Abstract
A methicillin-resistant mecB-positive Macrococcus caseolyticus (strain KM45013) was isolated from the nares of a dog with rhinitis. It contained a novel 39-kb transposon-defective complete mecB-carrying staphylococcal cassette chromosome mec element (SCCmecKM45013). SCCmecKM45013 contained 49 coding sequences (CDSs), was integrated at the 3′ end of the chromosomal orfX gene, and was delimited at both ends by imperfect direct repeats functioning as integration site sequences (ISSs). SCCmecKM45013 presented two discontinuous regions of homology (SCCmec coverage of 35%) to the chromosomal and transposon Tn6045-associated SCCmec-like element of M. caseolyticus JCSC7096: (i) the mec gene complex (98.8% identity) and (ii) the ccr-carrying segment (91.8% identity). The mec gene complex, located at the right junction of the cassette, also carried the β-lactamase gene blaZm (mecRm-mecIm-mecB-blaZm). SCCmecKM45013 contained two cassette chromosome recombinase genes, ccrAm2 and ccrBm2, which shared 94.3% and 96.6% DNA identity with those of the SCCmec-like element of JCSC7096 but shared less than 52% DNA identity with the staphylococcal ccrAB and ccrC genes. Three distinct extrachromosomal circularized elements (the entire SCCmecKM45013, ΨSCCmecKM45013 lacking the ccr genes, and SCCKM45013 lacking mecB) flanked by one ISS copy, as well as the chromosomal regions remaining after excision, were detected. An unconventional circularized structure carrying the mecB gene complex was associated with two extensive direct repeat regions, which enclosed two open reading frames (ORFs) (ORF46 and ORF51) flanking the chromosomal mecB-carrying gene complex. This study revealed M. caseolyticus as a potential disease-associated bacterium in dogs and also unveiled an SCCmec element carrying mecB not associated with Tn6045 in the genus Macrococcus.
INTRODUCTION
The genus Macrococcus is composed of seven species of Gram-positive bacteria closely related to staphylococci, including Macrococcus caseolyticus (formerly identified as Staphylococcus caseolyticus) (1). Unlike staphylococci, macrococci do not usually cause human or animal diseases and are typically isolated from animal skin and food products, such as milk and meat (1, 2). The only association of M. caseolyticus with an infection was observed in abscesses from slaughtered lambs in 1992 (3). Even though M. caseolyticus is not primarily targeted by antibiotic treatment as an infectious agent, a few strains have acquired antibiotic resistance mechanisms identical or similar to those found in staphylococci, such as cfr-mediated multidrug resistance (4) and mecB-mediated methicillin resistance (5), respectively.
In staphylococci, methicillin resistance is caused by the synthesis of a modified penicillin binding protein (PBP2a) with low affinity to virtually all β-lactams. This protein is encoded by either the mecA or the mecC gene (6, 7), whose expression is often regulated by the presence of MecR1 (sensor/signal transducer mecR1 gene) and MecI (mec transcription repressor mecI gene). These genes are arrayed in an operon designated the mec gene complex, which is located within the staphylococcal cassette chromosome mec (SCCmec) element. Cassette chromosome recombinases (Ccr), the second essential component of the SCCmec element, encoded by different allotypes of the ccrAB and ccrC genes, are responsible for site-specific integration and excision of the element at the integration site sequence (ISS) of SCCmec located at the 3′ end of the chromosomal orfX gene. The combination of the different allotypes defines the ccr gene complex. SCCmec elements are flanked by characteristic direct repeats (DRs) containing the ISSs that define the transferable unit (8).
The mecA and mecC homologue mecB genes have been identified in M. caseolyticus in two plasmids as well as on a macrococcal chromosomal primordial form of the SCCmec element, designated a SCCmec-like element due to the location of the mecB gene complex (mecR1m-mecIm-mecB-blaZm) within a transposon-driven genetic element (5). In this element, transposon Tn6045 was shown to be responsible for the excision of the region peripheral to mecB (5). While the SCCmec-like element exhibited characteristic 18-bp DRs that potentially also enable spontaneous excisions of ΨSCCmec7096 (SCCmec lacking the ccr genes) and SCC7096 (SCC lacking the mecB gene) as independent units in strain JCSC7096, the two mecB-containing plasmids lacked ccr genes (5).
In 2013, a 9-year-old male neutered Bernese mountain dog was presented several times to a veterinary practice with coughing and signs of rhinitis, including sneezing, nasal and ocular discharge, and swelling of the tonsils and regional lymph nodes. Bacteriological analysis of a nasal sample revealed massive growth of hemolytic Gram-positive cocci which exhibited resistance to penicillin as well as to oxacillin and cefoxitin, which are used for the prediction of the mec genes in staphylococci (9, 10). This prompted us to further identify this bacterium and characterize the genetic background of the β-lactam resistance, revealing a novel mecB-containing SCCmec element not associated with a transposon in a hemolytic Macrococcus.
MATERIALS AND METHODS
Bacterial identification.
Strain KM45013, obtained from our diagnostic unit, was identified as M. caseolyticus by 16S rRNA gene PCR amplification of cell lysates and sequence analysis (11). M. caseolyticus was routinely grown on either Trypticase soy agar containing 5% sheep blood (TSA-S; Becton, Dickinson and Company, Franklin Lakes, NJ) or in LB broth at 37°C with aeration.
Determination of antimicrobial resistance profile.
MICs were measured in Mueller-Hinton broth by the microdilution technique using custom-made Sensititre susceptibility plates (NLEUST; Trek Diagnostics Systems, East Grinstead, United Kingdom) and following the Clinical and Laboratory Standards Institute (CLSI) guidelines (9). The production of β-lactamase was tested on nitrocefin dry slides (Becton, Dickinson and Company).
DNA extraction and determination of mecB location.
Genomic DNA was isolated using a phenol-chloroform method with the following modifications for improved cell lysis (12). Five milliliters of overnight culture in LB broth was centrifuged for 10 min at 15,000 rpm, and cells were resuspended in 100 μl Tris-EDTA buffer containing 2 mg/ml lysozyme and 0.5 mg/ml lysostaphin and incubated for 20 min at 37°C. Plasmid DNA was obtained by phenol-chloroform extraction as described by Anderson and McKay (13), also including a lysis step with lysozyme and lysostaphin. The integrity and concentration of the extracted DNA were assessed by agarose gel electrophoresis and spectrophotometric measurement (Qubit; Invitrogen), respectively.
Southern blot hybridization was performed on both genomic and plasmid DNA of strain KM45013 using a digoxigenin-labeled mecB probe obtained using primers mecB-Fw and mecB-Rv (see Table S1 in the supplemental material) following the manufacturer's protocol (Roche, Switzerland). Hybridization signals were visualized on the membrane using a LAS-3000 imaging system (Fujifilm), anti-DIG-AP Fab fragments, and the CDP-Star chemiluminescence substrate (Roche, Switzerland). Genomic and plasmid DNA of M. caseolyticus strain JCSC5402 (5) served as a positive control for the determination of the location of the mecB gene.
Whole-genome sequencing, assembly, and annotation of the novel SCCmec element.
High-throughput whole-genome sequencing (WGS) of M. caseolyticus KM45013 was performed with Roche 454 GS Titanium chemistry according to the manufacturer's standard protocols (GS Junior System; Roche Diagnostics, Switzerland). Resulting contigs were analyzed for the presence of characteristic genes of the SCCmec-like element (mecB, mecIm, mecRm, ccrAm, ccrBm) and surrounding chromosomal segments (orfX, transposase gene of Tn6045, MCCL_0033, and MCCL_0034) of M. caseolyticus JCSC7096 (5) (GenBank accession no. AB498756) using a BLAST search (http://www.ncbi.nlm.nih.gov) and specific PCRs (see Table S1 in the supplemental material). Sanger sequencing of PCR products was performed using BigDye Terminator cycle sequencing and an ABI Prism 3100 genetic analyzer (Applied Biosystems, Foster City, CA) to fill the gaps between SCCmec-containing contigs and to define their specific orientations. Open reading frames (ORFs) were defined with the help of Prodigal (14), GeneMark (15), and ORF Finder (NCBI) software. Only those with plausible ribosomal binding sites were considered. Annotation of the ORFs was performed by BLAST homology, and motif analysis of the translated reading frames was performed against the ScanProsite database (16), the Pfam database (only significant matches were considered for annotation) (17), and the National Center for Biotechnology Information conserved domain database (CDD) (18).
Detection of ISSs was achieved by searching for the consensus sequence GA[A/G][TCG/ATG]TATCATAAGTGA (positions with possible alternative nucleotides are indicated within square brackets, and the possible nucleotides are separated by slashes) in all contigs. Characteristic inverted repeats (IRs), typically found after element insertion catalyzed by Ccrs, were also examined directly upstream and downstream of the ISSs.
Characterization of the chromosomal SCCmec structure and detection of spontaneous extrachromosomal excisions.
Primers designed for the detection of the mec and ccr gene complexes of SCCmecKM45013 and the SCCmec-like element of JCSC7096 are shown in Table S1 in the supplemental material. In addition, primers used for identification of the chromosomal orfX gene of M. caseolyticus strain KM45013, as well as the conserved reading frames MCCL_0033 and MCCL_0034 located at the 3′ end of orfX in the previously described mecB-carrying macrococcal strains (5), are also described (see Table S1). BamHI and BglII restriction analysis of different regions of the SCCmec element amplified by long-range PCR (GoTaq long PCR master mix; Promega) was performed for assembling and scaffolding confirmation (see Table S1). The presence of potential circular intermediates (CIs) of SCCmec segments delimited by ISS sequences, as well as other possible extrachromosomal circularized structures, was tested by specific PCR and sequenced using primers reading outward from the ISSs or a corresponding region (for an ISS-independent excision event). The chromosomal region, where segment excision was expected to have occurred, was also amplified by PCR using adapted elongation times and was sequenced (see Table S1).
Phylogenetic relationship of ccr genes and blaZ-containing mec gene complexes.
The phylogenetic relationships of one representative of each type of ccr gene (19) and the mec gene complexes containing the blaZ gene (5–7, 20) were investigated by the construction of a maximum likelihood phylogenetic tree using the SeaView program, version 4.4.0 (21), with nucleotide sequences deposited in the ENA/GenBank databases. Sequences were aligned using MUSCLE, and the trees were built with PhyML using a general time-reversible (GTR) model.
Nucleotide sequence accession number.
The 41,563-bp nucleotide sequence of M. caseolyticus strain KM45013 containing the complete 38,941-bp SCCmecKM45013 and its 602-bp upstream and 2,020-bp downstream chromosomal regions has been deposited in the GenBank/ENA/DDBJ databases under the accession number HG970732.
RESULTS AND DISCUSSION
Identification of M. caseolyticus KM45013.
Strain KM45013 was identified as M. caseolyticus based on the 16S rRNA gene sequence, which exhibited 99.7% nucleotide identity with that of M. caseolyticus type strain ATCC 13548T and M. caseolyticus JCSC5402, the only macrococcal strain whose genome has been completely sequenced (2). Decreased susceptibility to β-lactams was confirmed by the determination of the MICs for penicillin (MIC, >2 μg/ml), oxacillin (MIC, >8 μg/ml), and cefoxitin (MIC, >8 μg/ml). M. caseolyticus KM45013 differed from other members of Macrococcus species by the formation of a complete hemolysis on a sheep blood plate. Hemolysins are known virulence factors in staphylococci which have been associated with different types of infections (22, 23). Whether the hemolytic property of strain KM45013, which is so far unique among Macrococcus caseolyticus, represents a virulence factor in dogs still remains to be clarified. Nevertheless, the massive presence of M. caseolyticus in the nasal sample may be indicative of an association with the disease, even if other causes cannot be excluded. Since this discovery, 2 additional hemolytic M. caseolyticus isolates were obtained in our laboratory from 2 dogs diagnosed with otitis and dermatitis, indicating that more attention should be paid to this microorganism.
Characterization of the novel SCCmecKM45013 element and comparison with other mecB-carrying elements.
WGS of strain KM45013 resulted in 92,893 filter reads and coverage equivalent to 14.3×. Sequence reads were de novo assembled using Newbler 2.6 (Roche) at the Vital-IT Center for High-Performance Computing at the Swiss Institute of Bioinformatics (http://www.vital-it.ch), yielding 116 contigs (86 contigs >500 bp) with an N50 (length-weighted median) of 46,794 bp, a mean contig size of 19,287 bp, a maximum contig length of 227,824 bp, and a contig sum of 2,275,932 bp. WGS of M. caseolyticus KM45013 as well as Southern blot hybridization experiments allowed the identification and characterization of the novel 38,941-bp chromosomally located SCCmecKM45013 element. A total of 49 coding sequences (CDSs) were identified in SCCmecKM45013. The genome of strain KM45013 presented a GC content of 37.0%, while that of SCCmecKM45013 was 31.5%, suggesting that SCCmecKM45013 was integrated as an exogenous element. SCCmecKM45013 was located at the 3′ end of the chromosomal orfX gene and was demarcated at both extremities by DRs with the following ISSs: 5′-GAATCGTATCATAAGTGA-3′ (ISS1) and 5′-GAGTCGTATCATAAGTGA-3′ (ISS3) (Fig. 1). An additional ISS, ISS2 (5′-GAAAGTTATCATAAGTGA-3′), was detected 26,883 bp downstream of the orfX gene and 6,842 bp upstream of the mecB complex. Imperfect inverted repeats (IRs), which have been shown to play a role in the excision but not the integration of SCCmec (8), were detected adjacent to the three ISS elements (data not shown). These IRs had similar sequences to those detected in staphylococcal SCCmec elements and in the SCCmec-like element of M. caseolyticus JCSC7096 (5, 24).
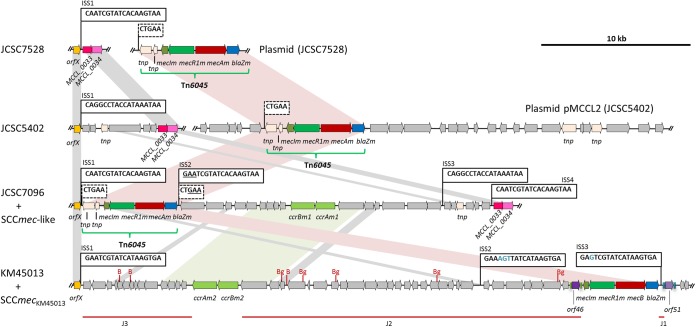
FIG 1.
Schematic presentation of the orfX downstream region in the M. caseolyticus KM45013 chromosome, including the novel SCCmecKM45013 (ENA accession no. HG970732), and a comparison with the previous mecB-carrying genetic structures detected in macrococci: M. caseolyticus strain JCSC7528 (GenBank accession no. AB498758), JCSC5402 (GenBank accession no. AP009486), and JCSC7096 (GenBank accession no. AB498756) and their correspondent chromosomal orfX downstream regions (GenBank accession no. AB498757, AP009484, and AB498756, respectively). The arrows indicate the extent and direction of transcription of the open reading frames. All annotated regions of M. caseolyticus KM45013 are colored as follows: yellow (orfX), pale pink (tnp), pale green (ccrAm, ccrBm), dark green (mecIm), green (mecRm), red (mecB, mecAm), blue (blaZm), purple (orf46), pale purple (orf51), magenta (MCCL_0033) and pink (MCCL_0034). The different integration site sequences (ISSs) for SCC (ISS1 to ISS4) are shown within boxes. The direct repeats (CTGAA) of transposon Tn6045 in strains JCSC7528, JCSC5402, and JCSC7096 are shown within dashed boxes. Shadowed areas indicate regions with more than 84% nucleotide sequence identity. Tn6045 in strains JCSC7528, JCSC5402, and JCSC7096 (green horizontal curly brackets) and the joining regions J1 to J3 (red horizontal bar) in KM45013 are also shown. B and Bg indicate the BamHI and BglII restriction sites, respectively, within SCCmecKM45013. A size scale in kb is displayed in the upper right-hand corner.
SCCmecKM45013 also shared the highest identity with the SCCmec-like element of JCSC7096 (BLAST hit of 35% query cover and 92% sequence identity) (Fig. 1). The high nucleotide identity value was mainly due to the presence of two discontinuous regions: the mec gene complex and a ccr-carrying segment. The mec gene complex shared 98.8% DNA identity with the corresponding segments of the three mecB-carrying macrococcal strains, and the ccr-carrying region shared 91.8% DNA identity with that detected in the SCCmec-like element of JCSC7096 (5). The ccr genes were absent in the mecB-carrying plasmids of M. caseolyticus strains JCSC5402 and JCSC7528, which instead contained transposon-associated transfer mechanisms (5) (Fig. 1).
The mec gene complex was located at the right-end junction of the cassette and carried a functionally active β-lactamase resistance gene, blaZm, as determined by the nitrocefin test. No other β-lactamase gene was detected in the remaining genomic sequence. In strains JCSC7096, JCSC5402, and JCSC7528, the mec gene complex (mecR1m-mecIm-mecB-blaZm) formed part of transposon Tn6045, which contains two adjacent transposase genes immediately upstream of the mec gene complex and is flanked by a set of short DRs (5′-CTGAA-3′), presumably generated by transposon integration (5). Neither transposons nor transposase genes were detected in the entire SCCmecKM45013 element. In contrast, the mec gene complex was flanked by two 775-bp to 777-bp duplicated sequence fragments that shared 93.2% identity. This duplicated DNA fragment comprised two CDSs (ORF46, ORF51), with 94.6% amino acid identity, and their flanking regions (121 bp upstream of both genes in addition to 81 and 83 bp downstream of orf46 and orf51, respectively). orf51 was located downstream of ISS3 and was therefore outside SCCmecKM45013. Analysis of the putative functional domains of the ORF46 and ORF51 proteins revealed a helix-turn-helix (HTH) domain of the XRE family (CDD accession no. cd00093) and a HipB domain profile (CDD accession no. COG1396), both associated with transcriptional regulators. The transposase of Tn6045 (GenPept accession no. BAI83381) also exhibits an HTH-like domain of the family HTH_21 (CDD accession no. pfam13276) but has two additional integrase domains of the rve superfamily (rve CDD accession no. pfam00665 and rve_3 CDD accession no. pfam13683). These domains, which are necessary for transposition, were not detected in ORF46 or ORF51. The presence of a mecB gene complex independent of Tn6045 and the detection of an entire SCCmec are novel characteristics for this bacterial genus. The high sequence and structure similarity levels between the mecB complex of JCSC7096 and KM45013, both driven by a completely different circularization machinery (see below), illustrate different possible mechanisms for the appearance of SCCmec in this species.
The ccr-carrying region was located 5.3 kb downstream of orfX and was comprised of five additional CDSs, three of which encoded proteins with domains (pfam07799 [DUF1643], pfam06124 [DUG960], and pfam04002 [PF04002]) that were also detected in the mecC-containing staphylococcal SCCmec XI (6).
In addition to the mec gene complex and the ccr-carrying region, SCCmecKM45013 contained three joining (J) regions (Fig. 1). Two of them (J3 and J2) carried additional CDSs, encoding hypothetical proteins for the vast majority but also proteins with putative metabolic functions (GenBank accession no. HG970732). Neither additional antimicrobial nor heavy-metal resistance genes were detected within SCCmecKM45013.
Variable regions downstream of the integration site of SCCmec have been previously observed in staphylococci (25, 26). In M. caseolyticus strains JCSC7096, JCSC5402, and JCSC7528, the orfX downstream region contains two adjacent conserved ORFs, named MCCL_0033 and MCCL_0034, coding for proteins of unknown function. These ORFs were not detected downstream of SCCmecKM45013 nor in the entire KM45013 genome. Instead, orf51 was present, sharing 72% identity with a sequence downstream of orfX of the methicillin-resistant Staphylococcus pseudintermedius strain 57395 (comprising a CDS named mrsp-29) (24) and of methicillin-susceptible S. pseudintermedius ED99 and HKU10-03 (27, 28), all encoding putative transcriptional regulators.
Phylogenetic analysis of ccr and mec gene complexes.
The mecB complex of SCCmecKM45013 presented structural similarities to the mecC complexes detected in SCCmec elements of Staphylococcus aureus LGA251 (mecI-mecR1-mecC-blaZ) (6) and of Staphylococcus xylosus S04009 (mecI-mecR1-mecC1-blaZ) (20), both belonging to the class E mec complex. Phylogenetic comparison of the mec gene complex from macrococci with the class E mec gene complexes revealed 57.4% nucleotide identity to that of S. xylosus and 56.8% to that of S. aureus (Fig. 2A). However, since the current nomenclature of the International Working Group on Staphylococcal Cassette Chromosome elements (IWG-SCC) is set for staphylococcal species, in particular for S. aureus, this mecB complex was not assigned to a specific class (19).
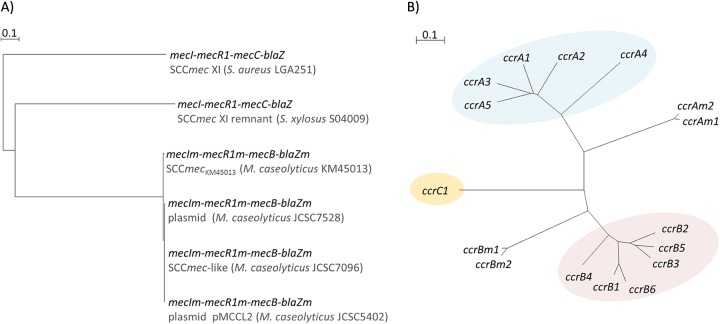
FIG 2.
Phylogenetic relationships of the mec gene complex and serine recombinase ccr genes. Bar length indicates the number of substitutions per site. (A) Phylogenetic relationships of the mec gene complex (mecI-mecR1-mecC-blaZ) detected in SCCmec XI of S. aureus strain LGA251 (GenBank accession no. FR821779) and SCCmec XI remnant of S. xylosus S04009 (GenBank accession no. HE993884) and those (mecIm-mecR1m-mecB [formerly mecAm]-blaZm) detected in M. caseolyticus strains JCSC7528 (GenBank accession no. AB498758), JCSC5402 (GenBank accession no. AP009486), and JCSC7096 (GenBank accession no. AB498756). (B) Phylogenetic relationship of the ccr genes currently described in macrococci (ccrAm1 and ccrBm1 [SCCmec-like element of M. caseolyticus strain JCSC7096, GenBank accession no. AB498756] and ccrAm2 and ccrBm2 [SCCmecKM45013 of M. caseolyticus strain KM45013, GenBank accession no. HG970732]) and one representative staphylococcal ccr per type (ccrA1 and ccrB1 [SCCmec I of S. aureus strain NCTC10442, GenBank accession no. AB033763], ccrA2 and ccrB2 [SCCmec II of S. aureus strain N315, GenBank accession no. BA000018], ccrA3 and ccrB3 [SCCmec III of S. aureus strain 85/2082, GenBank accession no. AB037671], ccrA4 and ccrB4 [SCCmec VI of S. aureus strain HDE288, Genbank accession no. AF411935], ccrA5 and ccrB5 [SCCmec VII-241 of S. pseudintermedius strain KM241, GenBank accession no. AM904731], ccrB6 [SCCmec X of S. aureus strain JCSC6945, GenBank accession no. AB505630], and ccrC1 [SCCmec VII of S. aureus strain JCSC6082, GenBank accession no. AB373032]).
Integration and excision of SCCmec at the orfX gene is mediated by CcrAB or CcrC, which are responsible for catalyzing DNA cleavage, strand exchange, and recombination between the two attachment sites, one within the SCC element (attSCC) and the other on the bacterial chromosome (attB) (8). The ccr genes detected in SCCmecKM45013, showed 94.3% and 95.6% identity with the ccrAm1 and ccrBm1 genes, respectively, from the SCCmec-like element of M. caseolyticus JCSC7096 and were designated ccrAm2 and ccrBm2 according to the nomenclature first described by Tsubakishita et al. (5) with the agreement of the members of the IWG-SCC (see reference 19 for a list of the members). The ccrAm2 and ccrBm2 genes showed the closest identity to the staphylococcal ccr genes from methicillin-resistant S. aureus strain HDE288 (GenBank accession no. AF411935), with an overall nucleotide identity of 51.6% to ccrA4 and 47.3% to ccrB4, respectively (Fig. 2B). Phylogenetic comparative analysis of the ccr genes from the macrococcal SCCmec elements with the other ccr types revealed that the macrococcal ccr genes formed two separate branches outside the staphylococcal ccrA, ccrB, and ccrC clades (Fig. 2B).
Analysis of spontaneous chromosomal excision of different SCCmecKM45013 element units.
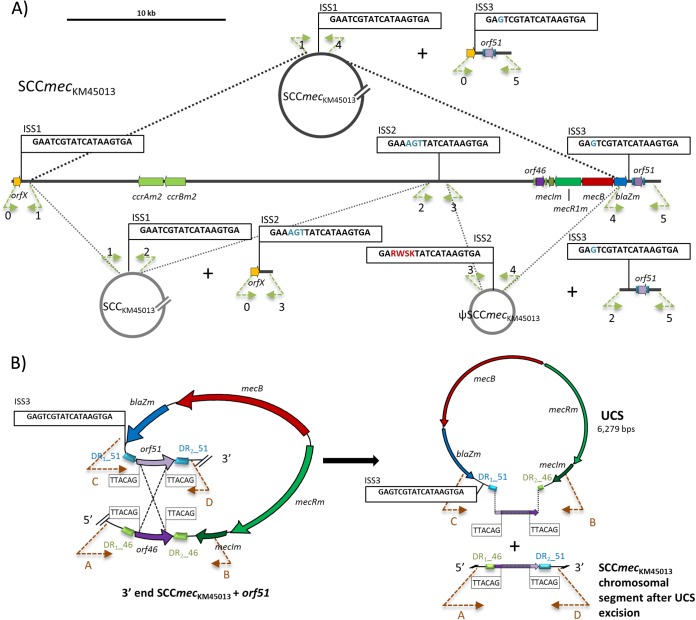
PCR and sequence analysis detected four distinct extrachromosomal CIs, three of them carrying one ISS copy as joining regions, which is characteristic of Ccr-mediated excision. These three ISS-associated CIs have been named with the same nomenclature as that described for the SCCmec-like element of JCSC7096 (5): (i) SCCmecKM45013 (entire cassette), (ii) SCCKM45013 (SCC lacking the mec gene) (26.9 kbp, 37 CDSs, GC content of 32.7%), and (iii) ΨSCCmecKM45013 (SCCmec lacking the ccr genes) (12 kbp, 12 CDSs, GC content of 29%) (Fig. 3A). This excision ability was also observed for the SCCmec-like element of JCSC7096 (5), indicating a high functional activity of the ccrABm gene complex. In addition, all corresponding chromosomal segments remaining after excision were detected (Fig. 3).
FIG 3.
Graphical representation of the spontaneous circular chromosomal excisions detected in SCCmecKM45013. (A) Display of the circular intermediates (CIs) designated to those with integration site sequences (ISSs) as delimiting region (SCCmecKM45013, SCCKM45013, and ΨSCCmecKM45013), and the resulting chromosomal regions after spontaneous loss. The arrows indicate the extent and direction of transcription of orfX, orf46, and orf51, the site-specific recombinase genes (ccrAm2, ccrBm2), and the genes comprising the mecB operon (mecIm, mecRm, mecB and blaZm). ISS1 to ISS3 are boxed. Bases in blue indicate divergences from ISS1, while letters in red indicate the presence of double peaks (Sanger sequencing) in the sequence chromatograms: R (G or A), W (A or T), S (G or C), and K (T or G). A size scale in kb is displayed in the upper left-hand corner. (B) Spontaneous chromosomal excision of an unconventional circularizable structure (UCS) carrying the mecB complex. The left panel shows the potential homologous recombination event between orf46 and orf51 in M. caseolyticus strain KM45013. The right panel shows the resulting UCS and the chromosomal region after excision. The recombined area is colored in purple with white dots. The imperfect direct repeats (DR1, DR2) flanking orf46 and orf51 are indicated as blue (DR_51) or green (DR_46) blocks. The suggested recombination sites (5′-TTACAG-3′) and the ISS3 are indicated within black boxes. Primers used to detect the different circularized elements and the excision sites are indicated as dashed arrows, with the arrowhead indicating the direction of amplification. They are named 0 to 5 for the named CIs and A to D for the designated UCS. See Table S1 in the supplemental material for nomenclature.
The fourth CI (6,279 bp, GC content of 28.1%) consisted of the mec gene complex (mecR1m-mecIm-mecB-blaZm) joined by one recombined copy of the putative transcriptional regulator genes orf46 and orf51 (Fig. 3B). The GC content of this CI was remarkably lower than that in the genome of M. caseolyticus KM45013 (37.0%), indicating that it probably originated from another bacterial species with a lower GC content. Additionally, the presence of long DRs as a joining region instead of ISSs suggested a ccrABm2-independent mechanism for excision, categorizing this CI as an unconventional circularized structure (UCS). UCSs have been recently described as particular genetic structures, mostly carrying antimicrobial resistance determinants (29) which, despite the lack of their own recombinase genes, are able to be excised in circular forms thanks to extensive flanking DRs (29). Moreover, UCSs are frequently carried by conventional mobile genetic elements. Mobilization via site-specific recombination and usage of host trans-acting functions has been suggested for UCSs; however, an active role of the ccrABm2 genes in the excision of this UCS cannot be excluded.
Nucleotide sequence alignment of the recombined region located in the UCS element, the recombined copy that remains in the chromosome after excision, and the individual orf46 and orf51 genes revealed that the CDS located in the UCS and the one that remained in the chromosome after excision resulted from a recombination event between orf46 and orf51, with a 6-bp sequence DR (5′-TTACAG-3′) present at the 5′ and 3′ ends of both CDSs as a presumptive homologous recombination site (see Fig. S1 in the supplemental material). Both extensive repeated regions on each side of mecB may still play a role in the UCS integration/excision. Additionally, this UCS contained ISS3 (Fig. 3B) and thus retained the potential to be integrated by Ccrs.
In conclusion, a mecB-carrying SCCmec element was discovered in a clinical hemolytic M. caseolyticus strain of canine origin. The mecB gene complex was not associated with transposases of Tn6045, revealing for the first time a true SCCmec element in Macrococcus. The high sequence and structure similarity between Tn6045 and the mecB complex of KM45013 within two structurally different elements gives new insight into the acquisition of mecB and the birth of SCCmec in Macrococcus. The detection of several excised circularized elements may also contribute to the further diversification of SCCmec elements in M. caseolyticus. This study underlines the role of commensal bacteria both as potential opportunistic animal pathogens and as reservoirs for novel and primordial forms of SCCmec with high potential genomic plasticity.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by grant 21/2012 awarded to Elena Gómez-Sanz by the Spanish Society of Infectious Diseases and Clinical Microbiology (SEIMC) and by grant 35-539 of the Institute of Veterinary Bacteriology, University of Bern, Bern, Switzerland.
We thank Teruyo Ito and Keiichi Hiramatsu (Department of Bacteriology, Juntendo University, Tokyo) for providing M. caseolyticus JCSC5402. We thank Teruyo Ito and the members of the IWG-SCC for advice on the SCCmec nomenclature. We also thank Ursina Nufer (Tierarztpraxis AG, Meiringen) for providing clinical data and Alexandra Collaud and Isabelle Brodard (Institute of Veterinary Bacteriology, University of Bern) for excellent technical assistance.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.05064-14.
REFERENCES
- 1.Götz F, Bannerman T, Schleifer K-H. 2006. The genera Staphylococcus and Macrococcus, p 5–75. In Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (ed), The prokaryotes, vol 4. Bacteria: firmicutes, cyanobacteria, 3rd ed Springer, New York, NY. [Google Scholar]
- 2.Baba T, Kuwahara-Arai K, Uchiyama I, Takeuchi F, Ito T, Hiramatsu K. 2009. Complete genome sequence of Macrococcus caseolyticus strain JCSCS5402, reflecting the ancestral genome of the human-pathogenic staphylococci. J Bacteriol 191:3429. doi: 10.1128/JB.00366-09 (Erratum.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de la Fuente R, Suarez G, Ruiz Santa Quiteria JA, Meugnier H, Bes M, Freney J, Fleurette J. 1992. Identification of coagulase-negative staphylococci isolated from lambs as Staphylococcus caseolyticus. Comp Immunol Microbiol Infect Dis 15:47–52. doi: 10.1016/0147-9571(92)90101-V. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y, Wang Y, Schwarz S, Shen Z, Zhou N, Lin J, Wu C, Shen J. 2012. Detection of the staphylococcal multiresistance gene cfr in Macrococcus caseolyticus and Jeotgalicoccus pinnipedialis. J Antimicrob Chemother 67:1824–1827. doi: 10.1093/jac/dks163. [DOI] [PubMed] [Google Scholar]
- 5.Tsubakishita S, Kuwahara-Arai K, Baba T, Hiramatsu K. 2010. Staphylococcal cassette chromosome mec-like element in Macrococcus caseolyticus. Antimicrob Agents Chemother 54:1469–1475. doi: 10.1128/AAC.00575-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.García-Álvarez L, Holden MT, Lindsay H, Webb CR, Brown DF, Curran MD, Walpole E, Brooks K, Pickard DJ, Teale C, Parkhill J, Bentley SD, Edwards GF, Girvan EK, Kearns AM, Pichon B, Hill RL, Larsen AR, Skov RL, Peacock SJ, Maskell DJ, Holmes MA. 2011. Methicillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the United Kingdom and Denmark: a descriptive study. Lancet Infect Dis 11:595–603. doi: 10.1016/S1473-3099(11)70126-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shore AC, Deasy EC, Slickers P, Brennan G, O'Connell B, Monecke S, Ehricht R, Coleman DC. 2011. Detection of staphylococcal cassette chromosome mec type XI carrying highly divergent mecA, mecI, mecR1, blaZ, and ccr genes in human clinical isolates of clonal complex 130 methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 55:3765–3773. doi: 10.1128/AAC.00187-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shore AC, Coleman DC. 2013. Staphylococcal cassette chromosome mec: recent advances and new insights. Int J Med Microbiol 303:350–359. doi: 10.1016/j.ijmm.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 9.Clinical and Laboratory Standards Institute. 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 8th ed, vol 29, no. 2. Approved standard M07-A8. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 10.Clinical and Laboratory Standards Institute. 2014. Performance standards for antimicrobial susceptibility testing; 23rd informational supplement. CLSI M100-S24, vol 33, no. 1. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 11.Kuhnert P, Capaul S, Nicolet J, Frey J. 1996. Phylogenetic positions of Clostridium chauvoei and Clostridium septicum based on 16S rRNA gene sequences. Int J Syst Bacteriol 46:1174–1176. doi: 10.1099/00207713-46-4-1174. [DOI] [PubMed] [Google Scholar]
- 12.Pitcher DG, Saunders NA, Owen RJ. 1989. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol 8:151–156. doi: 10.1111/j.1472-765X.1989.tb00262.x. [DOI] [Google Scholar]
- 13.Anderson DG, McKay LL. 1983. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol 46:549–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hyatt D, Chen GL, LoCascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borodovsky M, McIninch J. 1993. GeneMark: parallel gene recognition for both DNA strands. Comput Chem 17:123–133. doi: 10.1016/0097-8485(93)85004-V. [DOI] [Google Scholar]
- 16.de Castro E, Sigrist CJA, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, Bairoch A, Hulo N. 2006. ScanProsite: detection of Prosite signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res 34:W362–W365. doi: 10.1093/nar/gkl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt R, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer EL, Tate J, Punta M. 2014. Pfam: the protein families database. Nucleic Acids Res 42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH. 2011. CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res 39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito T, Hiramatsu K, Tomasz A, de Lencastre H, Perreten V, Holden MT, Coleman DC, Goering R, Giffard PM, Skov RL, Zhang K, Westh H, O'Brien F, Tenover FC, Oliveira DC, Boyle-Vavra S, Laurent F, Kearns AM, Kreiswirth B, Ko KS, Grundmann H, Sollid JE, John JF Jr, Daum R, Soderquist B, Buist G, International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC). 2012. Guidelines for reporting novel mecA gene homologues. Antimicrob Agents Chemother 56:4997–4999. doi: 10.1128/AAC.01199-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harrison EM, Paterson GK, Holden MT, Morgan FJ, Larsen AR, Petersen A, Leroy S, De Vliegher S, Perreten V, Fox LK, Lam TJ, Sampimon OC, Zadoks RN, Peacock SJ, Parkhill J, Holmes MA. 2013. A Staphylococcus xylosus isolate with a new mecC allotype. Antimicrob Agents Chemother 57:1524–1528. doi: 10.1128/AAC.01882-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic treebuilding. Mol Biol Evol 27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 22.Otto M. 2014. Staphylococcus aureus toxins. Curr Opin Microbiol 17:32–37. doi: 10.1016/j.mib.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vandenesch F, Lina G, Henry T. 2012. Staphylococcus aureus hemolysins, bi-component leukocidins, and cytolytic peptides: a redundant arsenal of membrane-damaging virulence factors? Front Cell Infect Microbiol 2:12. doi: 10.3389/fcimb.2012.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perreten V, Chanchaithong P, Prapasarakul N, Rossano A, Blum SE, Elad D, Schwendener S. 2013. Novel pseudo-staphylococcal cassette chromosome mec element (ψSCCmec57395) in methicillin-resistant Staphylococcus pseudintermedius CC45. Antimicrob Agents Chemother 57:5509–5515. doi: 10.1128/AAC.00738-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bouchami O, Ben Hassen A, de Lencastre H, Miragaia M. 2012. High prevalence of mec complex C and ccrC is independent of SCCmec type V in Staphylococcus haemolyticus. Eur J Clin Microbiol Infect Dis 31:605–614. doi: 10.1007/s10096-011-1354-3. [DOI] [PubMed] [Google Scholar]
- 26.Zong Z. 2013. Characterization of a complex context containing mecA but lacking genes encoding cassette chromosome recombinases in Staphylococcus haemolyticus. BMC Microbiol 13:64. doi: 10.1186/1471-2180-13-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ben Zakour NL, Bannoehr J, van den Broek AH, Thoday KL, Fitzgerald JR. 2011. Complete genome sequence of the canine pathogen Staphylococcus pseudintermedius. J Bacteriol 193:2363–4236. doi: 10.1128/JB.00137-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tse H, Tsoi HW, Leung SP, Urquhart IJ, Lau SK, Woo PC, Yuen KY. 2011. Complete genome sequence of the veterinary pathogen Staphylococcus pseudintermedius strain HKU10-03, isolated in a case of canine pyoderma. J Bacteriol 193:1783–1784. doi: 10.1128/JB.00023-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palmieri C, Mingoia M, Varaldo PE. 2013. Unconventional circularizable bacterial genetic structures carrying antibiotic resistance determinants. Antimicrob Agents Chemother 57:2440–2441. doi: 10.1128/AAC.02548-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.