Abstract
We report the first OXA-181-producing strain in China. blaOXA-181 was found in sequence type 410 (ST410) Escherichia coli strain WCHEC14828 from a Chinese patient without recent travel history. Genome sequencing and conjugation experiments were performed. blaOXA-181 was carried on a 51-kb self-transmissible IncX3 plasmid and was linked with qnrS1, a quinolone resistance gene. blaOXA-181 was introduced onto the IncX3 plasmid from a ColE2-type plasmid, and IncX3 plasmids have the potential to mediate the dissemination of blaOXA-181.
TEXT
OXA-181 is an OXA-48-type carbapenemase conferring resistance to penicillins and carbapenems, and its encoding gene blaOXA-181 originates from Shewanella xiamenensis, an environmental bacterium (1). The blaOXA-181 gene was initially identified in Enterobacter cloacae and Klebsiella pneumoniae isolates that were recovered in 2007 at several locations in India (2). Since then, blaOXA-181 has been sporadically found in several species of Enterobacteriaceae in a few countries, but it has not been identified in China previously. Unlike other major carbapenemase genes, e.g., blaNDM and blaKPC, found in the Enterobacteriaceae, the genetic context of blaOXA-181 and the plasmid and host strain carrying this gene remain largely uninvestigated. This study characterizes a carbapenem-resistant Escherichia coli clinical isolate that was found to carry blaOXA-181 in China.
Strain WCHEC14828 was obtained from the blood culture of a leukemia patient receiving stem cell transplantation on 2014. It was identified as E. coli and was resistant to imipenem (MIC, 16 μg/ml), meropenem (MIC, 32 μg/ml), ceftazidime (MIC, 128 μg/ml), and ciprofloxacin (MIC, 64 μg/ml) but was susceptible to amikacin (MIC, 16 μg/ml), colistin (MIC, 2 μg/ml), and tigecycline (MIC, 1 μg/ml) as determined using the microdilution method following recommendations of the Clinical and Laboratory Standards Institute (CLSI) (3). Strain WCHEC14828 was also resistant to ampicillin-sulbactam, aztreonam, cefazolin, cefepime, cefotaxime, cefoxitin, ceftriaxone, ertapenem, gentamicin, levofloxacin, nitrofurantoin, piperacillin-tazobactam, tobramycin, and trimethoprim-sulfamethoxazole as determined by Vitek II and to cefoperazone-sulbactam as determined by disk diffusion.
The strain was screened for the acquired carbapenemase-encoding genes blaGES, blaKPC, blaIMP, blaNDM, blaOXA-48-like, and blaVIM using PCR as described previously (4–7). The blaOXA-48-like gene was the only carbapenemase-encoding gene that was detected in strain WCHEC14828. Sequencing the complete coding sequence of the blaOXA-48-like gene amplified with an additional pair of primers (8) revealed the presence of blaOXA-181. To our knowledge, this is the first report of OXA-181 in China. Since the identification of OXA-181 in India in 2007, OXA-181-producing Enterobacteriaceae has been reported from several other countries in the Indian subcontinent, i.e., Bangladesh (9) and Sri Lanka (10) and potentially Nepal, as a Nepalese patient hospitalized in New Zealand was found to be carrying an OXA-181-producing K. pneumoniae (11). Outside the subcontinent, Enterobacteriaceae isolates producing OXA-181 have been found in Canada (12), France (13), the Netherlands (14), New Zealand (in a patient from Nepal) (11), Norway (in a patient from Romania) (15), Oman (16), Romania (17), Singapore (9), South Africa (18), and United Kingdom (in a patient from India) (19). In most cases (9, 11–14, 19, 20), the patients from whom OXA-181-producing Enterobacteriaceae were recovered had a recent travel history to the Indian subcontinent. In addition, blaOXA-181 has been identified in the chromosome of an S. xiamenensis isolate recovered in India (1). All of the above findings suggest that the Indian subcontinent is likely the origin place of OXA-181. Nonetheless, the patient in this study had no recent travel history to the Indian subcontinent, and it remains unclear how and where she acquired this OXA-181-producing strain.
Genome DNA of strain WCHEC14828 was prepared using the QIAamp DNA minikit (Qiagen, Hilden, Germany) and was subjected to whole-genome sequencing with a ca. 100× coverage using the HiSeq 2500 Sequencer (Illumina, San Diego, CA, USA) following the manufacturer's protocol at the Beijing Genomics Institute. A total of 6,051,062 clean reads were obtained from the genome sequencing for strain WCHEC14626. The GC content was 50.24%. Reads were assembled to 154 contigs, of which 98 were ≥500 bp in length using the SPAdes program (21). The Prokka program (22) was employed for annotating the genome sequence.
Strain WCHEC14828 was assigned to sequence type 410 (ST410) using the assembled genome sequence (see below) to query the seven alleles of the multilocus sequence typing scheme for Escherichia coli (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli) (23). ST410 is of the ST23 clonal complex, and it has been proposed as the founder ST of the ST23 complex (24). E. coli of ST410 has a worldwide distribution, as it has been recovered from humans and animals in Europe (Belgium, France, Germany, Greece, Ireland, Italy, Norway, Portugal, Spain, Switzerland, and United Kingdom), Africa (Congo, Ghana, Mauritania and Tunisia), Asia (Taiwan and Vietnam), North America (Canada), and South America (Brazil) (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli) (25–27). In addition, strain WCHEC14828 belonged to the phylogenetic group A as determined by phylogenetic group (A, B1, B2, and D) typing performed as described previously (28).
In strain WCHEC14828, blaOXA-181 was carried on a 51-kb plasmid, pOXA181_EC14828, which was completely circularized with intervals between contigs being filled by PCR with primers designed based on available sequences and Sanger sequencing. pOXA181_EC14828 was a self-transmissible plasmid, as it was able to be transferred to the recipient strain, azide-resistant E. coli strain J53, in the conjugation experiment and belonged to the IncX3 type. blaOXA-181 has not been found on an IncX3 plasmid before but has been detected on a 84-kb IncT plasmid (29) or on a 7.6-kb ColE2-type plasmid (1). The presence of blaOXA-181 on a self-transmissible IncX3 plasmid is of significance, as IncX3 plasmids have been found as a common vehicle mediating the dissemination of blaNDM-1 and blaNDM-5 among the Enterobacteriaceae in China (30, 31). Although this is the first case of blaOXA-181, IncX3 plasmids have the potential to facilitate the wide dissemination of this carbapenemase gene.
The IncX3 plasmid backbone (26 kb in size) of pOXA181_EC14828 was nearly identical (with only two nucleotide differences) to those of two IncX3 plasmids carrying blaNDM-1, of which one was recovered from another local ST3835 (the ST10 complex) E. coli strain (unpublished data), and the other was plasmid pNDM-HF727 from an E. cloacae strain from Guangdong, another province in China (31). The presence of blaOXA-181 on an IncX3 plasmid with a nearly identical backbone to IncX3 plasmids carrying blaNDM-1 suggests that blaOXA-181 might have coexisted with blaNDM-1 in the same host strain. Indeed, the coexistence of blaOXA-181 and blaNDM-1 has been found in a few strains previously (9, 10, 12, 15–17, 29). However, plasmids within almost all of those strains carrying blaNDM-1 and blaOXA-181 have not been characterized in previous studies, and therefore it remains largely unclear whether blaNDM-1 and blaOXA-181 have been carried by a single plasmid or by different ones in those strains. The only exception was in a Citrobacter freundii strain; blaNDM-1 was not detected on the plasmid carrying blaOXA-181, suggesting that blaNDM-1 is likely located on a different plasmid. It would be interesting to explore whether blaNDM-1 was located on an IncX3 plasmid within those strains carrying blaNDM-1 and blaOXA-181 (9, 10, 12, 15–17, 29). On the other hand, as WCHEC14828 has no blaNDM-1, it may also be possible that an IncX3 plasmid carrying blaOXA-181 has replaced the IncX3 plasmid carrying blaNDM-1 due to plasmid incompatibility.
In addition to blaOXA-181, strain WCHEC14828 had a few resistance genes, including blaCTX-M-15 (an extended-spectrum β-lactamase [ESBL] gene widely distributed in the world), blaCMY-2 (a plasmid-borne AmpC gene), blaOXA-1 (a non-ESBL oxacillinase gene), blaTEM-1b (a non-ESBL β-lactamase gene), blaampC (a chromosome-based AmpC gene), aac(6′)-Ib-cr (encoding an aminoglycoside acetyltransferase with low-level activity against fluoroquinolones), qnrS1 (conferring low-level resistance to fluoroquinolones), and tetA (a tetracycline resistance gene). The vast majority of these genes were bounded by IS elements, making their positioning very difficult. Among these resistance genes, only qnrS1 was located on pOXA181_EC14828 (Fig. 1). qnrS1 was flanked by an IS2-like insertion sequence upstream, which was truncated by IS26, and by a Tn3-like transposon downstream, which was truncated by ISKpn19. The IS26-qnrS1-ISKpn19 region is also present on several plasmids carrying the carbapenemase genes blaNDM-1 and blaKPC-2 in China (GenBank accession numbers KC958437 and KF914891) (32) but none was of the IncX3 type. The qnrS1 region on pOXA181_EC14828 was, therefore, introduced from another plasmid, which was likely due to the action of a transposable element.
FIG 1.
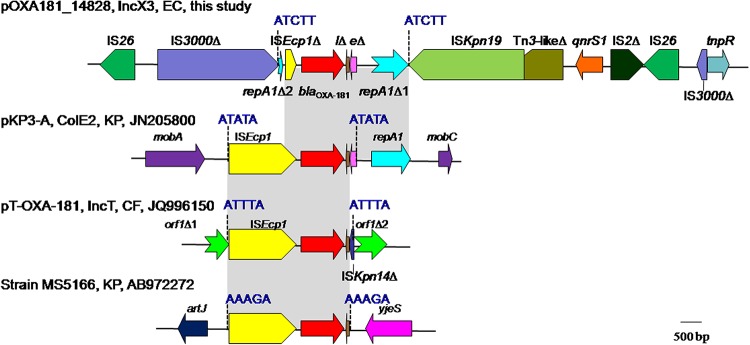
Genetic contexts of blaOXA-181. The name, the type, the host strain, and the GenBank accession number of plasmids carrying blaOXA-181 are shown. In strain MS5166, it remains unknown whether blaOXA-181 is on a plasmid, as only a partial sequence is available. EC, E. coli; KP, K. pneumoniae; CF, C. freundii; Δ, Incomplete; lΔ, lysRΔ; eΔ, ereAΔ. The 5-bp AT-rich DRs due to the transposition of ISEcp1 are blue. Identical regions of different contexts are gray. On plasmid pOXA181_EC14828, repA1 (encoding the ColE2-type replication initiation protein) is interrupted into two parts, which are shown as repA1Δ1 and repA1Δ2 here. repA1Δ2 was originally at the 3′ end of repA1, i.e., downstream of repA1Δ1 but is located close to blaOXA-181 on pOXA181_EC14828. On plasmid pKP3-A, mobA and mobC are genes encoding proteins for plasmid mobilization. On plasmid pT-OXA-181, a gene of unknown function, which is shown as an open reading frame (ORF) here, is interrupted into two parts by the insertion of ISEcp1 and the neighboring sequences (blaOXA-181, lysRΔ, and a truncated ISKpn14). In strain MS5166, artJ is a lysine-arginine-ornithine-binding periplasmic protein-encoding gene, and yjeS encodes an Fe-S electron transport protein.
Based on the limited currently available data, blaOXA-181 is always adjacent to ISEcp1. It is known that a single copy of ISEcp1 is able to mobilize neighboring resistance genes including blaOXA-181 by misidentifying various sequences as its alternative right-hand inverted repeat (IR) (1, 33). Indeed, in all of three contexts of blaOXA-181 available in the GenBank, the 5-bp AT-rich direct target repeats (DRs) that are the characteristics of the transposition of ISEcp1 have always been identified flanking the left-hand IR and downstream of blaOXA-181 (Fig. 1). This suggests that the mobilization of blaOXA-181 resulted from the action of ISEcp1 in the three cases. However, unlike those in previous contexts of blaOXA-181, ISEcp1 was truncated on pOXA181_EC14828. On plasmid pKP3-A, a ColE2-type replication initiation protein-encoding gene repA1 is located downstream of blaOXA-181. The presence of repA1 on pOXA181_EC14828 suggests that blaOXA-181 was introduced from a ColE2-type plasmid onto the IncX3 scaffold and formed the plasmid pOXA181_EC14828. Surprisingly, repA1 was interrupted into two parts on pOXA181_EC14828 with the small part present upstream of blaOXA-181 instead (Fig. 1). A 5-bp AT-rich duplicate sequence (ATCTT) was identified at the end of the large part and the beginning of the small part of the interrupted repA1 gene. This suggests that the interruption of repA1 was likely due to the insertion of an additional ISEcp1. The truncation of the ISEcp1 abutting blaOXA-181 and translocation of a part of repA1 from the downstream of blaOXA-181 to the upstream might be explained by subsequent homologous recombination between two copies of ISEcp1 as proposed before (34). blaOXA-181 was on pOXA181_EC14828, and the generation of such a complicated context of blaOXA-181 is likely a result of the direct insertion and subsequent recombination of multiple transposable elements. In addition, the two copies of IS26 in the region containing blaOXA-181 and qnrS1 can form a composite transposon, which has the potential to mobilize blaOXA-181 independent of the action of ISEcp1.
In conclusion, we report here the first OXA-181-producing strain in China, which was an ST410 E. coli strain causing blood stream infection (BSI) in a patient with leukemia. blaOXA-181 was likely introduced from a ColE2-type plasmid onto the IncX3 scaffold. The association of blaOXA-181 and an IncX3 plasmid is worrying as the IncX3 plasmid has the potential to serve as a common vehicle for mediating the further dissemination of blaOXA-181 among the Enterobacteriaceae like it is currently doing for blaNDM-1 in China.
Nucleotide sequence accession number.
Reads of WCHEC14828 genome sequence were deposited into the NCBI database under BioProject number PRJNA270793 and BioSample number SAMN03268883. The complete sequence of pOXA181_EC14828 was deposited into GenBank under the accession number KP400525.
ACKNOWLEDGMENTS
This work was supported by a grant from the National Natural Science Foundation of China (project 81222025), the New Century Excellent Talents Program, Ministry of Education, China (project NCET-13-0399), and a grant from Sichuan Bureau of Science, China (project 2013JQ0042).
REFERENCES
- 1.Potron A, Poirel L, Nordmann P. 2011. Origin of OXA-181, an emerging carbapenem-hydrolyzing oxacillinase, as a chromosomal gene in Shewanella xiamenensis. Antimicrob Agents Chemother 55:4405–4407. doi: 10.1128/AAC.00681-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Castanheira M, Deshpande LM, Mathai D, Bell JM, Jones RN, Mendes RE. 2011. Early dissemination of NDM-1- and OXA-181-producing Enterobacteriaceae in Indian hospitals: report from the SENTRY Antimicrobial Surveillance Program, 2006-2007. Antimicrob Agents Chemother 55:1274–1278. doi: 10.1128/AAC.01497-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clinical and Laboratory Standards Institute. 2013. Performance standards for antimicrobial susceptibility testing; 23rd informational supplement. CLSI M100-S23. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 4.Zong Z, Zhang X. 2013. blaNDM-1-carrying Acinetobacter johnsonii detected in hospital sewage. J Antimicrob Chemother 68:1007–1010. doi: 10.1093/jac/dks505. [DOI] [PubMed] [Google Scholar]
- 5.Mendes RE, Kiyota KA, Monteiro J, Castanheira M, Andrade SS, Gales AC, Pignatari AC, Tufik S. 2007. Rapid detection and identification of metallo-β-lactamase-encoding genes by multiplex real-time PCR assay and melt curve analysis. J Clin Microbiol 45:544–547. doi: 10.1128/JCM.01728-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poirel L, Le Thomas I, Naas T, Karim A, Nordmann P. 2000. Biochemical sequence analyses of GES-1, a novel class A extended-spectrum β-lactamase, and the class 1 integron In52 from Klebsiella pneumoniae. Antimicrob Agents Chemother 44:622–632. doi: 10.1128/AAC.44.3.622-632.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradford PA, Bratu S, Urban C, Visalli M, Mariano N, Landman D, Rahal JJ, Brooks S, Cebular S, Quale J. 2004. Emergence of carbapenem-resistant Klebsiella species possessing the class A carbapenem-hydrolyzing KPC-2 and inhibitor-resistant TEM-30 β-lactamases in New York City. Clin Infect Dis 39:55–60. doi: 10.1086/421495. [DOI] [PubMed] [Google Scholar]
- 8.Zong Z. 2012. Discovery of blaOXA-199, a chromosome-based blaOXA-48-like variant, in Shewanella xiamenensis. PLoS One 7:e48280. doi: 10.1371/journal.pone.0048280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balm MN, La MV, Krishnan P, Jureen R, Lin RT, Teo JW. 2013. Emergence of Klebsiella pneumoniae co-producing NDM-type and OXA-181 carbapenemases. Clin Microbiol Infect 19:E421–E423. doi: 10.1111/1469-0691.12247. [DOI] [PubMed] [Google Scholar]
- 10.Hall JM, Corea E, Sanjeewani HD, Inglis TJ. 2014. Molecular mechanisms of β-lactam resistance in carbapenemase-producing Klebsiella pneumoniae from Sri Lanka. J Med Microbiol 63:1087–1092. doi: 10.1099/jmm.0.076760-0. [DOI] [PubMed] [Google Scholar]
- 11.Williamson DA, Heffernan H, Sidjabat H, Roberts SA, Paterson DL, Smith M, Freeman JT. 2011. Intercontinental transfer of OXA-181-producing Klebsiella pneumoniae into New Zealand. J Antimicrob Chemother 66:2888–2890. doi: 10.1093/jac/dkr396. [DOI] [PubMed] [Google Scholar]
- 12.Peirano G, Ahmed-Bentley J, Fuller J, Rubin JE, Pitout JD. 2014. Travel-related carbapenemase-producing Gram-negative bacteria in Alberta, Canada: the first 3 years. J Clin Microbiol 52:1575–1581. doi: 10.1128/JCM.00162-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruppe E, Armand-Lefevre L, Estellat C, El-Mniai A, Boussadia Y, Consigny PH, Girard PM, Vittecoq D, Bouchaud O, Pialoux G, Esposito-Farese M, Coignard B, Lucet JC, Andremont A, Matheron S. 2014. Acquisition of carbapenemase-producing Enterobacteriaceae by healthy travellers to India, France, February 2012 to March 2013. Euro Surveill 19: pii=20768 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20768. [DOI] [PubMed] [Google Scholar]
- 14.Kalpoe JS, Al Naiemi N, Poirel L, Nordmann P. 2011. Detection of an Ambler class D OXA-48-type β-lactamase in a Klebsiella pneumoniae strain in The Netherlands. J Med Microbiol 60:677–678. doi: 10.1099/jmm.0.028308-0. [DOI] [PubMed] [Google Scholar]
- 15.Samuelsen O, Naseer U, Karah N, Lindemann PC, Kanestrom A, Leegaard TM, Sundsfjord A. 2013. Identification of Enterobacteriaceae isolates with OXA-48 and coproduction of OXA-181 and NDM-1 in Norway. J Antimicrob Chemother 68:1682–1685. doi: 10.1093/jac/dkt058. [DOI] [PubMed] [Google Scholar]
- 16.Dortet L, Poirel L, Al Yaqoubi F, Nordmann P. 2012. NDM-1, OXA-48 and OXA-181 carbapenemase-producing Enterobacteriaceae in Sultanate of Oman. Clin Microbiol Infect 18:E144–E148. doi: 10.1111/j.1469-0691.2012.03796.x. [DOI] [PubMed] [Google Scholar]
- 17.Szekely E, Damjanova I, Janvari L, Vas KE, Molnar S, Bilca DV, Lorinczi LK, Toth A. 2013. First description of blaNDM-1, blaOXA-48, blaOXA-181 producing Enterobacteriaceae strains in Romania. Int J Med Microbiol 303:697–700. doi: 10.1016/j.ijmm.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 18.Brink AJ, Coetzee J, Corcoran C, Clay CG, Hari-Makkan D, Jacobson RK, Richards GA, Feldman C, Nutt L, van Greune J, Deetlefs JD, Swart K, Devenish L, Poirel L, Nordmann P. 2013. Emergence of OXA-48 and OXA-181 carbapenemases among Enterobacteriaceae in South Africa and evidence of in vivo selection of colistin resistance as a consequence of selective decontamination of the gastrointestinal tract. J Clin Microbiol 51:369–372. doi: 10.1128/JCM.02234-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dimou V, Dhanji H, Pike R, Livermore DM, Woodford N. 2012. Characterization of Enterobacteriaceae producing OXA-48-like carbapenemases in the UK. J Antimicrob Chemother 67:1660–1665. doi: 10.1093/jac/dks124. [DOI] [PubMed] [Google Scholar]
- 20.Potron A, Nordmann P, Lafeuille E, Al Maskari Z, Al Rashdi F, Poirel L. 2011. Characterization of OXA-181, a carbapenem-hydrolyzing class D β-lactamase from Klebsiella pneumoniae. Antimicrob Agents Chemother 55:4896–4899. doi: 10.1128/AAC.00481-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 23.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turrientes MC, Gonzalez-Alba JM, del Campo R, Baquero MR, Canton R, Baquero F, Galan JC. 2014. Recombination blurs phylogenetic groups routine assignment in Escherichia coli: setting the record straight. PLoS One 9:e105395. doi: 10.1371/journal.pone.0105395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vervoort J, Gazin M, Kazma M, Kotlovsky T, Lammens C, Carmeli Y, Goossens H, Malhotra-Kumar S, SATURN WP1 and MOSAR WP2 study groups. 2014. High rates of intestinal colonisation with fluoroquinolone-resistant ESBL-harbouring Enterobacteriaceae in hospitalised patients with antibiotic-associated diarrhoea. Eur J Clin Microbiol Infect Dis 33:2215–2221. doi: 10.1007/s10096-014-2193-9. [DOI] [PubMed] [Google Scholar]
- 26.Chen YT, Lin JC, Fung CP, Lu PL, Chuang YC, Wu TL, Siu LK. 2014. KPC-2-encoding plasmids from Escherichia coli and Klebsiella pneumoniae in Taiwan. J Antimicrob Chemother 69:628–631. doi: 10.1093/jac/dkt409. [DOI] [PubMed] [Google Scholar]
- 27.Huber H, Zweifel C, Wittenbrink MM, Stephan R. 2013. ESBL-producing uropathogenic Escherichia coli isolated from dogs and cats in Switzerland. Vet Microbiol 162:992–996. doi: 10.1016/j.vetmic.2012.10.029. [DOI] [PubMed] [Google Scholar]
- 28.Clermont O, Bonacorsi S, Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol 66:4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Villa L, Carattoli A, Nordmann P, Carta C, Poirel L. 2013. Complete sequence of the IncT-type plasmid pT-OXA-181 carrying the blaOXA-181 carbapenemase gene from Citrobacter freundii. Antimicrob Agents Chemother 57:1965–1967. doi: 10.1128/AAC.01297-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang P, Xie Y, Feng P, Zong Z. 2014. blaNDM-5 carried by an IncX3 plasmid in Escherichia coli of the sequence type 167. Antimicrob Agents Chemother 58:7548–7552. doi: 10.1128/AAC.03911-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ho P, Li Z, Lo W, Cheung Y, Lin C, Sham P, Cheng V, Ng T, Que T, Chow K. 2012. Identification and characterization of a novel incompatibility group X3 plasmid carrying blaNDM-1 in Enterobacteriaceae isolates with epidemiological links to multiple geographical areas in China. Emerg Microbes and Infect 1:e39. doi: 10.1038/emi.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen L, Hu H, Chavda KD, Zhao S, Liu R, Liang H, Zhang W, Wang X, Jacobs MR, Bonomo RA, Kreiswirth BN. 2014. Complete sequence of a KPC-producing IncN multidrug-resistant plasmid from an epidemic Escherichia coli sequence type 131 strain in China. Antimicrob Agents Chemother 58:2422–2425. doi: 10.1128/AAC.02587-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poirel L, Decousser JW, Nordmann P. 2003. Insertion sequence ISEcp1B is involved in expression and mobilization of a blaCTX-M β-lactamase gene. Antimicrob Agents Chemother 47:2938–2945. doi: 10.1128/AAC.47.9.2938-2945.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zong Z, Partridge SR, Iredell JR. 2010. ISEcp1-mediated transposition and homologous recombination can explain the context of blaCTX-M-62 linked to qnrB2. Antimicrob Agents Chemother 54:3039–3042. doi: 10.1128/AAC.00041-10. [DOI] [PMC free article] [PubMed] [Google Scholar]


