Abstract
We evaluated markers of artemisinin resistance in Plasmodium falciparum isolated in Kampala in 2014. By standard in vitro assays, all isolates were highly sensitive to dihydroartemisinin (DHA). By the ring-stage survival assay, after a 6-h DHA pulse, parasitemia was undetectable in 40 of 43 cultures at 72 h. Two of 53 isolates had nonsynonymous K13-propeller gene polymorphisms but did not have the mutations associated with resistance in Asia. Thus, we did not see evidence for artemisinin resistance in Uganda.
TEXT
Artemisinin-based combination therapy is now the standard for treating P. falciparum malaria. However, this regimen is threatened by resistance to artemisinins, which manifests as delayed clearance of parasitemia after therapy, in Southeast Asia (1, 2). Recently, resistance has been associated with increased parasitemia in culture, compared to that in sensitive parasites, 72 h after a 6-h pulse with dihydroartemisinin (DHA) and has been associated with polymorphisms in propeller-encoding domains of the Plasmodium falciparum kelch (K13; UniProt number PF3D7_1343700) gene (3–6). Although artemisinin resistance is evident to date only in Southeast Asia, the bulk of P. falciparum malaria occurs in sub-Saharan Africa. Clinical resistance has not been noted in Africa, as rapid parasite clearance has been the rule in many trials, including those in Uganda (7–9). In recent surveys in Uganda (10) and other locations in Africa (11–15), K13-propeller gene polymorphisms were identified, but these were not the mutations clearly associated with artemisinin resistance in Asian isolates. Results for a new correlate of artemisinin resistance, the ex vivo ring-stage survival assay (RSA) (3), have not been reported previously for P. falciparum isolates from Africa. We characterized artemisinin sensitivity by this assay and assessed K13 polymorphisms in isolates from Uganda. Parasites were collected from patients diagnosed with malaria from May to August 2014 at Mulago Hospital, Kampala. Samples were delinked from patient information, so clinical data were unavailable. After malaria diagnosis, excess blood collected in a heparinized tube as part of clinical care was promptly delivered to the laboratory. Giemsa-stained thin smears were made, and samples containing only P. falciparum isolates with parasitemia of ≥0.1% were placed in culture after washing of erythrocytes, as previously described (16); blood was also spotted onto filter paper. Samples with parasitemia of >1% were diluted with uninfected erythrocytes for a final parasitemia of 1%.
The susceptibility of fresh isolates to DHA and chloroquine was assessed by a standard ex vivo histidine-rich protein 2-based enzyme-linked immunosorbent microplate assay, as previously described (17), except that drugs were freshly diluted from frozen stocks prior to each assay. Results from single assays were fitted to a variable-slope sigmoidal function using GraphPad Prism 6.0f to generate 50% inhibitory concentrations (IC50s). All data were visually assessed for parasite growth above the background and integrity of curve fits.
Parasite susceptibility to DHA was also assessed using the new ex vivo RSA (3). First, 20-μl erythrocyte pellets were added to 1.0 ml RPMI 1640 medium supplemented with 25 mM HEPES, 0.2% NaHCO3, 0.1 mM hypoxanthine, 100 μg/ml gentamicin, and 0.5% Albumax I (Invitrogen) containing 700 nM DHA or, for controls, 0.1% dimethyl sulfoxide (DMSO) at a hematocrit of 2%. Cultures were incubated at 37°C under 3% O2, 5% CO2, and 92% N2 and, after 6 h, washed 3 times with 10 ml RPMI prewarmed to 37°C; then, they were placed in drug-free culture medium. At 72 h after assay initiation, Giemsa-stained thin smears were prepared. Cultures were considered viable and appropriate for assessment if control parasitemia increased since the culture initiation. Parasitemias in the control cultures were determined by counting parasites per 1,000 to 2,000 erythrocytes. In DHA-pulsed cultures, 10,000 to 20,000 cells were scanned for parasites. RSA survival rates were expressed as the proportion of parasites in the DHA-treated cultures relative to controls at the end of the 72-h assay. For some samples, cultures were maintained for up to 4 weeks after initiation of the experiment, with smears assessed on alternating days. For positive cultures, blood spots were collected on filter paper.
DNA was extracted from filter paper blood spots using Chelex-100, and gene fragments spanning loci of interest were amplified by nested PCR (10, 18). K13-propeller-encoding domains (codons 440 to 726) were dideoxy sequenced, as previously described (4). To detect polymorphisms in pfcrt and pfmdr1, multiplex ligase detection reaction-fluorescent microsphere assays were performed, as previously described (19, 20).
We collected 53 isolates from patients with malaria from May to July 2014. Giemsa-stained thin smears from all blood samples demonstrated P. falciparum only, at parasitemias of 0.1% to 24.3%. We determined ex vivo IC50 data for 15 isolates. Values ranged from 0.4 to 2.7 nM (geometric mean, 1.6 nM) for DHA and 9.8 to 1,060 nM for chloroquine (Table 1).
TABLE 1.
Ex vivo measures of drug sensitivity and molecular features of Ugandan parasitesa
| Sample no. | CQ IC50 (nM [95% CI])b | DHA IC50 (nM [95% CI]) | RSA at 72 h (%)c | K13 | PfCRT aa 76d | PfMDR1 aad |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| 86 | 184 | 1034 | 1024 | 1246 | ||||||
| 1 | 15.6 (14.6–16.6) | 2.0 (1.7–2.4) | 0 | WTe | K | N | F | S | N | D |
| 2 | 123 (84.4–180) | 2.0 (1.5–2.6) | 0 | WT | T | N/Y | Y/F | S | N | Y |
| 3 | 38.8 (23.5–64.3) | 1.1 (1.0–1.3) | 0 | WT | K/T | N/Y | Y/F | S | N | D |
| 4 | 11.2 (9.7–13) | 1.4 (1.2–1.5) | 0 | WT | K | N | F | S | N | Y/D |
| 5 | 260 (154–440) | 2.3 (1.6–3.4) | 0 | WT | T | N | Y | S | N | Y |
| 6 | 196 (127–300) | 1.8 (1.5–2.0) | 0 | WT | T | N | F | S | N | D |
| 7 | 9.8 (9.2–10.5) | 1.3 (0.9–1.7) | 0 | WTf | K | N | F | S | N | D |
| 8 | 155 (133–182) | 2.3 (2.0–2.6) | 0 | WT | T | N | Y/F | S | N | Y |
| 9 | 141 (86.7–228) | 1.5 (1.2–1.9) | 0 | WT | T | N | Y/F | S | N | D |
| 10 | 139 (116–166) | 2.0 (1.8–2.2) | 0.7 | WT | T | N | F | S | N | D |
| 11 | 1060 (733–1520) | 2.2 (2.0–2.5) | 0 | WT | T | N | Y | S | N | D |
| 12 | 222 (169–291) | 1.6 (1.5–1.7) | 0 | A578Sg | T | N | F | S | N | D |
| 13 | 204 (147–283) | 2.7 (2.3–3.2) | 0 | WT | T | N | Y/F | S | N | D |
| 14 | 240 (183–313) | 0.47 (0.45–0.49) | ND | WT | T | N | Y | S | N | D |
| 15 | 182 (144–231) | 0.90 (0.81–0.93) | ND | WT | T | N/Y | Y | S | N | D |
Data are shown for isolates for which IC50 and K13 sequence data are available. WT, wild type; ND, no data collected.
CQ, chloroquine; CI, confidence interval; the 95% CI describes precision of the IC50 determined from the curve fit. Ex vivo IC50 tests are single assays.
Percentage of parasites in DHA-treated cultures relative to controls 72 h after initiation of a 6-h DHA pulse.
aa, amino acid. Wild-type PfCRT is K76, and wild-type PfMDR1 is NYSND.
Synonymous single nucleotide polymorphism (SNP) t1512c.
Synonymous SNP c1434g.
As standard drug sensitivity assays have not identified artemisinin resistance in Asian isolates, we also assessed DHA susceptibility using the ex vivo RSA (3). Ten isolates were lost to contamination or failed to grow. For the remaining 43 isolates, assessed 72 h after culture initiation, most DHA-pulsed cultures contained pyknotic forms, and only 3 of 43 demonstrated healthy-appearing parasites, all with parasitemia that was ≤0.025%. Of the 3 positive cultures, a single parasite was detected per ∼15,000 erythrocytes in 2 cultures, and 3 parasites were detected in the third (RSA survival rates, 0.7% to 1.9%). All 43 untreated control cultures demonstrated increased parasitemia and healthy-appearing parasites over the course of the assay. Because of the challenges of quantifying extremely low parasitemia, we assessed the viability of 14 DHA-pulsed cultures that did not demonstrate parasites after 72 h. These cultures were maintained for up to 30 days after initiation of the experiment. Twelve of these cultures demonstrated parasites, beginning 15 to 29 days after culture initiation; 2 cultures remained negative at 30 days. Thus, we did not observe enhanced growth of DHA-pulsed parasites after 72 h in culture, arguing against the presence of the artemisinin-resistant phenotype, although most cultures survived the DHA pulse.
Sequencing the K13-propeller-encoding domains from the 53 isolates revealed two nonsynonymous mutations, S552C (mixed with the wild-type sequence) and A578S, in propeller domains 2 and 4, respectively (Fig. 1). Ten isolates that grew in culture 15 to 29 days after the DHA pulse all had wild-type K13 sequences. Two of these samples were from isolates that were positive by microscopy 72 h after the DHA pulse. Considering the putative drug transporters encoded by pfcrt and pfmdr1, polymorphism prevalences were similar to those described recently from other regions of Uganda, with an increased prevalence of wild-type sequences at pfcrt K76, pfmdr1 N86, and pfmdr1 D1246 compared to older studies (Fig. 1) (20, 21). Sensitivity to chloroquine correlated well with the pfcrt K76T sequence, with IC50s that were >120 nM for all mutant parasites, <16 nM for all wild-type parasites, and intermediate for one mixed sample (Table 1).
FIG 1.
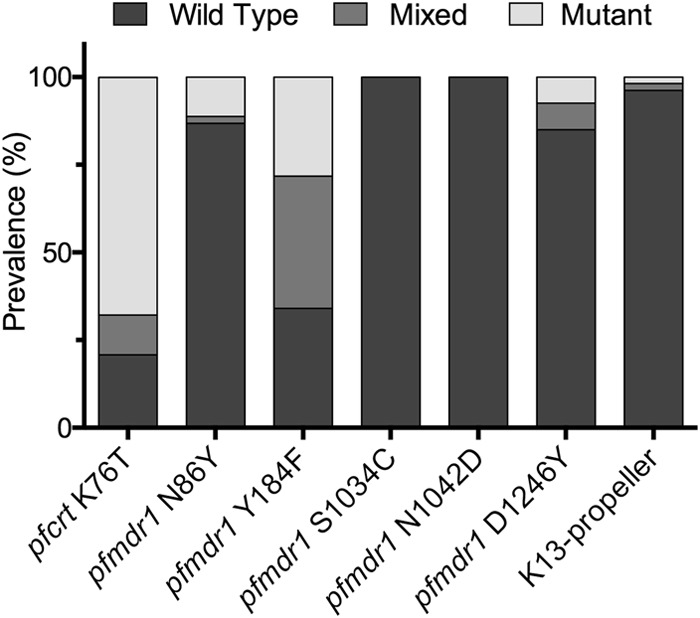
Prevalences of wild-type, mixed, and mutant sequences at the indicated positions.
Artemisinin resistance, with delayed clearance of parasites after treatment with artemisinin monotherapy or artemisinin combination therapies (ACTs), is of great concern but not clearly present in Africa, where clearance of parasites after treatment with ACTs has generally been rapid (2, 13). Parasites from Southeast Asia with delayed clearance typically do not show decreased sensitivity with standard in vitro assays but demonstrate decreased sensitivity to a DHA pulse in the RSA (3) and have specific mutations in K13-propeller-encoding domains (4). We screened fresh P. falciparum isolates from Uganda for these artemisinin resistance markers. Using the RSA, nearly all freshly isolated Ugandan parasites had undetectable parasitemia 72 h after exposure to a pulse of DHA. In contrast, DHA-treated Southeast Asian parasites had measurable parasitemia at 72 h, with parasitemia greater in those with delayed clearance in clinical trials and the presence of K13-propeller gene polymorphisms (3). A small percentage of Ugandan parasites had K13-propeller gene mutations, but these were not the mutations previously associated with drug resistance. Thus, our results suggest that artemisinin resistance is not yet a problem in Uganda.
Recent evidence suggests spreading of artemisinin resistance beyond the initial reports from Cambodia (1), with the delayed clearance phenotype and K13 mutations detected in parts of Thailand, Vietnam, Myanmar, and southern China (2, 6, 22). Additional K13-propeller gene mutations have been identified from other regions of Asia (23) and Africa (11-13, 15), but these have not been among the >10 mutations linked with delayed clearance in Southeast Asia (4, 22). The K13 mutations detected in our study, S522C and A578S, have been found elsewhere in Africa and in Bangladesh, but they were not associated with a resistance phenotype (14, 15, 23). Considering our ring-stage survival assay results in which 40 of 43 DHA-pulsed samples were smear-negative, it appeared that those parasites were more susceptible to a DHA pulse than even susceptible parasites from Southeast Asia (3). With prolonged observation, most samples showed regrowth of parasites, indicating that, although its effect was marked, the 6-h DHA pulse generally did not kill all parasites.
Our results are reassuring concerning artemisinin resistance in Uganda, but they must be considered together with results showing important changes in P. falciparum in the country in recent years. With adoption of artemether-lumefantrine as standard therapy for malaria within the last decade, parasites have demonstrated increasing prevalence of pfcrt and pfmdr1 alleles that mediate decreased lumefantrine sensitivity (20) and, in ex vivo assays, decreasing sensitivity to lumefantrine (17). Thus, even without the artemisinin resistance phenotype, changes in parasite drug sensitivity may threaten the antimalarial efficacies of ACTs, in particular artemether-lumefantrine, in Uganda.
Our study had some limitations. We studied a convenience sample of isolates, and consideration of host factors that may have impacted on parasite biology or genetics was not possible. The total number of isolates studied was small, and we were able to perform some assays only in subsets of these isolates. We also could not determine how an elevated survival phenotype would manifest in our ex vivo assay, since resistance-conferring K13 mutations were not evident in our study. Further, identification of unusual polymorphisms may have been difficult in samples with high complexity of infection, as is common in Uganda. For these reasons we may have missed uncommon resistance markers, but our results nonetheless argue against significant artemisinin resistance in Uganda at this time.
Taken together, results from our parasitological and molecular assessments and from recent clinical trials suggest that artemisinin resistance is not yet an important problem in Uganda. However, the recent spread of resistance in Asia suggests that the introduction or de novo emergence of resistance in Africa, the major world focus of P. falciparum malaria, is likely. Further, altered sensitivity to artemisinin partner drugs already threatens the efficacy of ACTs for the treatment of P. falciparum malaria in Africa (17, 20). Thus, regular surveillance for markers of artemisinin resistance is needed across Africa.
Nucleotide sequence accession numbers.
Nucleotide sequence data are available in the GenBank database under accession numbers KR055739 to KR055804.
ACKNOWLEDGMENTS
We thank Jenny Legac, Michelle Verghese, and Hadijah Nalubega for expert technical assistance.
This work was supported by research grants (AI1R01AI075045 and U19AI089674) and training programs (Training in Malaria Research in Uganda [D43TW007375] and University of California, Berkeley, Minority Health & Health Disparities International Research Training [T37MD003407]), all from the National Institutes of Health. R.A.C. was supported by a travel grant from Dominican University of California.
We have no commercial or other associations that may pose a conflict of interest regarding this research.
REFERENCES
- 1.Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, Lwin KM, Ariey F, Hanpithakpong W, Lee SJ, Ringwald P, Silamut K, Imwong M, Chotivanich K, Lim P, Herdman T, An SS, Yeung S, Singhasivanon P, Day NP, Lindegardh N, Socheat D, White NJ. 2009. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 361:455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashley EA, Dhorda M, Fairhurst RM, Amaratunga C, Lim P, Suon S, Sreng S, Anderson JM, Mao S, Sam B, Sopha C, Chuor CM, Nguon C, Sovannaroth S, Pukrittayakamee S, Jittamala P, Chotivanich K, Chutasmit K, Suchatsoonthorn C, Runcharoen R, Hien TT, Thuy-Nhien NT, Thanh NV, Phu NH, Htut Y, Han KT, Aye KH, Mokuolu OA, Olaosebikan RR, Folaranmi OO, Mayxay M, Khanthavong M, Hongvanthong B, Newton PN, Onyamboko MA, Fanello CI, Tshefu AK, Mishra N, Valecha N, Phyo AP, Nosten F, Yi P, Tripura R, Borrmann S, Bashraheil M, Peshu J, Faiz MA, Ghose A, Hossain MA, Samad R, Rahman MR, Hasan MM, Islam A, Miotto O, Amato R, MacInnis B, Stalker J, Kwiatkowski DP, Bozdech Z, Jeeyapant A, Cheah PY, Sakulthaew T, Chalk J, Intharabut B, Silamut K, Lee SJ, Vihokhern B, Kunasol C, Imwong M, Tarning J, Taylor WJ, Yeung S, Woodrow CJ, Flegg JA, Das D, Smith J, Venkatesan M, Plowe CV, Stepniewska K, Guerin PJ, Dondorp AM, Day NP, White NJ, Tracking Resistance to Artemisinin Collaboration (TRAC) . 2014. Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 371:411–423. doi: 10.1056/NEJMoa1314981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Witkowski B, Amaratunga C, Khim N, Sreng S, Chim P, Kim S, Lim P, Mao S, Sopha C, Sam B, Anderson JM, Duong S, Chuor CM, Taylor WR, Suon S, Mercereau-Puijalon O, Fairhurst RM, Menard D. 2013. Novel phenotypic assays for the detection of artemisinin-resistant Plasmodium falciparum malaria in Cambodia: in vitro and ex vivo drug-response studies. Lancet Infect Dis 13:1043–1049. doi: 10.1016/S1473-3099(13)70252-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois AC, Khim N, Kim S, Duru V, Bouchier C, Ma L, Lim P, Leang R, Duong S, Sreng S, Suon S, Chuor CM, Bout DM, Menard S, Rogers WO, Genton B, Fandeur T, Miotto O, Ringwald P, Le Bras J, Berry A, Barale JC, Fairhurst RM, Benoit-Vical F, Mercereau-Puijalon O, Menard D. 2014. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature 505:50–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghorbal M, Gorman M, Macpherson CR, Martins RM, Scherf A, Lopez-Rubio JJ. 2014. Genome editing in the human malaria parasite Plasmodium falciparum using the CRISPR-Cas9 system. Nat Biotechnol 32:819–821. doi: 10.1038/nbt.2925. [DOI] [PubMed] [Google Scholar]
- 6.Straimer J, Gnadig NF, Witkowski B, Amaratunga C, Duru V, Ramadani AP, Dacheux M, Khim N, Zhang L, Lam S, Gregory PD, Urnov FD, Mercereau-Puijalon O, Benoit-Vical F, Fairhurst RM, Menard D, Fidock DA. 2015. Drug resistance: K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science 347:428–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dorsey G, Staedke S, Clark TD, Njama-Meya D, Nzarubara B, Maiteki-Sebuguzi C, Dokomajilar C, Kamya MR, Rosenthal PJ. 2007. Combination therapy for uncomplicated falciparum malaria in Ugandan children: a randomized trial. JAMA 297:2210–2219. doi: 10.1001/jama.297.20.2210. [DOI] [PubMed] [Google Scholar]
- 8.Arinaitwe E, Sandison TG, Wanzira H, Kakuru A, Homsy J, Kalamya J, Kamya MR, Vora N, Greenhouse B, Rosenthal PJ, Tappero J, Dorsey G. 2009. Artemether-lumefantrine versus dihydroartemisinin-piperaquine for falciparum malaria: a longitudinal, randomized trial in young Ugandan children. Clin Infect Dis 49:1629–1637. doi: 10.1086/647946. [DOI] [PubMed] [Google Scholar]
- 9.Muhindo MK, Kakuru A, Jagannathan P, Talisuna A, Osilo E, Orukan F, Arinaitwe E, Tappero JW, Kaharuza F, Kamya MR, Dorsey G. 2014. Early parasite clearance following artemisinin-based combination therapy among Ugandan children with uncomplicated Plasmodium falciparum malaria. Malar J 13:32. doi: 10.1186/1475-2875-13-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conrad MD, Bigira V, Kapisi J, Muhindo M, Kamya MR, Havlir DV, Dorsey G, Rosenthal PJ. 2014. Polymorphisms in K13 and falcipain-2 associated with artemisinin resistance are not prevalent in Plasmodium falciparum isolated from Ugandan children. PLoS One 9:e105690. doi: 10.1371/journal.pone.0105690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Torrentino-Madamet M, Fall B, Benoit N, Camara C, Amalvict R, Fall M, Dionne P, Ba Fall K, Nakoulima A, Diatta B, Dieme Y, Menard D, Wade B, Pradines B. 2014. Limited polymorphisms in k13 gene in Plasmodium falciparum isolates from Dakar, Senegal in 2012 to 2013. Malar J 13:472. doi: 10.1186/1475-2875-13-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Escobar C, Pateira S, Lobo E, Lobo L, Teodosio R, Dias F, Fernandes N, Arez AP, Varandas L, Nogueira F. 2015. Polymorphisms in Plasmodium falciparum K13-propeller in Angola and Mozambique after the introduction of the ACTs. PLoS One 10:e0119215. doi: 10.1371/journal.pone.0119215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ouattara A, Kone A, Adams M, Fofana B, Maiga AW, Hampton S, Coulibaly D, Thera MA, Diallo N, Dara A, Sagara I, Gil JP, Bjorkman A, Takala-Harrison S, Doumbo OK, Plowe CV, Djimde AA. 2015. Polymorphisms in the K13-propeller gene in artemisinin-susceptible Plasmodium falciparum parasites from Bougoula-Hameau and Bandiagara, Mali. Am J Trop Med Hyg 92:1202–1206. doi: 10.4269/ajtmh.14-0605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamau E, Campino S, Amenga-Etego L, Drury E, Ishengoma D, Johnson K, Mumba D, Kekre M, Yavo W, Mead D, Bouyou-Akotet M, Apinjoh T, Golassa L, Randrianarivelojosia M, Andagalu B, Maiga-Ascofare O, Amambua-Ngwa A, Tindana P, Ghansah A, MacInnis B, Kwiatkowski D, Djimde AA. 2015. K13-propeller polymorphisms in Plasmodium falciparum parasites from sub-Saharan Africa. J Infect Dis 211:1352–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taylor SM, Parobek CM, DeConti DK, Kayentao K, Coulibaly SO, Greenwood BM, Tagbor H, Williams J, Bojang K, Njie F, Desai M, Kariuki S, Gutman J, Mathanga DP, Martensson A, Ngasala B, Conrad MD, Rosenthal PJ, Tshefu AK, Moormann AM, Vulule JM, Doumbo OK, Ter Kuile FO, Meshnick SR, Bailey JA, Juliano JJ. 2015. Absence of putative artemisinin resistance mutations among Plasmodium falciparum in sub-Saharan Africa: a molecular epidemiologic study. J Infect Dis 211:680–688. doi: 10.1093/infdis/jiu467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nsobya SL, Kiggundu M, Nanyunja S, Joloba M, Greenhouse B, Rosenthal PJ. 2010. In vitro sensitivities of Plasmodium falciparum to different antimalarial drugs in Uganda. Antimicrob Agents Chemother 54:1200–1206. doi: 10.1128/AAC.01412-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tumwebaze P, Conrad MD, Walakira A, LeClair N, Byaruhanga O, Nakazibwe C, Kozak B, Bloome J, Okiring J, Kakuru A, Bigira V, Kapisi J, Legac Gut JJ, Cooper RA, Kamya M, Havlir DV, Dorsey G, Greenhouse B, Nsobya SL, Rosenthal PJ. 2015. Impact of antimalarial treatment and chemoprevention on the drug sensitivity of malaria parasites isolated from Ugandan children. Antimicrob Agents Chemother 59:3018–3030. doi: 10.1128/AAC.05141-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plowe CV, Djimde A, Bouare M, Doumbo O, Wellems TE. 1995. Pyrimethamine and proguanil resistance-conferring mutations in Plasmodium falciparum dihydrofolate reductase: polymerase chain reaction methods for surveillance in Africa. Am J Trop Med Hyg 52:565–568. [DOI] [PubMed] [Google Scholar]
- 19.LeClair NP, Conrad MD, Baliraine FN, Nsanzabana C, Nsobya SL, Rosenthal PJ. 2013. Optimization of a ligase detection reaction-fluorescent microsphere assay for characterization of resistance-mediating polymorphisms in African samples of Plasmodium falciparum. J Clin Microbiol 51:2564–2570. doi: 10.1128/JCM.00904-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Conrad MD, LeClair N, Arinaitwe E, Wanzira H, Kakuru A, Bigira V, Muhindo M, Kamya MR, Tappero JW, Greenhouse B, Dorsey G, Rosenthal PJ. 2014. Comparative impacts over 5 years of artemisinin-based combination therapies on Plasmodium falciparum polymorphisms that modulate drug sensitivity in Ugandan children. J Infect Dis 210:344–353. doi: 10.1093/infdis/jiu141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mbogo GW, Nankoberanyi S, Tukwasibwe S, Baliraine FN, Nsobya SL, Conrad MD, Arinaitwe E, Kamya M, Tappero J, Staedke SG, Dorsey G, Greenhouse B, Rosenthal PJ. 2014. Temporal changes in prevalence of molecular markers mediating antimalarial drug resistance in a high malaria transmission setting in Uganda. Am J Trop Med Hyg 91:54–61. doi: 10.4269/ajtmh.13-0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang F, Takala-Harrison S, Jacob CG, Liu H, Sun X, Yang H, Nyunt MM, Adams M, Zhou S, Xia Z, Ringwald P, Bustos MD, Tang L, Plowe CV. 24 April 2015. A single mutation in K13 predominates in southern China and is associated with delayed clearance of Plasmodium falciparum following artemisinin treatment. J Infect Dis doi: 10.1093/infdis/jiv249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mohon AN, Alam MS, Bayih AG, Folefoc A, Shahinas D, Haque R, Pillai DR. 2014. Mutations in Plasmodium falciparum K13-propeller gene from Bangladesh (2009 to 2013). Malar J 13:431. doi: 10.1186/1475-2875-13-431. [DOI] [PMC free article] [PubMed] [Google Scholar]


