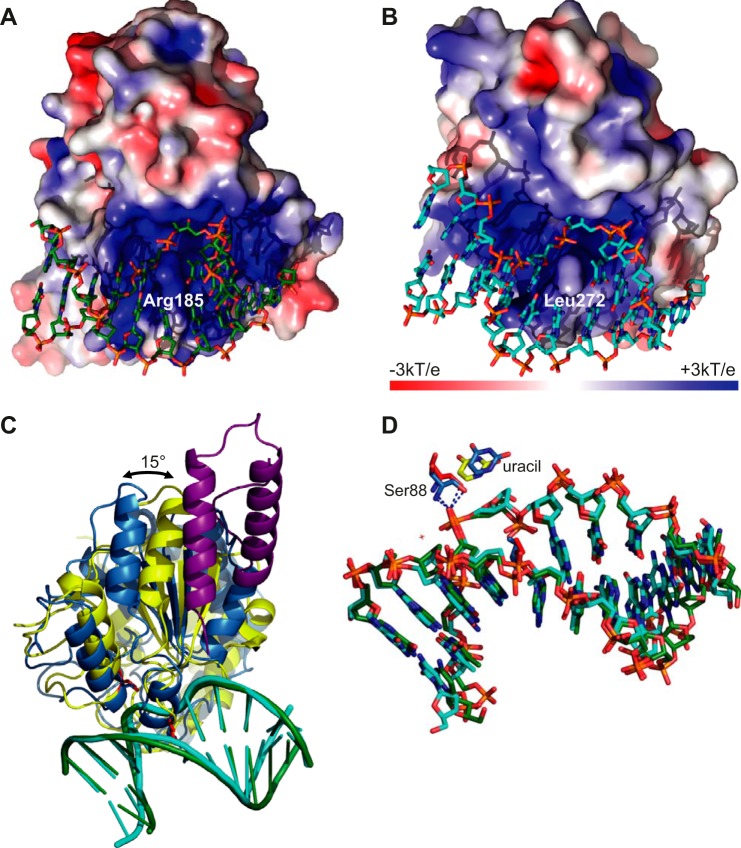
A detailed analysis shows that Ser-88 (Ser-169 in hUNG), which is part of the Pro-rich loop motif, makes two hydrogen bonds to the phosphate 5 of the abasic site through its amide nitrogen atom and the hydroxyl group of the side chain (Fig. 1B). A backbone phosphate on the 3′ side of the expelled base (7 in Fig. 1B) forms hydrogen bonds with the amide nitrogen of His-181 and two amides of the Gly-Ser loop (Lys-160 and Thr-161; Figs. 1B and 2A). In contrast, the corresponding phosphate of the hUNG·DNA complex is engaged in only one hydrogen bond with residue Ser-247 of the Gly-Ser loop, in addition to the interaction with the amide of conserved His-268 (Fig. 2A). The glycine residue of the Gly-Ser loop (Gly-159 in D4) contacts the backbone and is conserved for steric reasons. In the case of D4, the interaction on the 3′ side of the abasic site is strengthened by an additional hydrogen bond between the hydroxyl of Tyr-180 and a backbone phosphate (8 in Fig. 1B), an interaction that is absent in its human counterpart. The intercalating residue Arg-185 forms a salt bridge with the backbone phosphate (6 in Fig. 1B) on the 3′ side of the extrahelical base. The nucleotides involved in the protein/DNA contacts are shown in Fig. 2B. The contact surface between D4 and the DNA appears more positively charged than its human counterpart (Fig. 3, A and B). The positively charged residue intercalating into the DNA at the position of the expulsed base (Arg-185 versus Leu-272 in hUNG) is largely responsible for this difference. When viral and hUNG·DNA complexes are compared, we observed that the orientation of the enzyme on the DNA changes by 15° (Fig. 3C). The pivot of the rotation is located in the vicinity of the above mentioned Ser-88 residue (Fig. 3, C and D). However, the conformation of the DNA duplex is extremely similar in both viral and human structures, which superpose with an rms of 0.9 Å (Fig. 3D), and essentially the same nucleotides are involved in the contact (Fig. 2B).
FIGURE 3.
Interactions of D4 and hUNG with DNA. A, electrostatic potential of the solvent-accessible surface (49). Note the positive charge of D4 at the level of the DNA-binding site. The intercalating Arg-185 residue contributes to this charge. B, the corresponding view on the human enzyme in complex with DNA. C, orientation of D4 (yellow) and hUNG (blue) on the DNA. A201–50 is shown in violet. The phosphate backbone of the DNA has been superposed (VACV in green and human in cyan). The intercalating residue and the conserved serine of the 4Pro loop are shown in red. D, superposition of the dsDNA oligomers containing an abasic product site of the complex structures (green carbon atoms for VACV and cyan carbon atoms for human). The cleaved uracil molecule and the conserved interaction with Ser-88, the pivot of the rotation, are shown.
Structure of His-D4·A201–50 with Bound Uracil
The structure of the complex with uracil determined at a resolution of 1.85 Å was obtained after a soak of His-D4·A201–50 crystals (20) with uracil. The base is stacked onto Phe-79, and the face of the aromatic ring of Tyr-70 contacts the C5 carbon on the side of the uracil ring and assures the discrimination against thymine (Fig. 1C). Asn-120 and the amide nitrogen of Ile-67 form a specific hydrogen bond pattern. This interaction is identical to the one observed in the structure of the complex with DNA (Fig. 1C), as well as the interaction of uracil with free D4 (33) (not shown). Interestingly, a comparison between the uracil complex and the DNA-bound structure showed a structural rearrangement next to the uracil binding site induced by the bound DNA. The movement of the intercalation loop is linked to a movement of His-181 by 1.6 Å (Fig. 1C, red arrow) and to a rotation of the Ile-67 side chain.
Rigidity of the D4 Structure
When the different available structures of D4 (apo-His-D4·A201–50 (20), His-D4·A201–50 in complex with uracil, and the DNA-bound enzyme of the present work) were compared, the Cα atoms superposed with less than 0.38 Å rms when the mobile intercalation loop (residues 182–189) were excluded from the superposition. This shows that D4 is rigid apart from the intercalation loop and does not move upon uracil and DNA binding (Fig. 4A). A comparison of D4 from the His-D4·A201–50·DNA complex structure with free D4 or free D4 bound to uracil (Protein Data Bank entries 4DOF, 4DOG, and 4LZB (33)) gave the same result in addition to the greater flexibility of D4 in vicinity of the missing A20 molecule affecting in particular helices α7 and α8 of D4 (not shown). In contrast, conformational changes observed for hUNG upon DNA binding (24) were much more important (1.44 Å rms for all Cα atoms excluding the intercalation loop) (Fig. 4B) and are generally discussed in terms of an open to closed transition (23–25).
FIGURE 4.
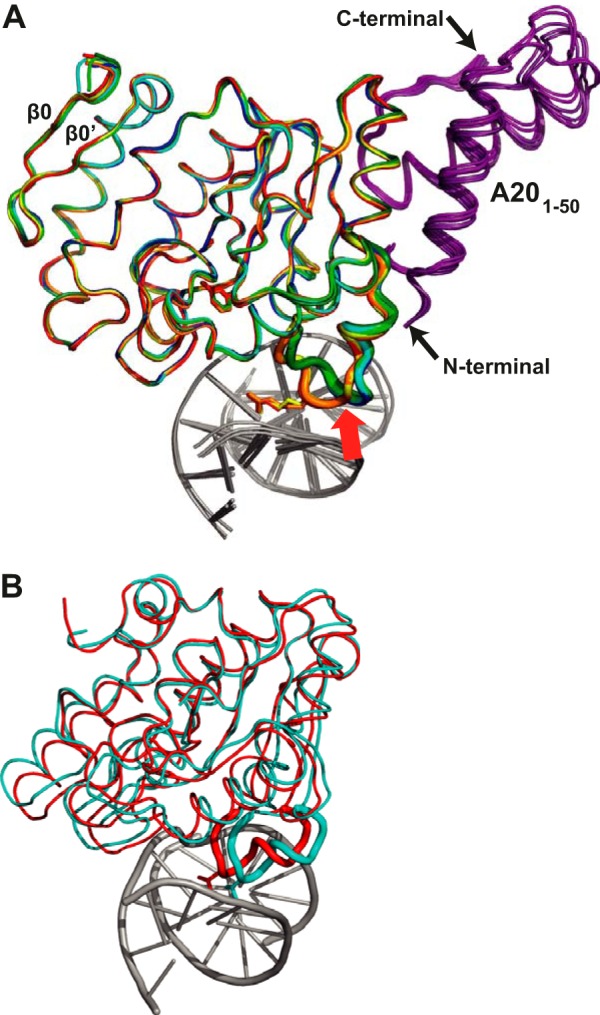
Rigidity of the D4 structure. A, alignment of the backbone of the two distinct D4 apoenzyme structures (dark blue, cyan) (20), of the two D4 structures in complex with uracil (dark green, light green), and of the three D4 structures in complex with DNA (red, orange, and yellow). A201–50 is depicted in violet, and the DNA is in gray. The intercalation loop is highlighted as a thick tube, and the Arg-185 residue is shown in stick representation in the DNA bound structure. This part of the structure is the only part of D4 to show significant rearrangements upon DNA binding (red arrow). The two strands of a D4-specific β-sheet are indicated. B, the corresponding superposition of hUNG in complex with DNA (Protein Data Bank entry 1SSP, red) and the structure of the human apoenzyme (Protein Data Bank entry 1AKZ, cyan).
Study of His-D4·A201–50 Binding to DNA by SPR
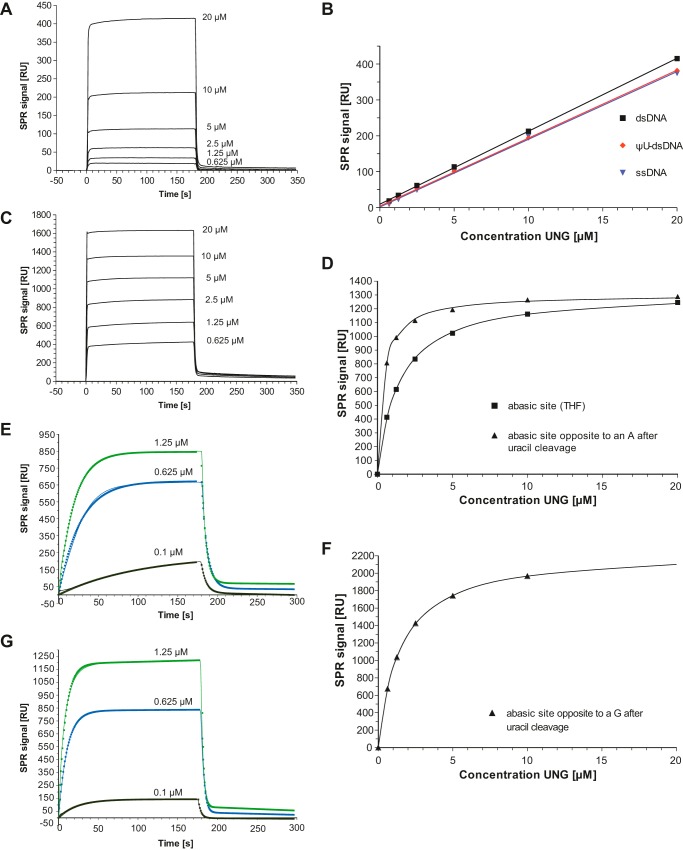
Binding of His-D4 (in complex with A201–50) to different 30-mer DNA substrates was examined at physiological ionic strength using a buffer containing 150 mm NaCl (Fig. 5). Unmodified dsDNA oligomers (Fig. 5A), ΨU-containing dsDNA, and single-stranded DNA (ssDNA) showed a similar linear response of ∼19 resonance units·μm−1 to increasing His-D4·A201–50 concentrations (Fig. 5B and Table 2), indicating Kd values above 50 μm. As shown in Fig. 5A, both association and dissociation are very fast events so that the corresponding rates could not be fitted. The DNA substrate analogue containing ΨU did not contribute to binding over unmodified DNA. On the other hand, substrates containing abasic sites either obtained by chemical synthesis where the deoxyribose has been replaced by THF (Fig. 5C) or obtained by the glycosylase action of His-D4·A201–50 on a DNA oligomer containing uracil (not shown) showed a much higher response with an indication of saturation. When the average contribution of nonspecific binding was subtracted from the plateau values of the SPR traces, an approach possible because of similar densities of immobilized DNA (Table 2), an equilibrium binding model could be fitted corresponding to the specific contribution of the abasic site (Fig. 5D). His-D4·A201–50 binds clearly better to abasic site-containing DNA than nonmodified ssDNA or dsDNA. The THF site showed a Kd of 1.4 μm, whereas the abasic site formed by UNG activity showed a Kd of 0.39 μm (Table 2). The preference of the enzymatically formed site containing an α-deoxyribose over the synthesized THF site can be rationalized by the interactions observed in the His-D4·A201–50·DNA complex (Fig. 1B). Indeed, THF lacks the hydroxyl group at position 1, abolishing a number of favorable interactions. Off rate kd values could be determined for the substrates containing abasic sites using single exponential fits and a linear baseline correction (Table 2). On rates (kobs) had a tendency to show a biphasic behavior with a dominant fast component when fitted directly (not shown) so that the ka was rather calculated from the Kd value and the off rate (kd).
FIGURE 5.
Study of His-D4·A201–50 binding to DNA by SPR. Kinetic parameters are given in Table 2. A, sensorgrams of increasing His-D4·A201–50 concentrations binding to immobilized unmodified 30-mer dsDNA. B, binding of His-D4·A201–50 to immobilized 30-mer: unmodified dsDNA, ΨU-containing dsDNA and ssDNA. A linear fit to the concentration-dependent binding is shown. C, sensorgrams of increasing His-D4·A201–50 concentrations binding to immobilized 30-mers dsDNA containing a THF abasic site. D, fit of an equilibrium binding model to the plateau values of His-D4·A201–50 binding to THF abasic sites (see C) and to abasic sites produced from uracil containing dsDNA. E, binding of His-D4·A201–50 to a uracil containing 30-mer dsDNA (continuous thin lines). Experiments use different flow cells of a CM5 chip. Sensorgrams have been fitted with a single exponential for the association phase and an exponential decay combined with a linear function for the decay phase. The points of the fitted functions are shown. F, fit of an equilibrium binding model to the plateau values obtained for His-D4·A201–50 binding to abasic sites produced from dsDNA with a U:G mismatch. G, binding of His-D4·A201–50 to a uracil containing 30-mer dsDNA with a U:G mismatch analyzed as in E.
When His-D4·A201–50 was injected on a chip with bound uracil-containing DNA present as U:A base pair, the initial response remained very low (Fig. 5E), indicating that the uracil-containing substrate interacted differently with the UNG than the substrates containing abasic sites, which would have given a rapid and strong response. This suggests an initial weak interaction similar to that of unmodified substrate DNA followed by the slow formation of a high affinity site, which could be described by a single exponential fit yielding a rate constant kobs (Fig. 5E and Table 2). This SPR signal is due to the catalytic action forming an abasic site, which binds His-D4·A201–50 with fast kinetics. Independent of the individual rates of the reaction of uracil cleavage, all intermediates should remain bound to the DNA and yield similar SPR signals. The dissociation phase, which can be fitted with a single exponential (Fig. 5E), is in agreement with this model because the observed value agrees with the Kd of the abasic site formed after uracil cleavage (Table 2). We then addressed the effect of a U:G mismatch on VACV UNG. Surprisingly, we found that the affinity for the formed abasic site is lower than for an U:A base pair (Fig. 5F and Table 2), although catalysis proceeds more rapidly (Fig. 5G and Table 2). The interaction with uracil-containing ssDNA was also analyzed; the reaction proceeded even more rapidly with a rather weak interaction with the produced abasic site (Kd ∼ 8 μm; Table 2).
Discussion
D4 is the most divergent enzyme among members from family I UNGs. Within this family, hUNG shares almost 60% amino acid identity with E. coli, whereas it drops to less than 20% with VACV UNG (5). Although the UNG overall structures are very similar (1.0 Å rms deviation with 207 aligned residues) when E. coli (Protein Data Bank entry 3EUG) (34) and hUNG (Protein Data Bank entry 1AKZ) (25) are aligned, D4 from His-D4·A201–50·DNA shows an rms deviation of 2.3 Å when compared with the different structures of hUNG bound to DNA (22–24).
Functionally one of the most noticeable features of D4 is its lack of sensitivity to Ugi (Uracil glycosylase inhibitor) proteins from bacteriophages PBS1 and PSB2 (21). In contrast, Ugi specifically binds and inhibits UNGs from human (35), E. coli (36), and various viruses such as Epstein-Barr virus (37) and HSV-1 (38). The structural basis for UNG inhibition is well characterized and reveals that Ugi targets the DNA binding surface of the UNG mimicking the DNA backbone interaction (35, 39). Binding was shown to be mediated by hydrogen bonding and packing contacts involving the conserved leucine residue (Leu-272 for hUNG) from the intercalation loop that perfectly fits into a hydrophobic pocket of Ugi (35). The structure of the intercalation loop in the free and the DNA-bound state of His-D4·A201–50 nicely explain the presumed lack of interaction between D4 and Ugi. As shown in Fig. 6A, whereas the leucine residue of the intercalation loop of the human enzyme is buried within a hydrophobic pocket of the inhibitor, the longer Arg-185 from D4 clashes with this pocket (Fig. 6B). In addition, the absence of hydrogen bond partners for the charged guanidinium group of the arginine residue within the fully hydrophobic binding pocket is very detrimental for a D4/Ugi interaction.
FIGURE 6.
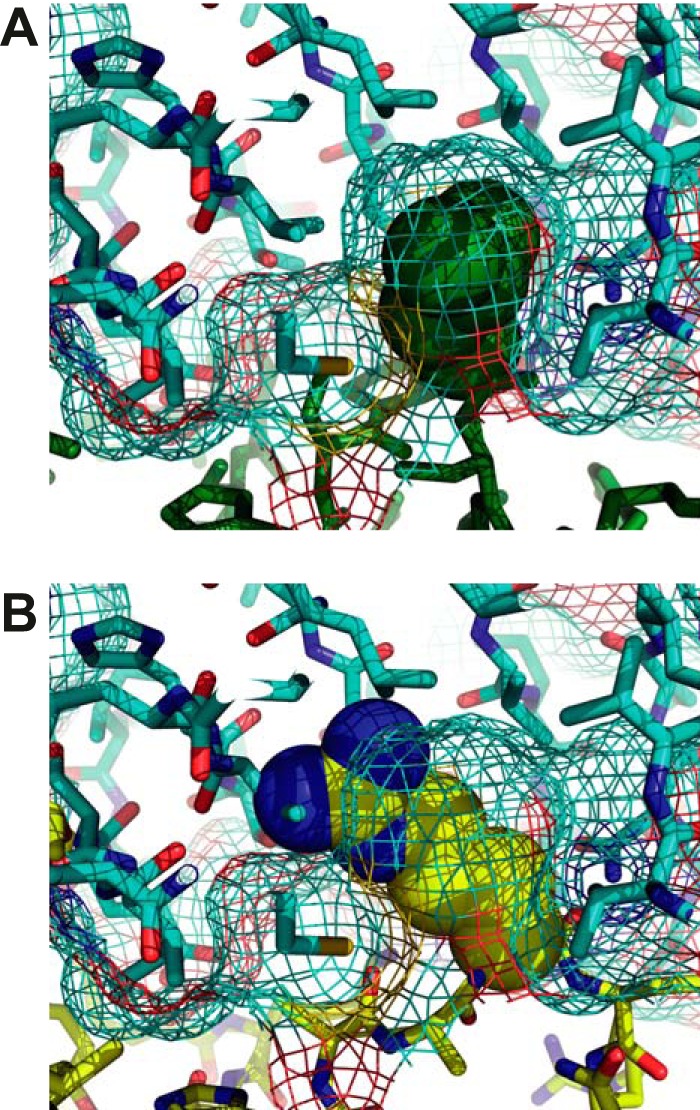
UNG/Ugi interaction. A, the structure of the hUNG/Ugi complex is shown (35). Ugi is shown in stick representation together with its surface in mesh representation (cyan carbon atoms). Residue Leu-272 of the intercalation loop is shown in space-filling representation (green). hUNG is shown in stick representation with dark green carbon atoms. B, Ugi is represented as in A. Arg-185 of His-D4-A201–50 (yellow) is shown in space-filling representation after manual superposition of equivalent atoms of the intercalation loop with its hUNG counterpart. The rest of D4 is shown in yellow stick representation. Note the clash of the larger arginine residue with the leucine-binding pocket of Ugi and the absence of hydrogen bond partners for the charged guanidinium group within the hydrophobic binding pocket.
This second structure of a family I UNG·DNA complex shows a surprising divergence of the DNA binding interface compared with its human counterpart. The divergent evolution of the contact interface includes the intercalation loop located between Pro-182 and Phe-189 carrying Arg-185 (Fig. 2A). However, the extent and nature of the contacts of the intercalation loop appear rather similar when the human and the VACV enzyme are compared with 480 versus 450 Å2 of total buried surface area. The replacement of the Leu-272 residue in hUNG by Arg-185 in VACV still allows a very similar conformation of the DNA in vicinity of the inserted residue (Fig. 3D). Thus, it appears that evolution led to divergence in the contacts between human and viral UNGs with DNA (Fig. 2A) and even in the relative orientation of the molecule onto the DNA template (Fig. 3C). In sharp contrast, the conformation of the DNA duplex in both complexes, in particular the central part around the abasic site, matches perfectly (Fig. 3D). Therefore, the well defined deformation of the DNA structure seems to be required for UNG function rather than specific protein/DNA contacts. It is noteworthy that catalytic residues and interactions of the cleaved uracil base in the active site of the glycosylase are strictly conserved between the human and the VACV enzyme.
Site-directed mutagenesis in the D4 subunit identified residues important for the processivity of the E9·A20·D4 holoenzyme (40). These mutations changing positively charged residues into valine can be localized on the structure of the His-D4·A201–50·DNA complex (Fig. 7). Lys-131, Arg-187, and Lys-160 mutations showed a negative effect on DNA synthesis and impaired, reduced, or preserved UNG activity, respectively. These lysine residues do not form well defined hydrogen bonds with the DNA backbone; however, they contribute to the positively charged environment observed at the protein/DNA contact surface (Fig. 3A). Valine residues at these positions are likely to reduce affinity of D4 for the DNA. Finally, Lys-126, which seemed critical for processive activity (40), is far away from the DNA (12.5 Å; Fig. 7), and its effect can only be explained by a long range structural change induced by the introduction of the hydrophobic valine residue or by the participation of this specific residue in a transient contact with the DNA.
FIGURE 7.
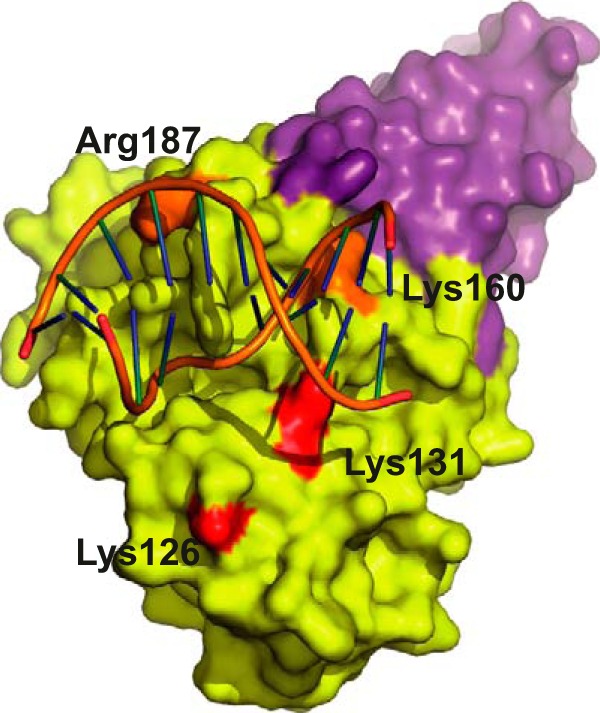
D4 point mutants defective in processivity. The mutated residues (40) are mapped onto the D4 surface (yellow). A201–50 is shown in violet. Mutants shown in orange have ∼35% activity in a DNA replication assay, whereas the ones in red have only ∼10%.
Despite a wealth of studies on the enzymatic action of UNGs, few of them have been carried out in physiological ionic strength condition. Studies using low salt concentration conditions artificially increase the contribution of electrostatic interactions with DNA. For VACV UNG, a steep dependence of activity and Km on the salt concentration has been reported (41). Our first observation was a weak interaction of His-D4·A201–50 with unmodified ssDNA or dsDNA (Fig. 5A), which showed no sign of saturation at a concentration of 20 μm so that only a lower margin of the Kd could be estimated (Table 2). This weak binding contributes also to the interaction with oligomers carrying modified bases. Unexpectedly, binding to ΨU containing substrates was not increased over binding to unmodified DNA, suggesting that this base is not bound preferentially. It is questionable whether ΨU is a good substrate analogue because its hydrogen bond donor nitrogen in position 5 of the base may interact unfavorably with the hydrophobic residue Tyr-70 (Fig. 1C). Surprisingly, the interaction with a hydrolyzable uracil-containing substrate appeared to be also very weak as shown by the absence of fast substrate binding (Fig. 5E), suggesting that the interaction is similar to that with unmodified ssDNA or dsDNA. On the other hand, there was a marked affinity for abasic sites produced after uracil cleavage (Fig. 5, C and D).
The binding profile of His-D4·A201–50 contrasts from the one of HSV-1 UNG (42). Using SPR under similar conditions HSV-1 UNG bound weakly to unmodified DNA substrates or abasic sites but had a strong affinity (in the nanomolar range) for uracil in ssDNA and dsDNA. Hydrolysis by wild-type HSV-1 UNG appeared to be too fast for the detection of an interaction with uracil-containing substrates in SPR experiments. Krusong et al. (43) showed that the activity profile of the human enzyme and the HSV-1 enzyme are similar. The activity profile of VACV His-D4·A201–50 is more similar to the C:T/U mismatch specific glycosylase (MUG), which shows a higher affinity for abasic sites in dsDNA than for the substrate, C:T/U mismatches in dsDNA (44).
However, unlike the situation in mismatch specific glycolylase, the interaction of VACV UNG with DNA containing a G opposite to the abasic site is weaker (i.e. reduced by a factor 4 compared with the presence of an A). When the structure of the DNA complex is used to model the effect of an A → G substitution, it becomes obvious that the interaction of guanine with a hydrophobic environment formed by Arg-185 and the neighboring base is rather unfavorable (not shown). On the other hand the catalytic rate is increased by a factor of ∼7 for the G:U mismatch as expected because the extrahelical position of the base required for catalysis would be favored in the absence of specific hydrogen bonding. In summary, D4 activity against uracil in an A:U or a G:U context seems similar as generally observed for UNGs, for example, hUNG (45).
When aligned with PDBeFold (46), the His-D4·A201–50 structure is slightly more similar to the closed DNA-bound structures of the human enzyme (22–24) (2.3 Å rms deviation of equivalent Cα positions) than when compared with the apo form of hUNG (25) with 2.6 Å rms, but as structure of VACV UNG differs in particular in the mobile parts of the hUNG structure, the assimilation of the VACV UNG structure to a closed form of the human enzyme is not unambiguous. Some flexible parts of hUNG actually appear buttressed in VACV UNG by A20 on one side of the enzyme and by the N-terminal extension of D4 carrying a small β-sheet of strands β0 and β0′ (Fig. 4A) on the other side.
Because all the existing D4 structures are superimposable, the structural information alone does not hint a transition from an open to a closed state upon DNA binding as observed for the human enzyme (22–25, 39). This might suggest that UNG activity does not always require a conformational change upon DNA binding. The only significant main chain movement of D4 upon DNA binding affects the intercalation loop (Fig. 4A). Additional side chain movements affect the active site (Fig. 1C), in particular a rotation of the Ile-67 side chain induced by the movement of His-181 upon binding to a backbone phosphate of the DNA (7 in Fig. 1B), which could be a mechanism to prevent activity against free deoxyuridine phosphates.
Altogether our data suggest that His-D4·A201–50 acts through a hit and run mechanism: a brief encounter with the DNA substrate could lead to the expulsion of the base followed by cleavage if uracil is present. There should not be any preference for uracil in the initial binding events; however, catalysis might be favored by a mispaired uracil base facilitating its expulsion from the base stack. We propose that the absence of the formation of a tight enzyme-substrate complex could be a consequence of the rigidity of the DNA binding site on His-D4·A201–50.
D4 appears to interact preferentially with uracil in the context of dsDNA narrowing down potential models of position and orientation of the polymerase holoenzyme within the replication fork (47). The current model of D4 tethering the polymerase holoenzyme to the DNA may be too simplistic because of its weak interaction with DNA at physiological ionic strength, although mutants affecting polymerase processivity point to a role of the DNA-binding site of D4 (40). The simultaneous presence of two DNA interacting domains, the polymerase active site in one hand and the D4 DNA binding site on the other hand, may still lead to an increased avidity, but this effect is hard to predict because of the long distance (∼150 Å) between the two DNA interacting domains (47). It is likely that the bridging A20 protein contributes also to the interaction with DNA. Such a role of A20 for processivity cannot be studied directly because A20 cannot be produced on its own but only in complex with D4. In the context of the holoenzyme, the progression of the polymerase will lead to a systematic scan of the DNA by the UNG. The fast off rate of the enzyme when it encounters abasic sites speaks against ideas that UNG stays as a mark on newly formed abasic sites.
The structure of His-D4·A201–50·DNA complex shows that a remarkable variability of structure and flexibility of UNG is still compatible with its function. So far the adaptation of D4 as an essential subunit of the VACV DNA processivity factor remains enigmatic, and its understanding has to await more structural and functional information on the polymerase holoenzyme and in particular concerning the role of A20.
Author Contributions
F. I. and W. P. B. designed the study and wrote the paper. C. C.-R. produced and purified His-D4·A201–50. F. I. crystallized the His-D4·A201–50·DNA complex. N. T. determined the x-ray structures. P. F. performed the SPR experiments shown in Fig. 5. C. N. P. contributed reagents and materials. F. I., W. P. B., P. F., and N. T. analyzed the experiments. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank the members of the High-Throughput Crystallisation Laboratory (HTX Lab, EMBL Grenoble, France). We are grateful to the European Synchrotron Radiation Facility for access to beamtime and to the local contacts for help.
This work was supported by ANR Grant ANR-13-BSV8-0014 (REPLIPOX) and by research grants from the Service de Santé des Armées and the Délégation Générale pour l'Armement. The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (codes 4YGM and 4YIG) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- UNG
- uracil-DNA glycosylase
- hUNG
- human UNG
- VACV
- vaccinia virus
- ssDNA
- single-stranded DNA
- ΨU
- noncleavable pseudo-uracil base
- rms
- root mean square
- HSV-1
- herpes simplex virus type 1
- SPR
- surface plasmon resonance
- Bicine
- N,N-bis(2-hydroxyethyl)glycine
- THF
- tetrahydrofuran
- EDC-NHS
- ethyl(dimethylaminopropyl) carbodiimide/N-hydroxysuccinimide.
References
- 1. Zharkov D. O., Mechetin G. V., Nevinsky G. A. (2010) Uracil-DNA glycosylase: structural, thermodynamic and kinetic aspects of lesion search and recognition. Mutat. Res. 685, 11–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Parikh S. S., Putnam C. D., Tainer J. A. (2000) Lessons learned from structural results on uracil-DNA glycosylase. Mutat. Res. 460, 183–199 [DOI] [PubMed] [Google Scholar]
- 3. Lindahl T. (1974) An N-glycosidase from Escherichia coli that releases free uracil from DNA containing deaminated cytosine residues. Proc. Natl. Acad. Sci. U.S.A. 71, 3649–3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kim Y.-J., Wilson D. M., 3rd (2012) Overview of base excision repair biochemistry. Curr. Mol. Pharmacol. 5, 3–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schormann N., Ricciardi R., Chattopadhyay D. (2014) Uracil-DNA glycosylases-Structural and functional perspectives on an essential family of DNA repair enzymes. Protein Sci. 23, 1667–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Porecha R. H., Stivers J. T. (2008) Uracil DNA glycosylase uses DNA hopping and short-range sliding to trap extrahelical uracils. Proc. Natl. Acad. Sci. U.S.A. 105, 10791–10796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schonhoft J. D., Kosowicz J. G., Stivers J. T. (2013) DNA translocation by human uracil DNA glycosylase: role of DNA phosphate charge. Biochemistry 52, 2526–2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schonhoft J. D., Stivers J. T. (2013) NA translocation by human uracil DNA glycosylase: the case of single-stranded DNA and clustered uracils. Biochemistry 52, 2536–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen R., Wang H., Mansky L. M. (2002) Roles of uracil-DNA glycosylase and dUTPase in virus replication. J. Gen. Virol. 83, 2339–2345 [DOI] [PubMed] [Google Scholar]
- 10. Mullaney J., Moss H. W., McGeoch D. J. (1989) Gene UL2 of herpes simplex virus type 1 encodes a uracil-DNA glycosylase. J. Gen. Virol. 70, 449–454 [DOI] [PubMed] [Google Scholar]
- 11. Reddy S. M., Williams M., Cohen J. I. (1998) Expression of a uracil DNA glycosylase (UNG) inhibitor in mammalian cells: varicella-zoster virus can replicate in vitro in the absence of detectable UNG activity. Virology 251, 393–401 [DOI] [PubMed] [Google Scholar]
- 12. Prichard M. N., Duke G. M., Mocarski E. S. (1996) Human cytomegalovirus uracil DNA glycosylase is required for the normal temporal regulation of both DNA synthesis and viral replication. J. Virol. 70, 3018–3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ward T. M., Williams M. V., Traina-Dorge V., Gray W. L. (2009) The simian varicella virus uracil DNA glycosylase and dUTPase genes are expressed in vivo, but are non-essential for replication in cell culture. Virus Res. 142, 78–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Millns A. K., Carpenter M. S., DeLange A. M. (1994) The vaccinia virus-encoded uracil DNA glycosylase has an essential role in viral DNA replication. Virology 198, 504–513 [DOI] [PubMed] [Google Scholar]
- 15. Holzer G. W., Falkner F. G. (1997) Construction of a vaccinia virus deficient in the essential DNA repair enzyme uracil DNA glycosylase by a complementing cell line. J. Virol. 71, 4997–5002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. De Silva F. S., Moss B. (2003) Vaccinia virus uracil DNA glycosylase has an essential role in DNA synthesis that is independent of its glycosylase activity: catalytic site mutations reduce virulence but not virus replication in cultured cells. J. Virol. 77, 159–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stanitsa E. S., Arps L., Traktman P. (2006) Vaccinia virus uracil DNA glycosylase interacts with the A20 protein to form a heterodimeric processivity factor for the viral DNA polymerase. J. Biol. Chem. 281, 3439–3451 [DOI] [PubMed] [Google Scholar]
- 18. Boyle K. A., Stanitsa E. S., Greseth M. D., Lindgren J. K., Traktman P. (2011) Evaluation of the role of the vaccinia virus uracil DNA glycosylase and A20 proteins as intrinsic components of the DNA polymerase holoenzyme. J. Biol. Chem. 286, 24702–24713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schormann N., Grigorian A., Samal A., Krishnan R., DeLucas L., Chattopadhyay D. (2007) Crystal structure of vaccinia virus uracil-DNA glycosylase reveals dimeric assembly. BMC Struct. Biol. 7, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Contesto-Richefeu C., Tarbouriech N., Brazzolotto X., Betzi S., Morelli X., Burmeister W. P., Iseni F. (2014) Crystal structure of the vaccinia virus DNA polymerase holoenzyme subunit D4 in complex with the A20 N-terminal domain. PLoS Pathog. 10, e1003978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ellison K. S., Peng W., McFadden G. (1996) Mutations in active-site residues of the uracil-DNA glycosylase encoded by vaccinia virus are incompatible with virus viability. J. Virol. 70, 7965–7973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Parker J. B., Bianchet M. A., Krosky D. J., Friedman J. I., Amzel L. M., Stivers J. T. (2007) Enzymatic capture of an extrahelical thymine in the search for uracil in DNA. Nature 449, 433–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Parikh S. S., Walcher G., Jones G. D., Slupphaug G., Krokan H. E., Blackburn G. M., Tainer J. A. (2000) Uracil-DNA glycosylase-DNA substrate and product structures: conformational strain promotes catalytic efficiency by coupled stereoelectronic effects. Proc. Natl. Acad. Sci. U.S.A. 97, 5083–5088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Slupphaug G., Mol C. D., Kavli B., Arvai A. S., Krokan H. E., Tainer J. A. (1996) A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA. Nature 384, 87–92 [DOI] [PubMed] [Google Scholar]
- 25. Parikh S. S., Mol C. D., Slupphaug G., Bharati S., Krokan H. E., Tainer J. A. (1998) Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA. EMBO J. 17, 5214–5226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dimasi N., Flot D., Dupeux F., Márquez J. A. (2007) Expression, crystallization and X-ray data collection from microcrystals of the extracellular domain of the human inhibitory receptor expressed on myeloid cells IREM-1. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 63, 204–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Krissinel E., Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 33. Schormann N., Banerjee S., Ricciardi R., Chattopadhyay D. (2013) Structure of the uracil complex of vaccinia virus uracil DNA glycosylase. Acta Crystallograph. Sect. F Struct. Biol. Cryst. Commun. 69, 1328–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xiao G., Tordova M., Jagadeesh J., Drohat A. C., Stivers J. T., Gilliland G. L. (1999) Crystal structure of Escherichia coli uracil DNA glycosylase and its complexes with uracil and glycerol: structure and glycosylase mechanism revisited. Proteins 35, 13–24 [PubMed] [Google Scholar]
- 35. Mol C. D., Arvai A. S., Sanderson R. J., Slupphaug G., Kavli B., Krokan H. E., Mosbaugh D. W., Tainer J. A. (1995) Crystal structure of human uracil-DNA glycosylase in complex with a protein inhibitor: protein mimicry of DNA. Cell. 82, 701–708 [DOI] [PubMed] [Google Scholar]
- 36. Handa P., Roy S., Varshney U. (2001) The role of leucine 191 of Escherichia coli uracil DNA glycosylase in the formation of a highly stable complex with the substrate mimic, ugi, and in uracil excision from the synthetic substrates. J. Biol. Chem. 276, 17324–17331 [DOI] [PubMed] [Google Scholar]
- 37. Géoui T., Buisson M., Tarbouriech N., Burmeister W. P. (2007) New insights on the role of the gamma-herpesvirus uracil-DNA glycosylase leucine loop revealed by the structure of the Epstein-Barr virus enzyme in complex with an inhibitor protein. J. Mol. Biol. 366, 117–131 [DOI] [PubMed] [Google Scholar]
- 38. Savva R., Pearl L. H. (1995) Nucleotide mimicry in the crystal structure of the uracil-DNA glycosylase-uracil glycosylase inhibitor protein complex. Nat. Struct. Biol. 2, 752–757 [DOI] [PubMed] [Google Scholar]
- 39. Putnam C. D., Shroyer M. J., Lundquist A. J., Mol C. D., Arvai A. S., Mosbaugh D. W., Tainer J. A. (1999) Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase. J. Mol. Biol. 287, 331–346 [DOI] [PubMed] [Google Scholar]
- 40. Druck Shudofsky A. M., Silverman J. E., Chattopadhyay D., Ricciardi R. P. (2010) Vaccinia virus D4 mutants defective in processive DNA synthesis retain binding to A20 and DNA. J. Virol. 84, 12325–12335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Scaramozzino N., Sanz G., Crance J. M., Saparbaev M., Drillien R., Laval J., Kavli B., Garin D. (2003) Characterisation of the substrate specificity of homogeneous vaccinia virus uracil-DNA glycosylase. Nucleic Acids Res. 31, 4950–4957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Panayotou G., Brown T., Barlow T., Pearl L. H., Savva R. (1998) Direct measurement of the substrate preference of uracil-DNA glycosylase. J. Biol. Chem. 273, 45–50 [DOI] [PubMed] [Google Scholar]
- 43. Krusong K., Carpenter E. P., Bellamy S. R., Savva R., Baldwin G. S. (2006) A comparative study of uracil-DNA glycosylases from human and herpes simplex virus type 1. J. Biol. Chem. 281, 4983–4992 [DOI] [PubMed] [Google Scholar]
- 44. Barrett T. E., Savva R., Panayotou G., Barlow T., Brown T., Jiricny J., Pearl L. H. (1998) Crystal structure of a G:T/U mismatch-specific DNA glycosylase: mismatch recognition by complementary-strand interactions. Cell. 92, 117–129 [DOI] [PubMed] [Google Scholar]
- 45. Kavli B., Sundheim O., Akbari M., Otterlei M., Nilsen H., Skorpen F., Aas P. A., Hagen L., Krokan H. E., Slupphaug G. (2002) hUNG2 is the major repair enzyme for removal of uracil from U:A matches, U:G mismatches, and U in single-stranded DNA, with hSMUG1 as a broad specificity backup. J. Biol. Chem. 277, 39926–39936 [DOI] [PubMed] [Google Scholar]
- 46. Krissinel E., Henrick K. (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 47. Sèle C., Gabel F., Gutsche I., Ivanov I., Burmeister W. P., Iseni F., Tarbouriech N. (2013) Low-resolution structure of vaccinia virus DNA replication machinery. J. Virol. 87, 1679–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Robert X., Gouet P. (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]