Abstract
Two new natural CphA metallo-β-lactamases, the CphA4 and CphA5 enzymes, were identified in water samples from municipal sewage in central Italy. Compared to CphA, the CphA4 and CphA5 enzymes showed numerous point mutations. These enzymes have a narrow spectrum of substrates focused on carbapenems only. CphA5 showed kcat values about 40-, 12-, and 97-fold higher than those observed for CphA4 versus imipenem, ertapenem, and biapenem, respectively.
TEXT
Even though Aeromonas species are generally isolated from rivers, lakes, drinking water, groundwater, wastewater natural soil, foods, and animals, some species (Aeromonas hydrophila, A. veronii, and A. caviae) are also involved in not very frequent but serious human infections, such as gastroenteritis, respiratory infections, and septicemia, in both immunocompetent and immunocompromised persons (1). Members of the genus Aeromonas have developed a high level of resistance to antibiotics, in particular to β-lactams and fluoroquinolones (2, 3). Several classes of β-lactamases are identified in Aeromonas spp., even though metallo-β-lactamases (MBLs), which belong to class B, are the usually identified most often (4, 5). In particular, in A. hydrophila, the most frequently encountered β-lactamase is the CphA enzyme, a metallo-β-lactamase included in subclass B2, which is characterized by a narrow specificity profile (especially carbapenems), and it is active with one zinc ion (6–8). In the present study, wastewater samples were collected from urban sewage effluents before the municipal treatment plant in L'Aquila, Italy. The collection was carried out from December 2011 to November 2012. The samples were treated in a laboratory, as previously reported (9). Briefly, samples (approximately 50 ml) were routinely collected the first Monday of each month from five sampling sites: site A, water from L'Aquila Hospital; site B, domestic sewage of the city; site C, upstream of the treatment plant, which collects water from A and B; site D, activated sludge; and site E, water collected after passage through the chlorination system. The dried sludge was collected (weighed in grams) and dissolved under shaking conditions in 10 ml of sterile water. A 100-μl volume of several dilutions (from 10−1 to 10−4) of the samples was plated on MacConkey and cystine-lactose-electrolyte-deficient (CLED) agar media supplemented with meropenem (4 μg/ml) to select potential resistance and carbapenemase producers. The plates were incubated at 37°C for 16 h. The bacteria were identified by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry with the Biotyper 3.0 (Bruker Daltonik, Germany).
Twenty-six environmental Aeromonas isolates (21 isolates of A. hydrophila and 5 isolates of A. veronii) resistant to meropenem were collected from five sampling sites at an urban sewage plant in L'Aquila.
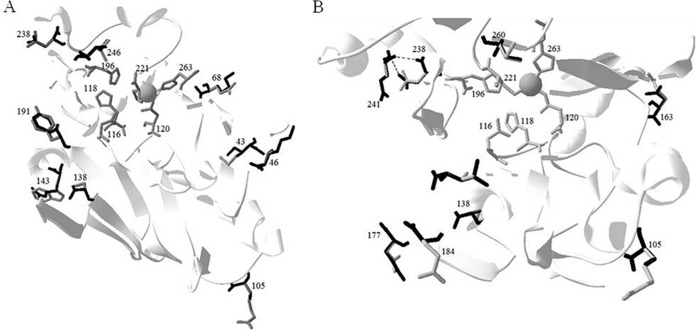
These strains were analyzed for the presence of plasmids, and no plasmids were found in A. hydrophila and A. veronii strains. All strains were investigated by PCR for the presence of blaKPC, blaNDM, blaVIM, blaIMP, blaOXA, and blaCphA determinants. The PCR screening with specific primers for CphA metallo-β-lactamase (CphABam_for, 5′-GGGGGATCCATGATGAAAGGTTGGATG, and CphAXho_rev, 5′-GGGGCTCGAGTTATGACTGGGGTGCGG) showed that all strains of A. veronii and A. hydrophila possess the CphA gene, whereas the remaining probes gave negative results. The positive amplicons were directly sequenced using ABI Prism 310 automated monocapillary sequencer (Life Technologies, Italy), and only two new variants were found. The nucleotide sequence of the amplimer carried a 750-bp open reading frame encoding a CphA enzyme of 227 amino acid residues, which showed all the conserved structural elements typical of the subclass B2 metallo-β-lactamase CphA (6). The new variants identified in this study were compared with the CphA prototype enzyme that was extensively characterized in the last 20 years (6–8). With respect to the CphA enzyme, CphA4 and CphA5 variants present 10 and 9 amino acid substitutions, respectively (Fig. 1). Compared with CphA, in the CphA4 enzyme, the positions with mutations are as follows: G30T, M43I, T46K, Q68K, R105S, S138A, R143W, Y191F, D238N, and L246I. In CphA5, the mutations found are R105S, S138A, E163D, D177E, E184G, V187L, D238N, A241E, and I260V (BBL numbering). Molecular modeling, using the program Swiss-PdbViewer, was carried out using the crystal structure of CphA in complex with biapenem (Protein Data Bank code 1X8G), described by Garau et al. (8). As can be seen in Fig. 2, in both CphA4 and CphA5, some mutated residues are in the proximity of the catalytic zinc and active site residues.
FIG 1.
Amino acid sequence of CphA (AE036), CphA4, and CphA5 enzymes. There is a natural mutation in CphA4 and CphA5 with respect to the CphA prototype.
FIG 2.
Ribbon representation of CphA4 and CphA5 superimposed onto CphA structure (Protein Data Bank 1X8G) described by Garau et al. (8). Shown are overviews with all amino acid substitutions labeled for CphA4 (A) and CphA5 (B). In the structure, we have drawn attention to the zinc ion and the residues of Zn1 (N116-H118-H196) and Zn2 (D120-C221-H263) that interact with the zinc ion. The residue in the CphA enzyme is highlighted in gray, whereas the replaced amino acid residues in CphA4 (A) and CphA5 (B) are highlighted in black.
The genes blaCphA4 and blaCphA5 were digested by KpnI/XhoI and BamHI/XhoI restriction enzymes and cloned into pBC-SK and pET-26(b) vectors. The recombinant plasmids pBC-CphA4 and pBC-CphA5 were inserted by transformation into Escherichia coli strain XL-1 Blue, whereas pET26-CphA4 and pET26-CphA5 were inserted into E. coli strain BL21(DE3). The phenotypic profiles of E. coli XL-1/pBC-CphA4 and E. coli XL-1 Blue/pBC-CphA5 versus a large panel of antibiotics were characterized by a microdilution method using a bacterial inoculum of 5 × 105 CFU/ml, according to Clinical and Laboratory Standards Institute (CLSI) performance standards (10). The CphA enzymes confer resistance to carbapenems but not to cefazolin, cefotaxime, cefepime, aztreonam, or amoxicillin (Table 1). The maximum concentration used for imipenem, meropenem, and ertapenem was 2,048 μg/ml. At high carbapenem concentrations, differences between the three strains were observed (Table 1).
TABLE 1.
Antibiotic resistance phenotype of E. coli XL-1 Blue/pBC-CphA, XL-1 Blue/pBC-CphA4, XL-1 Blue/pBC-CphA5, and XL-1 Blue/pBC-SK
| Antibiotic | MIC (μg/ml) for E. coli: |
|||
|---|---|---|---|---|
| XL-1/pBC-CphA | XL-1/pBC-CphA4 | XL-1/pBC-CphA5 | XL-1/pBC-SK | |
| Amoxicillin | 16 | 8 | 8 | 0.125 |
| Imipenem | 256 | 128 | 512 | 0.5 |
| Meropenem | 512 | 256 | 128 | 0.5 |
| Ertapenem | 128 | 256 | 1,024 | 0.5 |
| Aztreonam | 0.125 | 0.125 | 0.125 | 0.125 |
| Cefepime | 1 | 1 | 1 | 1 |
| Cefotaxime | 0.5 | 0.5 | 0.5 | 0.125 |
| Cefazolin | 2 | 1 | 2 | 0.5 |
The CphA4 and CphA5 MBLs were purified from E. coli BL21(DE3)/pET26-CphA4 and E. coli BL21(DE3)/pET26-CphA5 strains grown overnight at 37°C in 1 liter of brain heart infusion broth (BHI). The crude extract was prepared in 30 ml of 15 mM sodium cacodylate buffer (pH 6.3) (buffer A) by sonication (five times for 30 s each time at 60 W), followed by high-speed centrifugation (105,000 × g for 30 min at 4°C). The cleared extract was loaded onto an SP-Sepharose fast-flow (FF) column equilibrated with buffer A, and the β-lactamase was eluted with a linear gradient of NaCl (0 to 0.5 M) in buffer A. The fractions containing imipenem-hydrolyzing activity were pooled, dialyzed against buffer A, and loaded onto a Sephacryl S-100 equilibrated with the same buffer. The active fractions were pooled, concentrated 20-fold with an Amicon concentrator (YM-10 membrane; Millipore, Bedford, MA), and dialyzed against buffer A. At the end of two chromatographic steps, the CphA4 and CphA5 enzymes were >95% pure, as evaluated by SDS-PAGE analysis. Kinetic parameters were determined by monitoring the hydrolysis of each substrate at 25°C in 15 mM sodium cacodylate buffer (pH 6.3) with a Lambda 25 spectrophotometer (PerkinElmer, Italy). Steady-state kinetic parameters were determined on pure enzymes by analyzing either the complete hydrolysis time courses or under initial rate conditions (11). The effect of zinc content on CphA4 and CphA5 enzyme activity was evaluated by a determination of residual activity for imipenem at 0 to 500 μM ZnCl2. The hydrolysis of ZnCl2 alone at the same concentrations was also measured as with the control. When ZnCl2 concentration increased, the activity of the CphA4 and CphA5 enzymes decreased (data not shown).
CphA4 and CphA5 enzymes hydrolyzed carbapenems very efficiently (Table 2), but benzylpenicillin, cefazolin, cefotaxime and cefepime were poor substrates for them (data not shown). The catalytic efficiency values (kcat/Km) of CphA4 and CphA5 for carbapenems were less than those calculated for the CphA enzyme, with the exception of ertapenem, for which a slight enhancement of the kcat/Km value was observed for CphA5 only. Remarkable differences among the three enzymes were also observed in the Km and kcat values for meropenem. In particular, compared with CphA, reductions of 82- and 115-fold were noted in the meropenem kcat values for the CphA4 and CphA5 enzymes, respectively. The CphA5 enzyme showed kcat values about 2.5-, 4-, and 14-fold higher than those calculated for CphA. Moreover, significant differences between CphA4 and CphA5 in terms of kcat and kcat/Km values were noticed. The CphA4 and CphA5 enzymes showed the same behavior versus meropenem, as their kcat and Km values are similar. The behavior of CphA5 versus imipenem, ertapenem, and biapenem is different from that of the CphA4 natural mutant. In fact, the CphA5 enzyme has kcat values that are 40-, 12-, and 97-fold higher than those observed for CphA4 for the three drugs, respectively.
TABLE 2.
Kinetic parameters of the wild-type CphA and CphA4 and CphA5 enzymesa
| Substrate | CphA |
CphA4 |
CphA5 |
kcat/Km ratio for CphA5/CphA4 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Km (μM) | kcat (s−1) | kcat/Km (μM s−1) | Km (μM) | kcat (s−1) | kcat/Km (μM s−1) | Km (μM) | kcat (s−1) | kcat/Km (μM s−1) | ||
| Imipenem | 110 | 460 | 4.18 | 209 ± 4 | 28 | 0.13 | 453 ± 8 | 1,112 | 2.45 | 18.8 |
| Meropenem | 1,600 | 3,100 | 1.94 | 314 ± 6 | 38 | 0.12 | 374 ± 5 | 27 | 0.07 | 0.58 |
| Ertapenem | 230 | 700 | 3.04 | 688 ± 12 | 250 | 0.36 | 569 ± 15 | 2,963 | 5.21 | 14.5 |
| Biapenem | 1,700 | 120 | 0.07 | 870 ± 50 | 17 | 0.019 | >3,000 | 1,645 | NDb | ND |
Each kinetic value is the mean of five different measurements, with an error of <5%. Plus/minus values indicate the range from the five measurements.
ND, not determined.
In this study, two natural CphA metallo-β-lactamases, isolated from environmental A. veronii and A. hydrophila, were well characterized. Among the amino acid substitutions, the CphA variants showed three identical mutations, R105S, S138A, and D238N. The CphA4 and CphA5 enzymes exhibited a narrow spectrum of substrates compared to that of the CphA prototype enzyme. However, remarkable differences between CphA4 and CphA5 in terms of substrate binding and hydrolysis were observed, in particular for imipenem, ertapenem, and biapenem. The kinetic data are in agreement with the MIC values shown in Table 1. CphA5 very efficiently hydrolyzed imipenem, ertapenem, and biapenem. In particular, the CphA5 enzyme showed a kcat value about 97-fold higher than that calculated for CphA4. Despite CphA prototype and CphA4, in CphA5, the residues N238 and E241 are able to form two hydrogen bonds that might perturb the enzyme structure. This is an interesting example of how the environment can select new β-lactamase variants.
Nucleotide sequence accession numbers.
The nucleotide sequence data of these enzymes appear in the GenBank database under accession numbers KM609958 (CphA4 gene in A. veronii) and KP771880 (CphA5 gene in A. hydrophila).
ACKNOWLEDGMENT
We thank Anna Toso (Toronto Catholic District School Board, Toronto, Canada) for revising the language of the manuscript.
REFERENCES
- 1.Parker JL, Shaw JG. 2011. Aeromonas spp. clinical microbiology and disease. J Infect 62:109–118. doi: 10.1016/j.jinf.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 2.Carvalho MJ, Martínez-Murcia A, Esteves AC, Correia A, Saavedra MJ. 2013. Phylogenetic diversity, antibiotic resistance and virulence traits of Aeromonas spp. from untreated waters from human consumption. Int J Food Microbiol 159:230–239. doi: 10.1016/j.ijfoodmicro.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 3.Skwor T, Shinko J, Augustyniak A, Gee C, Andraso G. 2014. Aeromonas hydrophila and Aeromonas veronii predominate among potentially pathogenic ciprofloxacin- and tetracycline-resistant Aeromonas isolates from Lake Erie. Appl Environ Microbiol 80:841–848. doi: 10.1128/AEM.03645-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rossolini GM, Walsh T, Amicosante G. 1996. The Aeromonas metallo-β-lactamases: genetics, enzymology, and contribution to drug resistance. Microb Drug Resist 2:245–252. doi: 10.1089/mdr.1996.2.245. [DOI] [PubMed] [Google Scholar]
- 5.Neuwirth C, Siebor E, Robin F, Bonnet R. 2007. First occurrence of an IMP metallo-β-lactamase in Aeromonas cavie: IMP-19 in an isolate from France. Antimicrob Agents Chemother 51:4486–4488. doi: 10.1128/AAC.01462-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Segatore B, Massidda O, Satta G, Setacci D, Amicosante G. 1993. High specificity of CphA-encoded metallo-β-lactamase from Aeromonas hydrophila AE036 for carbapenems and its contribution to β-lactam resistance. Antimicrob Agents Chemother 37:1324–1328. doi: 10.1128/AAC.37.6.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hernandez Valladares M, Kiefer M, Heinz U, Soto RP, Meyer-Klaucke W, Nolting HF, Zeppezauer M, Galleni M, Frère JM, Rossolini GM, Amicosante G, Adolph HW. 2000. Kinetic and spectroscopic characterization of native and metal-substituted β-lactamase from Aeromonas hydrophila AE036. FEBS Lett 467:221–225. doi: 10.1016/S0014-5793(00)01102-9. [DOI] [PubMed] [Google Scholar]
- 8.Garau G, Bebrone C, Anne C, Galleni M, Frère JM, Dideberg O. 2005. A metallo-β-lactamase enzyme in action: crystal structures of the monozinc carbapenemase CphA and its complex with biapenem. J Mol Biol 345:785–795. doi: 10.1016/j.jmb.2004.10.070. [DOI] [PubMed] [Google Scholar]
- 9.Perilli M, Bottoni C, Pontieri E, Segatore B, Celenza G, Setacci D, Bellio P, Strom R, Amicosante G. 2013. Emergence of blaKPC-3-Tn4401a in Klebsiella pneumoniae ST512 in the municipal wastewater treatment plant and in the university hospital of a town in central Italy. J Global Antimicrob Resist 1:217–220. doi: 10.1016/j.jgar.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 10.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 7th ed CLSI document M7-A7. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 11.De Meester F, Joris B, Reckinger G, Bellefroid-Bourguignon C, Frère JM, Waley SG. 1987. Automated analysis of enzyme inactivation phenomena. Application to β-lactamases and dd-peptidases. Biochem Pharmacol 36:2393–2403. [DOI] [PubMed] [Google Scholar]