Abstract
The monoterpene carvacrol, the major component of oregano and thyme oils, is known to exert potent antifungal activity against the pathogenic yeast Candida albicans. This monoterpene has been the subject of a considerable number of investigations that uncovered extensive pharmacological properties, including antifungal and antibacterial effects. However, its mechanism of action remains elusive. Here, we used integrative chemogenomic approaches, including genome-scale chemical-genetic and transcriptional profiling, to uncover the mechanism of action of carvacrol associated with its antifungal property. Our results clearly demonstrated that fungal cells require the unfolded protein response (UPR) signaling pathway to resist carvacrol. The mutants most sensitive to carvacrol in our genome-wide competitive fitness assay in the yeast Saccharomyces cerevisiae expressed mutations of the transcription factor Hac1 and the endonuclease Ire1, which is required for Hac1 activation by removing a nonconventional intron from the 3′ region of HAC1 mRNA. Confocal fluorescence live-cell imaging revealed that carvacrol affects the morphology and the integrity of the endoplasmic reticulum (ER). Transcriptional profiling of pathogenic yeast C. albicans cells treated with carvacrol demonstrated a bona fide UPR transcriptional signature. Ire1 activity detected by the splicing of HAC1 mRNA in C. albicans was activated by carvacrol. Furthermore, carvacrol was found to potentiate antifungal activity of the echinocandin antifungal caspofungin and UPR inducers dithiothreitol and tunicamycin against C. albicans. This comprehensive chemogenomic investigation demonstrated that carvacrol exerts its antifungal activity by altering ER integrity, leading to ER stress and the activation of the UPR to restore protein-folding homeostasis.
INTRODUCTION
Fungal pathogens represent a serious risk to the growing population of immunocompromised individuals resulting from the increasing success of organ and bone marrow transplantation, immune-suppressive cancer chemotherapy, premature births, and the AIDS pandemic. Candida albicans is a diploid ascomycete yeast that is an important commensal and opportunistic pathogen in humans. Systemic infections resulting from C. albicans are on the rise and are associated with mortality rates of 50% or greater despite currently available antifungal therapy (1–3). Current therapeutic options are limited to treatment with three longstanding antifungal classes, the polyenes, azoles, and echinocandins (4). These compounds target the specific fungal biological process of ergosterol metabolism (azoles and polyenes) and cell wall β-1,3-glucan synthesis (echinocandins). However, these drugs have serious side effects such as nephrotoxicity and/or create complications such as resistance due to their fungistatic rather than fungicidal characteristics (4–6). There is, thus, an urgent need for new strategies to identify novel protein targets and bioactive molecules for antifungal therapeutic intervention.
Plants are an interesting reservoir of secondary metabolites with an attractive and broad spectrum of antimicrobial properties. Carvacrol is a monoterpene phenol and a major component of essential oil extract from oregano and other plants belonging to the Labiatae family (7). This monoterpene is considered nontoxic to humans and is commonly used as a flavoring substance. Carvacrol has been the subject of a considerable number of investigations that uncovered extensive pharmacological proprieties, including antifungal and antibacterial effects (8). Previous investigations demonstrated that carvacrol is one of the potent monoterpenes against C. albicans, impeding the growth of different morphological forms, including yeast, hyphae, and the highly drug-resistant biofilm (9–11). Recent studies have shown that the monoterpenes carvacrol and eugenol, but not thymol, synergize with the azole antifungal fluconazole and inhibit planktonic growth and biofilm in clinical resistant strains (12). Interestingly, carvacrol has been proved to be an effective treatment against vaginal candidiasis in an immunosuppressed rat model (13).
Despite the growing interest in using carvacrol in antifungal therapy, the mechanism of action (MoA) of this phytomolecule and other antimicrobial monoterpenes or sesquiterpenes remains unclear. Prior investigations suggested that carvacrol acts as a membrane-disrupting agent by targeting and binding ergosterol (11, 14, 15). Transcription profiling in the model yeast Saccharomyces cerevisiae exposed to carvacrol revealed a transcriptional signature similar to that experienced under calcium stress (16), suggesting that the antifungal activity of carvacrol is probably the consequence of the perturbation of Ca+ or H+ ion homeostasis. In the current study, we have used state-of-the-art chemical genomic approaches, including chemical-genetic profiling using the complete pool of bar-coded S. cerevisiae haploid deletion strains, in addition to genome-wide transcriptional profiling to accurately determine the MoA of carvacrol that is relevant to its antifungal activity. Similar chemogenomic approaches have been successfully used to confirm the known MoA of clinically approved antifungals such as fluconazole and also to uncover the MoA of novel antifungal compounds (17–19). We demonstrate that carvacrol acts as an antifungal by causing endoplasmic reticulum (ER) stress and by inducing the unfolded protein response (UPR).
MATERIALS AND METHODS
Inhibition and synergism assay.
A growth assay of Candida cells treated with carvacrol was performed in a 96-well plate using the Sunrise plate reader (Tecan). C. albicans clinical strain SC5314 (20) and ire1, mkc1, and bck1 mutants were grown overnight in yeast extract-peptone-dextrose (YPD) medium at 30°C in a shaking incubator. Candida cells were then resuspended in fresh YPD at an optical density at 595 nm (OD595) of 0.05. A total volume of 99 μl of cells was added to each well in the 96-well plate in addition to 1 μl of the corresponding stock solution of carvacrol (W224511; Sigma). The plates were incubated at 30°C under agitation, and OD readings were taken every 10 min over 20 h. Samples were done in triplicate, and the average was used for analysis. C. albicans ire1/ire1, mkc1/mkc1, and bck1/bck1 mutants were from the kinase collection of A. Mitchell (Carnegie Mellon University) (21). Carvacrol and other monoterpenes used in this study were dissolved in dimethyl sulfoxide (DMSO). As a control, an equal volume of DMSO was added (1% [vol/vol] final concentration). The MIC was determined by the first well with a growth reduction of 10% as referred to OD595 values in the presence of the tested compounds compared to untreated cells.
For the spot serial dilution assay, the S. cerevisiae wild-type (WT) strain BY4741 and the indicated deletion mutant strains were grown in YPD overnight at 30°C. Cells were diluted to a concentration of 107 cells/ml, and then 10-fold serial dilutions of the indicated strains were spotted on media containing the indicated compounds. Plates were incubated at 30°C for 2 days.
Carvacrol synergistic interactions with tunicamycin (T7765; Sigma), dithiothreitol (DTT) (BP172; Fisher), fluconazole (F8929; Sigma), caspofungin (SML0425; Sigma), and amphotericin B (A488; Sigma) were tested as described by Epp et al. (22). The fractional inhibitory concentration (FIC) index was determined as follows: (MIC of carvacrol in combination/MIC of carvacrol alone) plus (MIC of a drug in combination/MIC of a drug alone). Tunicamycin, amphotericin B, and fluconazole were dissolved in DMSO and added from stock solutions of 10, 30, and 300 mg/ml, respectively. Caspofungin was dissolved in water to a stock concentration of 10 mg/ml.
RNA extractions and microarray profiling.
Cultures of C. albicans strain SC5314 were inoculated from a fresh colony and grown overnight in YPD at 30°C. Cultures were then diluted to an OD595 of 0.05 in 100 ml of fresh YPD and grown at the same initial temperature until an OD595 of 0.8. The culture was divided into two volumes of 50 ml; one sample was maintained as the control where DMSO was added, and the other treated with 0.2 mM carvacrol or 0.3 mM thymol (MIC of each monoterpene). Candida cells were exposed to carvacrol for 5 and 30 min and to thymol for 30 min. Cells were then centrifuged 2 min at 3,500 rpm, the supernatants were removed, and the samples were quick-frozen and stored at −80°C. RNA was extracted using the Qiagen RNeasy kit as described previously by Sellam et al. (23). RNA quality and integrity were checked using an Agilent 2100 bioanalyzer. cDNA labeling and microarray experiments were performed as described by Nantel et al. (24). Briefly, 18 μg of total RNA was reverse transcribed using 9 ng of oligo(dT)21 in the presence of Cy3 or Cy5-dCTP (GE Healthcare) and 400 U of SuperScript III reverse transcriptase (Life Technologies) at 42°C for 3 h. After cDNA synthesis, template RNA was removed by adding 2.5 units RNase H (Promega) and 1 mg RNase A (Pharmacia) followed by incubation for 15 min at 37°C. The labeled cDNAs were purified with a QIAquick PCR purification kit (Qiagen). DNA microarrays were processed and analyzed as previously described by Nantel et al. (24). Data handling and analysis were carried out using Genespring v.7.3 (Agilent Technologies, Palo Alto, CA). Statistical analysis used Welch's t test with a false-discovery rate (FDR) of 5% and 1.5-fold enrichment cutoff. Hierarchical clustering of the expression profiling data was performed using Genespring v.7.3. Gene ontology (GO) annotation was performed using the Cytoscape (25) plug-in BiNGO (26).
Haploid deletion chemical-genetic profiling.
Screens of the haploid deletion pool were performed as described by Parsons et al. (18) with 0.64 mM carvacrol. Enrichment of GO terms was performed using Gene Ontology Finder (http://www.yeastgenome.org/cgi-bin/GO/goTermFinder.pl). The P value was calculated using a hypergeometric distribution.
HAC1 mRNA splicing assay.
The HAC1 splicing assay was performed by reverse transcription-PCR (RT-PCR). RNAs were extracted from C. albicans cells challenged with tunicamycin (4.7 μM), either alone or in combination with carvacrol (0.2 mM) as described for microarray experiments. cDNAs were obtained using Superscript II reverse transcriptase (Life Technologies) as recommended by the supplier. The obtained cDNA was used as a template to amplify the spliced and unspliced forms of HAC1 using the primer pair TGAGGATGAACACCAAGAAGAA (forward primer) and TCAAAGTCCAACTGAAATGAT (reverse primer). The PCR products were resolved on 1.5% agarose gel.
Evaluation of ER integrity by confocal microscopy.
The S. cerevisiae Sec61-green fluorescent protein (GFP) strain used for fluorescence microscopy is from the Yeast-GFP clone collection (27). An overnight culture was diluted in YPD supplemented with 1 mM carvacrol to an OD595 of 0.05 and grown for four generations at 30°C under agitation. Images of fluorescence microscopy were acquired with a 63×, 1.3-numerical-aperture (NA) objective on a Leica DMI6000B inverted microscope connected to a Hamamatsu C9100-13 camera.
RESULTS
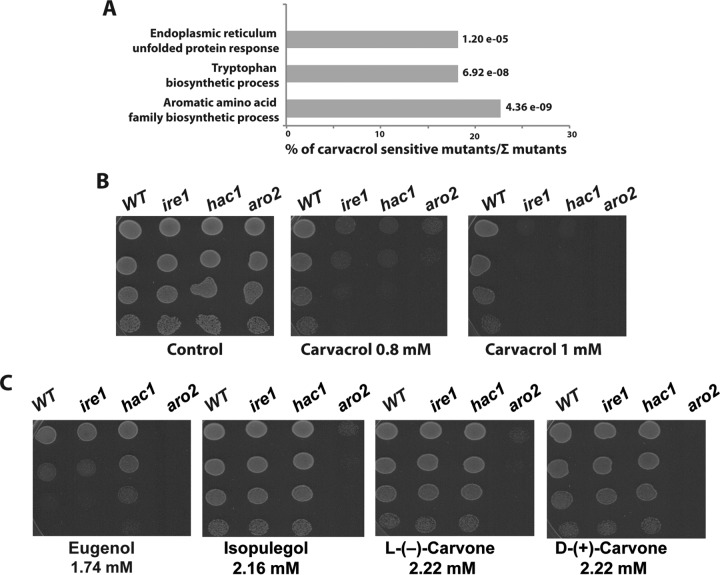
Chemogenomic fitness assay identifies key UPR regulators as required for carvacrol tolerance.
Chemical-genetic profiling is a powerful tool that has been widely used to uncover the MoA of many bioactive compounds (28). In order to determine the MoA of the monoterpene carvacrol, we used S. cerevisiae haploid deletion chemical-genetic profiling (HCGP) to identify gene deletions that confer sensitivity to carvacrol (Table 1; see also Table S1 in the supplemental material). GO terms associated with carvacrol-sensitive strains were determined, and relevant functional categories are summarized in Fig. 1A. The GO terms ER-mediated UPR and tryptophan amino acid biosynthesis were significantly enriched in the HCGP profile. Strains with mutations of the endonuclease Ire1 (ire1; Z-score = 3.03) and the transcription factor Hac1 (hac1; Z-score = 2.83), which are conserved components of the eukaryotic UPR signaling (29), were the most sensitive strains to carvacrol (Table 1). In response to ER stress, the endonuclease Ire1 mediates the splicing of a nonconventional intron from the 3′ region of HAC1 mRNA, which in turn activates the UPR transcriptional program to restore protein-folding homeostasis (30, 31). Strains with mutations of the cell wall integrity (CWI) signaling pathway, including the mitogen-activated protein kinase kinase kinase (MAPKKK) Bck1, the MAPK Slt2, and the transcription factor Swi6, were also hypersensitive to carvacrol. Previous investigations demonstrated that, in addition to its role in cell wall maintenance, the CWI pathway is also required for ER stress response and protein-folding homeostasis (32, 33). These data suggest that the UPR pathway is required for cells to tolerate carvacrol.
TABLE 1.
Chemical-genetic profiling of carvacrola
| Gene name | Description of product | Z-score |
|---|---|---|
| IRE1 | Serine-threonine kinase and endoribonuclease; mediates the unfolded protein response by regulating Hac1p synthesis through HAC1 mRNA splicing | 3.03 |
| HAC1 | Transcription factor; regulates the unfolded protein response | 2.83 |
| TRP3 | Indole-3-glycerol-phosphate synthase | 1.92 |
| ARO2 | Chorismate synthase and flavin reductase; catalyzes the conversion of 5-enolpyruvylshikimate 3-phosphate to form chorismate, which is a precursor to aromatic amino acids | 1.87 |
| ARO1 | Pentafunctional arom protein; catalyzes steps 2 through 6 in the biosynthesis of chorismate | 1.86 |
| BCK1 | MAPKKK acting in the protein kinase C signaling pathway | 1.79 |
| PHO84 | Hig high-affinity inorganic phosphate transporter | 1.63 |
| YPT6 | Ras-like GTP binding protein involved in the secretory pathway, required for fusion of endosome-derived vesicles with the late Golgi complex | 1.62 |
| TRP2 | Anthranilate synthase; catalyzes the initial step of tryptophan biosynthesis | 1.56 |
| TLG2 | Syntaxin-like t-SNARE; mediates fusion of endosome-derived vesicles with the late Golgi complex | 1.48 |
| CWH41 | ER type II integral membrane N-glycoprotein involved in assembly of cell wall β-1,6-glucan and asparagine-linked protein glycosylation | 1.47 |
| GTR2 | Putative GTP binding protein; negatively regulates Ran/Tc4 GTPase cycle | 1.36 |
| SLT2 | Serine/threonine MAP kinase involved in regulating maintenance of cell wall integrity, cell cycle progression, and nuclear mRNA retention | 1.32 |
| TRP4 | Anthranilate phosphoribosyl transferase; transferase of the tryptophan biosynthetic pathway | 1.30 |
| MDM31 | Mitochondrial protein that may have a role in phospholipid metabolism | 1.22 |
| SMI1 | Protein involved in the regulation of cell wall synthesis | 1.14 |
| SWI6 | Transcription cofactor; forms complexes with Swi4p and Mbp1p to regulate transcription at the G1/S transition; required for the unfolded protein response | 1.13 |
| COG6 | Component of the conserved oligomeric Golgi complex; functions in protein trafficking to mediate fusion of transport vesicles to Golgi complex | 1.12 |
| YPS7 | Putative GPI-anchored aspartic protease; member of the yapsin family of proteases involved in cell wall growth and maintenance; located in the cytoplasm and endoplasmic reticulum | 1.11 |
| RPS8A | Protein component of the small (40S) ribosomal subunit | 1.01 |
| TRS85 | Component of transport protein particle complex III; regulates endosome-Golgi complex traffic and required for membrane expansion | 1.01 |
| TRP1 | Phosphoribosylanthranilate isomerase; catalyzes the third step in tryptophan biosynthesis | 1.00 |
Identification by HCGP assay of gene deletion mutants that confer sensitivity to carvacrol. Fitness defect scores were calculated based on bar code microarray hybridization, and the top 22 sensitive deletion strains sorted by Z-score are shown.
FIG 1.
Chemical-genetic profiling using HCGP assay identified key UPR regulators as required for carvacrol tolerance. (A) GO term enrichment of carvacrol sensitive mutants. The P value was calculated using the hypergeometric distribution. (B, C) Individual confirmations of the chemical-genetic screen by spot serial dilution assay. A total of three deletion mutants, the ire1, hac1, and aro2 mutants, were selected and spotted on YPD with DMSO (control), YPD containing 0.8 mM or 1 mM carvacrol (B) or 1.74 mM eugenol, 2.16 mM isopulegol, 2.22 mM l-(−)-carvone (vol/vol), or 2.22 mM d-(+)-carvone (C). Plates were incubated at 30°C for 2 days.
The two mutants of the UPR pathway, the ire1 and hac1 mutants, and the mutant of tryptophan biosynthesis, the aro2 mutant, were selected, and their sensitivity to carvacrol was confirmed using serial dilution assay (Fig. 1B). The three mutants were also tested for their sensitivity to four other monoterpenes: eugenol, isopulegol, and the two enantiomers l-(−)-carvone and d-(+)-carvone (Fig. 1B). As shown in Fig. 1C, the aro2 mutant was sensitive to the four-tested monoterpenes. However, ire1 and hac1 mutants were sensitive only to carvacrol, suggesting that tolerance of the other monoterpenes does not require the UPR pathway (Fig. 1B). Taken together, these data suggest that UPR pathway signaling is specifically required for the tolerance of carvacrol.
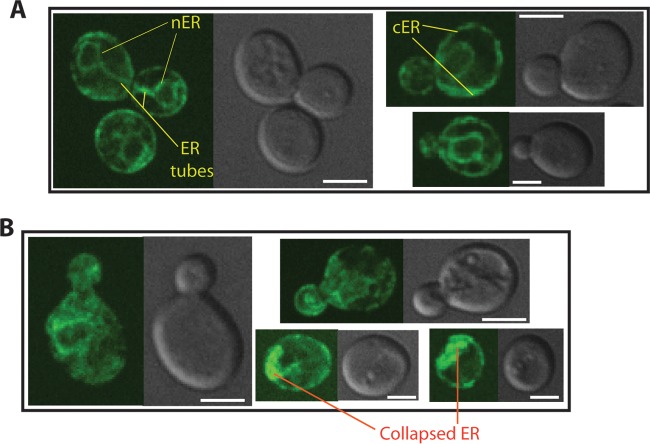
Carvacrol disrupts the morphology and the integrity of ER.
The UPR pathway requirement for carvacrol tolerance supports the hypothesis that carvacrol might act as an ER stressor, perhaps by altering ER integrity and/or its protein-folding capacity. To check if the UPR requirement is related to a direct effect of carvacrol on cellular organization of ER, we have used a Sec61-GFP fusion as an ER marker (34). Sec61 is an essential ER translocation channel required for protein import to ER and localizes to nuclear ER (nER) and cortical ER (cER). ER organization as judged by Sec61-GFP fluorescence was assessed in cells treated with 0.8 mM carvacrol and in untreated cells. The control cells exhibited a clear and well-defined ER distribution with nER surrounding the nucleus, cER at the periphery of the cell adjacent to plasma membrane, and few cytoplasmic ER tubes (Fig. 2A). However, in cells challenged with carvacrol, the ER became fragmented, and the GFP signal was diffuse in the cytoplasm. The nER structure was partially or completely disrupted in some cells (Fig. 2B). Cells treated with carvacrol accumulated cytoplasmic foci, likely representing collapsed ER (Fig. 2B). Taken together, these observations indicate that carvacrol disrupts ER organization.
FIG 2.
Carvacrol disrupts the morphology and the integrity of ER. Sec61-GFP fusion was used as an ER marker. (A) Fluorescence micrographs of Sec61 localization in control cells treated with DMSO. Cortical (cER) and nuclear ERs (nER) are labeled. (B) Reorganization of Sec61 localization in cells treated with 0.8 mM carvacrol. Bars, 4 μm.
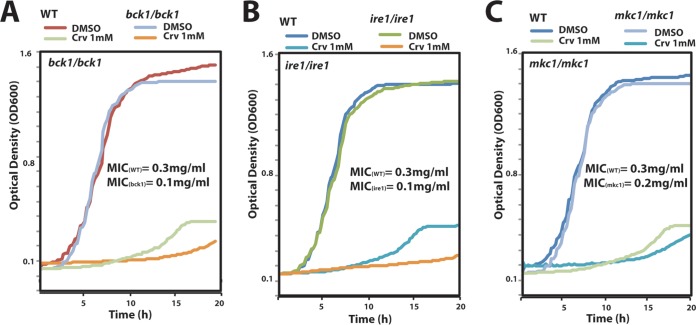
UPR pathway is required for carvacrol tolerance in the pathogenic yeast C. albicans.
Carvacrol has been widely investigated for its antifungal activity mainly against the pathogenic yeast C. albicans (9–11). To check whether the conserved eukaryotic UPR signaling pathway is also required for carvacrol tolerance in C. albicans, sensitivity of ire1, mkc1 (Mkc1 is the ortholog of Slt2), and bck1 homozygous mutants to carvacrol was assessed. Our data revealed that all tested mutants had increased sensitivity to carvacrol compared to their parental strains (Fig. 3). Consistent with the HCGP assay in S. cerevisiae, these data demonstrate that C. albicans UPR is also required for tolerance of carvacrol.
FIG 3.
The UPR pathway is important for carvacrol tolerance in the pathogenic yeast C. albicans. Growth assays of C. albicans bck1 (A), ire1 (B), and mkc1 (C) mutants and the WT strain SC5314 challenged with 1 mM carvacrol. Cells were grown in YPD at 30°C, and OD595 readings were taken every 10 min. MIC values for each mutant and WT strain are indicated.
Genome-wide transcriptional profiling reveals that carvacrol induces the unfolded protein response in C. albicans.
We undertook microarray transcriptional profiling to uncover cellular responses to carvacrol. The C. albicans clinical strain SC5314 was treated with 0.2 mM (MIC of carvacrol) carvacrol for 5 or 30 min. Using a statistical significance analysis with an estimated FDR of 5%, in addition to a 1.5-fold cutoff, 499 and 317 transcripts were differentially expressed after 5 min and 30 min exposure to carvacrol, respectively (see Table S2 in the supplemental material). GO term enrichment analysis of upregulated transcripts demonstrated that carvacrol activates genes involved in proteolysis, amino acid metabolism, phospholipid translocation, response to oxidative stress, and DNA repair mechanisms (Fig. 4A and B; see also Table S3 in the supplemental material). Transcripts related to GO terms ribosome biogenesis, glycosylation, sugar transport, drug export, and nuclear import were repressed. The carvacrol transcriptional signature in C. albicans was reminiscent of the unfolded protein stress response expressed in eukaryotic organisms (35, 36). In response to UPR inducers such as DTT or tunicamycin, C. albicans and other fungi, including S. cerevisiae, Aspergillus niger, and Aspergillus fumigatus, activate genes involved in vesicle trafficking, protein folding, amino acid metabolism, proteolysis, glycosylation, lipid metabolism, and cell wall biogenesis (35, 37–39). All these UPR-associated GO terms are represented in our data set (Fig. 4 and Table 2; see also Table S3 in the supplemental material). In agreement with the HCGP assay, our data suggest that carvacrol generates ER stress and induces UPR response in C. albicans.
FIG 4.
Genome-wide transcriptional profiling reveals that the monoterpene carvacrol induces the UPR in C. albicans. GO analysis of transcripts differentially regulated in C. albicans cells treated with carvacrol for 5 min (A) or 30 min (B) using BiNGO software (26). Results were charted using Cytoscape (25) and the Enrichment Map plug-in (52). (C) Heat map and two-dimensional hierarchical clustering of the transcriptional profiles of carvacrol- and thymol-treated cells. Upregulated and downregulated genes are indicated by red and green, respectively. Molecular structures of carvacrol and thymol are shown to emphasize the unique difference, which is the position of the hydroxyl group.
TABLE 2.
Different manually curated GO terms related to the unfolded protein response and their associated transcripts that are activated in response to carvacrol
| GO term category | Genes in response to: |
|
|---|---|---|
| Carvacrol (5 min) | Carvacrol (30 min) | |
| Vesicle-mediated transport | ENT2, YSC84, UBP15, PEP3, ECM21, OBPA, RVS167, SDS24, SEC18, SRO77, VPS15, VPS21, VPS35, VPS4, CSP37 | LSB5, SEM11, DDI1, GDI1, SEC18, TRX1, DID2, VPS8, EFM4 |
| Proteolytic degradation | NPL4, PR26, PRE3, PRE5, PUP1, RPN10, RPN2, RPT1 VPS4, UBP15, LAP41, DOA4, DOA1, CYM1, UBP2, CDC48, PBN1, PRE4, EAR1, SAN1, RQC1, PNG1, MNL1, UBP14, UBX5, UBP12, UBP16, PRB1, LAP4, RPN9, RPN12, HOD1, UBP2, ASI3, DDI1, PIM1, PRE1, PRE3, PRE9, PUP2, RPT2 | RPN9, RPN12, SEM1, CDC48, UBP2, ASI3, DDI1, JEM1, PIM1, PRE1, PRE3, PRE9, PUP2, RPT2 |
| Protein folding | FMO1, HSP104, HSP70, HSP78, TRX1, RBP1, JEM1, CYP5 | CYP5, JEM1, RBP1, TRX1, HSP12, orf19.4216 |
| Amino acid metabolism | SHM2, SER33, STR2, TRP5, LYS4, LYS1, LYS2, SER1 MET15, MET10, ARO8, ARO3, ARG1, AAT1 | MUP1, ALP1, AGP2, AGP3, HIP1, MET3, MET6 |
| Lipid metabolism | LCB4, PLC2, TGL1, ROG1, OBPA, ORM1, YDC1 VPS4, GPI15, ALG13, SMP2, HAL22, POX1, NPR1, PBN1, GPI13, PDR16, PEX5, SCT1, HFD1, YJU3, YFT2, YEH1, GPI15, IFA38, PEX6, PLC1, PLC2, RTA2 MIT1, MCR1, ERG4, ERG2 | HFD1, YJU3, YFT2, YEH1, GPI15, IFA38, ERG2, ERG4, MCR1, MIT1, MLS1, PEX6, PLC1, PLC2, RTA2 |
| Cellular transport | CDR1, GAP2, GNP1, AQY1, ITR1, AGP3, HGT20, ATM1, SUL2, MEP1, HIP1, HAK1, CRP1 | ALP1, ATX1, CDR1 CRP1, YET3, DDI1, GDI1, MEP1, HAK1, MIM1, RTA2, SUL2, SIT1 |
| Cell wall proteins and biogenesis | ALS1, ECM29, MNN4, ECM15, DDR48, PHR1, BMT4 | CAS5, PHR1, WSC4, RHD3, ENG1, CSH1, CSP37, ALS1, BMT4 |
In order to assess whether the UPR response uncovered here is specific to carvacrol, the transcriptional profile of C. albicans cells challenged with thymol, a monoterpene structurally related to carvacrol, was evaluated. Thymol is a positional isomer of carvacrol and has a phenolic hydroxyl at a different position on the phenolic ring. As shown in Fig. 4C, hierarchical clustering distinguished clearly the transcriptional signature exhibited by cells treated with carvacrol from that displayed by cells exposed to thymol (see Table S2 in the supplemental material). We conclude that the mechanism of action of carvacrol is different from that of thymol.
Carvacrol induces unconventional splicing of the transcription factor Hac1.
Our data demonstrated that Ire1 and Hac1, key players of UPR signaling, were required for fungal tolerance of carvacrol. This prompted us to assess whether the UPR signaling pathway is activated following exposure to carvacrol. The activation of the UPR was assayed by detecting the splicing of HAC1 mRNA using RT-PCR (40). As shown in Fig. 5A, Candida cells treated with carvacrol displayed UPR activation as evidenced by increased splicing of HAC1 mRNA.
FIG 5.
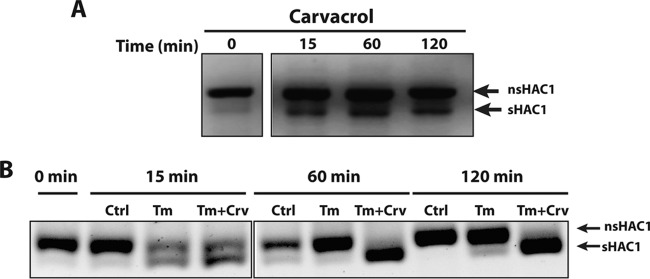
Carvacrol induces splicing of the transcription factor gene HAC1 mRNA. (A) Effect of carvacrol on HAC1 mRNA splicing in WT C. albicans. Cells were treated with carvacrol and at the indicated time samples were harvested and splicing of HAC1 was assessed using RT-PCR. nsHAC1, nonspliced HAC1; sHAC1, spliced HAC1. (B) Effect of tunicamycin on HAC1 splicing in the presence or absence of carvacrol. Cells were treated with tunicamycin (Tm) or with tunicamycin and carvacrol (Tm + Crv) and sampled at the indicated times to asses HAC1 splicing. As a control, splicing of HAC1 mRNA was also monitored in nontreated cells (Ctrl).
In contrast to what was previously reported with the UPR inducers tunicamycin and DTT (37), the nonspliced form of HAC1 (nsHAC1) predominated over the spliced form (sHAC1) following treatment with carvacrol. Since the HAC1 mRNA splicing factor ire1 was one of the most sensitive mutants to carvacrol, we wanted to check whether carvacrol itself directly compromised Ire1 activity. Therefore, we assessed the splicing of HAC1 mRNA in response to tunicamycin, a well-known UPR stressor, alone or in combination with carvacrol. As shown in Fig. 5B, cells treated with tunicamycin alone or in combination with carvacrol for 15 min were able to splice the cryptic intron at the 3′ region of HAC1, suggesting that Ire1 activity was not compromised by carvacrol. Interestingly, after 60 min, cells treated with tunicamycin exhibited predominantly the unspliced form of HAC1, possibly reflecting an adaptive response, while cells treated with tunicamycin and carvacrol had exclusively the spliced form of HAC1. This finding suggests that carvacrol exacerbates the effect of tunicamycin on HAC1 splicing and sustained UPR signaling.
Synergistic interaction of carvacrol with ER stressors and caspofungin.
Drug combination treatments are powerful strategies that have been used to increase the efficacy and reduce the toxicity of preexisting single-drug therapies. Synergistic action can result from complementary action of the synergized drugs, which target different parts along the same biological pathway or protein (41). A well-known example in anticancer therapy is the combination of aplidin and cytarabine, which target the same apoptotic pathway (42). In C. albicans, combination of the azole fluconazole with either ketoconazole or terbinafine, each targeting the ergosterol biosynthesis pathway, led to a synergistic antifungal activity (43). Here, we wanted to test whether other well-known UPR inducers and ER stressors, such as the reducing agent DTT and the N-linked glycosylation inhibitor tunicamycin, potentiate the antifungal activity of carvacrol. As shown in Table 3, combination of carvacrol with DTT or tunicamycin resulted in a potent antifungal synergy in the C. albicans clinical strain SC5314, while either compound alone had minor inhibitory effect. We also confirmed the synergistic interaction of carvacrol with the antifungal fluconazole as reported previously (12) and uncovered a potent synergism with the echinocandin caspofungin (Table 3). However, carvacrol did not synergize with the polyene antifungal amphotericin B.
TABLE 3.
Synergistic interaction of carvacrol with ER stressors and the antifungals caspofungin and fluconazole
| Drug(s) (alone or in combination)a | MIC (μg/ml)b | FIC index |
|---|---|---|
| FCZ | 48 | |
| CRV | 28.12 | |
| FCZ-CRV | 12/3.51 | 0.37 |
| TNC | 1 | |
| CRV | 28.12 | |
| TNC-CRV | 0.06/7.03 | 0.31 |
| DTT | 93.7 | |
| CRV | 28.12 | |
| DTT-CRV | 23.4/3.51 | 0.36 |
| CSP | 0.5 | |
| CRV | 28.12 | |
| CSP-CRV | 0.06/3.51 | 0.24 |
| AmpB | 1 | |
| CRV | 28.12 | |
| AmpB-CRV | 0.5/14.06 | 1 |
CRV, carvacrol; FCZ, fluconazole; TNC, tunicamycin; DTT, dithiothreitol; CSP, caspofungin; AmpB, amphotericin B.
The MIC values for the individual drugs in a combination are separated by a slash.
DISCUSSION
In the current investigation, we have used state-of-the-art chemogenomic approaches to uncover the MoA of the monoterpene carvacrol in the pathogenic yeast C. albicans. UPR is a cytoprotective response that is engaged as a consequence of the accumulation of unfolded or misfolded proteins following stress affecting the ER. Our chemical-genetic profiling assay, supported by the transcriptional profiling data, led to the hypothesis that carvacrol might target and compromise ER integrity and perturb protein-folding capacity, which in turn activates the UPR pathway. Cellular investigation of the ER demonstrated clearly that carvacrol affected the integrity and the organization of nER and cER. In accordance with this result, many S. cerevisiae mutants that exhibited defective ER morphology or organization express a constitutive UPR response and depend tightly on it for their survival (34). Thus, our results demonstrated that carvacrol exerts its antifungal activity by disrupting ER integrity, which in turn causes ER stress and leads to Ire1-mediated UPR to restore protein-folding homeostasis in C. albicans. In our HCGP assay, deletion of genes involved in different trafficking pathways such as ER-to-Golgi (trs85), Golgi-to-ER (ypt6), and intra-Golgi transport (cog6) were also required for carvacrol tolerance. This supposes that, in addition to ER, carvacrol might target other intracellular vesicular trafficking. Another possible explanation, and taking into consideration that ER is the main cellular membrane source for many trafficking systems (44, 45), is that disrupting ER by carvacrol might result in a collapse of the ER-dependent cellular vesicle trafficking network.
While previous studies suggested that carvacrol exerts its antifungal activity by disrupting calcium homeostasis (16), ergosterol biosynthesis (14), and the plasma membrane (15), our HCGP and transcriptional profiling results were not supportive of such MoAs. These presumed MoAs might be an indirect consequence of ER stress triggered by carvacrol. In fact, calcium in the cell is stored in the ER, and many studies report that calcium homeostasis is significantly perturbed under UPR and ER stress (46–49). In fungi, the ER is also the site for the synthesis of ergosterol and lipids as well as cell wall components (50). Thus, ER perturbations might disturb many aspects of membrane biology, such as permeability and ergosterol or other lipid content.
Mutants uncovered by the HCGP assay often reflect mechanisms that buffer the impact of the target compromised by a bioactive compound (17). Our HCGP assay showed that, in addition to UPR signaling mutants, deletion of genes involved in tryptophan biosynthesis, including trp1, trp2, trp3, trp4, aro1, and aro2, resulted in a hypersensitivity to carvacrol. Interestingly, recent investigations showed that the monoterpene eugenol interferes with aromatic amino acid uptake, including tryptophan in S. cerevisiae (51). This suggests that, in addition to targeting ER, carvacrol might also interfere with tryptophan uptake.
The newly revisited MoA of carvacrol uncovered in this study was exploited to predict and validate complementary synergistic drug interactions with other ER stressors and with well-known antifungals. Overall, our data suggest that pharmacological perturbation of ER function results in increased sensitivity to fluconazole and caspofungin. In agreement with this, Epp et al. demonstrated that compromising ER function genetically (mutation of the ARF protein, Age3) or pharmacologically (by brefeldin A, an inhibitor of the retrograde Golgi-to-ER transport and UPR inducer) resulted in a potentiation of the activity of many azoles as well as the echinocandins against C. albicans and other human fungal pathogens (22). Interestingly, our HAC1 splicing assay reflected synergistic interaction of carvacrol and the UPR stressor, tunicamycin. Addition of the two compounds caused complete splicing of the HAC1 mRNA, while treating cells with each compound separately resulted in incomplete splicing.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Aaron Mitchel (Carnegie Mellon University) and Christian Landry (IBIS, Université Laval) for providing mutants used in this work. We thank J. L. Lévesque for technical assistance. M.B. is grateful to A. Azzouzi for helping to set up the Medical Biology Unit at the Faculty of Medicine and Pharmacy of Oujda.
This work was supported by the Faculty of Medicine, Université Laval, and CHUQ Startup funding to A.S. J.C. received a Faculty of Medicine Ph.D. scholarship (Université Laval). A.S. is supported by the Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant (06625). A.S. is a recipient of the Fonds de Recherche du Québec-Santé (FRQS) J1 salary award. G.W.B. is supported by grants from the Canadian Institutes of Health Research (MOP-84305 and MOP-79368).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00551-15.
REFERENCES
- 1.Pfaller MA, Diekema DJ. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev 20:133–163. doi: 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gudlaugsson O, Gillespie S, Lee K, Vande Berg J, Hu J, Messer S, Herwaldt L, Pfaller M, Diekema D. 2003. Attributable mortality of nosocomial candidemia, revisited. Clin Infect Dis 37:1172–1177. doi: 10.1086/378745. [DOI] [PubMed] [Google Scholar]
- 3.Pappas PG, Kauffman CA, Andes D, Benjamin DK Jr, Calandra TF, Edwards JE Jr, Filler SG, Fisher JF, Kullberg BJ, Ostrosky-Zeichner L, Reboli AC, Rex JH, Walsh TJ, Sobel JD, Infectious Diseases Society of America. 2009. Clinical practice guidelines for the management of candidiasis: 2009 update by the Infectious Diseases Society of America. Clin Infect Dis 48:503–535. doi: 10.1086/596757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shapiro RS, Robbins N, Cowen LE. 2011. Regulatory circuitry governing fungal development, drug resistance, and disease. Microbiol Mol Biol Rev 75:213–267. doi: 10.1128/MMBR.00045-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perlin DS. 2007. Resistance to echinocandin-class antifungal drugs. Drug Resist Updat 10:121–130. doi: 10.1016/j.drup.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akins RA. 2005. An update on antifungal targets and mechanisms of resistance in Candida albicans. Med Mycol 43:285–318. doi: 10.1080/13693780500138971. [DOI] [PubMed] [Google Scholar]
- 7.Buchbauer G, Ilic A. 2013. Biological activities of selected mono- and sesquiterpenes: possible uses in medicine, p 4109–4159. In Ramawat KG, Ḿerillon J-M (ed), Natural products. Springer, Berlin, Germany. [Google Scholar]
- 8.Baser KH. 2008. Biological and pharmacological activities of carvacrol and carvacrol bearing essential oils. Curr Pharm Des 14:3106–3119. doi: 10.2174/138161208786404227. [DOI] [PubMed] [Google Scholar]
- 9.Raut JS, Shinde RB, Chauhan NM, Karuppayil SM. 2013. Terpenoids of plant origin inhibit morphogenesis, adhesion, and biofilm formation by Candida albicans. Biofouling 29:87–96. doi: 10.1080/08927014.2012.749398. [DOI] [PubMed] [Google Scholar]
- 10.Inouye S, Takahashi M, Abe S. 2009. Inhibitory activity of hydrosols, herbal teas and related essential oils against filament formation and the growth of Candida albicans. Nihon Ishinkin Gakkai Zasshi 50:243–251. doi: 10.3314/jjmm.50.243. [DOI] [PubMed] [Google Scholar]
- 11.Lima IO, de Oliveira Pereira F, de Oliveira WA, de Oliveira Lima E, Menezes EA, Cunha FA, de Fátima Formiga Melo Diniz M. 2013. Antifungal activity and mode of action of carvacrol against Candida albicans strains. J Essential Oil Res 25:138–142. doi: 10.1080/10412905.2012.754728. [DOI] [Google Scholar]
- 12.Doke SK, Raut JS, Dhawale S, Karuppayil SM. 2014. Sensitization of Candida albicans biofilms to fluconazole by terpenoids of plant origin. J Gen Appl Microbiol 60:163–168. doi: 10.2323/jgam.60.163. [DOI] [PubMed] [Google Scholar]
- 13.Chami F, Chami N, Bennis S, Trouillas J, Remmal A. 2004. Evaluation of carvacrol and eugenol as prophylaxis and treatment of vaginal candidiasis in an immunosuppressed rat model. J Antimicrob Chemother 54:909–914. doi: 10.1093/jac/dkh436. [DOI] [PubMed] [Google Scholar]
- 14.Ahmad A, Khan A, Akhtar F, Yousuf S, Xess I, Khan LA, Manzoor N. 2011. Fungicidal activity of thymol and carvacrol by disrupting ergosterol biosynthesis and membrane integrity against Candida. Eur J Clin Microbiol Infect Dis 30:41–50. doi: 10.1007/s10096-010-1050-8. [DOI] [PubMed] [Google Scholar]
- 15.Chami N, Bennis S, Chami F, Aboussekhra A, Remmal A. 2005. Study of anticandidal activity of carvacrol and eugenol in vitro and in vivo. Oral Microbiol Immunol 20:106–111. doi: 10.1111/j.1399-302X.2004.00202.x. [DOI] [PubMed] [Google Scholar]
- 16.Rao A, Zhang Y, Muend S, Rao R. 2010. Mechanism of antifungal activity of terpenoid phenols resembles calcium stress and inhibition of the TOR pathway. Antimicrob Agents Chemother 54:5062–5069. doi: 10.1128/AAC.01050-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho CH, Piotrowski J, Dixon SJ, Baryshnikova A, Costanzo M, Boone C. 2011. Combining functional genomics and chemical biology to identify targets of bioactive compounds. Curr Opin Chem Biol 15:66–78. doi: 10.1016/j.cbpa.2010.10.023. [DOI] [PubMed] [Google Scholar]
- 18.Parsons AB, Lopez A, Givoni IE, Williams DE, Gray CA, Porter J, Chua G, Sopko R, Brost RL, Ho CH, Wang J, Ketela T, Brenner C, Brill JA, Fernandez GE, Lorenz TC, Payne GS, Ishihara S, Ohya Y, Andrews B, Hughes TR, Frey BJ, Graham TR, Andersen RJ, Boone C. 2006. Exploring the mode-of-action of bioactive compounds by chemical-genetic profiling in yeast. Cell 126:611–625. doi: 10.1016/j.cell.2006.06.040. [DOI] [PubMed] [Google Scholar]
- 19.Hoepfner D, Helliwell SB, Sadlish H, Schuierer S, Filipuzzi I, Brachat S, Bhullar B, Plikat U, Abraham Y, Altorfer M, Aust T, Baeriswyl L, Cerino R, Chang L, Estoppey D, Eichenberger J, Frederiksen M, Hartmann N, Hohendahl A, Knapp B, Krastel P, Melin N, Nigsch F, Oakeley EJ, Petitjean V, Petersen F, Riedl R, Schmitt EK, Staedtler F, Studer C, Tallarico JA, Wetzel S, Fishman MC, Porter JA, Movva NR. 2014. High-resolution chemical dissection of a model eukaryote reveals targets, pathways and gene functions. Microbiol Res 169:107–120. doi: 10.1016/j.micres.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 20.Gillum AM, Tsay EY, Kirsch DR. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet 198:179–182. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- 21.Blankenship JR, Fanning S, Hamaker JJ, Mitchell AP. 2010. An extensive circuitry for cell wall regulation in Candida albicans. PLoS Pathog 6:e1000752. doi: 10.1371/journal.ppat.1000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Epp E, Vanier G, Harcus D, Lee AY, Jansen G, Hallett M, Sheppard DC, Thomas DY, Munro CA, Mullick A, Whiteway M. 2010. Reverse genetics in Candida albicans predicts ARF cycling is essential for drug resistance and virulence. PLoS Pathog 6:e1000753. doi: 10.1371/journal.ppat.1000753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sellam A, Hogues H, Askew C, Tebbji F, van Het Hoog M, Lavoie H, Kumamoto CA, Whiteway M, Nantel A. 2010. Experimental annotation of the human pathogen Candida albicans coding and noncoding transcribed regions using high-resolution tiling arrays. Genome Biol 11:R71. doi: 10.1186/gb-2010-11-7-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nantel A, Rigby T, Hogues H, Whiteway M. 2006. Microarrays for studying pathogenicity in Candida Albicans, p 181–205. In Kavanagh K. (ed), Medical mycology: cellular and molecular techniques. John Wiley & Sons, Ltd., Chichester, UK. [Google Scholar]
- 25.Saito R, Smoot ME, Ono K, Ruscheinski J, Wang PL, Lotia S, Pico AR, Bader GD, Ideker T. 2012. A travel guide to Cytoscape plugins. Nat Methods 9:1069–1076. doi: 10.1038/nmeth.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maere S, Heymans K, Kuiper M. 2005. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 27.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. 2003. Global analysis of protein localization in budding yeast. Nature 425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 28.Smith AM, Ammar R, Nislow C, Giaever G. 2010. A survey of yeast genomic assays for drug and target discovery. Pharmacol Ther 127:156–164. doi: 10.1016/j.pharmthera.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hetz C. 2012. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol 13:89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 30.Chapman RE, Walter P. 1997. Translational attenuation mediated by an mRNA intron. Curr Biol 7:850–859. doi: 10.1016/S0960-9822(06)00373-3. [DOI] [PubMed] [Google Scholar]
- 31.Kaufman RJ. 1999. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev 13:1211–1233. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- 32.Chen Y, Feldman DE, Deng C, Brown JA, De Giacomo AF, Gaw AF, Shi G, Le QT, Brown JM, Koong AC. 2005. Identification of mitogen-activated protein kinase signaling pathways that confer resistance to endoplasmic reticulum stress in Saccharomyces cerevisiae. Mol Cancer Res 3:669–677. doi: 10.1158/1541-7786.MCR-05-0181. [DOI] [PubMed] [Google Scholar]
- 33.Levin DE. 2011. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: the cell wall integrity signaling pathway. Genetics 189:1145–1175. doi: 10.1534/genetics.111.128264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manford AG, Stefan CJ, Yuan HL, Macgurn JA, Emr SD. 2012. ER-to-plasma membrane tethering proteins regulate cell signaling and ER morphology. Dev Cell 23:1129–1140. doi: 10.1016/j.devcel.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 35.Back SH, Schroder M, Lee K, Zhang K, Kaufman RJ. 2005. ER stress signaling by regulated splicing: IRE1/HAC1/XBP1. Methods 35:395–416. doi: 10.1016/j.ymeth.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 36.Arensdorf AM, Diedrichs D, Rutkowski DT. 2013. Regulation of the transcriptome by ER stress: non-canonical mechanisms and physiological consequences. Front Genet 4:256. doi: 10.3389/fgene.2013.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wimalasena TT, Enjalbert B, Guillemette T, Plumridge A, Budge S, Yin Z, Brown AJ, Archer DB. 2008. Impact of the unfolded protein response upon genome-wide expression patterns, and the role of Hac1 in the polarized growth, of Candida albicans. Fungal Genet Biol 45:1235–1247. doi: 10.1016/j.fgb.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 38.Guillemette T, van Peij N, Goosen T, Lanthaler K, Robson GD, van den Hondel CA, Stam H, Archer DB. 2007. Genomic analysis of the secretion stress response in the enzyme-producing cell factory Aspergillus niger. BMC Genomics 8:158. doi: 10.1186/1471-2164-8-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101:249–258. doi: 10.1016/S0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 40.Guillemette T, Ram AF, Carvalho ND, Joubert A, Simoneau P, Archer DB. 2011. Methods for investigating the UPR in filamentous fungi. Methods Enzymol 490:1–29. doi: 10.1016/B978-0-12-385114-7.00001-5. [DOI] [PubMed] [Google Scholar]
- 41.Sun X, Vilar S, Tatonetti NP. 2013. High-throughput methods for combinatorial drug discovery. Sci Transl Med 5:205rv201. doi: 10.1126/scitranslmed.3006667. [DOI] [PubMed] [Google Scholar]
- 42.Humeniuk R, Menon LG, Mishra PJ, Saydam G, Longo-Sorbello GS, Elisseyeff Y, Lewis LD, Aracil M, Jimeno J, Bertino JR, Banerjee D. 2007. Aplidin synergizes with cytosine arabinoside: functional relevance of mitochondria in Aplidin-induced cytotoxicity. Leukemia 21:2399–2405. doi: 10.1038/sj.leu.2404911. [DOI] [PubMed] [Google Scholar]
- 43.Spitzer M, Griffiths E, Blakely KM, Wildenhain J, Ejim L, Rossi L, De Pascale G, Curak J, Brown E, Tyers M, Wright GD. 2011. Cross-species discovery of syncretic drug combinations that potentiate the antifungal fluconazole. Mol Syst Biol 7:499. doi: 10.1038/msb.2011.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Viotti C, Kruger F, Krebs M, Neubert C, Fink F, Lupanga U, Scheuring D, Boutte Y, Frescatada-Rosa M, Wolfenstetter S, Sauer N, Hillmer S, Grebe M, Schumacher K. 2013. The endoplasmic reticulum is the main membrane source for biogenesis of the lytic vacuole in Arabidopsis. Plant Cell 25:3434–3449. doi: 10.1105/tpc.113.114827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mullins C. 2005. The biogenesis of cellular organelles. Springer, New York, NY. [Google Scholar]
- 46.Zhang K, Kaufman RJ. 2004. Signaling the unfolded protein response from the endoplasmic reticulum. J Biol Chem 279:25935–25938. doi: 10.1074/jbc.R400008200. [DOI] [PubMed] [Google Scholar]
- 47.Tessitore A, del P Martin M, Sano R, Ma Y, Mann L, Ingrassia A, Laywell ED, Steindler DA, Hendershot LM, d'Azzo A. 2004. GM1-ganglioside-mediated activation of the unfolded protein response causes neuronal death in a neurodegenerative gangliosidosis. Mol Cell 15:753–766. doi: 10.1016/j.molcel.2004.08.029. [DOI] [PubMed] [Google Scholar]
- 48.Torres M, Encina G, Soto C, Hetz C. 2011. Abnormal calcium homeostasis and protein folding stress at the ER: a common factor in familial and infectious prion disorders. Commun Integr Biol 4:258–261. doi: 10.4161/cib.4.3.15019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoyer-Hansen M, Jaattela M. 2007. Connecting endoplasmic reticulum stress to autophagy by unfolded protein response and calcium. Cell Death Differ 14:1576–1582. doi: 10.1038/sj.cdd.4402200. [DOI] [PubMed] [Google Scholar]
- 50.Parks LW, Casey WM. 1995. Physiological implications of sterol biosynthesis in yeast. Annu Rev Microbiol 49:95–116. doi: 10.1146/annurev.mi.49.100195.000523. [DOI] [PubMed] [Google Scholar]
- 51.Darvishi E, Omidi M, Bushehri AA, Golshani A, Smith ML. 2013. The antifungal eugenol perturbs dual aromatic and branched-chain amino acid permeases in the cytoplasmic membrane of yeast. PLoS One 8:e76028. doi: 10.1371/journal.pone.0076028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. 2010. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One 5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.