Abstract
Enzymes of the OXA-48 family have become some of the most important beta-lactamases in the world. A new OXA-48 variant (OXA-370) was first described for an Enterobacter hormaechei strain isolated in Rio Grande do Sul (southern region of Brazil) in 2013. Here we report detection of the blaOXA-370 gene in 24 isolates belonging to three Enterobacteriaceae species (22 Klebsiella pneumoniae isolates, 1 Enterobacter cloacae isolate, and 1 Enterobacter aerogenes isolate) collected from five hospitals in Rio de Janeiro, Brazil, in 2013 and 2014. The isolates showed a multidrug resistance profile, and 12.5% were resistant to polymyxin B. Besides blaOXA-370, no other carbapenemase genes were observed by PCR, whereas blaOXA-1 was found in all isolates and 22 isolates (91.6%) possessed blaCTX-M-15. Molecular typing of the K. pneumoniae isolates by pulsed-field gel electrophoresis (PFGE) showed the presence of two clonal groups, i.e., KpA (21 isolates) and KpB (1 isolate). KpA was characterized as sequence type 16 (ST16) and KpB as ST1041 by multilocus sequence typing (MLST). ST16 has been observed for KPC-producing K. pneumoniae in Rio de Janeiro. Plasmid analysis performed with six representative OXA-370-producing isolates showed plasmids harboring the blaOXA-370 gene in all strains, ranging from 25 kb to 150 kb. This study suggests that there is an urgent need to investigate the presence of OXA-370 and dissemination of the K. pneumoniae ST16 clone carrying this gene in Brazil.
INTRODUCTION
The Enterobacteriaceae are ubiquitous Gram-negative bacteria that are commonly associated with diverse types of infections and are increasingly showing resistance, especially to beta-lactams (1). The most commonly acquired mechanism of resistance against carbapenems, which are the widest-spectrum beta-lactams available, is the production of carbapenemases (2). The most important carbapenemases are serine carbapenemases (including KPC-type enzymes); metallo beta-lactamases such as IMP, VIM, and NDM; and the OXA type (like OXA-48), which includes enzymes showing lower hydrolytic capacities against carbapenems than enzymes of other classes. OXA-48 carbapenemase was first isolated in Turkey in 2001; since then, this enzyme has been frequently observed in different parts of the world (3). To date, nine other allelic variants have been noted in a few individual reports (3). The phenotypic detection of OXA-type enzymes is a problem for clinical laboratories, since there is no specific inhibitor for those enzymes; consequently, they may be underreported. In Brazil, a new allelic variant, namely, OXA-370, was recently observed in an Enterobacter hormaechei strain isolated in Rio Grande do Sul (4). Here we describe the detection of OXA-370 in three Enterobacteriaceae species and the clonal dissemination of OXA-370-producing Klebsiella pneumoniae in five hospitals in Rio de Janeiro.
MATERIALS AND METHODS
Clinical strains.
The Laboratório de Pesquisa em Infecção Hospitalar (LAPIH) located at Oswaldo Cruz Institute (Rio de Janeiro, Brazil) routinely receives clinical bacterial isolates from hospitals that are part of the Bacterial Nosocomial Infection Resistance Surveillance network. We routinely perform multiplex PCR assays with carbapenem-resistant or intermediate isolates, to identify some of the more important epidemiological genes currently associated with Gram-negative bacilli (KPC and NDM). For the Enterobacteriaceae isolates that show negative results, we usually carry out PCR assays to detect any OXA-48-like genes (5).
Between August 2013 and January 2014, our laboratory received 24 carbapenem-resistant Enterobacteriaceae isolates (22 K. pneumoniae isolates, one Enterobacter aerogenes isolate, and one Enterobacter cloacae complex isolate) that were negative for KPC and NDM and positive for OXA-48-like carbapenemase; they had been obtained from different clinical specimens from five hospitals in Rio de Janeiro state. Bacterial isolates were identified by conventional techniques (6), and the identification of OXA-48-like allele variants was performed by PCR (5) and sequencing.
Antimicrobial susceptibility tests.
Antimicrobial susceptibility tests were performed and interpreted using the disk diffusion method described by the Clinical and Laboratory Standards Institute (CLSI) (7). The following antimicrobials (Oxoid) were tested: ertapenem (10 μg), imipenem (10 μg), meropenem (10 μg), cefoxitin (30 μg), cefepime (30 μg), ceftazidime (30 μg), cefotaxime (30 μg), aztreonam (30 μg), gentamicin (10 μg), amikacin (30 μg), ciprofloxacin (5 μg), and sulfamethoxazole-trimethoprim (25 μg). MICs were determined for meropenem, imipenem, ertapenem, and polymyxin B using the Etest method (AB Biodisk, Solna, Sweden). The results were interpreted according to CLSI breakpoints except for polymyxin B, for which the EUCAST 2014 breakpoints were used (http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/Breakpoint_table_v_4.0.pdf).
Detection of beta-lactamase genes.
Despite the previous detection of OXA-370, screening for other beta-lactamase genes (blaCTX-M, blaCTX-M-15, blaOXA-1, blaIMP, blaVIM, blaNDM, blaGES, and blaKPC) was performed by PCR using previously reported conditions and primers (8).
Molecular typing.
For epidemiological analysis, pulsed-field gel electrophoresis (PFGE) was used for K. pneumoniae, whereas multilocus sequence typing (MLST) analysis was performed on the representative isolates of the PFGE clones of this species. For PFGE (9), the plug containing the genomic DNA was digested with XbaI for 3 h and the fragments were separated in a 1.0% Seakem Gold agarose gel (Lonza) in a CHEF-DRIII system (Bio-Rad, Richmond, CA), under the following conditions: 13°C, 120° field angle, 6 V/cm, 0.5- to 35-s pulse times, for 15.5 h. Band patterns were analyzed using Bionumerics 6.6 software (Applied Maths, Sint-Martens-Latem, Belgium), and isolates displaying ≥85% similarity (Dice coefficient) were considered to belong to the same clone.
For MLST, seven housekeeping genes of K. pneumoniae (infB, tonB, pgi, gapA, phoE, rpoB, and mdh) were amplified and sequenced according to the protocol described on the K. pneumoniae MLST website (http://bigsdb.web.pasteur.fr/klebsiella/klebsiella.html). The sequence analyses were performed using BioEdit software (version 7.0.5.3).
Plasmid analysis.
Plasmid analysis by restriction digestion was performed with S1 nuclease (10). The blaOXA-370-specific probe labeled with digoxigenin (DIG) was generated with the PCR DIG detection system (Roche Diagnostics). Hybridization experiments were performed as reported by Sambrook and Russell (11).
RESULTS AND DISCUSSION
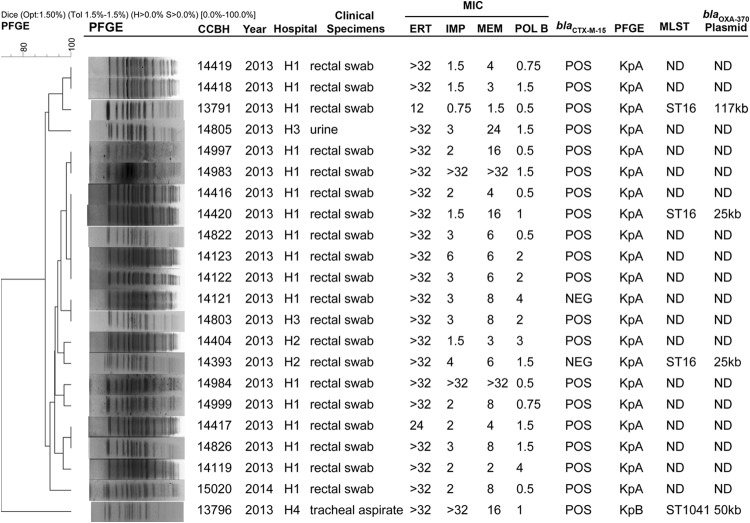
The K. pneumoniae isolates (n = 22) were recovered from four hospitals (hospital 1 [H1], H2, H3, and H4) (Fig. 1). Most of those isolates (n = 17 [77.2%]) were obtained from rectal swabs from patients at H1 between September 2013 and January 2014, during routine surveillance for carbapenem-resistant Enterobacteriaceae. Of the remaining isolates, two were obtained from rectal swabs from patients at H2 in October 2013, two were obtained from urine and rectal swabs from patients at H3 in November 2013, and one was recovered from the tracheal aspirate of a patient at H4 in August 2013 (Fig. 1). During the same period, an E. aerogenes isolate was recovered from a rectal swab from another patient at H4. Also, an E. cloacae isolate was obtained from the urine of a patient at H5.
FIG 1.
Characteristics of 22 OXA-370-producing K. pneumoniae isolates obtained in the Rio de Janeiro state, Brazil. All MIC values are presented in milligrams per liter. The banding patterns were compared by using the unweighted pair group method with arithmetic averages (UPGMA) with the Dice similarity coefficient using the following parameters: optimization (Opt), 1.5%; position tolerance (Tol), 1.5%; minimal height (H) > 0.0%; and minimal surface (S) > 0.0%. CCBH, Coleção de Culturas de Bactérias de Origem Hospitalar; H1, hospital 1; H2, hospital 2; H3, hospital 3; H4, hospital 4; ERT, ertapenem; IMP, imipenem; MEM, meropenem; POLB, polymyxin B; PFGE, pulsed-field gel electrophoresis; MLST, multilocus sequence typing; POS, positive; NEG, negative; ND, not determined.
The high prevalence of OXA-48-like-carbapenemase-producing K. pneumoniae isolates from rectal swabs (n = 20 [90.9%]) was also observed in a study performed from 2001 to 2011 in various Mediterranean countries in Europe and North Africa by Potron et al. and published in 2013 (12). This phenomenon may contribute to the silent dissemination of this gene in bacterial populations.
In performing PCR and sequencing, we observed that all of the isolates possessed the OXA-370 allelic variant, which differs from blaOXA-48 by three nucleotide changes, resulting in one amino acid substitution (4). This variant was recently identified in an Enterobacter hormaechei strain in the southern region of Brazil (4). This species is one of the five species belonging to the Enterobacter cloacae complex that cannot be differentiated by conventional identification techniques (13).
OXA-48-like-carbapenemase-producing bacteria may show different patterns of resistance to beta-lactams. Some strains show susceptibility to broad-spectrum cephalosporins and carbapenems, while others are susceptible to broad-spectrum cephalosporins and resistant to carbapenems and others are resistant to both broad-spectrum cephalosporins and carbapenems (3). These differences represent a challenge for identifying the production of these OXA-48-producing bacteria. Thus, the criteria used in this study to select OXA-48-producing bacteria (resistant or intermediate to carbapenems) might have underestimated the real occurrence of OXA-48-like carbapenemase in our set of isolates.
In the present study, most of the K. pneumoniae isolates were resistant to β-lactams, such as cefotaxime (95.4%), cefepime (95.4%), ceftazidime (95.4%), aztreonam (95.4%), ertapenem (100%), imipenem (95,4%), and meropenem (100%), according to CLSI breakpoints. The E. aerogenes and E. cloacae isolates were resistant to all of the beta-lactams tested.
Some OXA-48 variants, such as OXA-163 and OXA-247, do not have carbapenem-hydrolyzing activity (3). Although the hydrolytic profile of OXA-370 has not yet been determined, Sampaio et al. showed that the OXA-370-producing E. hormaechei strain was resistant to ertapenem (MIC = 4 mg/liter) and exhibited reduced susceptibility to imipenem (MIC = 1.5 mg/liter) but was susceptible to meropenem (MIC = 0.5 mg/liter) and the Escherichia coli transformant was susceptible to carbapenems (4).
Assessing the MICs of K. pneumoniae isolates, our study showed high MIC50 values for ertapenem (>32 mg/liter), with a MIC range of 12 to >32 mg/liter. For meropenem and imipenem, the MIC50 values were lower (6.0 mg/liter and 2.0 mg/liter, respectively), with MIC ranges of 1.5 to >32 mg/liter for meropenem and 0.75 to >32 mg/liter for imipenem. The E. aerogenes isolate showed MICs of >32 mg/liter for all carbapenems tested, while the E. cloacae isolate showed MICs of >32 mg/liter for ertapenem, 1.5 mg/liter for meropenem, and 2 mg/liter for imipenem. These findings showed higher levels of resistance to carbapenems in the isolates included in this study, compared with the previously detected OXA-370-producing E. hormaechei isolate (4).
All isolates were resistant to sulfamethoxazole-trimethoprim and ciprofloxacin, with the exception of the E. aerogenes isolate, which was considered to have intermediate resistance to ciprofloxacin. For the aminoglycosides, 95.4% of the K. pneumoniae isolates were considered nonsusceptible to amikacin (63.6% resistant) and 54.5% nonsusceptible to gentamicin (18.1% resistant). The E. aerogenes and E. cloacae isolates were resistant to both drugs.
The MICs for polymyxin B placed 12.5% of the isolates in the resistant category; all of them were K. pneumoniae strains. In Brazil, previous reports showed similar rates of resistance to this drug among KPC-producing K. pneumoniae isolates (14). This result is alarming, since polymyxin B is considered one of the last resources in the fight against carbapenemase-producing pathogens.
Apart from OXA-370, no other carbapenemase genes were detected by PCR. However, genes encoding narrow-spectrum beta-lactamases and extended-spectrum beta-lactamases (ESBLs) were detected. The blaOXA-1 gene was found in all isolates, and 22 (91.6%) possessed the blaCTX-M-15 gene (including the two Enterobacter isolates). Many different studies have shown the association of the OXA-48-like carbapenemase with other beta-lactamases, mainly CTX-M-15, the most disseminated ESBL reported worldwide (5, 12). Differently from the isolates included in our study, blaOXA-370 was found to be associated with the blaTEM-1 and blaCTX-M-8 variants in the first OXA-370-producing E. hormaechei isolate in Brazil (4). The association of OXA-48-like carbapenemase production with ESBLs probably contributes to increased MICs against third-generation cephalosporins, against which the oxacillinases of this family are not very effective (3).
Molecular typing, performed first by PFGE of K. pneumoniae isolates, showed the presence of two clonal groups (KpA and KpB) (Fig. 1). KpA was represented by 21 isolates (95.4%) and KpB by only one isolate, which was recovered from the tracheal aspirate from a patient at H4. We selected three representative KpA isolates and the one KpB isolate for MLST analysis. The isolate belonging to pulsotype KpB was characterized as sequence type 1041 (ST1041), which was deposited in the Pasteur Klebsiella pneumoniae MLST database in 2012 from other isolates recovered in Rio de Janeiro in 2008.
All KpA isolates tested belonged to ST16. This clone has already been associated with different carbapenemases and blaCTX-M-15 in diverse countries around the world (for example, NDM-1 in Canada and CTX-M-15 in Taiwan and Copenhagen, Denmark) (15–17). Furthermore, ST16 was also described for an OXA-48-producing K. pneumoniae strain that caused outbreaks in two hospitals in different regions of Spain (18). In Brazil, ST16 has already been observed for two KPC-producing isolates that were recovered from blood, in 2008 and 2010 (14, 19), in Rio de Janeiro state. In this work, the K. pneumoniae isolates belonging to KpA ST16 were found in three of the four hospitals studied (Fig. 1). This spread in Rio de Janeiro of a clone of K. pneumoniae carrying OXA-370 is very worrisome, because this species has plasticity to acquire different mechanisms of resistance and has great dispersion capacity.
Plasmid analysis was performed for six OXA-370-producing isolates, i.e., three K. pneumoniae isolates belonging to ST16, the K. pneumoniae ST1041 isolate, and the E. cloacae and E. aerogenes isolates. Plasmids harboring the blaOXA-370 gene were observed in all strains, ranging from 25 kb to 150 kb (Fig. 1). In the K. pneumoniae strains, three different plasmids were observed. The K. pneumoniae ST1041 isolate (CCBH13796) had blaOXA-370 in a plasmid of ∼50 kb. In two ST16 isolates (CCBH14393 and CCBH14420), this gene was observed in an ∼25-kb plasmid; however, in the CCBH13791 isolate, which also belongs to ST16, the plasmid was ∼117 kb. In the E. cloacae isolate CCBH14402, the gene was observed in a plasmid of approximately 40 kb. Only the E. aerogenes isolate presented a plasmid of ∼150 kb, the same plasmid size as described for the E. hormaechei strain detected in Rio Grande do Sul state (4). However, it is necessary to sequence these plasmids to compare the similarities between them. This study showed that the blaOXA-370 gene was detected in plasmids of different sizes in representative isolates. Other genes from the OXA-48 family have also been detected in different genetic platforms (12).
This study showed the presence of OXA-370 in different Enterobacteriaceae species in Rio de Janeiro, Brazil, and dissemination of the K. pneumoniae ST16 clone carrying this gene. Since phenotypic detection of the OXA-48-like carbapenemase is a serious concern in clinical laboratories, we deduce that the extent of the spread of OXA-370 may well be underestimated. Thus, our findings call attention to an urgent need to investigate the presence of OXA-370, as well as studying the phenotypic and molecular characteristics of the blaOXA-370 gene.
ACKNOWLEDGMENTS
This work was funded by research grants from Conselho Nacional de Desenvolvimento Científico e Tecnológico, Fundação Carlos Chagas de Amparo a Pesquisa, and Instituto Oswaldo Cruz-Fiocruz.
We thank the Genomic Platform for DNA Sequencing (Instituto Oswaldo Cruz).
REFERENCES
- 1.Nordmann P, Naas T, Poirel L. 2011. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 17:1791–1798. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gupta N, Limbago BM, Patel JB, Kallen AJ. 2011. Carbapenem-resistant Enterobacteriaceae: epidemiology and prevention. Clin Infect Dis 53:60–67. doi: 10.1093/cid/cir202. [DOI] [PubMed] [Google Scholar]
- 3.Poirel L, Potron A, Nordmann P. 2012. OXA-48-like carbapenemases: the phantom menace. J Antimicrob Chemother 67:1597–1606. doi: 10.1093/jac/dks121. [DOI] [PubMed] [Google Scholar]
- 4.Sampaio JL, Ribeiro VB, Campos JC, Rozales FP, Magagnin CM, Falci DR, da Silva RC, Dalarosa MG, Luz DI, Vieira FJ, Antochevis LC, Barth AL, Zavascki AP. 2014. Detection of OXA-370, an OXA-48-related class D β-lactamase, in Enterobacter hormaechei from Brazil. Antimicrob Agents Chemother 58:3566–3567. doi: 10.1128/AAC.02510-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poirel L, Héritier C, Tolün V, Nordmann P. 2004. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother 48:15–22. doi: 10.1128/AAC.48.1.15-22.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abbott SL. 2011. Klebsiella, Enterobacter, Citrobacter, Serratia, Plesiomonas, and other Enterobacteriaceae, p 639–657. In Versalovic J, Carroll KC, Jorgensen JG, Funke G, Landry ML, Warnock DW (ed), Manual of clinical microbiology, 10th ed, vol 1 ASM Press, Washington, DC. [Google Scholar]
- 7.Clinical and Laboratory Standards Institute. 2014. Performance standards for antimicrobial susceptibility testing; 21st informational supplement. CLSI M100-S24. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 8.Dallenne C, Da Costa A, Decré D, Favier C, Arlet G. 2010. Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J Antimicrob Chemother 65:490–495. doi: 10.1093/jac/dkp498. [DOI] [PubMed] [Google Scholar]
- 9.Ribot EM, Fair MA, Gautom R. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis 3:59–67. doi: 10.1089/fpd.2006.3.59. [DOI] [PubMed] [Google Scholar]
- 10.Barton BM, Harding GP, Zuccarelli AJ. 1995. A general method for detecting and sizing large plasmids. Anal Biochem 226:235–240. doi: 10.1006/abio.1995.1220. [DOI] [PubMed] [Google Scholar]
- 11.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 12.Potron A, Poirel L, Rondinaud E, Nordmann P. 2013. Intercontinental spread of OXA-48 beta-lactamase-producing Enterobacteriaceae over a 11-year period, 2001 to 2011. Euro Surveill 18(31):pii=20549 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20549. [DOI] [PubMed] [Google Scholar]
- 13.Brady C, Cleenwerck I, Venter S, Coutinho T, De Vos P. 2013. Taxonomic evaluation of the genus Enterobacter based on multilocus sequence analysis (MLSA): proposal to reclassify E. nimipressuralis and E. amnigenus into Lelliottia gen. nov. as Lelliottia nimipressuralis comb. nov. and Lelliottia amnigena comb. nov., respectively, E. gergoviae and E. pyrinus into Pluralibacter gen. nov. as Pluralibacter gergoviae comb. nov. and Pluralibacter pyrinus comb. nov., respectively, E. cowanii, E. radicincitans, E. oryzae and E. arachidis into Kosakonia gen. nov. as Kosakonia cowanii comb. nov., Kosakonia radicincitans comb. nov., Kosakonia oryzae comb. nov. and Kosakonia arachidis comb. nov., respectively, and E. turicensis, E. helveticus and E. pulveris into Cronobacter as Cronobacter zurichensis nom. nov., Cronobacter helveticus comb. nov. and Cronobacter pulveris comb. nov., respectively, and emended description of the genera Enterobacter and Cronobacter. Syst Appl Microbiol 36:309–319. doi: 10.1016/j.syapm.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 14.Pereira PS, de Araujo CF, Seki LM, Zahner V, Carvalho-Assef AP, Asensi MD. 2013. Update of the molecular epidemiology of KPC-2-producing Klebsiella pneumoniae in Brazil: spread of clonal complex 11 (ST11, ST437 and ST340). J Antimicrob Chemother 68:312–316. doi: 10.1093/jac/dks396. [DOI] [PubMed] [Google Scholar]
- 15.Mulvey MR, Grant JM, Plewes K, Roscoe D, Boyd DA. 2011. New Delhi metallo-β-lactamase in Klebsiella pneumoniae and Escherichia coli, Canada. Emerg Infect Dis 17:103–106. doi: 10.3201/eid1701.101358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee MY, Ko KS, Kang CI, Chung DR, Peck KR, Song JH. 2011. High prevalence of CTX-M-15-producing Klebsiella pneumoniae isolates in Asian countries: diverse clones and clonal dissemination. Int J Antimicrob Agents 38:160–163. doi: 10.1016/j.ijantimicag.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 17.Nielsen JB, Skov MN, Jørgensen RL, Heltberg O, Hansen DS, Schønning K. 2011. Identification of CTX-M15-, SHV-28-producing Klebsiella pneumoniae ST15 as an epidemic clone in the Copenhagen area using a semi-automated Rep-PCR typing assay. Eur J Clin Microbiol Infect Dis 30:773–778. doi: 10.1007/s10096-011-1153-x. [DOI] [PubMed] [Google Scholar]
- 18.Oteo J, Hernández JM, Espasa M, Fleites A, Sáez D, Bautista V, Pérez-Vázquez M, Fernández-García MD, Delgado-Iribarren A, Sánchez-Romero I, García-Picazo L, Miguel MD, Solís S, Aznar E, Trujillo G, Mediavilla C, Fontanals D, Rojo S, Vindel A, Campos J. 2013. Emergence of OXA-48-producing Klebsiella pneumoniae and the novel carbapenemases OXA-244 and OXA-245 in Spain. J Antimicrob Chemother 68:317–321. doi: 10.1093/jac/dks383. [DOI] [PubMed] [Google Scholar]
- 19.Seki LM, Pereira PS, de Souza MP, Conceição MS, Marques EA, Porto CO, Colnago EM, Alves CF, Gomes D, Assef AP, Samuelsen Ø, Asensi MD. 2011. Molecular epidemiology of KPC-2-producing Klebsiella pneumoniae isolates in Brazil: the predominance of sequence type 437. Diagn Microbiol Infect Dis 70:274–247. doi: 10.1016/j.diagmicrobio.2011.01.006. [DOI] [PubMed] [Google Scholar]