Background: ERp44 tightly controls adiponectin assembly in the early secretory compartment.
Results: ERp44 exclusively recognizes and converts assembly-trapped adiponectin intermediates back to precursors of the biologically potent high molecular weight form.
Conclusion: ERp44 enhances the population of adiponectin intermediates with appropriate oxidative state for HMW assembly.
Significance: Our findings provide a mechanism for the regulation of adiponectin assembly and shed light on ERp44 function.
Keywords: adiponectin, disulfide, ER quality control, peptide interaction, thiol
Abstract
Adiponectin, a collagenous hormone secreted abundantly from adipocytes, possesses potent antidiabetic and anti-inflammatory properties. Mediated by the conserved Cys39 located in the variable region of the N terminus, the trimeric (low molecular weight (LMW)) adiponectin subunit assembles into different higher order complexes, e.g. hexamers (middle molecular weight (MMW)) and 12–18-mers (high molecular weight (HMW)), the latter being mostly responsible for the insulin-sensitizing activity of adiponectin. The endoplasmic reticulum (ER) chaperone ERp44 retains adiponectin in the early secretory compartment and tightly controls the oxidative state of Cys39 and the oligomerization of adiponectin. Using cellular and in vitro assays, we show that ERp44 specifically recognizes the LMW and MMW forms but not the HMW form. Our binding assays with short peptide mimetics of adiponectin suggest that ERp44 intercepts and converts the pool of fully oxidized LMW and MMW adiponectin, but not the HMW form, into reduced trimeric precursors. These ERp44-bound precursors in the cis-Golgi may be transported back to the ER and released to enhance the population of adiponectin intermediates with appropriate oxidative state for HMW assembly, thereby underpinning the process of ERp44 quality control.
Introduction
Adiponectin is a collagenous adipokine possessing direct antidiabetic, antiatherogenic, anti-inflammatory, and antitumor properties (1–13). It acts on a wide range of target tissues and is present at high plasma concentrations (14, 15). Adiponectin monomers consist of four domains: an N-terminal secretion-signaling segment followed by a variable domain, a triple helical collagenous domain, and a C-terminal globular head domain (16, 17). Adiponectin monomers spontaneously form trimers (low molecular weight (LMW)3) that are held together by intermonomer hydrophobic interactions within the globular head domain and stabilized by the trimeric collagen helix (18). Via the trimeric building block, adiponectin can further oligomerize into hexamers (middle molecular weight (MMW)) or 12–18-mers (high molecular weight (HMW)) (19–21). Polymorphism in its oligomeric state together with post-translational modifications appears to be intimately linked to the functional diversity of adiponectin (14, 22, 23). Studies provide strong evidence that the most biologically potent form of adiponectin is the HMW species, which is markedly reduced in obesity concomitant with loss of protective properties of adiponectin (24). Despite its importance, the maturation process of the various adiponectin oligomeric species is not fully understood.
Assembly of the MMW and HMW forms is mediated by the formation of intertrimer disulfide bonds by the conserved cysteine residue (Cys39) in the variable domain (16, 20). In vitro studies (14, 25) have demonstrated the importance of intratrimer disulfide bonds en route to the formation of HMW adiponectin from its precursors. In this context, our earlier study showed that the conserved tryptophan (Trp42) controls the oxidation state of Cys39 and promotes the formation of HMW adiponectin (26). In addition, this study further supported the previously reported (27) importance of the endoplasmic reticulum (ER) chaperone ERp44 in controlling oxidative maturation of HMW adiponectin (26).
ERp44 is a member of the protein-disulfide isomerase family of proteins. It belongs to an ensemble of ER chaperones, including Ero1L-α and DsblA (28–30), that mediate assembly of adiponectin and act on other cysteine-rich client proteins such as IgM antibodies and serotonin transporter (31, 32). Unlike other protein-disulfide isomerase family members, which are mainly found in the ER, the majority of ERp44 is localized in the ER-Golgi intermediate compartment/cis-Golgi region (31, 33–35). The x-ray crystal structure of ERp44 (36) and recent findings (37) have revealed new insights into ERp44 action.
ERp44 consists of an N-terminal thioredoxin domain (a) followed by two thioredoxin-like domains (b and b′) arranged in a clover-leaflike structure containing six distinct cysteine pairs, namely 29 and 63, 160 and 212, and 272 and 289. For instance, the Cys160/Cys212 pair has been implicated in the regulation of binding to the IP3R1 channel (38), whereas the biological roles of the Cys289/Cys272 pair and that of residue Cys63 have not yet been assigned. Cys29 was shown to be responsible for thiol-mediated retention and release of ERp44 substrates such as IgM antibodies and oxidoreductase Ero1-α and also suggested to be implicated in a similar process with adiponectin (34, 36, 37). Binding and release of ERp44 substrates is believed to be pH-dependent occurring in the following manner. A flexible tail at the C terminus of ERp44 shields Cys29 contained in the CRFS motif at the binding site in the a domain. At higher pH in the ER (pH 7.2), the buried Cys29 is then deprotonated, and in addition, an RDEL motif facilitates the transport of ERp44 substrates from the Golgi back to the ER upon binding to the KDEL receptors, making the active site inaccessible. It is believed that at lower pH in cis-Golgi (pH 6.7) Cys29 becomes protonated, a state that exhibits higher affinity to target Cys residues of ERp44 substrates, and the C-terminal tail rearranges to make the active site accessible. This exposes the RDEL motif at the end of the C-terminal tail, which can facilitate binding of the ERp44-client complex to KDEL receptors (37).
The discovery of pH-regulated ERp44 activity and shuttling of substrates between the ER and cis-Golgi is of marked significance for understanding the thiol-mediated retention and the quality control cycle, i.e. establishment of the correct disulfide-linked oligomers of client secretory proteins. However, several aspects of the underlying mechanism remain unclear. For example, what provides the oxidative power for ERp44 to form mixed disulfide bonds, and how does the ERp44-cargo complex dissociate when retrieved to the ER (39)? In this study, we investigated factors that underpin the role of ERp44 action in adiponectin assembly. To further our understanding of this process, we used short peptide mimetics derived from the variable domain of adiponectin for revealing the mode of adiponectin complexation with ERp44. Our findings provide a mechanism for the regulation of adiponectin assembly and shed light on ERp44 function.
Experimental Procedures
Production of Adiponectin
The production and purification of murine adiponectin was as described (26). The expression vector encoding murine adiponectin with a FLAG epitope tag at the C terminus was transfected into HEK293 cells. Single colonies overexpressing FLAG-tagged adiponectin were selected for large scale expansion. The cells were incubated in serum-free Dulbecco's modified Eagle's medium (DMEM) containing 0.2% vitamin C and 0.2% BSA for 48 h. The FLAG-tagged recombinant protein was purified from the conditioned medium using the monoclonal anti-FLAG affinity gel as described previously (40).
Production of Mouse ERp44
The cDNA encoding mouse ERp44 without signal sequence was amplified by PCR and cloned into vector pET28b (from Novagen) at the NheI and XhoI sites. The recombinant form of ERp44 was overexpressed in Escherichia coli and purified as described (37) with one additional purification step to separate ERp44 monomers and dimers. For this purpose, purified ERp44 was loaded onto a Superdex-200 10/300 GL column (GE Healthcare) pre-equilibrated with 20 mm MES, 150 mm NaCl, pH 6.5 (buffer A). The fractions containing monomeric ERp44 or dimeric ERp44 as the main component were pooled and confirmed by non-reducing SDS-PAGE (data not shown).
Production of 9-Amino Acid Residue Peptides
WT36–44, C39S36–44, and control peptides were synthesized manually using Fmoc/tert-butyl solid phase synthesis in a fritted glass reaction vessel (41) on aminomethyl polystyrene resin (42) equipped with the acid-labile 4-hydroxymethylphenoxyacetic acid linker (43). The N-Fmoc group was deprotected with 20% (v/v) piperidine in dimethyl formamide twice for 10 min, and coupling of individual amino acids was performed with 5.5 eq of Fmoc-protected amino acid in dimethyl formamide (0.2 m), 5 eq of O-(benzotriazol-1-yl)-N,N,N′,N″-tetramethyluroniumhexafluorophosphate in dimethyl formamide (0.45 m), and 10 eq of diisopropylethylamine in N-methylpyrrolidine (2 m) for 30 min. Upon completion of the synthesis, the peptide was released from the resin with concomitant removal of protecting groups by treatment with trifluoroacetic acid/triisopropylsilane/H2O/ethane dithiol (94:1:2.5:2.5, v/v/v/v) at room temperature for 3 h. The crude peptide was precipitated with ice-cold diethyl ether, isolated by centrifugation, washed with cold diethyl ether, dissolved in 1:1 (v/v) acetonitrile:water containing 0.1% trifluoroacetic acid, and lyophilized. Purification using a solvent system of A (0.1% TFA in H2O) and B (0.1% TFA in acetonitrile) was performed by semipreparative reverse phase HPLC (Dionex Ultimate 3000 equipped with a four-channel UV detector) at 210 nm using a Gemini C18 (5 μm; 10 × 250-mm) column (Phenomenex) at a 5.0 ml/min flow rate and eluting with an appropriate shallow gradient of increasing concentration of B. Fractions were analyzed for purity by HPLC and/or MS, pooled, lyophilized, and stored at −20 °C. Purified peptides were analyzed for purity by analytical HPLC (Dionex Ultimate 3000 equipped with a four-channel UV detector) at 214 nm using a Zorbax Eclipse XDB-C8 (5 μm; 4.6 × 150-mm) column (Agilent) at 1 ml/min flow rate using a linear gradient of 5–65% over 21 min at ∼3% B/min. WT36–44: NH2-KGTCAGWMA-CO2H (22.1 mg; yield, 24.0%; purity, >95%); observed mass, 925.3 Da (M + H)1+; calculated mass, 925.0 Da. C39S36–44: NH2-KGTSAGWMA-CO2H (16.5. mg; yield, 18.3%; purity, >95%); observed mass, 909.3 Da (M + H)1+; calculated mass, 909.02 Da. Control peptide: NH2-AGACGMWTK-CO2H (18.3 mg; yield, 19.9%; purity, >95%); observed mass, 925.2 Da (M + H)1+; calculated mass, 925.0 Da.
Adiponectin Hypervariable Domain (AHD) Peptide
The peptide was synthesized and purified as described previously (44).
Production of Dimeric WT36–44 Peptide
WT36–44 at a concentration of 2 mg/ml was dissolved in buffer A containing 5 mm H2O2 and incubated for 2 h on ice. Subsequently, the sample was extensively dialyzed against buffer A using a membrane with a molecular mass cutoff of 100 Da.
Production and Separation of Oxidized AHD Trimer and Hexamer
AHD peptide was dissolved in 20 mm Tris-HCl, 150 mm NaCl, pH 7.4 (buffer B) containing 5 mm H2O2 at a concentration of 20 mg/ml and incubated for 16 h at 4 °C. The sample was subsequently loaded onto a Superdex-200 10/300 GL column pre-equilibrated with buffer A, and the trimeric AHD was thereby separated from the hexameric form. The fractions containing trimeric AHD peptide or hexameric AHD peptide as a main component were pooled. In size exclusion chromatography coupled to multiangle laser light scattering (SEC-MALS) experiments, these pooled fractions exhibited single peaks with corresponding masses for the trimer and hexamer, respectively.
Co-immunoprecipitation (IP) and Western Blotting
For detection of different oligomeric adiponectin complexes, cell lysates or purified adiponectin was incubated with a non-reducing sample buffer (1% SDS, 5% glycerol, 10 mm Tris-HCl, pH 6.8) at ambient temperature for 10 min, separated by 4–20% gradient SDS-PAGE, and transferred to polyvinylidene difluoride (PVDF) membranes for immunoblotting with an in-house anti-adiponectin antibody (45) or anti-ERp44 antibody (Santa Cruz Biotechnology).
For the co-immunoprecipitation studies, cells were washed with PBS buffer (137 mm NaCl, 2.7 mm KCl, 4.3 mm Na2HPO4, 1.47 mm KH2PO4, pH 7.4) and solubilized in radioimmune precipitation assay buffer (25 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5 mm NaF, 1% sodium deoxycholate, 1% Nonidet P-40, 0.1% Triton X-100 plus protease inhibitor mixtures). 100 μg of cell lysate was precleared with protein G beads and then incubated with the antibody and protein G beads overnight at 4 °C with shaking. The beads were precipitated and washed with radioimmune precipitation assay buffer three times, and the immunoprecipitated complexes were eluted by incubation with non-reducing SDS-PAGE sample buffer and analyzed by Western blotting.
Far-Western Blotting
Purified adiponectin (1 μg) was first separated by non-reducing SDS-PAGE, transferred to PVDF membranes, and incubated with or without ERp44 (10 μg) in control buffer (50 mm Tris-HCl, 150 mm NaCl, 0.05% Tween 20, pH 7.5) for 24 h at 4 °C. After incubation, membranes were washed three times with control buffer and probed with anti-ERp44 antibody.
Co-incubation Experiments with Adiponectin and ERp44
Purified murine adiponectin (1 μg) was incubated with purified ERp44 (10 μg) in PBS buffer. As a control, adiponectin and ERp44 were incubated independently under the same condition. Subsequently, the samples were separated by non-reducing SDS-PAGE. In two independent experiments, either anti-adiponectin antibody or anti-ERp44 antibody was used for identification of the protein bands. For the co-incubation assay at varying pH, experiments were conducted as described above using buffer containing 50 mm HEPES, 150 mm NaCl at pH 6.5, pH 7.5, and pH 8.0. For the pH stability test, an aliquot of the sample incubated at pH 6.5 was taken, and pH was adjusted to 8.0 by adding NaOH. After incubation for 48 h, the sample was separated by non-reducing SDS-PAGE, and anti-adiponectin antibody was used for identification of the protein bands.
SEC-MALS Experiments
SEC-MALS was used to determine the solution molecular weight, stability, and composition of AHD peptide (oxidized and reduced trimer and hexamer) and to identify the formation of ERp44-peptide complexes. AHD peptide was dissolved in buffer B at a concentration of 20 mg/ml in the absence and presence of 5 mm H2O2. After incubation for 24 h, a 100-μl aliquot was subjected to SEC-MALS as described below.
The binding assay between ERp44 and AHD peptide oligomers was carried out at the stated peptide concentration in the presence of 0.5 (26.5 μm), 1 (53 μm), and 2 eq (106 μm) of ERp44 monomer or dimer. After incubation at 4 °C for 16 h, samples (100 μl) were applied to a Superdex-200 10/300 GL column mounted on a Dionex HPLC at a flow rate of 0.5 ml/min. The scattered light intensity and protein concentration in the column eluate were recorded using a PSS SLD7000 MALS detector and a Shodex RI-101 differential refractometer (dn/dc = 0.186), respectively. The weight-averaged molecular mass of material in chromatographic peaks was determined using PSS winGPC UniChrom software.
Electrospray Ionization Mass Spectroscopy (ESI-MS)
The mass spectroscopy assay was used to analyze the amount of disulfide bond formation between ERp44 and the various peptides. To this end, ERp44 (43 μm) was incubated with a 10-fold excess (430 μm) of peptide (WT36–44 monomer, WT36–44 dimer, C39S36–44, control peptide, AHD reduced trimer, AHD oxidized trimer, and AHD hexamer). All samples were incubated for 7 days in buffer A unless stated otherwise and applied to mass spectroscopy. Samples at pH 7.5 and 8.0 were incubated in 20 mm Tris-HCl, 150 mm NaCl, pH 7.5 or pH 8.0. To analyze the effects of a reducing condition, the WT36–44 sample was incubated in buffer A in the presence of 5 mm β-mercaptoethanol.
For LC-MS analysis, samples were diluted in 0.1% formic acid, and 10 μl was injected onto a 0.32 × 100-mm 3-μm Discovery Bio Wide pore C5 column (Supelco, Bellefonte, PA) and separated using the following gradient at 6 ml/min: 0–4 min 10% B; 24 min 70% B, 27 min 97% B, 30 min 97% B, 32 min 10% B, and 35 min 10% B where A was 0.1% formic acid in water and B was 0.1% formic acid in acetonitrile. The column eluate was ionized in the electrospray source of a QSTAR-XL quadrupole time-of-flight mass spectrometer (Applied Biosystems, Foster City, CA). A TOF-MS scan from 400 to 1600 m/z was performed. The resulting data were deconvoluted into protein molecular weights using the Bayesian Protein Reconstruct Tool within Analyst QS 1.1 (Applied Biosystems).
Tryptophan Fluorescence Quenching
Each peptide (30 μm) was incubated at ambient temperature in the presence or absence of 15 μm ERp44 in buffer A for 5 min at a total volume of 300 μl. Fluorescence was recorded between 340 and 450 nm on an EnSpire multimode plate reader (PerkinElmer). Data were analyzed using the EnSpire Manager software package. The ratio of the fluorescence intensities was used as a measure of quenching. The maximum of fluorescence intensities at 388 nm was used, and the ratio f/f0 was calculated as described (46) where f is fluorescence of the mixture (peptide + ERp44) and f0 is the fluorescence signal of the peptide alone.
Non-reducing SDS-PAGE
Disulfide bond formation between ERp44 monomer and AHD peptide oligomers was analyzed by non-reducing SDS-PAGE. ERp44 monomer (15 μm) was incubated with 1 (15 μm), 2 (30 μm), or 5 eq (75 μm) of the particular AHD peptide. After incubation for 24 h at 4 °C, samples were loaded onto non-reducing SDS-PAGE (12.5%). To determine pH dependence of disulfide bond formation between ERp44 and AHD peptide, ERp44 monomer (15 μm) was incubated with 1 (15 μm), 2 (30 μm), or 5 eq (75 μm) of the reduced trimeric AHD peptide for 5 days at 4 °C in buffer A at pH 6.5 and 20 mm Tris-HCl, 150 mm NaCl, pH 7.5 or 20 mm Tris-HCl, 150 mm NaCl, pH 8.0. Subsequently, the samples were analyzed by non-reducing SDS-PAGE (12.5%). To analyze pH stability of the ERp44-AHD peptide complex, ERp44 monomer (15 μm) was incubated separately with 30 μm reduced trimeric, oxidized trimeric, and hexameric AHD peptide. After incubation for 5 days at 4 °C, the pH of 10-μl aliquots of each sample was set to 8.0 by the addition of 100 mm Tris-HCl, 150 mm NaCl, pH 8.0 solution. The samples were subsequently incubated for a further 16 h at 4 °C prior to loading onto non-reducing SDS-PAGE (12.5%). GelQuant.NET software (BiochemLabSolutions) was used to quantify the intensity of the gel bands by densitometry.
Results
ERp44 Binds LMW and MMW but Not the HMW Form of Adiponectin
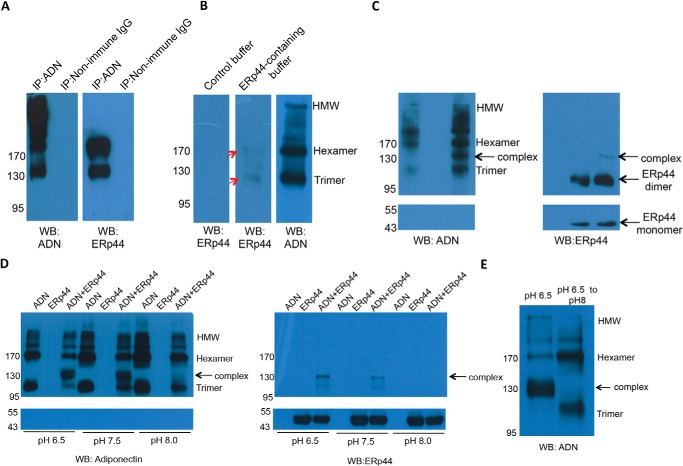
We investigated the adiponectin/ERp44 interaction by (i) co-IP using HEK293 cells stably expressing adiponectin, (ii) far-Western blot analysis, and (iii) Western blot analysis using purified recombinant ERp44 and adiponectin. In the immunoprecipitated complex milieu generated by using anti-adiponectin antibody, two species were recognized by anti-ERp44 antibody (Fig. 1A), one migrating at ∼130 kDa likely corresponding to the ERp44-adiponectin LMW complex and another migrating at around 190 kDa likely corresponding to the ERp44-adiponectin MMW complex. Far-Western blotting using purified recombinant adiponectin also showed complexes with ERp44 corresponding to LMW and MMW adiponectin (Fig. 1B), but again no signal corresponding to the HMW form was detected (Fig. 1B).
FIGURE 1.
Examination of interactions between ERp44 and different adiponectin oligomers. A, co-IP profiles of cell lysates of transfected HEK293 cells stably expressing murine adiponectin. The immune complexes, separated by non-reducing SDS-PAGE and probed using anti-adiponectin (left panel) and anti-ERp44 antibodies (right panel) for the presence of adiponectin and ERp44, are shown. Non-immune IgG was used as the control antibody. The species migrating at ∼130 kDa corresponds to the ERp44-adiponectin LMW complex, whereas the species migrating at around 190 kDa corresponds to the ERp44-adiponectin MMW complex. B, purified murine adiponectin (1 μg) was separated by non-reducing SDS-PAGE and transferred to PVDF membranes. After incubating with control buffer with (10 μg) or without ERp44 for 24 h at 4 °C, the membrane was washed with the control buffer and probed by anti-ERp44 antibody (first panel and second panel) and anti-adiponectin antibody to show the distribution of oligomers of adiponectin (third panel). The red arrows indicate the locations of LMW and MMW adiponectin (molecular masses of ∼110 and ∼170 kDa) that bind ERp44. C, ERp44 (10 μg), purified murine adiponectin (1 μg), or the mixture of two recombinant proteins was incubated for 24 h at 4 °C in PBS. After separation by non-reducing SDS-PAGE, the membranes were probed with antibodies against adiponectin and ERp44. Anti-adiponectin antibody recognizes the different adiponectin oligomers and a complex consisting of ERp44 and LMW adiponectin (at ∼130 kDa). The estimated amounts as reflected by densitometry of the band intensity in adiponectin-only control were 2% HMW, 89% MMW, and 9% LMW. These amounts for the ERp44-adiponectin co-incubated sample were 2% HMW, 62% MMW, 23% LMW, and 13% ERp44-adiponectin complex showing an increase of trimeric adiponectin after ERp44 co-incubation. Anti-ERp44 antibody recognizes ERp44-LMW adiponectin complex (at ∼130 kDa) and ERp44 monomers and dimers. D, the same experiment as in C carried out using HEPES buffer at pH 6.5, 7, and 8. The amount of complex formed decreased with increasing pH and was almost abolished at pH 8.0 as confirmed by densitometry, which showed complex formation at a level of 30% at pH 6.5, 18% at pH 7.5, and 6% at pH 8.0 when adiponectin antibody was used for detection. When detected with ERp44 antibody, the measured complex formation was at the level of 30% at pH 6.5, 5% at pH 7.5, and <1% at pH 8.0. E, the pH of an aliquot of the sample incubated at pH 6.5 as in D was adjusted to 8. Subsequently, the sample was analyzed as in C. After incubation at pH 8.0, no ERp44-LMW adiponectin was detected. Quantitation of the gel bands for the pH 8.0 experiment indicated 75% complex at pH 6.5 and <1% at pH 8.0, whereas the amount of the uncomplexed LMW adiponectin increased from 4 to 46%, and the amount of uncomplexed MMW adiponectin increased from 21 to 56%. WB, Western blotting; ADN, adiponectin.
Experiments i and ii suggest that ERp44 does not associate with the HMW form, instigating experiment iii. Here, we wanted to reveal what happens to the different adiponectin oligomers in solution after binding to ERp44 while not being constrained and localized on the PVDF membrane as is the case for far-Western blots. Toward this end, we performed a direct co-incubation of purified murine adiponectin and ERp44 and subjected this to Western blot analysis after separation by non-reducing SDS-PAGE (Fig. 1C). The Western blot showed no band corresponding to HMW or MMW adiponectin in complex with ERp44. When compared with the controls, irrespective of whether anti-ERp44 or anti-adiponectin antibody was used, an additional band was detected that may be assigned to the ERp44-LMW adiponectin complex (Fig. 1C). The co-incubated sample also features a change in the relative levels of the various adiponectin oligomers. As can be seen (Fig. 1C), when compared with the ratio of LMW to MMW in the control, the co-incubated sample displays an enhanced level of LMW adiponectin after ERp44 treatment (Fig. 1C). We also note that in addition to the ERp44 monomer band another band consistent with ERp44 existing as a dimer was also observed. This is due to the presence of both monomers and dimers in the purified ERp44 sample, which is in line with previously observed in vivo data (33).
Given that regulation of ERp44 activity has been indicated to be pH-dependent (37), we queried whether pH dependence also applies to the binding of full-length adiponectin to ERp44. Co-incubation of ERp44 and adiponectin at pH 6.5, 7.5, and 8.0 showed the largest complexation at pH 6.5 and very little at pH 8.0 with an intermediate amount of complex seen at pH 7.5 (Fig. 1D). In agreement with this observation, when the pH of an aliquot of such a sample incubated at 6.5 to generate the ERp44-LMW adiponectin complex was raised to 8.0, the complex disassembled. Western blot analysis of the sample showed that most of the released adiponectin was trimeric, whereas only a small increase of hexameric species was seen (Fig. 1E).
Taken together, these results using full-length adiponectin for complexation with ERp44 established distinctive binding profiles for the various oligomeric states of adiponectin. However, our results described above also raised the following questions. (a) Why was the ERp44/MMW adiponectin interaction observed by co-IP and far-Western blotting experiments but not by Western blotting, and (b) what factors lead to the apparent increase of the LMW adiponectin level after co-incubation in solution?
Investigation of ERp44 Interaction with Adiponectin N-terminal Model Peptides
To answer these questions and in the process reveal details of the mode of ERp44 interaction with the various adiponectin oligomers, we focused on small peptides derived from the variable domain of adiponectin. Our strategy of using peptide mimetics mitigates the complexity of a biophysical study as posed by the post-translational modifications and the various oligomeric states when working with the full-length adiponectin (14). We are informed by the knowledge that apart from Cys39 the aromatic residue Trp42 in the conserved 42WMA44 triplet is also involved in adiponectin assembly by influencing the redox state of Cys39 (19, 26, 27). Previously, we have shown that the N-terminal region of adiponectin clusters together into a compact mass in the HMW form (19, 26), signifying an important role for this region of the sequence in the oligomerization process.
Design of Adiponectin N-terminal Model Peptides
For this purpose, we utilized two types of model peptides. The first consist of a set of short peptides only comprising residues of the variable region. Peptides of the second set span the variable region and collagen-like repeats as a bona fide trimeric adiponectin mimetic for the N-terminal region (Fig. 2A).
FIGURE 2.
Illustration of the model peptide ensemble. A, top panel, the three domains of mature adiponectin and the secretion signal domain are shown for murine adiponectin. The N-terminal stretch of the primary sequence of mouse adiponectin encompassing the hypervariable domain and the first seven repeats of the collagenous domain is indicated below the various domains of the polypeptide chain. Also shown are the amino acid sequence of the WT36–44 peptide (middle panel) as well as that for the AHD peptide mimetic (O, hydroxyproline) (bottom panel). B, SEC-MALS analysis of the AHD peptide after incubation in buffer B in the presence (right) and absence (left) of 5 mm H2O2 for 24 h at 4 °C. The UV280 nm trace (black line) is reported for each sample. The average molecular weights across the protein peaks as determined by MALS (black circles) are shown in the plot. In the absence of 5 mm H2O2, the AHD peptide forms trimers eluting at 14.3 ml (left panel). In the presence of 5 mm H2O2, the AHD peptide forms oxidized hexamers eluting at 12.7 ml and oxidized trimers eluting at 14.3 ml (right panel).
As a minimal model for the ERp44 target region in adiponectin, we first synthesized a 9-amino acid-long peptide wherein Cys39 and Trp42 are flanked by 3 and 2 amino acids of the murine sequence at the N- and C-terminal ends, respectively (36KGTCAGWMA44; WT36–44) (Fig. 2A). For the full N-terminal adiponectin mimetic, we made use of the previously reported trimeric model for the N-terminal domain of adiponectin (44). This peptide, termed AHD hereafter, constitutes the entire variable domain and the N-terminal part of the collagen-like domain of murine adiponectin (residues 17–53) fused to nine canonical collagen repeats (GPO where O is hydroxyproline) at the C terminus (Fig. 2A) (44).
We first characterized the behavior of AHD under different redox conditions. SEC-MALS of oxidized AHD revealed trimeric and hexameric species as well as small amounts of higher oligomers (Fig. 2B). Conversely, reduced AHD formed only trimers (18 kDa) (Fig. 2B). This behavior is similar to that of the full-length adiponectin in vitro that is trimeric under reducing conditions and under uncontrolled oxidizing conditions (using H2O2 or oxidized glutathione) assembles mainly into the MMW form and oxidized trimers (LMW) with very little of the HMW form (25, 47). Oxidation of AHD followed by SEC enabled us to isolate both the oxidized trimer and hexamer of AHD and allowed us to query how the oligomeric and oxidative state of AHD may affect its interaction with ERp44.
ERp44 Binds WT36–44 in a Sequence-specific Manner
Optimal complex formation between ERp44 and the WT36-44 peptide appears to occur at pH 6.5 as evidenced by ESI-MS. Thus, for exploring the mode of interaction of ERp44 with various model peptides, the following experiments were carried out at pH 6.5 (20 mm MES, 150 mm NaCl) using ERp44 monomers (see below) unless stated differently.
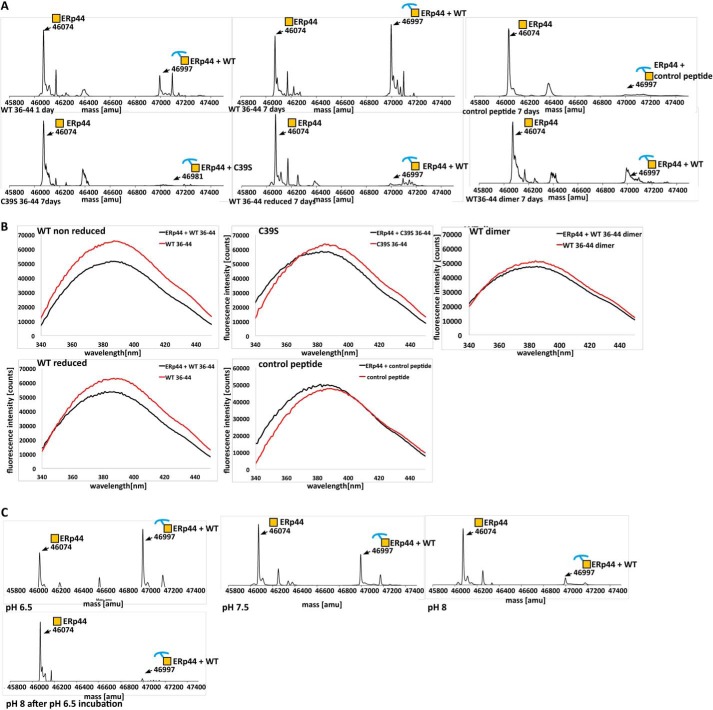
To test whether WT36–44 peptide is an informative model in binding ERp44, we used ESI-MS and tryptophan fluorescence quenching experiments. After incubation of ERp44 with a 10-fold excess of WT36–44 for 16 h, ESI-MS detected ∼15% of ERp44 to be covalently linked to the peptide, whereas after 7 days of incubation, this value had increased to ∼65% (Fig. 3A). The mass for the complex was increased by 923 Da, which is the mass of the model peptide less one hydrogen atom, giving evidence that the peptide has formed a disulfide bond with ERp44 (Fig. 3A). The slow formation of disulfide bond suggests that oxidation is mainly driven by atmospheric oxygen. Because the peptide contains a tryptophan (corresponding to Trp42 in adiponectin), this residue can be used as probe for fluorescence quenching experiments. However, Trp28 in ERp44, located next to Cys29, also contributes to the fluorescence signal albeit at a much smaller level. Despite this “background” signal, a significant decrease in fluorescence was still observed, giving evidence of binding (Fig. 3B). Fluorescence quenching occurs on a much shorter time scale compared with disulfide bond formation monitored by ESI-MS (i.e. minutes versus days), suggesting that electrostatic and/or hydrophobic factors are likely to be responsible for initiating the peptide-protein recognition. This was confirmed by tryptophan quenching experiments in the presence of a reducing agent (Fig. 3A). Furthermore, we also observed interaction of ERp44 with an analogous peptide construct in which Cys39 was substituted by serine(C39S36–44; 36KGTSAGWMA44) (Fig. 3B). Compared with WT36–44, the quenching for C39S36–44 and WT36–44 under reducing conditions was less pronounced but still detectable, whereas ESI-MS experiments did not detect covalently linked complexes (Fig. 3A). Taken together, these results indicate that disulfide bond formation between Cys39 of adiponectin and a thiol group of ERp44, likely Cys29, is the key driver for binding, but additional, possibly sequence-specific factors are also likely at play. We were unable to detect non-covalent interactions by ESI-MS, which may be due to dissociation of a complex upon ionization.
FIGURE 3.
Binding experiments of ERp44 with the 9-amino acid model peptide. A, ERp44 was incubated with WT36–44 for 1 and 7 days, and the mixture was analyzed by ESI-MS (top panel). The spectra show two peaks, one at 46,074 atomic mass units (amu) representing free ERp44 and another at 49,997 atomic mass units representing the disulfide-linked ERp44-peptide complex. The corresponding spectra for C39S36–44, the control peptide, WT36–44 dimer, and WT36–44 under reducing condition are also shown (top and bottom panels). The satellite peaks of the primary ERp44 peak that appear in some of the ERp44 overexpression experiments are likely due to adducts with the recombinantly purified ERp44. The existing ESI-MS database failed to detect identities for instance for the extra ∼100- and ∼150-Da species in the primary satellite peaks. B, ERp44 was incubated with a 2-fold access of WT36–44 (under reducing and non-reducing conditions), C39S36–44, WT36–44 dimer, and the control peptide. Fluorescence emission spectra from 340 to 450 nm were recorded. In each case, a spectrum for the peptide alone was recorded in the same wavelength range. Quenching was determined as described (46) with f/f0 = 0.78 for WT, f/f0 = 0.91 for C39S, f/f0 = 0.84 for WT under reducing condition, f/f0 = 1.04 for the control peptide, and f/f0 = 0.92 for WT36–44 dimer. C, ERp44 was co-incubated with WT36–44 at pH 6.5, 7.5, and 8.0 and analyzed by ESI-MS (first three plots as indicated). The pH of an aliquot of the sample incubated at pH 6.5 was set to 8.0 using 100 mm Tris-HCl, pH 8.0. After incubation for 16 h, the sample was analyzed by ESI-MS. The peaks for ERp44 (mass, 46,074 atomic mass units) and those for the ERp44-WT36–44 complex (mass, 46,997 atomic mass units) are indicated in the deconvoluted spectrum.
Next, we asked whether non-covalent interactions between ERp44 and WT36–44 are influenced by the sequence of the peptide. For this purpose, we scrambled the sequence of WT36–44 but maintained the length and composition to be used as control (control peptide; WGAMAKGCT). ESI-MS and the tryptophan quenching assay for this control peptide showed a markedly reduced level of complex (<10%) and no detectable quenching, respectively (Fig. 3, A and B). This result suggests that indeed ERp44 recognition is sensitive to the sequence with the location of the Cys39 residue in the peptide being crucial for proper binding. As an aside, we noted that the monomeric form of the peptide binds more readily to ERp44 than its dimeric counterpart (produced by treatment with 5 mm H2O2) as assayed by ESI-MS (data not shown). This was confirmed by fluorescence quenching, which also showed higher affinity of reduced peptide monomers relative to oxidized dimers (Fig. 3, A and B).
ERp44 Binds WT36–44 in a pH-dependent Manner
Next, we queried whether similar pH-dependent binding as demonstrated for full-length adiponectin (see above) applies to the binding of our model peptides to ERp44. ESI-MS analysis of the formation of ERp44-WT36–44 complex for a range of pH (6.5, 7.5, and 8) showed 65% complex at pH 6.5, 40% complex at pH 7.5, and a very minor amount of complex at pH 8.0 (Fig. 3C). The ERp44-WT36–44 complex can be disassembled by raising the pH to 8 with an efficiency of over 95% (Fig. 3C). This is analogous to what we observed for the ERp44-LMW adiponectin complex.
ERp44 Can Distinguish between the Oligomeric and Oxidative States of AHD
Given the binding results for the full-length adiponectin (described above) and to gain possible molecular level insight into ERp44-mediated adiponectin assembly, we queried whether ERp44 can distinguish between different oligomeric states and disulfide-linked configurations of AHD.
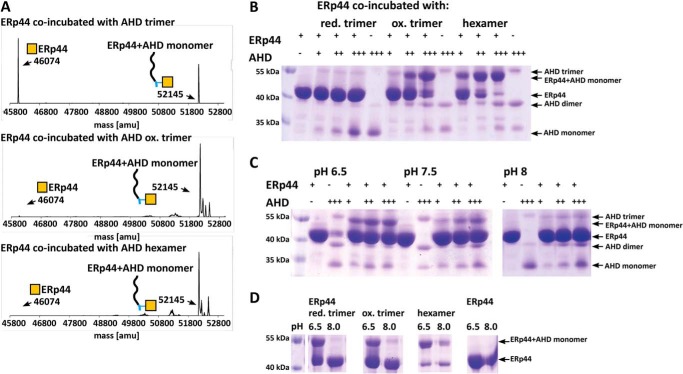
We generated and subjected reduced and oxidized trimers as well as hexamers of AHD to the same treatment and ESI-MS binding assay as outlined above. In the case of the reduced AHD trimers, 50% of the peptide was found to be complexed with ERp44 after 1 week of incubation (Fig. 4A). In contrast, in the case of the oxidized forms of trimer and hexamer, nearly 100% of the peptide was complexed with ERp44 (Fig. 4A). Due to the sizable mass difference of 6072 Da, ERp44 and ERp44-AHD complexes could be resolved by non-reducing SDS-PAGE unlike in the case of the short WT36–44 peptides. Non-reducing SDS-PAGE analysis of a titration series using ERp44 and AHD at ratios of 1:1, 1:2, and 1:5 revealed the largest level of binding for the oxidized hexamers followed by oxidized trimers with the reduced trimers binding the least (Fig. 4B).
FIGURE 4.
ESI-MS and non-reducing SDS-PAGE experiments showing different binding affinity of ERp44 to AHD peptide mimetics. A, reduced trimeric AHD, oxidized trimeric AHD, and hexameric AHD peptides were co-incubated with ERp44 at a 10:1 ratio (peptide:protein) and analyzed by ESI-MS measurements. The peak for ERp44 (mass, 46,074 atomic mass units (amu)) and that for the AHD-ERp44 complex (mass, 52,145 atomic mass units; 46,074 + 6071 Da) are indicated in the deconvoluted mass spectra; the latter indicates the formation of S–S link. The “harsh” ionization process applied in this method disassembles all but the covalently linked complexes. Hence, only complex consisting of ERp44 and peptide monomer was detected even though ERp44 was incubated with the AHD trimer or hexamer. B, non-reducing SDS-PAGE experiment of ERp44 co-incubated with different amounts of reduced AHD trimer, oxidized AHD trimer, and AHD hexamer (protein:peptide ratios, 1:1, 1:2, and 1:5), respectively. SDS denatures and disassembles all but non-covalently linked complexes. As a result, we detected ERp44-peptide monomer complex for all samples irrespective of the nature of the peptide complex (trimer, oxidized trimer, or hexamer) used in the incubation experiments. However, uncomplexed AHD appeared to be somewhat SDS-resistant so that AHD trimers, dimers, and monomers are all detected in the gel. Because of its linear shape, the peptide apparently moves slower through the gel and migrates effectively at higher than the true molecular weight (as indicated by the marker). Based on quantitation by densitometry, ERp44 co-incubated with AHD hexamer resulted in the largest level of complexation (22% at a 1:1 ratio; 49% at a 1:2 ratio, and 63% at a 1:5 ratio), a somewhat lower level of complexation was observed with the oxidized trimer (11% at a 1:1 ratio; 22% at a 1:2 ratio, and 54% at a 1:5 ratio), and a very low level of complexation was observed with the reduced trimer (4% at a 1:1 ratio, 4% at a 1:2 ratio, and 3% at a 1:5 ratio). C, ERp44 and reduced AHD peptide were co-incubated at different ratios (1:1, 1:2, and 1:5) at pH 6.5, 7.5, and 8.0, respectively. After 5 days, the samples were analyzed by non-reducing SDS-PAGE. The amount of complex formed decreased with increasing pH. Thus the amount of complex detected was 17% at a 1:1 ratio; 25% at a 1:2 ratio, and 26% at a 1:5 ratio at pH 6.5; 11% at a 1:1 ratio, 13% at 1:2 ratio, and 19% at a 1:5 ratio at pH 7.5; and 3% at a 1:1 ratio, 7% at a 1:2 ratio, and 9% at a 1:5 ratio at pH 8.0. D, ERp44 was incubated for 24 h with a 5-fold excess of reduced trimer, oxidized trimer, and hexamer of the AHD peptide at 4 °C. The pH of an aliquot of the sample incubated at pH 6.5 was set to 8.0 using 100 mm Tris-HCl, pH 8.0. After further incubation for 16 h, the samples were analyzed by non-reducing SDS-PAGE. An aliquot of the sample before and after treatment with 100 mm Tris-HCl, pH 8.0 was examined. At pH 8.0, the complex disassembled into free ERp44 and AHD peptide. The effect was most pronounced for the reduced AHD trimer with a decrease in the amount of complex from 48 to 3%. For the oxidized AHD trimer, the amount of complex decreased from 50 to 6%, and for the AHD hexamer, the amount of complex decreased from 70 to 38%.
Analogous to the experiments above, we assessed any pH dependence of AHD binding to ERp44. First, we confirmed that the influence of pH on the oligomeric state of the AHD peptide was minimal and therefore did not notably influence the binding assays (data not shown). Non-reducing SDS-PAGE analysis of ERp44-AHD complexes showed most efficient complex formation at pH 6.5 with lesser amounts of complex formed at pH 7.5 and almost no detectable complex at pH 8.0 (Fig. 4C). These results are in line with those of the WT36–44 peptides. Likewise, the pH stability assay showed that ERp44-AHD complexes formed at low pH disassemble (at a level of ∼90%) when incubated at pH 8.0 for 12 h (Fig. 4C). In the case of reduced trimers, the dissociation of the complex at pH 8.0 was most pronounced and went to completion.
ERp44 Disrupts AHD Hexamers to Form ERp44-AHD Trimer Complex
SEC-MALS was used for determining the molecular weights of ERp44-AHD complexes. Prior to these experiments, we confirmed that the different AHD oligomers (reduced and oxidized trimer as well as hexamer) were stable over time (data not shown) and that the exchange of monomeric to dimeric ERp44 had only a very minor effect on the interpretation of our results (data not shown).
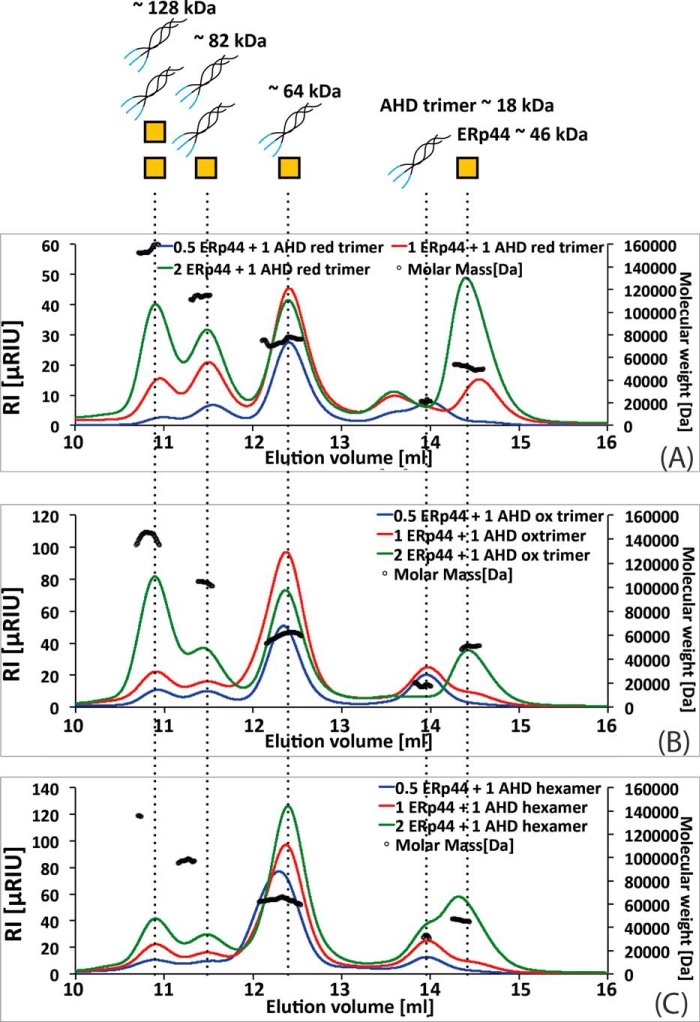
When ERp44 was mixed with hexameric AHD, surprisingly, SEC-MALS showed predominantly an ERp44-AHD trimer complex and only a small amount of ERp44-AHD hexamer complex (Fig. 5A). In fact, the SEC-MALS elution profile consisted of multiple peaks. The largest peak was an overlap of the peaks for two species, one attributed to the ERp44-AHD trimer complex (57kDa) and the other attributed to an unbound AHD hexamer. This peak resolved entirely to the ERp44-AHD trimer complex in a titration experiment where the amount of ERp44 used was varied. The three other smaller peaks could be attributed to 1) a free AHD trimer, 2) ERp44-AHD hexamer complex, and 3) an AHD hexamer in complex with two ERp44 molecules (Fig. 5A).
FIGURE 5.
SEC-MALS assay of ERp44 binding to various AHD peptide oligomers. The AHD hexamer (A), oxidized AHD trimer (B), and reduced AHD trimer (C) were incubated with varying relative amounts of ERp44 (peptide:ERp44, 1:0.5 (in blue), 1:1 (in red), and 1:2 (in green)) for 16 h at 4 °C and analyzed by SEC-MALS. The refractive index (RI) trace (colored line as indicated) is reported for each sample. The average molecular weight for one representative measurement across each peak as determined by MALS (black circles) is shown in the plot. The identity of each peak is illustrated in the top panel, and theoretical molecular weight of each “complex” is indicated. A, the location and therefore the mass for the major peak changed as a function of the ratio of ERp44 to AHD hexamer used in the experiment. The titration experiment showed that this major peak is an overlap of the peak associated with the free AHD hexamer and a species attributed to the ERp44-AHD trimer complex. The relatively small peaks in the SEC-MALS run eluting at 10.9, 11.4, and 13.9 ml correspond to two ERp44 molecules in complex with AHD hexamer, one ERp44 molecule in complex with AHD hexamer, and free AHD trimer, respectively. B and C, the SEC-MALS elution profiles of oxidized (B) and reduced (C) AHD trimer consist of multiple peaks eluting at 10.9, 11.4, 12.4, 13.9, and 14.4 ml corresponding to two ERp44 molecules in complex with AHD hexamer, one ERp44 molecule in complex with AHD hexamer, ERp44 in complex with AHD trimer, free AHD trimer, and monomeric ERp44, respectively. RIU, refractive index unit.
Next, we repeated the same set of experiments using oxidized AHD trimers instead. Here also we observed the formation of ERp44-AHD trimer complexes, but in addition and interestingly, we found the generation of species comprising two AHD trimers complexed with either one or two ERp44 molecules (Fig. 5B) even though in the incubated sample AHD hexamers were initially absent. In fact, the relative amounts of these species were actually larger than those observed in the case of the AHD hexamer experiment described above.
Lastly, we performed SEC-MALS experiments using reduced AHD trimers. The elution profiles in these experiments were identical to that of the oxidized AHD trimer with somewhat lesser amounts of complexes formed and a slight shift in mass toward the ERp44-AHD trimer complex (Fig. 5C). The complex formation appeared to be driven by non-covalent interactions rather than through disulfide bonds as evidenced by non-reducing SDS-PAGE (Fig. 4B).
Discussion
Very little is currently known about how ERp44 exerts quality control on the oligomeric assembly of adiponectin, one of its client proteins. Based on our current results, we propose a model of how ERp44 executes this quality control that we describe below.
ERp44 Binding Depends on the Relative Location of the Cys39 in Peptide Mimetics
We observed that recognition of peptide WT36–44 by ERp44 is sequence-specific, especially the relative location of the Cys in this peptide (analogous to Cys39 of adiponectin), which also established the fidelity of the designed 9-amino acid model peptide system. Additionally, we observed that the ERp44/peptide interaction is also driven by electrostatic and hydrophobic interactions.
We compared adiponectin sequences, especially the N-terminal region, with other ERp44 clients such as Ero1-α, IgM antibody, and serotonin transporter to examine whether we could reveal any consensus ERp44 binding motif. However, there appears to be no apparent consensus binding motif with only the 3-amino acid stretch (GTC) in the C terminus of IgM showing an overlap with the corresponding region in adiponectin (32, 33, 48, 49). It is interesting to note that most of these binding cysteine residues are found to be proximal to an aromatic residue, Trp, Tyr, or Phe, in the sequence.
Electron Transfer Drives Discriminative Binding of ERp44 to Oxidized AHD
Binding experiments with the AHD peptide in various redox states demonstrated that ERp44 could interfere with and redistribute the pattern of the disulfide bonds in the model peptide in a manner as follows. Binding of ERp44 to AHD hexamer disrupts the intermolecular disulfide bond between Cys39 residues linking the two trimeric AHD moieties of the AHD hexamer by donating an electron to one of the trimeric units. Concurrently, ERp44 covalently links to the other trimeric moiety by forming a disulfide bond between presumably Cys29 and Cys39 in AHD. This process results in a disulfide-linked ERp44-AHD trimer complex and a free trimeric unit with a thiol group. Conversely, when ERp44 interacts with the oxidized AHD trimer, ERp44 reduces the trimer by donating an electron to the trimer. We also observed that ERp44 binds reduced AHD trimer albeit at a considerably slower rate compared with the oxidized AHD trimers and hexamers. In this case, however, no intermolecular disulfide bonds are formed, suggesting that binding to the reduced trimer is triggered by hydrophobic and/or electrostatic interactions. We therefore surmise that the oxidation equivalent (i.e. the electron acceptor) necessary for the formation of a disulfide bond between ERp44 and the oxidized AHD trimer is supplied by the oxidized peptide itself. In contrast, when ERp44 was incubated with the oxidized, dimeric WT36–44, disulfide bond formation between ERp44 and the peptide was substantially reduced when compared with that in the case of the monomeric species. This observation and the fact that as mentioned above there is an increased disulfide bond formation between ERp44 and the oxidized AHD peptides compared with the reduced AHD trimer suggest that residues further up- or downstream of the 9 amino acids of the WT36–44 peptide also contribute to the process of exchange of disulfide bonds. This needs to be clarified by further examination.
ERp44 Promotes HMW Formation by Converting the Pool of Oxidized LMW and MMW to Reduced LMW Adiponectin
A key question remains as to the mechanism behind the promotion of HMW adiponectin formation by ERp44. Previous studies (25, 47) have reported that fully oxidized LMW adiponectin and MMW adiponectin fail to assemble into the HMW form, whereas reduced trimers could assemble into the HMW form through oxidative formation of disulfide bonds in vitro and in vivo (25, 47, 50). These observations indicate that the MMW form comprises oxidized trimers that are assembly-trapped. Our SEC-MALS data with AHD suggest that ERp44 transforms the AHD hexamer and the oxidized AHD trimer into reduced AHD trimers. These results are in line with the in vitro ERp44 and adiponectin co-incubation experiment. When ERp44 was co-incubated with the full-length adiponectin, only a complex of ERp44 with adiponectin trimer was detected (Fig. 1, C and D). In addition, the level of the LMW form was increased upon co-incubation. These results strongly suggest that the action of ERp44 on AHD mimetics parallels that for the full-length adiponectin hexamers and trimers. Thus, the observed disulfide bond exchange between the AHD and ERp44 may in part be similar to that happening for the full-length adiponectin in the transition from the LMW or MMW forms to the HMW form through an analogous disulfide bond exchange. Far Western blotting and co-IP experiments demonstrated that ERp44 binds MMW and LMW adiponectin, whereas the HMW form does not interact with ERp44. We hypothesize that ERp44 is not able to disrupt/modify the compact structure at the N terminus that has been visualized for the HMW form (19). In the case of MMW and LMW adiponectin, the N terminus is likely to be more flexible and/or accessible for ERp44 binding.
The pH Environment Tightly Regulates Binding and Release of AHD by ERp44
We observed a pH dependence for ERp44 binding to our model peptides and full-length adiponectin. This situation is analogous to the results reported in a recent study (37). We also investigated the pH stability of the ERp44-peptide complexes to answer the question of “how the ERp44-cargo complexes dissociate when retrieved to the ER” (39). Remarkably, raising the pH from 6.5 to 8.0 ruptures the covalently linked complex consisting of ERp44 and the binding moiety. This result is particularly interesting because a disulfide bond between two polypeptide segments requires a reducing equivalent, i.e. an electron, to transform the disulfide bond into two sulfhydryl groups, which cannot be achieved by just a pH change. The crystal structure of ERp44 (36) has shown that the cysteine pair Cys289/Cys272 in the b′ domain is disulfide-bonded, whereas the two cysteines in the b domain (Cys160 and Cys212) are reduced but are in close proximity. Furthermore, ERp44 contains a reduced cysteine (Cys63) in the a domain that is proximal to Cys29 (36). It is possible that the observed pH-influence on the disulfide bond stability is due to either the donation of electrons from the aforementioned cysteine pair in the ERp44 b domain or the disruption of the ERp44-reduced trimer complex leading to a disulfide bond between Cys29 and Cys63. This phenomenon, however, needs further examination.
Model for ERp44 Quality Control of Adiponectin Oligomerization
On the basis of our results discussed above and those from earlier studies (36, 37), we developed a model for the ERp44-mediated quality control of adiponectin oligomerization as described below (Fig. 6). As in the case of all secretory proteins, adiponectin is synthesized in the ER where an array of ER-resident chaperones and oxidoreductases assist the adiponectin monomers to fold into a trimeric unit. The trimeric building blocks further oligomerize in transit along the early secretion pathway from the ER to the Golgi and are monitored by the quality control mechanisms of the ER. Further downstream in the ER-Golgi intermediate compartment/cis-Golgi region, ERp44 is thought to scrutinize the assembly of HMW adiponectin (31). In the acidic pH environment of the cis-Golgi, the hydrophobic active site around the free cysteine (Cys29) of ERp44 is exposed and allows ERp44 to bind to its substrates (37). Here ERp44 scans for incorrect disulfide-linked adiponectin intermediates, namely oxidized trimers and hexamers, preventing these from subsequent secretion. Concomitantly, completely assembled HMW adiponectin is not recognized and continues down the secretion pathway to the Golgi where maturation through post-translational modification occurs. Our model posits that ERp44 traps incorrectly oxidized intermediates through the donation of an electron to the oxidized trimeric building block and by the simultaneous formation of a disulfide bond with the adiponectin intermediate (oxidized trimer or oxidized hexamer). In the case of the adiponectin hexamer, interaction with ERp44 results in a partly reduced trimer with one free cysteine and an ERp44-adiponectin trimer complex. The so-released, partly reduced trimeric unit may subsequently be prevented from secretion by another ERp44 oxidoreductase molecule. Subsequently, the covalently linked ERp44-adiponectin complex utilizes the RDEL sequence at the ERp44 C terminus to form an adiponectin-ERp44-KDEL receptor cargo complex that is brought back to the ER. In the ER, the neutral pH triggers the release of the reduced trimeric unit from ERp44, and this trimer may participate again for assembly into HMW adiponectin. Our results show that the pH change by itself may be sufficient to cause release of the adiponectin trimer from ERp44. However, for ERp44 to continue its role in adiponectin oligomerization, ERp44 needs to be reduced. Most likely Ero1-α, the foremost partner of ERp44, delivers the required reducing equivalents either by binding the ERp44-adiponectin complex directly, thereby setting free the reduced trimer, or by subsequently reducing oxidized ERp44 after the reduced trimer has been released. We noted that ERp44 could bind reduced adiponectin through an interaction that is likely to be hydrophobic in nature that is unlikely, however, to survive the ERp44-mediated transport from the cis-Golgi to the ER (37). Such a scenario raises the possibility that effectively retention of reduced adiponectin trimer in the cis-Golgi region caused by ERp44 allows the reduced trimer to participate again in the formation of the HMW form. These HMW assemblies can then traffic to the Golgi to undergo further maturation.
FIGURE 6.
Model for the quality control of adiponectin oligomerization executed by ERp44. Adiponectin is synthesized in the ER where it forms trimers. From the ER, adiponectin trimer is engaged in the secretion pathway and undergoes oxidative oligomerization into disulfide-linked trimers, hexamers, and HMW adiponectin. In the ER-Golgi intermediate compartment, ERp44 exists in its active state and screens for incorrectly assembled adiponectin intermediates, namely oxidized trimers and hexamers. Via its free cysteine (Cys29), ERp44 binds and reduces these intermediates. At the same time, ERp44 forms a mixed disulfide bond with its client, yielding a stable ERp44-adiponectin complex. Subsequently, the ERp44-adiponectin complex is retrieved to the ER by the KDLE receptor by engaging the C-terminal RDEL sequence. Once transported to the ER, the ERp44-adiponectin complex disassembles triggered by the neutral pH in the ER and potentially facilitated by ERo1-α. This process results in the release of a reduced adiponectin trimer, which in turn can participate in the cycle of the oxidative assembly pathway leading to the formation of HMW adiponectin.
In conclusion, in this study, we have explored the interaction of ERp44 with adiponectin through a combination of in vitro and in cellulo experiments with both wild-type adiponectin and representative, synthesized adiponectin mimetics in vitro. Our results elaborating a molecular level insight into the mechanism underpinning the observed differential binding of ERp44 to the various adiponectin oligomeric states reveal how ERp44 executes quality control of adiponectin assembly.
Author Contributions
L. H., M. R., and A. K. M. devised the project. L. H. did the experiments. P. W. R. H., L. H., and M. A. B. devised peptide synthesis experiments. G. B. gave advice on molecular biology and protein production. D. C. G. gave advice on the SEC-MALS experiment and data analysis. X. C. and Y. W. carried out the work with wild-type adiponectin. L. H. and A. K. M wrote the manuscript with contributions from the other authors.
Acknowledgments
We thank James Dickson for helpful hints in protein expression and purification, Ben Rushton with some of the protein preparations, Vivian Ward for helping to generate Fig. 6, and Meder Kamarov with peptide synthesis. We thank the Centre for Genomics, Proteomics, and Metabolomics at the University of Auckland for ESI-MS experiments.
This work was supported in part by a University of Auckland doctoral scholarship and the German Academic Exchange Service (DAAD) (Germany) (to L. H.), research funding from the Maurice Wilkins Centre for Molecular Biodiscovery (to A. K. M. and P. W. R. H.), and a New Zealand Health Research Council grant (to A. K. M.). The authors declare that they have no conflicts of interest with the contents of this article.
- LMW
- low molecular weight
- HMW
- high molecular weight
- MMW
- middle molecular weight
- ER
- endoplasmic reticulum
- Fmoc
- N-(9-fluorenyl)methoxycarbonyl
- AHD
- adiponectin hypervariable domain
- SEC-MALS
- size exclusion chromatography coupled to multiangle laser light scattering
- IP
- immunoprecipitation
- ESI-MS
- electrospray ionization mass spectroscopy.
References
- 1. Scherer P. E. (2006) Adipose tissue: from lipid storage compartment to endocrine organ. Diabetes 55, 1537–1545 [DOI] [PubMed] [Google Scholar]
- 2. Kishore U., Gaboriaud C., Waters P., Shrive A. K., Greenhough T. J., Reid K. B., Sim R. B., Arlaud G. J. (2004) C1q and tumor necrosis factor superfamily: modularity and versatility. Trends Immunol. 25, 551–561 [DOI] [PubMed] [Google Scholar]
- 3. Garaulet M., Hernández-Morante J. J., de Heredia F. P., Tébar F. J. (2007) Adiponectin, the controversial hormone. Public Health Nutr. 10, 1145–1150 [DOI] [PubMed] [Google Scholar]
- 4. Peake P. W., Shen Y., Walther A., Charlesworth J. A. (2008) Adiponectin binds C1q and activates the classical pathway of complement. Biochem. Biophys. Res. Commun. 367, 560–565 [DOI] [PubMed] [Google Scholar]
- 5. Wang Y., Lam K. S., Xu A. (2007) Adiponectin as a negative regulator in obesity-related mammary carcinogenesis. Cell Res 17, 280–282 [DOI] [PubMed] [Google Scholar]
- 6. Kelesidis I., Kelesidis T., Mantzoros C. S. (2006) Adiponectin and cancer: a systematic review. Br. J. Cancer 94, 1221–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Körner A., Pazaitou-Panayiotou K., Kelesidis T., Kelesidis I., Williams C. J., Kaprara A., Bullen J., Neuwirth A., Tseleni S., Mitsiades N., Kiess W., Mantzoros C. S. (2007) Total and high-molecular-weight adiponectin in breast cancer: in vitro and in vivo studies. J. Clin. Endocrinol. Metab. 92, 1041–1048 [DOI] [PubMed] [Google Scholar]
- 8. Shinoda Y., Yamaguchi M., Ogata N., Akune T., Kubota N., Yamauchi T., Terauchi Y., Kadowaki T., Takeuchi Y., Fukumoto S., Ikeda T., Hoshi K., Chung U. I., Nakamura K., Kawaguchi H. (2006) Regulation of bone formation by adiponectin through autocrine/paracrine and endocrine pathways. J. Cell. Biochem. 99, 196–208 [DOI] [PubMed] [Google Scholar]
- 9. Berner H. S., Lyngstadaas S. P., Spahr A., Monjo M., Thommesen L., Drevon C. A., Syversen U., Reseland J. E. (2004) Adiponectin and its receptors are expressed in bone-forming cells. Bone 35, 842–849 [DOI] [PubMed] [Google Scholar]
- 10. Berg A. H., Combs T. P., Scherer P. E. (2002) ACRP30/adiponectin: an adipokine regulating glucose and lipid metabolism. Trends Endocrinol. Metab. 13, 84–89 [DOI] [PubMed] [Google Scholar]
- 11. Berg A. H., Scherer P. E. (2005) Adipose tissue, inflammation, and cardiovascular disease. Circ. Res. 96, 939–949 [DOI] [PubMed] [Google Scholar]
- 12. Tilg H., Moschen A. R. (2006) Adipocytokines: mediators linking adipose tissue, inflammation and immunity. Nat. Rev. Immunol. 6, 772–783 [DOI] [PubMed] [Google Scholar]
- 13. Yamauchi T., Kadowaki T. (2008) Physiological and pathophysiological roles of adiponectin and adiponectin receptors in the integrated regulation of metabolic and cardiovascular diseases. Int. J. Obes. 32, Suppl. 7, S13–S18 [DOI] [PubMed] [Google Scholar]
- 14. Wang Y., Lam K. S., Yau M. H., Xu A. (2008) Post-translational modifications of adiponectin: mechanisms and functional implications. Biochem. J. 409, 623–633 [DOI] [PubMed] [Google Scholar]
- 15. Goldstein B. J., Scalia R. (2004) Adiponectin: a novel adipokine linking adipocytes and vascular function. J. Clin. Endocrinol. Metab. 89, 2563–2568 [DOI] [PubMed] [Google Scholar]
- 16. Tsao T.-S., Murrey H. E., Hug C., Lee D. H., Lodish H. F. (2002) Oligomerization state-dependent activation of NF-κB signaling pathway by adipocyte complement-related protein of 30 kDa (Acrp30). J. Biol. Chem. 277, 29359–29362 [DOI] [PubMed] [Google Scholar]
- 17. Tsao T.-S., Tomas E., Murrey H. E., Hug C., Lee D. H., Ruderman N. B., Heuser J. E., Lodish H. F. (2003) Role of disulfide bonds in Acrp30/adiponectin structure and signaling specificity: different oligomers activate different signal transduction pathways. J. Biol. Chem. 278, 50810–50817 [DOI] [PubMed] [Google Scholar]
- 18. Shapiro L., Scherer P. E. (1998) The crystal structure of a complement-1q family protein suggests an evolutionary link to tumor necrosis factor. Curr. Biol. 8, 335–338 [DOI] [PubMed] [Google Scholar]
- 19. Radjainia M., Wang Y., Mitra A. K. (2008) Structural polymorphism of oligomeric adiponectin visualized by electron microscopy. J. Mol. Biol. 381, 419–430 [DOI] [PubMed] [Google Scholar]
- 20. Pajvani U. B., Du X., Combs T. P., Berg A. H., Rajala M. W., Schulthess T., Engel J., Brownlee M., Scherer P. E. (2003) Structure-function studies of the adipocyte-secreted hormone Acrp30/adiponectin: implications for metabolic regulation and bioactivity. J. Biol. Chem. 278, 9073–9085 [DOI] [PubMed] [Google Scholar]
- 21. Suzuki S., Wilson-Kubalek E. M., Wert D., Tsao T.-S., Lee D. H. (2007) The oligomeric structure of high molecular weight adiponectin. FEBS Lett. 581, 809–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Simpson F., Whitehead J. P. (2010) Adiponectin—it's all about the modifications. Int. J. Biochem. Cell Biol. 42, 785–788 [DOI] [PubMed] [Google Scholar]
- 23. Schraw T., Wang Z. V., Halberg N., Hawkins M., Scherer P. E. (2008) Plasma adiponectin complexes have distinct biochemical characteristics. Endocrinology 149, 2270–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hara K., Horikoshi M., Yamauchi T., Yago H., Miyazaki O., Ebinuma H., Imai Y., Nagai R., Kadowaki T. (2006) Measurement of the high-molecular weight form of adiponectin in plasma is useful for the prediction of insulin resistance and metabolic syndrome. Diabetes Care 29, 1357–1362 [DOI] [PubMed] [Google Scholar]
- 25. Briggs D. B., Jones C. M., Mashalidis E. H., Nuñez M., Hausrath A. C., Wysocki V. H., Tsao T.-S. (2009) Disulfide-dependent self-assembly of adiponectin octadecamers from trimers and presence of stable octadecameric adiponectin lacking disulfide bonds in vitro. Biochemistry 48, 12345–12357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Radjainia M., Huang B., Bai B., Schmitz M., Yang S. H., Harris P. W., Griffin M. D., Brimble M. A., Wang Y., Mitra A. K. (2012) A highly conserved tryptophan in the N-terminal variable domain regulates disulfide bond formation and oligomeric assembly of adiponectin. FEBS J. 279, 2495–2507 [DOI] [PubMed] [Google Scholar]
- 27. Wang Z. V., Schraw T. D., Kim J. Y., Khan T., Rajala M. W., Follenzi A., Scherer P. E. (2007) Secretion of the adipocyte-specific secretory protein adiponectin critically depends on thiol-mediated protein retention. Mol. Cell. Biol. 27, 3716–3731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Qiang L., Wang H., Farmer S. R. (2007) Adiponectin secretion is regulated by SIRT1 and the endoplasmic reticulum oxidoreductase Ero1-Lα. Mol. Cell. Biol. 27, 4698–4707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang Z. V., Scherer P. E. (2008) DsbA-L is a versatile player in adiponectin secretion. Proc. Natl. Acad. Sci. U.S.A. 105, 18077–18078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu M., Zhou L., Xu A., Lam K. S., Wetzel M. D., Xiang R., Zhang J., Xin X., Dong L. Q., Liu F. (2008) A disulfide-bond A oxidoreductase-like protein (DsbA-L) regulates adiponectin multimerization. Proc. Natl. Acad. Sci. U.S.A. 105, 18302–18307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Anelli T., Ceppi S., Bergamelli L., Cortini M., Masciarelli S., Valetti C., Sitia R. (2007) Sequential steps and checkpoints in the early exocytic compartment during secretory IgM biogenesis. EMBO J. 26, 4177–4188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Freyaldenhoven S., Li Y., Kocabas A. M., Ziu E., Ucer S., Ramanagoudr-Bhojappa R., Miller G. P., Kilic F. (2012) The role of ERp44 in maturation of serotonin transporter protein. J. Biol. Chem. 287, 17801–17811 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33. Anelli T., Alessio M., Bachi A., Bergamelli L., Bertoli G., Camerini S., Mezghrani A., Ruffato E., Simmen T., Sitia R. (2003) Thiol-mediated protein retention in the endoplasmic reticulum: the role of ERp44. EMBO J. 22, 5015–5022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Anelli T., Alessio M., Mezghrani A., Simmen T., Talamo F., Bachi A., Sitia R. (2002) ERp44, a novel endoplasmic reticulum folding assistant of the thioredoxin family. EMBO J. 21, 835–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gilchrist A., Au C. E., Hiding J., Bell A. W., Fernandez-Rodriguez J., Lesimple S., Nagaya H., Roy L., Gosline S. J., Hallett M., Paiement J., Kearney R. E., Nilsson T., Bergeron J. J. (2006) Quantitative proteomics analysis of the secretory pathway. Cell 127, 1265–1281 [DOI] [PubMed] [Google Scholar]
- 36. Wang L., Wang L., Vavassori S., Li S., Ke H., Anelli T., Degano M., Ronzoni R., Sitia R., Sun F., Wang C. C. (2008) Crystal structure of human ERp44 shows a dynamic functional modulation by its carboxy-terminal tail. EMBO Rep. 9, 642–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vavassori S., Cortini M., Masui S., Sannino S., Anelli T., Caserta I. R., Fagioli C., Mossuto M. F., Fornili A., van Anken E., Degano M., Inaba K., Sitia R. (2013) A pH-regulated quality control cycle for surveillance of secretory protein assembly. Mol. Cell 50, 783–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pan C., Zheng J., Wu Y., Chen Y., Wang L., Zhou Z., Yin W., Ji G. (2011) ERp44 C160S/C212S mutants regulate IP3R1 channel activity. Protein Cell 2, 990–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Anelli T., Sannino S., Sitia R. (2015) Proteostasis and “redoxtasis” in the secretory pathway: tales of tails from ERp44 and immunoglobulins. Free Radic. Biol. Med. 83, 323–330 [DOI] [PubMed] [Google Scholar]
- 40. Wang Y., Xu A., Knight C., Xu L. Y., Cooper G. J. (2002) Hydroxylation and glycosylation of the four conserved lysine residues in the collagenous domain of adiponectin: potential role in the modulation of its insulin-sensitizing activity. J. Biol. Chem. 277, 19521–19529 [DOI] [PubMed] [Google Scholar]
- 41. Amblard M., Fehrentz J.-A., Martinez J., Subra G. (2006) Methods and protocols of modern solid phase peptide synthesis. Mol. Biotechnol. 33, 239–254 [DOI] [PubMed] [Google Scholar]
- 42. Harris P. W., Yang S. H., Brimble M. A. (2011) An improved procedure for the preparation of aminomethyl polystyrene resin and its use in solid phase (peptide) synthesis. Tetrahedron Lett. 52, 6024–6026 [Google Scholar]
- 43. Albericio F., Barany G. (1985) Improved approach for anchoring Nα-9-fluorenylmethyloxycarbonylamino acids as p-alkoxybenzyl esters in solid-phase peptide synthesis. Int. J. Pept. Protein Res. 26, 92–97 [DOI] [PubMed] [Google Scholar]
- 44. Harris P. W., Hampe L., Radjainia M., Brimble M. A., Mitra A. K. (2014) An investigation of the role of the adiponectin variable domain on the stability of the collagen-like domain. Biopolymers 101, 313–321 [DOI] [PubMed] [Google Scholar]
- 45. Xu A., Chan K. W., Hoo R. L., Wang Y., Tan K. C., Zhang J., Chen B., Lam M. C., Tse C., Cooper G. J., Lam K. S. (2005) Testosterone selectively reduces the high molecular weight form of adiponectin by inhibiting its secretion from adipocytes. J. Biol. Chem. 280, 18073–18080 [DOI] [PubMed] [Google Scholar]
- 46. Lakowicz J. R. (2006) in Principles of Fluorescence Spectroscopy (Lakowicz J., ed) pp. 277–330, Springer, New York [Google Scholar]
- 47. Briggs D. B., Giron R. M., Malinowski P. R., Nuñez M., Tsao T. S. (2011) Role of redox environment on the oligomerization of higher molecular weight adiponectin. BMC Biochem. 12, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Anelli T., van Anken E. (2013) Missing links in antibody assembly control. Int. J. Cell Biol. 2013, 606703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Otsu M., Bertoli G., Fagioli C., Guerini-Rocco E., Nerini-Molteni S., Ruffato E., Sitia R. (2006) Dynamic retention of Ero1α and Ero1β in the endoplasmic reticulum by interactions with PDI and ERp44. Antioxid. Redox Signal. 8, 274–282 [DOI] [PubMed] [Google Scholar]
- 50. Kim J. A., Nuñez M., Briggs D. B., Laskowski B. L., Chhun J. J., Eleid J. K., Quon M. J., Tsao T. S. (2012) Extracellular conversion of adiponectin hexamers into trimers. Biosci. Rep. 32, 641–652 [DOI] [PMC free article] [PubMed] [Google Scholar]