Abstract
A CMY-2-producing capsular type K2 Klebsiella pneumoniae strain (TVGHKP93) with multidrug resistance was isolated from a recurrent liver abscess in a patient who also carried a CMY-2-producing Escherichia coli strain (TVGHEC01) in the stool. TVGHKP93 retained its high virulence compared with that of the isogenic strain (TVGHKP60) with wild-type resistance from the first liver abscess. Our conjugation experiment showed the successful transfer of the blaCMY-2-carrying plasmid from TVGHEC01 into TVGHKP60. The transconjugant showed both high virulence and the multidrug-resistant phenotype, as did TVGHKP93.
TEXT
Klebsiella pneumoniae liver abscess (KPLA) has been increasingly reported in Asia and is considered to be endemic in Taiwan (1, 2). The capsular type of K. pneumoniae appears to be the major virulence factor (3, 4), and the K1 and K2 types were the most prevalent in KPLA (1). In KPLA, K. pneumoniae isolates demonstrate an almost unique antibiogram indicative of resistance to ampicillin only, and multidrug-resistant isolates have rarely been reported (5, 6). Currently, several investigations have provided evidence that KPLA is preceded by gastrointestinal colonization (7–10).
We identified an 84-year-old patient with diabetes who had recurrent KPLA. The strain (TVGHKP60) isolated from the first abscess in November 2012 was susceptible to all antibiotics tested, except for ampicillin, consistent with the natural resistance of K. pneumoniae. The patient received intravenous ceftriaxone for 3 weeks and recovered well. After discharge, he received oral cefuroxime for another 2 weeks. However, the patient had a recurrent liver abscess in January 2013, and a second K. pneumoniae strain with multidrug resistance (TVGHKP93) was isolated. The patient received intravenous ciprofloxacin for 3 weeks and was discharged uneventfully. Interestingly, a multidrug-resistant Escherichia coli strain (TVGHEC01) was isolated from the patient's stool during the recurrence episode. We further investigated the two K. pneumoniae strains and one E. coli strain from this case. The protocol was approved by the institutional review board of Taipei Veterans General Hospital.
Bacterial identification and antimicrobial susceptibility were determined using a Vitek2 system (bioMérieux, Marcy l'Etoile, France). The antimicrobial susceptibility was interpreted according to the guidelines of the CLSI (11) and is shown in Table 1. Pulsed-field gel electrophoresis DNA fingerprinting (12, 13) showed that the wild-type TVGHKP60 strain was nearly identical (only one band difference) to the multidrug-resistant TVGHKP93 strain. Capsular genotyping, detection of rmpA/rmpA2, molecular characterization of β-lactamases, and colony mucoviscosity were performed as previously described (14–16). Multilocus sequence typing (MLST) was performed on the TVGHKP60 and TVGHKP93 strains, and the results were analyzed as previously described (17). The TVGHKP60 and TVGHKP93 strains both belonged to capsular type K2 and sequence type (ST) 86. They showed hypermucoviscosity phenotypes and carried rmpA and rmpA2 genes. Regarding the detection of β-lactamases, blaSHV-1 was detected in the TVGHKP60 strain, and blaSHV-1 and blaCMY-2 were detected in the TVGHKP93 strain. Interestingly, blaCMY-2 was also detected in the TVGHEC01 strain.
TABLE 1.
Antimicrobial susceptibility test data for the strains in this study
| Antibiotic(s) | MIC (mg/liter)a |
|||
|---|---|---|---|---|
| K. pneumoniae TVGHKP60 | K. pneumoniae TVGHKP93 | E. coli TVGHEC01 | K. pneumoniae transconjugant: TVGHKP60::blaCMY-2 | |
| Ampicillin | ≥32 | ≥32 | ≥32 | ≥32 |
| Cefazolin | ≤4 | ≥64 | ≥64 | ≥64 |
| Cefuroxime | 2 | 16 | 16 | 16 |
| Cefoxitin | ≤4 | 32 | 32 | 32 |
| Ceftriaxone | ≤1 | 8 | 8 | 8 |
| Ceftazidime | ≤1 | 16 | 4 | 16 |
| Cefepime | ≤1 | ≤1 | ≤1 | ≤1 |
| Piperacillin-tazobactam | ≤4 | 8 | ≤4 | 8 |
| Gentamicin | ≤1 | ≤1 | ≤1 | ≤1 |
| Amikacin | ≤2 | ≤2 | ≤2 | ≤2 |
| Ciprofloxacin | ≤0.25 | ≤0.25 | ≤0.25 | ≤0.25 |
| Levofloxacin | ≤0.12 | ≤0.12 | ≤0.12 | ≤0.12 |
| Ertapenem | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 |
| Imipenem | ≤0.25 | ≤0.25 | ≤0.25 | ≤0.25 |
| Trimethoprim-sulfamethoxazole | 0.19 | 0.38 | 0.094 | 0.38 |
The values are MIC correlates determined by the Vitek2 system, except for trimethoprim-sulfamethoxazole, which was determined by an Etest.
The K. pneumoniae strains were cultured in LB broth at 37°C for 16 h to obtain bacterial growth curves as described previously (18). The isogenic blaCMY-2-producing strain (TVGHKP93) and the original strain (TVGHKP60) showed no significant differences in their growth rates. The in vivo virulence of the TVGHKP60 and TVGHKP93 strains was compared using a murine model of septicemia generated by intraperitoneal injection. Male 6- to 8-week-old C57/B6 mice were observed for 1 week after intraperitoneal inoculation of 100 CFU of K. pneumoniae. All animal care procedures and protocols were approved by the Institutional Animal Care and Use Committee of National Yang-Ming University. Upon intraperitoneal infection of mice, both strains showed hypervirulence with 50% lethal dose(LD50) values of <100 CFU (Fig. 1). The clinical K. pneumoniae strain (capsular type K64) isolated from blood was used as the control and its LD50 was 107 CFU.
FIG 1.
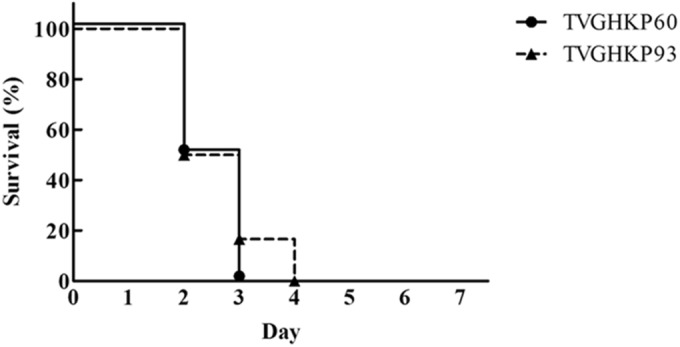
In vivo virulence study. Mouse lethality data following challenges with the TVGHKP60 and TVGHKP93 strains are presented. Male C57BL/6 mice (n = 6 from two independent experiments) were inoculated by intraperitoneal injection with 100 CFU of the TVGHKP60 and TVGHKP93 strains. Survival was assessed for 7 days following infection. The Kaplan-Meier method was used to evaluate the survival rate. Upon intraperitoneal infection, all mice from each group (TVGHKP60 versus TVGHKP93) were dead within 5 days (log rank test; P = 0.6285).
Plasmids were extracted from TVGHEC01 for high-throughput sequencing using the Illumina/Solexa GAII sequencing platform. Coding sequences were predicted and annotated with NCBI protein BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome). A circular map of a blaCMY-2-carrying plasmid, designated pCMY2, with 102,199 bp was obtained (GenBank accession no. LC019731). Nucleotide blast results revealed that pCMY2 exhibited a high level of similarity with a 108,660-bp E. coli plasmid, pC49-108 (GenBank accession no. KJ484638), which showed 94% coverage and shared 99% DNA sequence identity. Conjugation-related genes (associated with tra and trb) and type IV pili-associated genes were found in both plasmids. However, pC49-108 harbored the drug resistance genes addA5, dhfrA17, and blaCTX-M-1, while pCMY2 carried an AmpC-type β-lactamase, blaCMY-2, instead. The PCR results using four primer pairs located within the plasmid revealed that pCMY2 was present in both TVGHEC01 and TVGHKP93 but absent from the TVGHKP60. It is reasonable to consider that horizontal transfer of a blaCMY-2-carrying resistance determinant from E. coli to K. pneumoniae developed in the patient's bowel.
Our attempts to mimic the natural transfer of the pCMY2 plasmid from TVGHEC01 into TVGHKP60 using conjugation experiments described previously (19) were successful. The conjugation frequency was determined to be 5.2 × 10−6. The MICs for cefazolin, cefuroxime, cefoxitin, ceftriaxone, ceftazidime, and piperacillin-tazobactam increased in the transconjugant (TVGHKP60::blaCMY-2) and were the same as those for TVGHKP93 (Table 1). The in vivo virulence assessment also confirmed that K. pneumoniae TVGHKP60::blaCMY-2 retained the hypervirulence characteristics (data not shown). These results implied that TVGHKP60 had acquired pCMY2 from TVGHEC01 in the patient's gut, leading to the formation of the hypervirulent and multidrug-resistant TVGHKP93 in the recurrent liver abscess.
Capsular type K2 and ST86 are considered to be hypervirulent clones that can cause invasive diseases (20, 21). Our study first demonstrated that this virulent strain acquired the pCMY2 plasmid but still retained its virulence. The selective pressure by cephalosporins may predispose to the possible plasmid transfer in this case. Although increased antimicrobial resistance is generally associated with decreased virulence and fitness, evidence has also shown the opposite, and it is increasingly evident that the relationship is often of greater benefit to the pathogen, resulting in a growing public health problem (22).
A recent study showed that multidrug-resistant and hypervirulent populations of K. pneumoniae were mostly nonoverlapping, although two isolates with combined virulence and resistance features were detected (23). These results show that the threat of dual-risk K. pneumoniae strains, combining virulence and multidrug-resistance features, is becoming a reality. The multidrug-resistant and highly virulent TVGHKP93 strain derived from the wild-type TVGHKP60 strain serves as a good example to verify the relationship between virulence and resistance in K. pneumoniae.
With the increasing rate of drug-resistant Enterobacteriaceae colonizing the intestine, the possibility of interspecies transfer of drug resistance determinants into highly virulent K. pneumoniae increases. The acquisition of an important mechanism of antibiotic resistance, such as CMY-2, might suggest that virulent strains may be a potential cause of nosocomial infections in the future (24).
Nucleotide sequence accession number.
The complete nucleotide sequence of pCMY2 was deposited in GenBank under accession no. LC019731.
ACKNOWLEDGMENTS
This work was supported by grants from the Ministry of Science and Technology in Taiwan, Taipei Veterans General Hospital (V104B-001 and V104C-072), and Szu-Yuan Research Foundation of Internal Medicine.
We also thank Taipei Veterans General Hospital for providing experimental space and facilities in the Medical Science & Technology Building.
We declare no conflicts of interest.
REFERENCES
- 1.Siu LK, Yeh KM, Lin JC, Fung CP, Chang FY. 2012. Klebsiella pneumoniae liver abscess: a new invasive syndrome. Lancet Infect Dis 12:881–887. doi: 10.1016/S1473-3099(12)70205-0. [DOI] [PubMed] [Google Scholar]
- 2.Tsai FC, Huang YT, Chang LY, Wang JT. 2008. Pyogenic liver abscess as endemic disease, Taiwan. Emerg Infect Dis 14:1592–1600. doi: 10.3201/eid1410.071254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fung CP, Chang FY, Lee SC, Hu BS, Kuo BI, Liu CY, Ho M, Siu LK. 2002. A global emerging disease of Klebsiella pneumoniae liver abscess: is serotype K1 an important factor for complicated endophthalmitis? Gut 50:420–424. doi: 10.1136/gut.50.3.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fang CT, Chuang YP, Shun CT, Chang SC, Wang JT. 2004. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med 199:697–705. doi: 10.1084/jem.20030857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hsueh PR, Wu JJ, Teng LJ, Chen YC, Yang PC, Ho SW, Luh KT. 2002. Primary liver abscess caused by one clone of Klebsiella pneumoniae with two colonial morphotypes and resistotypes. Emerg Infect Dis 8:100–102. doi: 10.3201/eid0801.010167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su SC, Siu LK, Ma L, Yeh KM, Fung CP, Lin JC, Chang FY. 2008. Community-acquired liver abscess caused by serotype K1 Klebsiella pneumoniae with CTX-M-15-type extended-spectrum beta-lactamase. Antimicrob Agents Chemother 52:804–805. doi: 10.1128/AAC.01269-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tu YC, Lu MC, Chiang MK, Huang SP, Peng HL, Chang HY, Jan MS, Lai YC. 2009. Genetic requirements for Klebsiella pneumoniae-induced liver abscess in an oral infection model. Infect Immun 77:2657–2671. doi: 10.1128/IAI.01523-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung DR, Lee H, Park MH, Jung SI, Chang HH, Kim YS, Son JS, Moon C, Kwon KT, Ryu SY, Shin SY, Ko KS, Kang CI, Peck KR, Song JH. 2012. Fecal carriage of serotype K1 Klebsiella pneumoniae ST23 strains closely related to liver abscess isolates in Koreans living in Korea. Eur J Clin Microbiol Infect Dis 31:481–486. doi: 10.1007/s10096-011-1334-7. [DOI] [PubMed] [Google Scholar]
- 9.Fung CP, Lin YT, Lin JC, Chen TL, Yeh KM, Chang FY, Chuang HC, Wu HS, Tseng CP, Siu LK. 2012. Klebsiella pneumoniae in gastrointestinal tract and pyogenic liver abscess. Emerg Infect Dis 18:1322–1325. doi: 10.3201/eid1808.111053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin YT, Liu CJ, Yeh YC, Chen TJ, Fung CP. 2013. Ampicillin and amoxicillin use and the risk of Klebsiella pneumoniae liver abscess in Taiwan. J Infect Dis 208:211–217. doi: 10.1093/infdis/jit157. [DOI] [PubMed] [Google Scholar]
- 11.Clinical and Laboratory Standards Institute. 2012. Performance standards for antimicrobial susceptibility testing: 22nd informational supplement. CLSI document M100-S22. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 12.Lin YT, Wang FD, Chan YJ, Fu YC, Fung CP. 2014. Clinical and microbiological characteristics of tigecycline non-susceptible Klebsiella pneumoniae bacteremia in Taiwan. BMC Infect Dis 14:1. doi: 10.1186/1471-2334-14-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol 33:2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang CT, Lai SY, Yi WC, Hsueh PR, Liu KL, Chang SC. 2007. Klebsiella pneumoniae genotype K1: an emerging pathogen that causes septic ocular or central nervous system complications from pyogenic liver abscess. Clin Infect Dis 45:284–293. doi: 10.1086/519262. [DOI] [PubMed] [Google Scholar]
- 15.Hsu CR, Lin TL, Chen YC, Chou HC, Wang JT. 2011. The role of Klebsiella pneumoniae rmpA in capsular polysaccharide synthesis and virulence revisited. Microbiology 157:3446–3457. doi: 10.1099/mic.0.050336-0. [DOI] [PubMed] [Google Scholar]
- 16.Yu WL, Ko WC, Cheng KC, Lee HC, Ke DS, Lee CC, Fung CP, Chuang YC. 2006. Association between rmpA and magA genes and clinical syndromes caused by Klebsiella pneumoniae in Taiwan. Clin Infect Dis 42:1351–1358. doi: 10.1086/503420. [DOI] [PubMed] [Google Scholar]
- 17.Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. 2005. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol 43:4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin WH, Kao CY, Yang DC, Tseng CC, Wu AB, Teng CH, Wang MC, Wu JJ. 2014. Clinical and microbiological characteristics of Klebsiella pneumoniae from community-acquired recurrent urinary tract infections. Eur J Clin Microbiol Infect Dis 33:1533–1539. doi: 10.1007/s10096-014-2100-4. [DOI] [PubMed] [Google Scholar]
- 19.Chuang YP, Fang CT, Lai SY, Chang SC, Wang JT. 2006. Genetic determinants of capsular serotype K1 of Klebsiella pneumoniae causing primary pyogenic liver abscess. J Infect Dis 193:645–654. doi: 10.1086/499968. [DOI] [PubMed] [Google Scholar]
- 20.Bialek-Davenet S, Criscuolo A, Ailloud F, Passet V, Nicolas-Chanoine MH, Decré D, Brisse S. 2014. Development of a multiplex PCR assay for identification of Klebsiella pneumoniae hypervirulent clones of capsular serotype K2. J Med Microbiol 63:1608–1614. doi: 10.1099/jmm.0.081448-0. [DOI] [PubMed] [Google Scholar]
- 21.Lin YT, Wang YP, Wang FD, Fung CP. 2015. Community-onset Klebsiella pneumoniae pneumonia in Taiwan: clinical features of the disease and associated microbiological characteristics of isolates from pneumonia and nasopharynx. Front Microbiol 9:122. doi: 10.3389/fmicb.2015.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beceiro A, Tomas M, Bou G. 2013. Antimicrobial resistance and virulence: a successful or deleterious association in the bacterial world? Clin Microbiol Rev 26:185–230. doi: 10.1128/CMR.00059-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bialek-Davenet S, Criscuolo A, Ailloud F, Passet V, Jones L, Delannoy-Vieillard AS, Garin B, Le Hello S, Arlet G, Nicolas-Chanoine MH, Decré D, Brisse S. 2014. Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg Infect Dis 20:1812–1820. doi: 10.3201/eid2011.140206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shon AS, Bajwa RP, Russo TA. 2013. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence 4:107–118. doi: 10.4161/viru.22718. [DOI] [PMC free article] [PubMed] [Google Scholar]


