Abstract
The aim of this study was to assess the impact of ciprofloxacin, clindamycin, and placebo administration on culturable Gram-negative isolates and the antibiotic resistance genes they harbor. Saliva and fecal samples were collected from healthy human volunteers before and at intervals, up to 1 year after antibiotic administration. Samples were plated on selective and nonselective media to monitor changes in different colony types or bacterial species. Following ciprofloxacin administration, there was a decrease of Escherichia coli in feces and after clindamycin administration a decrease of Bacteroides in feces and Leptotrichia in saliva, which all returned to pretreatment levels within 1 to 4 months. Ciprofloxacin administration also resulted in an increase in ciprofloxacin-resistant Veillonella in saliva, which persisted for 12 months. Additionally, 949 aerobic and anaerobic isolates purified from ciprofloxacin- and clindamycin-containing plates were screened for the presence of resistance genes. Resistance gene carriage was widespread in isolates from all three treatment groups, and no association was observed between genes and antibiotic administration. Although the anaerobic component of the microbiota was not a major reservoir of aerobe-associated antimicrobial resistance (AMR) genes, we detected the sulfonamide resistance gene sul2 in anaerobic isolates. The longitudinal nature of the study allowed identification of distinct Escherichia coli clones harboring multiple resistance genes, including one carrying an extended-spectrum β-lactamase blaCTX-M group 9 gene, which persisted in the gut for up to 4 months. This study provided insight into the effects of antibiotic administration on healthy microbiota and the diversity of resistance genes harbored therein.
INTRODUCTION
The advent of antimicrobial chemotherapy for the treatment of bacterial disease has revolutionized medicine and made a significant contribution to reductions in human morbidity and mortality. However, antimicrobial use selects for resistance in bacteria, and multidrug resistance has become a significant public health issue worldwide (1). Furthermore, antibiotic treatment can alter the composition of the natural microbiota and the prevalence of antimicrobial resistance (AMR) genes (2–6). The human microbiota also serves as a reservoir of AMR genes (7–12) that may be acquired by susceptible pathogenic or commensal bacteria, contributing to the spread of multidrug-resistant bacteria.
In order to discover new means of limiting the development of resistance and the transmission of antibiotic-resistant strains, a greater understanding of how the administration of particular antibiotics affects the carriage of AMR genes in the individual bacteria present in the human microbiota is required. This is of particular concern for Gram-negative bacteria such as Escherichia coli that are currently the most common causative organisms in infections such as urinary tract infections (13). Both bacterial clonal expansion and acquisition of mobile elements such as plasmids have resulted in increased multidrug resistance (14), and the problem of multidrug resistance in Gram-negative bacteria is worrying as there are few applicable new antibiotics under advanced development for this group (1). Two antibiotics that are active and commonly used against certain Gram-negative bacteria are ciprofloxacin and clindamycin. Ciprofloxacin is a fluoroquinolone antibiotic, with a broad spectrum of activity targeting aerobic Gram-negative and Gram-positive bacteria but with low potency against many anaerobic bacteria. Reduced susceptibility to ciprofloxacin is mediated by mutations in the topoisomerase genes targeted by the drug, by alterations in drug efflux, and by resistance genes (15). Clindamycin is a lincosamide antibiotic that is active against many aerobic Gram-positive cocci and a range of anaerobic Gram-positive and Gram-negative bacteria (16). Resistance mechanisms in Gram-negative anaerobes include mutations in the 23S rRNA gene targeted by the drug, alterations in efflux, and erythromycin resistance methylase genes (16). Gram-negative aerobes such as Enterobacteriaceae are impervious to clindamycin and so are inherently resistant (16).
The aim of this study was to examine the effects of administration of ciprofloxacin or clindamycin on Gram-negative bacteria cultured from the indigenous microbiota of healthy humans and the diversity of AMR genes they harbor. Aerobic and anaerobic isolates were examined in this study as both have the potential to serve as reservoirs and routes for transmission of AMR genes in the human microbiota (9). We were particularly interested to know if the AMR genes commonly associated with aerobic bacteria can be present in anaerobes, which can then act as a reservoir and possible source for dissemination of these genes.
MATERIALS AND METHODS
Study participants, sampling, and bacterial isolation.
The study was approved by the Ethics Committee of the Karolinska Institutet (Stockholm, Sweden), according to Swedish law. Verbal and written consent to participate in this study was obtained from the volunteers. Volunteer recruitment, sample collection, and bacterial isolation and purification were performed at the Karolinska Institutet as part of the Antiresdev consortium (http://www.ucl.ac.uk/antiresdev). In brief, 30 healthy adult volunteers, who had not received antimicrobials in the previous 3 months, were divided randomly into three groups of 10 volunteers. One group was administered ciprofloxacin (500 mg twice a day [b.i.d.]) for a 10-day period, one group was administered clindamycin (150 mg 4 times a day [q.i.d.]) for a 10-day period, and one group was a control group that received placebo only. All volunteers in the placebo group successfully completed the study. Nine volunteers from both the ciprofloxacin and clindamycin groups successfully completed the study (one volunteer left the study due to personal reasons and the other left due to treatment with other antibiotics). Fecal and saliva samples were collected from each volunteer at day 0, before administration; at day 11, immediately after treatment completion; and then at 1, 2, 4, and 12 months following treatment completion.
Saliva (∼0.5 ml) and fecal (∼1 g) samples were suspended in prereduced peptone-yeast extract medium and diluted to 10−7, and 100 μl of each dilution was inoculated on selective and nonselective agar plates: blood agar for total aerobes and anaerobes, cysteine lactose electrolyte-deficient agar for the detection of Enterobacteriaceae, kanamycin-vancomycin-blood agar for the cultivation of Bacteroides and Prevotella species, neomycin-vancomycin-blood agar for the cultivation of fusobacteria, and veillonella agar for the cultivation of Veillonella cocci, as described previously (4). In addition, all samples were inoculated onto antibiotic plates, containing either ciprofloxacin at 1 mg/liter or clindamycin at 4 mg/liter, which according to the EUCAST Clinical Breakpoints (v. 3.1) would enable the recovery of intermediate and resistant isolates. After incubation, different colony types were counted from all plates to establish the number of CFU per gram of saliva or feces. The effect of antibiotic administration on each colony type was investigated by grouping volunteers by treatment and plotting the geometric mean of the CFU obtained at each time point (a count of zero was assigned a value of 1 when the geometric means were calculated).
Representative colony types from antibiotic-containing plates were subcultured to purity, and bacteria were identified to the genus or species level, according to the Manual of Clinical Microbiology (17); subsequently, they were screened for AMR gene content by microarray (see below). Aerobic Gram-negative isolates were further identified to the species/genus level using matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF) (18) and tested for antibiotic susceptibility by the Vitek 2 system. On average, 1 aerobic Gram-negative isolate and 4 anaerobic Gram-negative representative isolates were picked and purified for each volunteer, at each visit, and tested by microarray.
DNA extraction and microarray.
Aerobic bacteria were grown at 37°C on blood agar plates, and anaerobic bacteria were grown on fastidious anaerobe agar as described previously (19). For extraction of DNA, a 10-μl loopful of bacteria was lysed, as described previously (20). The DNA was labeled in a linear multiplex reaction using primers described previously (19, 20) and some newly designed primers (see below). For the anaerobic samples, the labeled DNA was pooled because previous work (19) with anaerobic bacteria had shown that carriage of resistance genes represented on the microarray was low; each pool comprised material from one volunteer at a single time point (one to six labeling reactions per pool). The effect of pooling on the microarray sensitivity was assessed using pools of labeled DNA from isolates of known gene content and shown to be minimal (data not shown); this method was used because it was more cost-effective and time-efficient but equally sensitive. Labeled DNA was hybridized to immobilized probes present on the microarray using the HybPlus kit buffers (Alere Technologies, Jena, Germany) with adaptation to the manufacturer's protocol (see File S1 in the supplemental material). Microarray signals were detected with the ArrayMate device (Alere Technologies) using IconoClust software (standard version; Alere Technologies). The mean signal intensities of two replicate spots per probe were used for analysis, and values of ≥0.5 were considered positive.
Most primers and probes included for this microarray work were described previously (19, 20), with the exception of those for blaOXY and cfxA, which were validated during this study. New primers and probes for the genes blaCTX-M group 1, blaCTX-M group 2, qepA, qnrB, qnrC, and qnrS were also included and validated in this study. The new probes and primers (see Table S1 in the supplemental material) were designed as described previously (20), and their specificities were tested using control strains, in which the presence of the probe targets was verified by PCR and sequencing, using the PCR primers described in Table S1. Several probes on the microarray gave false-positive results when the pools of anaerobic isolates were tested, as determined by PCR (data not shown), and these probes were excluded from analysis. The false-positive results may have arisen because the microarray probes had been designed primarily using gene sequences from aerobic bacteria.
Pulsed-field gel electrophoresis.
Eighteen isolates were selected for analysis by pulsed-field gel electrophoresis (PFGE) using XbaI restriction endonuclease according to the PulseNet protocol for E. coli O157 (Centers for Disease Control and Prevention, Atlanta, GA, USA; http://www.cdc.gov/pulsenet/pathogens/index.html). PFGE profiles were clustered with the Dice similarity coefficient and unweighted pair group method using average linkages (UPGMA) clustering analysis using BioNumerics software (v. 3.00; Applied Maths Inc.). Isolates with ≥85% similarity were considered the same clone.
RESULTS
Impact of antibiotic administration on isolate counts.
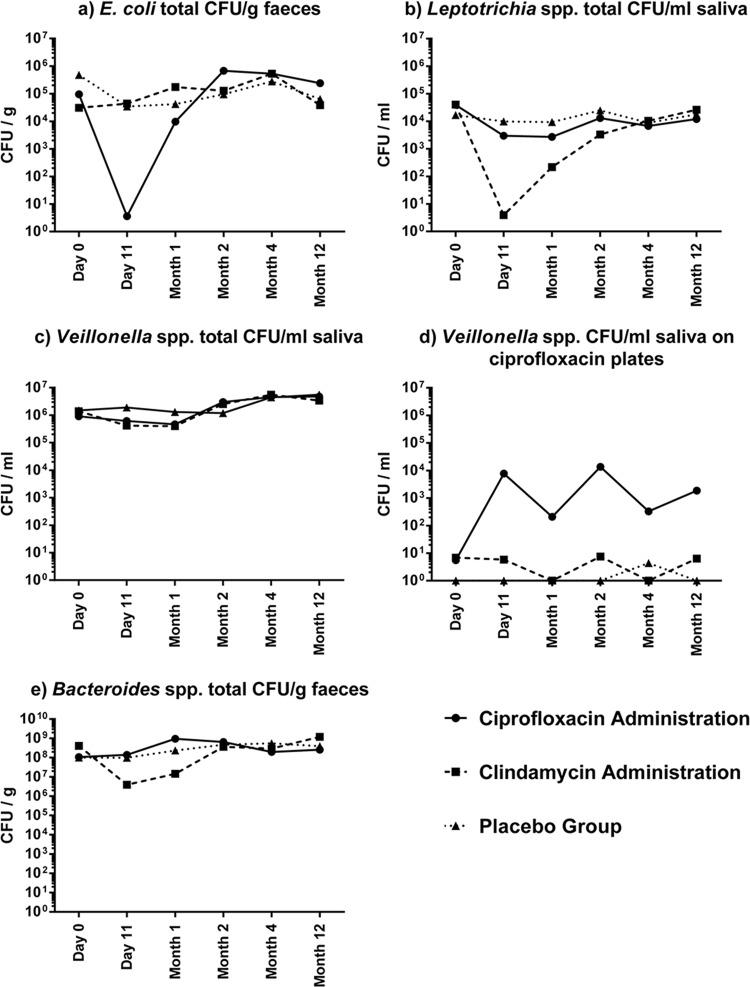
The impact of antibiotic administration on Gram-negative bacteria was determined from samples collected at six time points in a yearlong study using a range of selective, nonselective, and antibiotic-containing plates. The effect was assessed by grouping volunteers by treatment and plotting the geometric mean of the CFU obtained for each colony type at each visit. Only bacterial species showing notable changes, including those on antibiotic-containing plates, over the yearlong study, are reported.
This included a 5-log decrease in the geometric mean for E. coli CFU/g feces observed at day 11 in the ciprofloxacin group (i.e., immediately following the completion of ciprofloxacin administration), which was not observed in the clindamycin and placebo groups (Fig. 1a). Indeed, E. coli was present in only a single volunteer (volunteer P914) at day 11 in the ciprofloxacin group. The E. coli was ciprofloxacin resistant and was purified from the corresponding ciprofloxacin-containing plate for examination by DNA microarray and PFGE (see “Detection of antibiotic resistance genes by microarray” and “Persistence of AMR genes in the aerobic microbiota” below). By month 1, the E. coli population had largely recovered to preantibiotic administration levels in participants in this group (Fig. 1a), but throughout the year-long study, the geometric means of E. coli strains present on the ciprofloxacin plates remained very low (<20 CFU/g) in all three treatment groups (data not shown). Similarly, in the clindamycin group there was a 4-log decrease in Leptotrichia spp. at day 11 following clindamycin administration, which recovered to preadministration levels by month 4, and this effect was not observed in the ciprofloxacin and placebo groups (Fig. 1b). For all three treatment groups, Leptotrichia spp. were not present on clindamycin-containing plates at any time point.
FIG 1.
Impact of antibiotic administration on the geometric mean CFU of fecal E. coli (a), saliva Leptotrichia spp. (b), saliva Veillonella spp. (c), and saliva Veillonella spp. (d) on ciprofloxacin-containing plates and of fecal Bacteroides spp. (e).
Veillonella spp. were cultured from the saliva samples of all volunteers at every time point, and the geometric means of total CFU did not vary markedly between time points (Fig. 1c). Additionally, Veillonella spp. were rarely present on ciprofloxacin-containing plates before treatment and in the clindamycin and placebo groups following treatment, thus giving very low geometric means (Fig. 1d). However, following ciprofloxacin administration, the geometric means of Veillonella species CFU/g on ciprofloxacin-containing plates increased by up to 4 log, and this effect (with some variation) persisted for the duration of the 12-month study period (Fig. 1d).
In the fecal samples, following clindamycin administration there was a 2-log decrease in the counts of Bacteroides spp., which returned to preadministration levels by month 2, an effect not observed in the ciprofloxacin and placebo groups (Fig. 1e). Notable changes in the proportions of volunteers positive for the genera Prevotella and Fusobacteria were not observed following antibiotic administration (data not shown). Also, the impact of antibiotic administration on Klebsiella, Citrobacter, and Proteus isolates present in feces was not determined as too few volunteers were positive for these genera to enable analysis (data not shown).
Diversity of isolates purified from antibiotic-containing plates.
Representative isolates were purified from antibiotic-containing plates, based on colony morphology, for analysis of resistance gene carriage by microarray (see below) and species/genus identification by MALDI-TOF and biochemical characterization. These isolates were expected to be intermediate and fully resistant, according to EUCAST breakpoints. A total of 949 aerobic and anaerobic Gram-negative isolates were purified from antibiotic plates, with approximately one-third recovered from each administration group (see Table S2 in the supplemental material). From fecal samples, a total of 195 Gram-negative aerobic isolates were purified, of which 174 were recovered from clindamycin-containing plates. There was an average of 1.2 isolates per volunteer per visit, except at day 11 immediately following administration for the ciprofloxacin-administered group for which only one isolate was recovered from the only positive volunteer, P914. The most common Gram-negative aerobic species purified from feces was E. coli (166 isolates), while other species were recovered at low occurrence (see Table S3 in the supplemental material). Four Gram-negative aerobic isolates were recovered from saliva samples. All were recovered from the same volunteer and identified as Klebsiella pneumoniae.
A total of 509 anaerobic Gram-negative isolates were recovered from the fecal samples, the average number of isolates per volunteer per visit was 3.0 (range, 0 to 10), and all were identified as Bacteroides spp. For the saliva samples, the average number of Gram-negative anaerobic isolates per volunteer per visit was 1.4 (range, 0 to 5), and these were identified as members of the genera Prevotella, Fusobacteria, Veillonella, or Leptotrichia (29 isolates were not fully identified).
Detection of antibiotic resistance genes by microarray. (i) Antibiotic resistance genes in aerobic isolates.
All resistant and intermediate isolates purified from antibiotic-containing plates (see “Diversity of isolates purified from antibiotic containing plates” above) were screened for the carriage of AMR genes to identify ciprofloxacin and/or clindamycin resistance genes and to determine other resistance genes that are coselected with clindamycin and ciprofloxacin resistance. For this screen, we used a DNA microarray previously developed and updated by our group (19, 20) that targets a wide range of clinically relevant acquired resistance genes which are frequently located on mobile genetic elements such as plasmids, transposons, conjugative elements, and integrases.
The four K. pneumoniae isolates recovered from saliva samples were positive for the β-lactamase blaSHV gene only. For the fecal aerobes, at least one AMR gene was detected by microarray in 133 of 195 isolates, and in total 36 different genes were detected (see Table S4 in the supplemental material). The most commonly detected gene was blaTEM, present in 77 isolates, followed by sul2, strB, tet(A), tet(B), dfrA17, strA, sul1, blaOXA-2, and aadA4. The remaining AMR genes were detected in less than 10% of isolates (see Table S4) and included a blaCTX-M group 9 extended-spectrum β-lactamase (ESBL) which was detected in two isolates from consecutive visits from volunteer P910 (clindamycin-administered group) and a blaCTX-M group 1 gene which was detected in two isolates, each from a different volunteer (one of whom was P910). β-Lactamase genes were detected in many of the non-E. coli isolates, e.g., blaSHV in K. pneumoniae, blaCMY in Citrobacter freundii, and blaACC in Hafnia alvei. The two integrase genes intI1 and intI2, associated with class 1 and class 2 integrons, respectively, were also detected; intI1 was present in 49 isolates and intI2 in 11 isolates.
The AMR genes detected were grouped by their mode of activity (as classified previously [20]) and encoded resistances to eight antibiotic classes: aminoglycosides, β-lactams, macrolides, phenicols, quinolones, sulfonamides, tetracyclines, and trimethoprim (see Table S4 in the supplemental material). Sixty-one isolates encoded resistances to 1 or 2 antibiotic classes, and 72 isolates encoded resistances to 3 or more classes. Genes encoding resistances to β-lactam, tetracycline, and trimethoprim classes were present at every visit for all groups, as was the class 1 integrase gene intI1. Genes encoding resistances to the aminoglycoside and sulfonamide classes were also common, although not detected at day 0 and day 11 for the ciprofloxacin group. Genes encoding resistances to phenicol, macrolide, and quinolone classes were rarely detected in any treatment group.
Six plasmid-mediated quinolone resistance (PMQR) genes, qepA, qnrA, qnrB, qnrC, qnrS, and aac6-Ib, were represented on the microarray. Of these genes, only qnrB was detected in three C. freundii isolates, and each was susceptible to ciprofloxacin by the Vitek 2 system (data not shown). Additionally, 36/195 isolates (33 E. coli and 3 Comamonas kerstersii) were found to be ciprofloxacin resistant by the Vitek 2 system but showed no ciprofloxacin resistance gene by microarray; sequencing showed that all 33 E. coli isolates harbored a C248T point mutation in the gyrA gene, but the 3 Comamonas kerstersii isolates were not explored any further (data not shown). These isolates were recovered from all three treatment groups, with volunteer P914 (ciprofloxacin administration) providing 10 E. coli isolates and volunteer P929 (placebo group) providing 9 E. coli isolates; resistant isolates were recovered at day 0 from both volunteers. The clindamycin resistance genes ereA and erm(B) were represented on the microarray, but no Gram-negative aerobic isolate was positive for either gene.
(ii) Antibiotic resistance genes in anaerobic isolates.
The anaerobic isolates were tested in pools as microarray sensitivity was not compromised by pooling (data not shown), and the results were analyzed as percentages of pooled isolates positive for each gene (see Table S5 in the supplemental material). Four genes, cfxA, erm(B), sul2, and tet(Q), were detected in both fecal and saliva samples. An additional three genes, cblA, cepA, and tet(X), were detected only in fecal isolates. The sulfonamide resistance gene sul2 was detected by microarray (see Table S5) and verified by PCR (data not shown) in anaerobic isolates from three volunteers. In one volunteer, sul2-positive Bacteroides isolates were recovered at every visit except day 11. For clindamycin resistance, ereA was not detected in any pool of anaerobic isolates, while erm(B) was detected in all three treatment groups. Although there was a greater percentage of erm(B)-positive fecal samples in the clindamycin treatment group, there was no statistically significant difference in the number of erm(B)-positive volunteers following treatment using Fisher's exact test (not shown).
(iii) Persistence of AMR genes in the aerobic microbiota.
After the initial analysis of AMR genes present in fecal aerobes, we noted that in at least three volunteers there were core sets of AMR genes that were present at successive visits and these isolates clustered together in a dendrogram looking at the diversity of AMR genes. To determine whether this was due to the same E. coli clone being isolated at each visit, PFGE was performed on 18 representative isolates. Nine isolates from volunteer P914 from day 0 to day 120 had identical PFGE profiles and AMR gene content (blaTEM, dfrA17, tet(B), and intI1) (Fig. 2). The same four genes were also present in a single isolate from volunteer P918 (clindamycin treatment group; month 12), but this isolate had a different PFGE profile (Fig. 2). This isolate had a PFGE profile identical to that of a single E. coli isolate from volunteer P914 at month 12 but with a different AMR gene content [blaOXA-2, blaTEM, and tet(A)]. Seven isolates from volunteer P910 clustered into four groups by AMR gene content and PFGE profile. Each distinct AMR genotype could be matched to a distinct PFGE profile, indicating the presence of several different E. coli clones with discrete AMR gene content in this participant over the time period studied (Fig. 2). For one clone, two isolates were recovered at consecutive visits (months 1 and 2) and contained a blaCTX-M group 9 gene, along with blaOXA-2, strA, strB, sul2, and tet(A). This volunteer also possessed a single isolate that harbored 11 AMR genes that can confer resistance to six antibiotic classes (aadA4, strA, strB, blaCTX-group M1, blaTEM, intI1, mphA, sul1, sul2, tet(A), dfrA17, and dfrA19). In the placebo group, E. coli isolates containing sul2, tet(B), dfrA01, aadA1, and intl2 were recovered from volunteer P905 at five successive visits (day 0 to month 4); however, their clonal relationship was not examined by PFGE.
FIG 2.
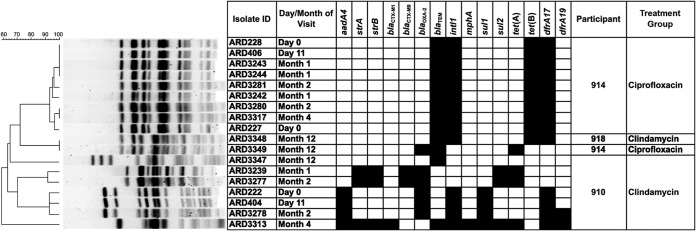
PFGE and antibiotic resistance gene profile of E. coli isolates recovered from the feces of three volunteers.
DISCUSSION
In this study, we have examined the effect of antibiotic administration on culturable Gram-negative bacterial populations and the carriage of AMR genes in isolates purified from antibiotic-containing plates from healthy human volunteers. Our data clearly showed that antibiotic administration resulted in changes to the Gram-negative component of the saliva and fecal microbiota. One notable change was that immediately following ciprofloxacin administration, E. coli was cultured from the feces of only one volunteer. This suggested that E. coli isolates present in all but one participant in the ciprofloxacin administration group were susceptible to ciprofloxacin and so were not recovered from other participants immediately following its administration. However, by month 1, E. coli cells in the gut flora had recovered significantly to pretreatment levels, although the numbers of ciprofloxacin-resistant E. coli isolates remained low throughout the study. Also, following ciprofloxacin administration, there was an increase in ciprofloxacin-resistant Veillonella spp., and the level remained elevated for the remainder of the study after antibiotic administration. The MIC range of Veillonella to ciprofloxacin can be broad (21), and it is possible that there was selection for more resistant strains as a consequence of ciprofloxacin treatment, which merits further work in the future. The significant decrease in Leptotrichia spp. in saliva samples following clindamycin administration was expected because these bacteria, commonly present in saliva, are generally susceptible to clindamycin (22). The decrease in Bacteroides spp. following clindamycin administration has been observed previously (3).
To gain insight into resistance gene carriage by clindamycin- and ciprofloxacin-resistant bacteria, microarray analysis was performed on representative Gram-negative aerobic and anaerobic isolates purified from antibiotic-containing plates from each group. The purified isolates represented many of the natural inhabitants of the human oral and fecal microbiota. All of the fecal anaerobes were identified as Bacteroides, one of the most abundant genera in the human gut (12, 23). The majority of the isolated fecal aerobes were E. coli strains, with other Gram-negative species recovered in low numbers, reflecting the normal microbial diversity in the human gut (5, 23). The saliva samples mainly yielded Gram-negative isolates that grew anaerobically and were from genera naturally resident in the human oral microbiota (12, 24). Only one aerobic isolate per participant per time point was selected based on colony morphology, but we believe this was representative of the most prevalent isolate at that time point, and the recovery of clonal isolates at different time points supports this view. Furthermore, in a similar study where up to 15 aerobic isolates were purified per participant per time point, we have shown that in many instances the same clones had been selected multiple times (6).
In this study, we found no association between the presence of AMR genes and administration of ciprofloxacin or clindamycin. This is similar to results we have reported recently for minocycline administration, which resulted in no alternation of gene carriage, but unlike the effect of amoxicillin administration, which resulted in an increase in levels of the corresponding resistance gene blaTEM (6). Instead, all treatment groups, including the placebo group, possessed a wide diversity of AMR genes. For example, the aerobic isolates contained AMR genes that spanned eight antibiotic classes and approximately 37% of these isolates encoded resistance to three or more different antibiotic classes. This indicates that multidrug resistance is possibly common in the microbiota of the healthy human population from Sweden, a finding which we previously noted from the United Kingdom (6). Commonly detected genes included blaTEM and sul2, both of which usually reside on plasmids or transposons and are widespread in the human microbiota (5, 6, 8, 10, 12, 25). Many of the other microarray-positive genes have been described previously in the microbiota of healthy humans (5, 6, 8, 10, 12, 25). The majority of the β-lactamase genes detected in non-E. coli aerobes are likely genes chromosomally located in these species, e.g., blaSHV in K. pneumoniae, blaCMY in C. freundii, and blaACC in H. alvei (26). In this study, 36 ciprofloxacin-resistant isolates were recovered, but none harbored any PMQR gene represented on the microarray; all 33 E. coli isolates possessed a mutation in the gyrA gene. The only potential PMQR gene detected by microarray was qnrB in three C. freundii isolates, but in this species qnrB can be chromosomally located (27). The majority of aerobic Enterobacteriaceae isolates were recovered from clindamycin plates; clindamycin is an antibiotic to which they are inherently resistant and so these are representative of the baseline population. The fact that none harbored any PMQR gene present on the microarray probably reflects the generally low prevalence of PMQR genes reported previously from epidemiological studies (28).
Two E. coli isolates possessed a blaCTX-M group 1 ESBL, and two carried a blaCTX-M group 9 ESBL. The prevalence of both these ESBL genes is increasing in clinical isolates (29), and healthy humans can be carriers (7, 30). The blaCTX-M group 9 positive isolates were clonal and recovered from the same volunteer on successive visits. Interestingly, the single isolate recovered at day 11 in the ciprofloxacin group was demonstrated by PFGE to be present at preadministration and persisted until month 4 of the study. In addition to the gyrA mutation (conferring ciprofloxacin resistance), this isolate harbored three other antibiotic resistance genes and had a multidrug-resistant phenotype. The persistence of these clones demonstrates that strains encoding multidrug resistance can be maintained in the gut of healthy humans for periods ranging from at least 1 to 4 months. The presence of resident E. coli strains that remain in the gut microbiota for months or years is well established (5, 31), but AMR gene carriage is rarely described. Persistence of AMR genes can also arise from the transfer of genes between bacteria via mobile genetic elements, such as plasmids. The presence of a class 1 and/or 2 integron in 29% of aerobic isolates indicates a potential for the transfer of genes via antibiotic resistance gene cassettes carried on integron-bearing transposons.
The anaerobic isolates were microarray positive for a smaller diversity of genes than aerobes, although this may reflect the smaller range of anaerobe AMR genes represented on the microarray. The most prevalent AMR gene detected in the anaerobes was tet(Q), which is the most common tetracycline resistance gene to be detected in Bacteroides spp. in healthy humans (9, 32) and is also reported in oral Prevotella species isolates (33). The tetracycline resistance gene tet(X) was detected only in Bacteroides isolates, where it can be located on mobile genetic elements (34). Three β-lactamase genes (cblA, cepA, and cfxA) were detected in Bacteroides species isolates and are common in this genus (32). cfxA was also present in some saliva isolates and has been previously reported in oral clinical isolates of Prevotella spp. (35). The clindamycin resistance gene erm(B) was detected in anaerobic isolates from fecal samples in all three treatment groups, but there was no significant association between the erm(B)-positive volunteers and treatment group. Löfmark et al. (3), reported an increase in the detection of erm(F) and erm(G) in Bacteroides spp. following clindamycin treatment but did not detect erm(B) in their study. However, erm(B) has been described in the microbiota of healthy humans previously (2, 12) and has a large Gram-negative host range, including Fusobacterium spp. and Bacteroides spp. (36). Therefore, the absence of any association in our study may be due to differences in the study method or the subjects.
The genes represented on the microarray encompass not only prevalent resistances present in anaerobic bacteria but also a large selection of AMR genes usually associated with aerobic Gram-negative bacteria. An additional purpose of this work was to determine if anaerobes can be a reservoir for these genes that are commonly present in Enterobacteriaceae. It is interesting to note, therefore, that in this study these genes were not detected in the anaerobic isolates, with the single exception of sul2 (encoding sulfonamide resistance). The sul2 gene was detected in anaerobic isolates from three volunteers and was present in Bacteroides isolates from one volunteer for a year. To date, sul2 has rarely been reported in Bacteroides species (11, 19), and its mode of carriage in this genus remains unknown. These data therefore support recent work (19) which suggests that the anaerobic component of the microbiota is not a major reservoir of aerobe-associated AMR genes.
An alternative method to investigate the diversity of AMR genes present in the microbiome is sequenced-based metagenomics using next-generation sequencing. These methods can provide a description of changes in bacterial composition using the 16S rRNA gene microbial profile and can also look at changes in AMR genes present in the microbiome following antibiotic administration. The general conclusions of one such study (2) is similar to ours in that certain bacterial species declined immediately after treatment and subsequently most recovered to pretreatment levels. The study also found elevated levels of ermB genes in treated groups but did not look at any other resistance gene. Identifying AMR genes by homology searches from shotgun sequences of the metagenome is still a challenging and expensive task to perform. In contrast, using AMR microarrays on either purified isolates (as in this study) or the total microbiome (12) to look for resistances is still much easier and cheaper, although has other limitations.
Therefore, in this study we have shown that even in the absence of recent exposure to antibiotics, AMR genes are commonly detected in isolates from healthy adults and probably reside in this milieu for months, if not longer. In the future, whether by microarrays or whole-genome sequencing, characterizing the molecular basis of resistance in these isolates will continue to prove vital for monitoring the dissemination of antibiotic-resistant Gram-negative bacteria in humans and the environment and controlling their rise.
Supplementary Material
ACKNOWLEDGMENTS
The skillful technical assistance of Ann-Chatrin Palmgren, Monica Sörensson, and Elisabeth Wahlund from Karolinska Institutet in performing the microbiological and biochemical analyses is gratefully acknowledged.
The research leading to these results has received funding from the European Union Seventh Framework Programme (FP7/2007-2013) under grant agreement 241446 (project Antiresdev). Funding was also provided by the Department for Innovation, University and Skills (project number DT0041; Public Sector Research Exploitation Fund: Fourth Round, MicroArray [Capacity Building]).
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00068-15.
REFERENCES
- 1.Freire-Moran L, Aronsson B, Manz C, Gyssens IC, So AD, Monnet DL, Cars O, ECDC-EMA Working Group. 2011. Critical shortage of new antibiotics in development against multidrug-resistant bacteria-Time to react is now. Drug Resist Updat 14:118–124. doi: 10.1016/j.drup.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 2.Jakobsson HE, Jernberg C, Andersson AF, Sjolund-Karlsson M, Jansson JK, Engstrand L. 2010. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS One 5:e9836. doi: 10.1371/journal.pone.0009836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Löfmark S, Jernberg C, Jansson JK, Edlund C. 2006. Clindamycin-induced enrichment and long-term persistence of resistant Bacteroides spp. and resistance genes. J Antimicrob Chemother 58:1160–1167. doi: 10.1093/jac/dkl420. [DOI] [PubMed] [Google Scholar]
- 4.Nord CE, Sillerstrom E, Wahlund E. 2006. Effect of tigecycline on normal oropharyngeal and intestinal microflora. Antimicrob Agents Chemother 50:3375–3380. doi: 10.1128/AAC.00373-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nyberg SD, Osterblad M, Hakanen AJ, Lofmark S, Edlund C, Huovinen P, Jalava J. 2007. Long-term antimicrobial resistance in Escherichia coli from human intestinal microbiota after administration of clindamycin. Scand J Infect Dis 39:514–520. doi: 10.1080/00365540701199790. [DOI] [PubMed] [Google Scholar]
- 6.Kirchner M, Mafura M, Hunt T, Abu-Oun M, Nunez-Garcia J, Hu Y, Weile J, Coates A, Card R, Anjum MF. 2014. Antimicrobial resistance characteristics and fitness of Gram negative faecal bacteria from volunteers treated with minocycline or amoxicillin. Front Microbiol 5:722. doi: 10.3389/fmicb.2014.00722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Machado E, Coque TM, Canton R, Sousa JC, Peixe L. 2013. Commensal Enterobacteriaceae as reservoirs of extended-spectrum beta-lactamases, integrons, and sul genes in Portugal. Front Microbiol 4:80. doi: 10.3389/fmicb.2013.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bailey JK, Pinyon JL, Anantham S, Hall RM. 2011. Distribution of the blaTEM gene and blaTEM-containing transposons in commensal Escherichia coli. J Antimicrob Chemother 66:745–751. doi: 10.1093/jac/dkq529. [DOI] [PubMed] [Google Scholar]
- 9.Salyers AA, Gupta A, Wang Y. 2004. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol 12:412–416. doi: 10.1016/j.tim.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 10.Sommer MO, Dantas G, Church GM. 2009. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science 325:1128–1131. doi: 10.1126/science.1176950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang L, Kinkelaar D, Huang Y, Li Y, Li X, Wang HH. 2011. Acquired antibiotic resistance: are we born with it? Appl Environ Microbiol 77:7134–7141. doi: 10.1128/AEM.05087-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Card RM, Warburton PJ, MacLaren N, Mullany P, Allan E, Anjum MF. 2014. Application of microarray and functional-based screening methods for the detection of antimicrobial resistance genes in the microbiomes of healthy humans. PLoS One 9:e86428. doi: 10.1371/journal.pone.0086428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pallett A, Hand K. 2010. Complicated urinary tract infections: practical solutions for the treatment of multiresistant Gram-negative bacteria. J Antimicrob Chemother 65(Suppl 3):iii25–iii33. doi: 10.1093/jac/dkq298. [DOI] [PubMed] [Google Scholar]
- 14.Woodford N, Turton JF, Livermore DM. 2011. Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol Rev 35:736–755. doi: 10.1111/j.1574-6976.2011.00268.x. [DOI] [PubMed] [Google Scholar]
- 15.Jacoby GA. 2005. Mechanisms of resistance to quinolones. Clin Infect Dis 41(Suppl 2):S120–S126. doi: 10.1086/428052. [DOI] [PubMed] [Google Scholar]
- 16.Gold HS, Moellering RC Jr. 1999. Macrolides and clindamycin, p 291− 297 In Root RK, Waldvogel F, Corey L, Stamm WE (ed), Clinical infectious diseases: a practical approach. Oxford University Press, New York, NY. [Google Scholar]
- 17.Fitzgerald C, Collins M, van Duyne S, Mikoleit M, Brown T, Fields P. 2007. Multiplex, bead-based suspension array for molecular determination of common Salmonella serogroups. J Clin Microbiol 45:3323–3334. doi: 10.1128/JCM.00025-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seng P, Drancourt M, Gouriet F, La Scola B, Fournier PE, Rolain JM, Raoult D. 2009. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis 49:543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- 19.Kirchner M, Mafura M, Hunt T, Card R, Anjum MF. 2013. Antibiotic resistance gene profiling of faecal and oral anaerobes collected during an antibiotic challenge trial. Anaerobe 23:20–22. doi: 10.1016/j.anaerobe.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 20.Card R, Zhang J, Das P, Cook C, Woodford N, Anjum MF. 2013. Evaluation of an expanded microarray for detecting antibiotic resistance genes in a broad range of gram-negative bacterial pathogens. Antimicrob Agents Chemother 57:458–465. doi: 10.1128/AAC.01223-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whiting JL, Cheng N, Chow AW. 1987. Interactions of ciprofloxacin with clindamycin, metronidazole, cefoxitin, cefotaxime, and mezlocillin against gram-positive and gram-negative anaerobic bacteria. Antimicrob Agents Chemother 31:1379–1382. doi: 10.1128/AAC.31.9.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Couturier MR, Slechta ES, Goulston C, Fisher MA, Hanson KE. 2012. Leptotrichia bacteremia in patients receiving high-dose chemotherapy. J Clin Microbiol 50:1228–1232. doi: 10.1128/JCM.05926-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. 2005. Diversity of the human intestinal microbial flora. Science 308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keijser BJ, Zaura E, Huse SM, van der Vossen JM, Schuren FH, Montijn RC, ten Cate JM, Crielaard W. 2008. Pyrosequencing analysis of the oral microflora of healthy adults. J Dent Res 87:1016–1020. doi: 10.1177/154405910808701104. [DOI] [PubMed] [Google Scholar]
- 25.Blahna MT, Zalewski CA, Reuer J, Kahlmeter G, Foxman B, Marrs CF. 2006. The role of horizontal gene transfer in the spread of trimethoprim-sulfamethoxazole resistance among uropathogenic Escherichia coli in Europe and Canada. J Antimicrob Chemother 57:666–672. doi: 10.1093/jac/dkl020. [DOI] [PubMed] [Google Scholar]
- 26.Philippon A, Arlet G, Jacoby GA. 2002. Plasmid-determined AmpC-type beta-lactamases. Antimicrob Agents Chemother 46:1–11. doi: 10.1128/AAC.46.1.1-11.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacoby GA, Griffin CM, Hooper DC. 2011. Citrobacter spp. as a source of qnrB alleles. Antimicrob Agents Chemother 55:4979–4984. doi: 10.1128/AAC.05187-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strahilevitz J, Jacoby GA, Hooper DC, Robicsek A. 2009. Plasmid-mediated quinolone resistance: a multifaceted threat. Clin Microbiol Rev 22:664–689. doi: 10.1128/CMR.00016-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coque TM, Baquero F, Canton R. 2008. Increasing prevalence of ESBL-producing Enterobacteriaceae in Europe. Euro Surveill 13(47):pii:19044 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19044. [PubMed] [Google Scholar]
- 30.Kirchner M, Abuoun M, Mafura M, Bagnall M, Hunt T, Thomas C, Weile J, Anjum MF. 2013. Cefotaxime resistant Escherichia coli collected from a healthy volunteer; characterisation and the effect of plasmid loss. PLoS One 8:e84142. doi: 10.1371/journal.pone.0084142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tenaillon O, Skurnik D, Picard B, Denamur E. 2010. The population genetics of commensal Escherichia coli. Nat Rev Microbiol 8:207–217. doi: 10.1038/nrmicro2298. [DOI] [PubMed] [Google Scholar]
- 32.Wexler HM. 2007. Bacteroides: the good, the bad, and the nitty-gritty. Clin Microbiol Rev 20:593–621. doi: 10.1128/CMR.00008-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tribble GD, Garza JJ, Yeung VL, Rigney TW, Dao DH, Rodrigues PH, Walker CB, Smith CJ. 2010. Genetic analysis of mobile tetQ elements in oral Prevotella species. Anaerobe 16:604–609. doi: 10.1016/j.anaerobe.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park BH, Levy SB. 1988. The cryptic tetracycline resistance determinant on Tn4400 mediates tetracycline degradation as well as tetracycline efflux. Antimicrob Agents Chemother 32:1797–1800. doi: 10.1128/AAC.32.12.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iwahara K, Kuriyama T, Shimura S, Williams DW, Yanagisawa M, Nakagawa K, Karasawa T. 2006. Detection of cfxA and cfxA2, the beta-lactamase genes of Prevotella spp., in clinical samples from dentoalveolar infection by real-time PCR. J Clin Microbiol 44:172–176. doi: 10.1128/JCM.44.1.172-176.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Hoek AH, Mevius D, Guerra B, Mullany P, Roberts AP, Aarts HJ. 2011. Acquired antibiotic resistance genes: an overview. Front Microbiol 2:203. doi: 10.3389/fmicb.2011.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.