Abstract
IMP-8 metallo-β-lactamase was identified in Klebsiella pneumoniae sequence type 252 (ST252), isolated in a Portuguese hospital in 2009. blaIMP-8 was the first gene cassette of a novel class 3 integron, In1144, also carrying the blaGES-5, blaBEL-1, and aacA4 cassettes. In1144 was located on a ColE1-like plasmid, pKP-M1144 (12,029 bp), with a replication region of limited nucleotide similarity to those of other RNA-priming plasmids, such as pJHCMW1. In1144 and pKP-M1144 represent an interesting case of evolution of resistance determinants in Gram-negative bacteria.
TEXT
Acquired carbapenem-hydrolyzing metallo-β-lactamases (MBLs) are resistance determinants of increasing clinical importance in Gram-negative pathogens. Of these, enzymes mainly of the VIM, IMP, and NDM types have been encountered in Klebsiella pneumoniae and other Enterobacteriaceae. In contrast to blaNDM genes, blaVIM and blaIMP occur as gene cassettes in class 1 integrons or, more rarely, integrons of class 2 or 3 (1–3). Here, we report on a novel class 3 integron, In1144, identified in a K. pneumoniae sequence type 252 (ST252) isolate from Portugal. In1144 codes for IMP-8 (4) and two other β-lactamases, GES-5 and BEL-1 (5, 6). It is located on a new ColE1-like plasmid, pKP-M1144, which was entirely sequenced in the study.
In 2009, K. pneumoniae strain Kpn-1144 was recovered from a patient in a Portuguese intensive care unit (ICU) of a hospital participating in the European Union (EU)-funded project MOSAR (7). During MOSAR, 17,945 patients in 18 clinical sites in Europe and Israel were screened for rectal carriage of expanded-spectrum cephalosporin-resistant Enterobacteriaceae and were tested also for carbapenem susceptibility (7, 8). Kpn-1144 was extended-spectrum β-lactamase (ESBL) positive by the double-disk synergy test (9) and showed reduced susceptibility to carbapenems, according to the EUCAST screening cutoffs (10). The isolate was positive in the MBL EDTA double-disk synergy test (11), and carbapenemase production was confirmed by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (12). Table 1 presents the antimicrobial susceptibility data for Kpn-1144 determined by broth microdilution (13) and interpreted using the EUCAST criteria (http://www.eucast.org/). Of note, the MICs of carbapenems were relatively low (0.75 to 1 μg/ml).
TABLE 1.
Antimicrobial susceptibility of K. pneumoniae Kp-1144 and the E. coli DH5α transformant harboring the IMP-8-encoding plasmid pKP-M1144
| Strain | MIC (μg/ml) ofa: |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMX | AMC | PIP | TZP | CTX | CAZ | FEP | ATM | IPM | MEM | ETP | GEN | AMK | CHL | CST | SXT | CIP | |
| K. pneumoniae Kpn-1144 | >256 | 24 | >64 | 32 | >8 | >32 | 16 | 48 | 0.75 | 1 | 0.5 | 4 | 8 | >32 | 0.25 | 8 | 0.5 |
| E. coli DH5α (pKP-M1144) | >256 | 24 | 64 | 8 | >8 | >32 | 16 | 2 | 0.5 | 0.5 | 0.25 | 0.5 | 1 | 2 | 0.25 | 1 | ≤0.12 |
| E. coli DH5α | 4 | 2 | 2 | 0.5 | ≤0.12 | ≤0.25 | ≤0.12 | ≤0.12 | ≤0.12 | ≤0.12 | ≤0.12 | 0.12 | 0.5 | ≤1 | ≤0.12 | 1 | ≤0.12 |
AMX, amoxicillin; AMC, amoxicillin-clavulanate (inhibitor fixed at 2 μg/ml); PIP, piperacillin; TZP, piperacillin-tazobactam (inhibitor fixed at 4 μg/ml); CTX, cefotaxime; CAZ, ceftazidime; FEP, cefepime; ATM, aztreonam; IPM, imipenem; MEM, meropenem; ETP, ertapenem; GEN, gentamicin; AMK, amikacin; CHL, chloramphenicol; CST, colistin; SXT, trimethoprim-sulfamethoxazole; CIP, ciprofloxacin.
PCR screening for various MBL genes (14, 15) followed by sequencing revealed the presence of the blaIMP-8 gene (4) in Kpn-1144. The multilocus sequence typing (MLST) analysis (16) classified the isolate into ST252. K. pneumoniae ST252 was originally identified in the United States in 2007 (http://bigsdb.web.pasteur.fr/klebsiella/klebsiella.html), but recently, it was also found among VIM-1-producing isolates from different health care institutions in Barcelona, Spain (8, 17).
Attempts to transfer β-lactam resistance from Kpn-1144 to rifampin-resistant (Rifr) Escherichia coli strain A15 by conjugation were unsuccessful, in contrast to electroporation of E. coli strain DH5α with purified plasmid DNA of Kpn-1144 (Qiagen maxi kit; Qiagen, Hilden, Germany). Transformants were selected on Luria-Bertani agar plates with ampicillin (50 μg/ml), confirmed to be IMP producers by PCR (14), and tested for antimicrobial susceptibility (Table 1). The plasmid location of the blaIMP-8 gene was demonstrated by the S1 nuclease analysis of Kpn-1144 and its transformant (18), followed by hybridization with a digoxigenin-labeled blaIMP probe. S1 profiling revealed multiple plasmids in Kpn-1144 comprising molecules of ∼15 kb, ∼100 kb, ∼170 kb, ∼230 kb, and ∼340 kb, of which only the ∼15-kb plasmid was also in the transformant and hybridized with the blaIMP probe (results not shown). This plasmid, designated pKP-M1144, was nontypeable by PCR-based replicon typing (PBRT) (19), and its whole nucleotide sequence was determined. Sequencing, assembling of the reads, filling of sequence gaps, and analysis and annotation of the plasmid sequence were performed as described previously (20).
Plasmid pKP-M1144 is 12,029 bp in size, with an average G+C content of 51.5%. Analysis of the sequence revealed that blaIMP-8 is the first gene cassette of a unique class 3 integron structure, In1144 (Fig. 1), composed of 4,679 bp (nucleotides [nt] 1 to 4679). Upstream of blaIMP-8, the intI3 gene encoding the class 3 integrase was identified (21). The pKP-M1144 intI3 exhibited 100% sequence identity to the intI3 associated with blaGES-1 in the IncQ-type plasmid pQ7 from E. coli strain TB7 from Switzerland (22) and 99% identity to that with blaIMP-1 from Serratia marcescens strain AK9373 from Japan (23). Downstream of blaIMP-8, three other cassettes were found. The second one was blaGES-5, encoding the ESBL GES-5 that exhibits weak carbapenemase activity (5). This was followed by blaBEL-1, specifying the prototype enzyme of the ESBL family BEL, observed only in Pseudomonas aeruginosa so far (6). The last cassette was aacA4, encoding the acetyltransferase AAC(6′)-Ib, which confers resistance to tobramycin, netilmicin, and amikacin (24). A putative promoter (TAGACA-N17-TAGGAT) was located within intI3 and significantly differed from common and relatively strong class 1 integron promoters (25).
FIG 1.
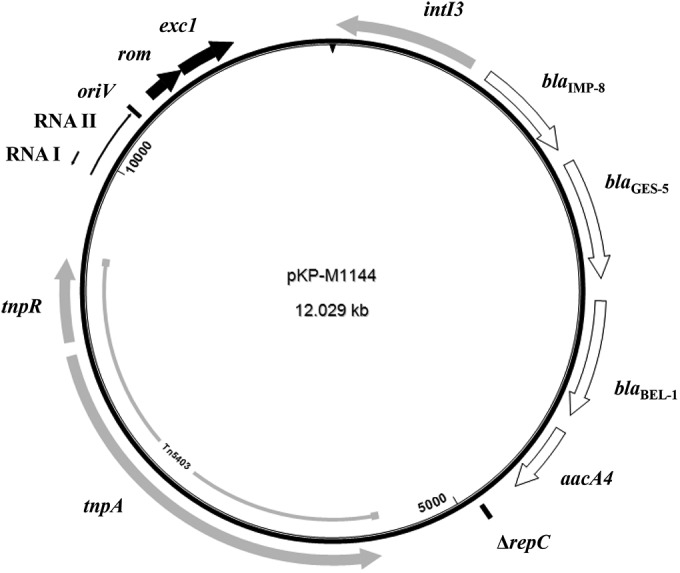
Circular genetic map of pKP-M1144. Arrows show directions of transcription of open reading frames and regulatory elements. Sequences characteristic of the ColE1-like plasmid backbone are indicated by black arrows. The intI3 gene and transposases are shaded gray. White arrows indicate the resistance genes. Position 1 is indicated by a vertical black arrow on strand ruler.
In1144 was inserted into a ColE1-like backbone of 2,737 bp (Fig. 1; nt 9293 to 12029). A DNA comparison showed that its 704-bp sequence (nt 9838 to 10541) had limited nucleotide similarity to replication regions of several RNA-priming plasmids (26–29). pKP-M1144 exhibited the highest similarity score with the replicon of pJHCMW1 (100% coverage, 75% identity) from K. pneumoniae strain JHCK1 from Argentina (28). A consensus sequence for the ColE1 replication origin (oriV) (30) was located at positions 10414 to 10416. Additionally, putative regions containing the RNA transcripts RNA II and RNA I that control the initiation of DNA replication and the plasmid copy number (31) were identified (Fig. 1). RNA II (nt 9882 to 10417) acts as a primer for the initiation of replication, while RNA I (nt 9984 to 9883) is an antisense molecule that controls replication initiation by binding to RNA II and preventing primer formation. The RNA II and RNA I of pKP-M1144 were 78% and 68% identical, respectively, to the corresponding transcripts from pJHCMW1 (28). Downstream of oriV, a 192-bp sequence resembling the rom gene (96% identity) of pNBL63 from Klebsiella oxytoca strain NBL63 (29) was present (nt 10589 to 10780). The Rom (or Rop) protein enhances the interaction of the RNA I inhibitor with its target, thus resulting in a reduction of the replication initiation frequency (32). Just next to rom, an exc1-like sequence of 423 bp, exhibiting 95% identity with that of pNE1280 from Enterobacter cloacae strain 1623 (33), was identified (nt 10780 to 11202). The Exc1 protein (entry exclusion protein 1) may reduce the formation of stable mating pairs (26). Of note is that an exc1-like gene, whose putative product showed low amino acid similarity (41%) with the putative Exc1 of pKP-M1144, has also been found downstream of the rom gene in pNBL63 (29). In the remaining part of pKP-M1144 (nt 4680 to 9292), a 37-bp segment similar to the replication region of IncQ plasmids (ΔrepC) (22) was found at the boundary of In1144, 258 bp downstream of aacA4. The repC gene at a similar position was identified in the IncQ plasmid pQ7 with the blaGES-1-carrying class 3 integron (22). Upstream of ΔrepC, the intact transposon Tn5403 was found (34).
Although no origin-of-transfer (oriT) and ColE1 mob genes (35) were identified in pKP-M1144, the mobilization capability of the plasmid was tested. pKP-M1144 was introduced into E. coli strain XL1-Blue that harbors the fertility factor F′, being an IncFIA-type conjugative plasmid. The resulting transformants were used in mating experiments with E. coli A15 Rifr (8). Transconjugants were selected on MacConkey agar plates with rifampin (150 μg/ml) and ampicillin (50 μg/ml) and confirmed to carry the ∼15-kb plasmid pKP-M1144. The transfer of pKP-M1144 was achieved at a relatively high frequency (2 × 10−5 blaIMP-positive recombinants per donor cell), indicating its capability to be mobilized by plasmids with apparently different conjugation systems. This finding might be explained by the presence of a sequence with limited identity with the ColE1 oriT (28); however, a further study is necessary to prove this hypothesis and characterize the putative oriT.
To our knowledge, this is the first description of a blaIMP-carrying class 3 integron identified in Europe. In1144 also carries the blaGES-5, blaBEL-1, and aacA4 gene cassettes. Of note was that Kpn-1144 showed only reduced susceptibility to carbapenems, even though it produced IMP-8 and GES-5 carbapenemases. It might be due to lower expression driven by a weaker promoter (25), low plasmid copy number, and/or good permeability of the outer membrane of the strain for antibiotics.
In general, IMP-like MBLs have been much more common in the Far East than in Europe (3), and class 3 integrons with MBL genes, exclusively blaIMP, have occurred rarely and only in Japan so far (23, 36). In 2010, the blaIMP-8 cassette was reported in K. oxytoca in Spain and Pseudomonas mendocina in Portugal, but it was located in class 1 integrons (37, 38); moreover, the cassettes blaGES-5 and blaBEL-1 have been found exclusively in such elements so far (5, 6). All these observations suggest that In1144 emerged by the exchange of resistance cassettes between class 1 and class 3 integrons. Moreover, the presence of blaBEL-1 in Kpn-1144 is the first report on a BEL-type ESBL in Enterobacteriaceae, suggesting its acquisition from P. aeruginosa. In1144 is carried by a new ColE1-like plasmid, pKP-M1144, but the presence of the IncQ-derived ΔrepC next to the integron suggests that it may have originated from an IncQ-like plasmid with a class 3 integron, like pQ7 (22). A plausible hypothesis is that this class 3 integron was acquired by a ColE1-like replicon from an IncQ-type plasmid at a certain step in pKP-M1144 evolution; however, it is not known whether this was In1144 already or any of its possible progenitors. This work confirms the significant role of ColE1-like plasmids in resistance dissemination (29, 39–41) and documents an interesting case of evolution of mobile genetic elements with resistance determinants in Gram-negative bacteria.
The nucleotide sequence of the plasmid pKP-M1144 has been assigned the GenBank accession no. KF745070.
ACKNOWLEDGMENTS
We thank A. Baraniak and J. Fiett for their assistance, and the curator team of the Institute Pasteur in Paris, France, for curating the MLST data of K. pneumoniae and making them publicly available.
This work was supported by funding from the European Community (MOSAR network contract LSHP-CT-2007-037941). It was also financed in part by grant NT11032-6/2010 from the Ministry of Health of the Czech Republic and grant P36 by the Charles University Research Fund.
We declare no conflicts of interest.
REFERENCES
- 1.Bebrone C. 2007. Metallo-β-lactamases (classification, activity, genetic organization, structure, zinc coordination) and their superfamily. Biochem Pharmacol 74:1686–1701. doi: 10.1016/j.bcp.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 2.Cornaglia G, Giamarellou H, Rossolini GM. 2011. Metallo-β-lactamases: a last frontier for β-lactams? Lancet Infect Dis 11:381–393. doi: 10.1016/S1473-3099(11)70056-1. [DOI] [PubMed] [Google Scholar]
- 3.Tzouvelekis LS, Markogiannakis A, Psichogiou M, Tassios PT, Daikos GL. 2012. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin Microbiol Rev 25:682–707. doi: 10.1128/CMR.05035-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan JJ, Ko WC, Wu JJ. 2001. Identification of a plasmid encoding SHV-12, TEM-1, and a variant of IMP-2 metallo-β-lactamase, IMP-8, from a clinical isolate of Klebsiella pneumoniae. Antimicrob Agents Chemother 45:2368–2371. doi: 10.1128/AAC.45.8.2368-2371.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vourli S, Giakkoupi P, Miriagou V, Tzelepi E, Vatopoulos AC, Tzouvelekis LS. 2004. Novel GES/IBC extended-spectrum β-lactamase variants with carbapenemase activity in clinical enterobacteria. FEMS Microbiol Lett 234:209–213. doi: 10.1016/j.femsle.2004.03.028. [DOI] [PubMed] [Google Scholar]
- 6.Poirel L, Brinas L, Verlinde A, Ide L, Nordmann P. 2005. BEL-1, a novel clavulanic acid-inhibited extended-spectrum β-lactamase, and the class 1 integron In120 in Pseudomonas aeruginosa. Antimicrob Agents Chemother 49:3743–3748. doi: 10.1128/AAC.49.9.3743-3748.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Derde LP, Cooper BS, Goossens H, Malhotra-Kumar S, Willems RJ, Gniadkowski M, Hryniewicz W, Empel J, Daurzenberg MJ, Annane D, Aragão I, Chalfine A, Dumpis U, Esteves F, Giamarellou H, Muzlovic I, Nardi G, Petrikkos GL, Tomic V, Martí AT, Stammet P, Brun-Buisson C, Bonten MJ; MOSAR WP3 Study Team. 2013. Interventions to reduce colonization and transmission of antimicrobial-resistant bacteria in intensive care units: an interrupted time series study and cluster randomised trial. Lancet Infect Dis 14:31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Papagiannitsis CC, Izdebski R, Baraniak A, Fiett J, Herda M, Hrabak J, Derde LPG, Bonten MJM, Carmeli Y, Goossens H, Hryniewicz W, Brun-Buisson C, Gniadkowski M; MOSAR WP2, WP3, and WP5 Study Groups. 2015. Survey of metallo-β-lactamase-producing Enterobacteriaceae colonizing patients in European ICUs and rehabilitation units, 2008–11. J Antimicrob Chemother, in press. [DOI] [PubMed] [Google Scholar]
- 9.Drieux L, Brossier F, Sougakoff W, Jarlier V. 2008. Phenotypic detection of extended-spectrum beta-lactamase production in Enterobacteriaceae. Clin Microbiol Infect 14(Suppl 1):90–103. [DOI] [PubMed] [Google Scholar]
- 10.European Committee on Antimicrobial Susceptibility Testing (EUCAST). 2013. EUCAST guideline for the detection of resistance mechanisms and specific resistances of clinical and/or epidemiological importance. European Society of Clinical Microbiology and Infectious Diseases (EUCAST), Basel, Switzerland: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Resistance_mechanisms/EUCAST_detection_of_resistance_mechanisms_v1.0_20131211.pdf. [Google Scholar]
- 11.Lee K, Lim YS, Yong D, Yum JH, Chong Y. 2003. Evaluation of the Hodge test and the imipenem-EDTA double-disk synergy test for differentiating metallo-β-lactamase producing isolates of Pseudomonas spp. and Acinetobacter spp. J Clin Microbiol 41:4623–4629. doi: 10.1128/JCM.41.10.4623-4629.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Papagiannitsis CC, Študentova V, Izdebski R, Oikonomou O, Pfeifer Y, Petinaki E, Hrabak J. 2015. Matrix-assisted laser desorption ionization–time of flight mass spectrometry meropenem hydrolysis assay with NH4HCO3, a reliable tool for the direct detection of carbapenemase activity. J Clin Microbiol 53:1731–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.European Committee on Antimicrobial Susceptibility Testing. 2003. Determination of minimum inhibitory concentrations (MICs) of antibacterial agents by broth dilution. Clin Microbiol Infect 9:1–7. doi: 10.1046/j.1469-0691.2003.00790.12691538 [DOI] [Google Scholar]
- 14.Ellington MJ, Kistler J, Livermore DM, Woodford N. 2007. Multiplex PCR for rapid detection of genes encoding acquired metallo-β-lactamases. J Antimicrob Chemother 59:321–322. [DOI] [PubMed] [Google Scholar]
- 15.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 12 from India. Antimicrob Agents Chemother 53:5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diancourt L, Passet V, Verhoef J, Grimont A, Brisse S. 2005. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolate. J Clin Microbiol 43:4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coelho A, Piedra-Carrasco N, Bartolome R, Quintero-Zarate JN, Larrosa N, Conejo-Sanchez T, Prats G, Garcillan-Barcia MP, de la Cruz F, Gonzalez-Lopez JJ. 2012. Role of IncHI2 plasmids harbouring blaVIM-1, blaCTX-M-9, aac(6′)-Ib and qnrA genes in the spread of multiresistant Enterobacter cloacae and Klebsiella pneumoniae strains in different units at Hospital Vall d'Hebron, Barcelona, Spain. Int J Antimicrob Agents 39:514–517 doi: 10.1016/j.ijantimicag.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 18.Barton BM, Harding GP, Zuccarelli AJ. 1995. A general method for the detecting and sizing large plasmids. Anal Biochem 226:235–240. [DOI] [PubMed] [Google Scholar]
- 19.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 20.Studentova V, Dobiasova H, Hedlova D, Dolejska M, Papagiannitsis CC, Hrabak J. 2015. Complete nucleotide sequences of two NDM-1-encoding plasmids from the same sequence type 11 Klebsiella pneumoniae strain. Antimicrob Agents Chemother 59:1325–1328. doi: 10.1128/AAC.04095-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collis CM, Kim MJ, Patridge SR, Stokes HW, Hall RM. 2002. Characterization of the class 3 integron and the site-specific recombination system it determines. J Bacteriol 184:3017–3026. doi: 10.1128/JB.184.11.3017-3026.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poirel L, Carattoli A, Bernabeu S, Bruderer T, Frei R, Nordmann P. 2010. A novel IncQ plasmid type harbouring a class 3 integron from Escherichia coli. J Antimicrob Chemother 65:1594–1598. doi: 10.1093/jac/dkq166. [DOI] [PubMed] [Google Scholar]
- 23.Arakawa Y, Murakami M, Suzuki K, Ito H, Wacharotayankin R, Ohsuka S, Kato N, Ohta M. 1995. A novel integron-like element carrying the metallo-β-lactamase gene blaIMP. Antimicrob Agents Chemother 39:1612–1615. doi: 10.1128/AAC.39.7.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shaw KJ, Rather PN, Hare RS, Miller GH. 1993. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol Rev 57:138–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Papagiannisis CC, Tzouvelekis LS, Miriagou V. 2009. Relative strengths of the class 1 integron promoter hybrid 2 and the combinations of strong and hybrid 1 with an active P2 promoter. Antimicrob Agents Chemother 53:277–280. doi: 10.1128/AAC.00912-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan PT, Ohmori H, Tomizawa J, Lebowitz J. 1985. Nucleotide sequence and gene organization of ColE1 DNA. J Biol Chem 260:8925–8935. [PubMed] [Google Scholar]
- 27.Nomura N, Murooka Y. 1994. Characterization and sequencing of the region required for replication of a non-selftransmissible plasmid pEC3 isolated from Erwinia carotovora subsp. carotovora. J Ferment Bioeng 78:250–254. doi: 10.1016/0922-338X(94)90299-2. [DOI] [Google Scholar]
- 28.Dery KJ, Chavideh R, Waters V, Chamoro R, Tomalsky LS, Tomalsky ME. 1997. Characterization of the replication and mobilization regions of the multiresistance Klebsiella pneumoniae plasmid pJHCMW1. Plasmid 38:97–105. doi: 10.1006/plas.1997.1303. [DOI] [PubMed] [Google Scholar]
- 29.Wu SW, Dornbusch K, Kronvall G, Norgen M. 1999. Characterization and nucleotide sequence of a Klebsiella oxytoca cryptic plasmid encoding a CMY-type beta-lactamase: confirmation that the plasmid-mediated cephamycinase originated from the Citrobacter freundii AmpC beta-lactamase. Antimicrob Agents Chemother 43:1350–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tomizawa JI, Ohmori H, Bird RE. 1977. Origin of replication of colicin E1 plasmid DNA. Proc Natl Acad Sci U S A 74:1865–1869. doi: 10.1073/pnas.74.5.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Polisky B. 1988. ColE1 replication control circuitry: sense from antisense. Cell 55:929–932. doi: 10.1016/0092-8674(88)90235-8. [DOI] [PubMed] [Google Scholar]
- 32.Cesareni G, Helmer-Citterich M, Castagnoli L. 1991. Control of ColE1 plasmid replication by antisense RNA. Trends Genet 7:230–235. doi: 10.1016/0168-9525(91)90370-6. [DOI] [PubMed] [Google Scholar]
- 33.Bryant KA, Van Schooneveld TC, Thapa I, Bastola D, Williams LO, Safranek TJ, Hinrichs SH, Rupp ME, Fey FD. 2013. KPC-4 is encoded within a truncated Tn4401 in an IncL/M plasmid, pNE1280, isolated from Enterobacter cloacae and Serratia marcescens. Antimicrob Agents Chemother 57:37–41. doi: 10.1128/AAC.01062-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rinkel M, Hubert JC, Roux B, Lett MC. 1994. Identification of a new transposon Tn5403 in a Klebsiella pneumoniae strain isolated from a polluted aquatic environment. Curr Microbiol 29:249–154. doi: 10.1007/BF01577436. [DOI] [PubMed] [Google Scholar]
- 35.Warren GJ, Saul MW, Sherratt DJ. 1979. ColE1 plasmid mobility: essential and conditional functions. Mol Gen Genet 170:103–107. [DOI] [PubMed] [Google Scholar]
- 36.Senda K, Arakawa Y, Ichiyama S, Nakashima K, Ito H, Ohsuka S, Shimokata K, Kato N, Ohta M. 1996. PCR detection of metallo-β-lactamase gene (blaIMP) in Gram-negative rods resistant to broad-spectrum β-lactams. J Clin Microbiol 34:2909–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Conejo MC, Dominguez MC, López-Cerero L, Serano L, Rodríguez-Baño J, Pascual A. 2010. Isolation of multidrug-resistant Klebsiella oxytoca carrying blaIMP-8, associated with OXY hyperproduction in the intensive care unit of a community hospital in Spain. J Antimicrob Chemother 65:1071–1073. doi: 10.1093/jac/dkq063. [DOI] [PubMed] [Google Scholar]
- 38.Santos C, Caetano T, Ferreira S, Mendo S. 2010. First description of blaIMP-8 in a Pseudomonas mendocina isolated at the Hospital Infante D. Pedro, Aveiro, Portugal. Res Microbiol 161:305–307. [DOI] [PubMed] [Google Scholar]
- 39.Cao V, Lambert T, Courvalin P. 2002. ColE1-like plasmid pIP843 of Klebsiella pneumoniae encoding extended-spectrum beta-lactamase CTX-M-17. Antimicrob Agents Chemother 46:1212–1217. doi: 10.1128/AAC.46.5.1212-1217.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sarno R, McGillivary G, Sherratt DJ, Actis LA, Tomalsky ME. 2002. Complete nucleotide sequence of Klebsiella pneumoniae multiresistance plasmid pJHCMW1. Antimicrob Agents Chemother 46:3422–3427. doi: 10.1128/AAC.46.11.3422-3427.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zioga A, Whichard JM, Kotsakis SD, Tzouvelekis LS, Tzelepi E, Miriagou V. 2009. CMY-31 and CMY-36 cephalosporinases encoded by ColE1-like plasmids. Antimicrob Agents Chemother 53:1256–1259. doi: 10.1128/AAC.01284-08. [DOI] [PMC free article] [PubMed] [Google Scholar]


