Abstract
The continuous emergence of multidrug-resistant pathogenic bacteria is compromising the successful treatment of serious microbial infections. GSK1322322, a novel peptide deformylase (PDF) inhibitor, shows good in vitro antibacterial activity and has demonstrated safety and efficacy in human proof-of-concept clinical studies. In vitro studies were performed to determine the frequency of resistance (FoR) to this antimicrobial agent in major pathogens that cause respiratory tract and skin infections. Resistance to GSK1322322 occurred at high frequency through loss-of-function mutations in the formyl-methionyl transferase (FMT) protein in Staphylococcus aureus (4/4 strains) and Streptococcus pyogenes (4/4 strains) and via missense mutations in Streptococcus pneumoniae (6/21 strains), but the mutations were associated with severe in vitro and/or in vivo fitness costs. The overall FoR to GSK1322322 was very low in Haemophilus influenzae, with only one PDF mutant being identified in one of four strains. No target-based mutants were identified from S. pyogenes, and only one or no PDF mutants were isolated in three of the four S. aureus strains studied. In S. pneumoniae, PDF mutants were isolated from only six of 21 strains tested; an additional 10 strains did not yield colonies on GSK1322322-containing plates. Most of the PDF mutants characterized from those three organisms (35/37 mutants) carried mutations in residues at or in close proximity to one of three highly conserved motifs that are part of the active site of the PDF protein, with 30 of the 35 mutations occurring at position V71 (using the S. pneumoniae numbering system).
INTRODUCTION
The continuous appearance and dissemination of multidrug resistance among pathogenic bacteria are compromising the successful treatment of serious microbial infections, and alternative therapies are urgently needed. The development of new agents against clinically unexploited targets provides the additional advantage of preventing potential cross-resistance with antibiotics already being marketed. One such target is peptide deformylase (PDF), a highly conserved bacterial metalloprotease that hydrolyzes the N-terminal formyl group from all nascent polypeptides (1–3) and plays an essential role in protein maturation. A large number of structurally diverse PDF inhibitors have been identified over the years, including several compounds with demonstrated in vivo efficacy and good safety profiles (4), some of which have progressed to clinical trials (5, 6). GSK1322322 is a novel nonpeptidic PDF inhibitor from the hydrazide class that shows good in vitro antibacterial activity (7) and has demonstrated safety and efficacy in human proof-of-concept clinical studies (8–11).
In bacteria where protein synthesis can be initiated with unformylated Met-tRNAi (therefore bypassing the need for a deformylation step), the main mechanism of resistance to PDF inhibitors involves loss-of-function mutations in genes involved in the formylation of the initiator tRNA; mutations occur mostly in the gene encoding formyl-methionyl transferase (FMT) (12–19) but mutations in FolD and GlyA, two enzymes involved in the synthesis of 10-formyl-tetrahydrofolate, have also been described (14, 16). Such mutants are highly resistant to PDF inhibitors, but they display compromised in vitro (12, 13, 15–18) and in vivo (12, 16, 20) growth. In Staphylococcus aureus, FMT mutants also show drastic reductions in the production of virulence factors and restricted ability to produce invasive infections (20). In organisms in which formylation seems to be essential, such as Streptococcus pneumoniae and Haemophilus influenzae, resistance to PDF inhibitors involves mutations in, or increased production of, the target gene (21–23) or efflux (24). This study was performed to determine the target-based frequency of resistance (FoR) to the PDF inhibitor GSK1322322 in major pathogens causing respiratory tract and skin infections, including H. influenzae, S. aureus, Streptococcus pyogenes, and S. pneumoniae.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All strains used in this study were clinical isolates obtained from the GlaxoSmithKline microbiology department culture collection. S. pneumoniae, S. pyogenes, and S. aureus strains were cultured on trypticase soy agar (TSA) or cation-adjusted Mueller-Hinton (CAMH) agar with 5% defibrinated sheep blood. H. influenzae strains were cultured on chocolate agar II plates. CAMH broth was used for S. aureus, with 3% lysed horse blood added for S. pneumoniae and S. pyogenes. Haemophilus test medium (HTM) was used for H. influenzae. For inoculum preparation for the resistance studies, S. pneumoniae and S. pyogenes strains were grown in Todd Hewitt broth with 0.5% yeast extract (THYB). A total of four strains of H. influenzae, S. aureus, and S. pyogenes and 21 strains of S. pneumoniae were evaluated in these studies.
Antimicrobial agents and susceptibility testing.
The PDF inhibitor GSK1322322 and the pleuromutilin tiamulin were obtained from GlaxoSmithKline Pharmaceuticals (Collegeville, PA) and dissolved in dimethyl sulfoxide (DMSO). MIC endpoints were determined with broth microdilution methodology according to Clinical and Laboratory Standards Institute (CLSI) guidelines (25).
Spontaneous FoR studies.
The frequency of spontaneous resistance was calculated by dividing the number of confirmed resistant colonies growing on antibiotic-containing plates by the total number of CFU in the initial test inoculum. Colonies were defined as resistant if their MICs were ≥4 times the MIC of the wild-type strain. When necessary, the total number of resistant colonies on the plates was extrapolated from a representative set tested.
(i) Preparation of plates.
GSK1322322 was added to the appropriate medium-containing molten agar at 50°C to yield 20 ml of agar at the correct multiple of the MIC for each organism. Plates containing GSK1322322 at 4× MIC and/or 10× MIC were poured and left to cool and to solidify. Plates containing no compound were also prepared, to be used for determination of viable counts and observation of growth.
(ii) Preparation of inocula.
Cultures were prepared by inoculating broth with a bacterial suspension in saline solution, made from plates derived from individual colonies (S. pneumoniae, S. aureus, and H. influenzae) or by dilution of overnight cultures (S. pyogenes). The cultures were incubated at 35°C, with (S. pneumoniae, S. aureus, and H. influenzae) or without (S. pyogenes) agitation, in the presence of CO2 (S. pneumoniae and H. influenzae), until the broth was visibly turbid. Cultures were centrifuged and cell pellets were resuspended to an appropriate cell concentration in fresh medium. Cell concentrations of 1 to 5 × 108 CFU/ml (S. pneumoniae and S. pyogenes) or 1 × 109 CFU/ml (S. aureus and H. influenzae) were targeted.
(iii) Plating of bacterial suspensions.
To determine the number of CFU present in the initial test inoculum, each organism suspension was serially diluted 1:10, and three 20-μl drops from the 10−4 to 10−8 dilutions were plated on the appropriate agar plates and incubated overnight at 35°C, in the presence of 5% CO2 (H. influenzae and S. pneumoniae). Counts were performed at the dilution that provided distinguishable colonies, and an average of the three samples was used to calculate the number of CFU in the original suspension. Colonies were counted after 48 h of incubation at 35°C in ambient air, with the aid of a magnifying glass when necessary.
(iv) Confirmation of resistance phenotype.
Colony size relative to that of wild-type colonies was noted, and single colonies from each drug-organism combination were streaked onto new plates containing identical drug concentrations or no drug. Plates were incubated at 35°C, and growth and resistance were evaluated at 24 and 48 h. In cases in which a large number of mutants were obtained, only a representative set of colonies was tested. In the cases of S. aureus and S. pyogenes, colonies were also analyzed for production of hemolysin in blood agar plates, as it is known that S. aureus fmt mutants are nonhemolytic (20).
Genetic characterization of isolates with reduced susceptibility to GSK1322322.
The PCR primers used for DNA amplification of the pdf and fmt genes were designed from the appropriate regions of the corresponding publically available genomic sequences by using Lasergene PrimerSelect software. Although S. pneumoniae and S. pyogenes contain a second pdf gene, i.e., def2, this was not amplified because it has been reported that def2 encodes an inactive protein (21). In addition, PCR amplification of folD and glyA, which encode bifunctional 5,10-methylene-tetrahydrofolate dehydrogenase/cyclohydrolase and serine hydroxymethyltransferase, respectively, was performed with selected S. aureus and S. pneumoniae mutants. The PCR templates were prepared by boiling cells collected from overnight plates. PCR products were purified using the QIAquick PCR purification kit and were sequenced using the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems). Sequencing reaction mixtures were purified using the Performa DTR V3 96-well short plate kit (Edge Bio) and were analyzed with a 3730XL genetic analyzer (Applied Biosystems). In order to identify specific mutations responsible for the resistance phenotype, alignment of the DNA sequences of the pdf, fmt, folD, and glyA genes and promoter regions from mutant strains and their corresponding parent organisms was carried out using Lasergene MegAlign software (DNAStar).
Growth curves for S. pyogenes strains.
S. pyogenes cells that had been grown on TSA with 5% sheep blood at 35°C for approximately 20 h were resuspended in 6 ml of THYB and adjusted to an optical density at 600 nm (OD600) (1-cm path length) of 0.01. The cultures were incubated at 35°C without agitation, and growth was monitored at different time points by assessing optical density at 600 nm and viable bacterial counts.
Competitive growth studies of parent and mutant S. pneumoniae strains.
For the preparation of early-log-phase cultures, a suspension equivalent to a 0.5 McFarland standard from an overnight agar plate was diluted 50-fold into 4 ml of THYB and incubated for approximately 4 h in the presence of 5% CO2, with shaking, to an OD600 of approximately 0.05. Competitive growth studies were carried out in THYB at 35°C in 5% CO2, with continuous shaking, using initial inocula from early-log-phase cultures to give a 1:1 mutant/wild-type ratio. At specific time points, samples were serially diluted and plated with or without selective concentrations of GSK1322322, in order to calculate mutant and wild-type CFU/ml values as described above. At the end of each growth cycle (4 h), cultures were diluted into fresh medium to start another cycle.
RESULTS
H. influenzae.
Studies were performed to determine both the overall and target-based FoR values for four strains of H. influenzae at 4 and 10 times the GSK1322322 MIC. The FoR was low in this species, and mutants with reduced susceptibility to GSK1322322 were not isolated for three of the four strains tested, resulting in a FoR of <1 × 10−9 (Table 1). Only one colony was identified from strain H. influenzae 222270, giving an overall FoR of 3 × 10−9. No phenotypic differences from the wild type were noted for this strain, which carried a mutation in the PDF protein (I45N) that was probably responsible for the >16-fold increase observed in the GSK1322322 MIC relative to the parent strain.
TABLE 1.
Frequency of resistance to GSK1322322 in H. influenzae strains and genetic characterization of mutants
| H. influenzae strain | Frequency of resistance |
Mutant characteristicsa |
|||||
|---|---|---|---|---|---|---|---|
| Overall |
Target-based |
GSK1322322 MIC (μg/ml) |
Genotype | ||||
| 4× MIC | 10× MIC | 4× MIC | 10× MIC | Wild-type | Mutant | ||
| H128 | <1 × 10−9 | <1 × 10−9 | <1 × 10−9 | <1 × 10−9 | NA | NA | NA |
| 222270 | <3 × 10−9 | 3 × 10−9 | <3 × 10−9 | 3 × 10−9 | 2 | >32 | PDF I45N |
| 195402 | <9 × 10−10 | <9 × 10−10 | <9 × 10−10 | <9 × 10−10 | NA | NA | NA |
| 216580 | <1 × 10−9 | <1 × 10−9 | <1 × 10−9 | <1 × 10−9 | NA | NA | NA |
NA, not applicable.
S. aureus.
Mutations in the S. aureus fmt gene are associated with a number of phenotypic features, including lack of hemolysin production, high-level resistance to PDF inhibitors, a small-colony phenotype, and hypersensitivity to pleuromutilins (20). Therefore, in order to easily identify FMT mutants, FoR studies were performed with four methicillin-resistant hemolytic S. aureus strains. The overall FoR values were high for all strains at both 4 and 10 times the GSK1322322 MIC, with similar frequencies ranging between 5 × 10−7 and 6 × 10−8 (Table 2). In order to determine the target-based FoR, colony size, susceptibility to GSK1322322 and tiamulin, and hemolytic phenotype were evaluated for a representative number of colonies. In every case, nonhemolytic isolates were highly resistant to GSK1322322 (MICs of >64 μg/ml), displayed a small-colony phenotype, and were classified as FMT-like mutants, although not all colonies were hypersensitive to tiamulin. Only FMT-like isolates were identified from the resistance study with S. aureus X31360. DNA sequencing analysis of a number of those isolates, including those that were not hypersensitive to tiamulin, confirmed that they carried mutations in the fmt gene that should lead to inactive FMT proteins, with premature stop codons, frameshifts, or amino acid substitutions known to affect the catalytic mechanism (26). Ten isolates from S. aureus WCUH29 and one each from S. aureus PVL-1 and T6466 were identified as having a hemolytic phenotype, elevated GSK1322322 MICs, and wild-type tiamulin MICs. Genotypic characterization of those isolates showed that most of them carried a mutation at amino acid V59 of the PDF protein. The GSK1322322 MIC increased 32-fold in isolates carrying the V59A substitution and >64-fold in those with a PDF V59D substitution (Table 3). Low-level resistance was associated with changes in the promoter region in two of the mutants, and one isolate had no obvious mutations in the fmt, folD, glyA, or pdf genes or in their promoter regions (Table 3). Resistance to GSK1322322 through target-based mutations occurred in those S. aureus strains at frequencies ranging from 2 × 10−8 to <1 × 10−9 at 4× MIC and from 9 × 10−9 to <8 × 10−10 at 10× MIC (Table 2).
TABLE 2.
Frequency of resistance to GSK1322322 in S. aureus, S. pyogenes, and S. pneumoniae strains and phenotypic characterization of mutants
| Organism | FoRa |
Phenotypic characteristics (no.)b |
|||||
|---|---|---|---|---|---|---|---|
| Overall |
Target-based |
||||||
| 4× MIC | 10× MIC | 4× MIC | 10× MIC | Total | Hemolytic | ≥4-fold decrease in tiamulin MIC | |
| S. aureus | |||||||
| WCUH29c | 3 × 10−7 | 3 × 10−7 | 2 × 10−8 | 9 × 10−9 | 76 | 10 | NA |
| PVL-1c | 6 × 10−8 | 7 × 10−8 | 2 × 10−9 | <9 × 10−10 | 58 | 1 | NA |
| T6466c | 2 × 10−7 | 2 × 10−7 | 5 × 10−9 | <8 × 10−10 | 80 | 1 | NA |
| X31360c | 4 × 10−7 | 5 × 10−7 | <1 × 10−9 | <1 × 10−9 | 80 | 0 | NA |
| S. pyogenes | |||||||
| 201308 | <2 × 10−8 | 1 × 10−7d | <2 × 10−8 | <2 × 10−8 | 6 | NA | 6 |
| 202130 | 3 × 10−7 | 5 × 10−9e | <5 × 10−8 | <5 × 10−9 | 6 | NA | 1 |
| 202370c | 9 × 10−8 | 7 × 10−7 | <3 × 10−8 | <4 × 10−8 | 9 | NA | 9 |
| 63294c | 1 × 10−6 | 7 × 10−7 | <3 × 10−8 | <3 × 10−8 | 16 | NA | 6 |
| S. pneumoniae | |||||||
| 1629 | 1 × 10−8 | 1 × 10−8 | 1 × 10−8 | 1 × 10−8 | 4 | NA | 0 |
| 403346 | 8 × 10−9 | <2 × 10−9 | 8 × 10−9 | <2 × 10−9 | 3 | NA | 0 |
| 1307006S | 3 × 10−9 | <1 × 10−9 | 3 × 10−9 | <1 × 10−9 | 3 | NA | 0 |
| 1302004Sc | 5 × 10−6 | <2 × 10−8 | <2 × 10−8 | <2 × 10−8 | 6 | NA | 0 |
| 340643 | ND | 3 × 10−8 | ND | 3 × 10−8 | 3 | NA | 0 |
| 1314005Sc | ND | 9 × 10−7 | ND | 3 × 10−8 | 24 | NA | 0 |
| 121175 | ND | 9 × 10−9 | ND | <1 × 10−9 | 7 | NA | 6 |
| 339881 | ND | 8 × 10−8 | ND | <2 × 10−8 | 4 | NA | 3 |
| 237442c | ND | 4 × 10−7 | ND | <4 × 10−8 | 11 | NA | 11 |
| 395259c | ND | 1 × 10−6 | ND | <6 × 10−8 | 16 | NA | 16 |
| 302305c | ND | 9 × 10−8 | ND | 2 × 10−9 | 18 | NA | 17 |
| 1309002S | ND | <1 × 10−9 | ND | <1 × 10−9 | NA | NA | NA |
| 164931 | ND | <4 × 10−9 | ND | <4 × 10−9 | NA | NA | NA |
| 193259 | ND | <4 × 10−9 | ND | <4 × 10−9 | NA | NA | NA |
| 239956 | ND | <1 × 10−8 | ND | <1 × 10−8 | NA | NA | NA |
| 245289 | ND | <1 × 10−8 | ND | <1 × 10−8 | NA | NA | NA |
| 245611 | ND | <5 × 10−9 | ND | <5 × 10−9 | NA | NA | NA |
| 300410 | ND | <2 × 10−8 | ND | <2 × 10−8 | NA | NA | NA |
| 335952 | ND | <5 × 10−9 | ND | <5 × 10−9 | NA | NA | NA |
| 456852 | ND | <7 × 10−9 | ND | <7 × 10−9 | NA | NA | NA |
| 428686 | ND | <1 × 10−8 | ND | <1 × 10−8 | NA | NA | NA |
ND, not done.
NA, not applicable.
The FoR was extrapolated from a representative set of colonies that were phenotypically characterized.
All resistant mutants were isolated from 1 of 3 plates (all had the same mutation in the fmt gene).
Results were obtained at 8 and 20 times the broth MIC.
TABLE 3.
Genetic characterization of S. aureus, S. pyogenes, and S. pneumoniae mutants with reduced susceptibility to GSK1322322
| Organism | GSK1322322 MIC (μg/ml) | Genotypic characteristicsa |
|
|---|---|---|---|
| PDF mutation | FMT mutation | ||
| S. aureus | |||
| Parent WCUH29 | 0.5 | NA | NA |
| WCUH29-M1 (n = 7) | 16 | V59A | None |
| WCUH29-M2 (n = 1) | 4 | +T at position −242 | None |
| WCUH29-M3 (n = 1) | 4 | A to G at position −240 | None |
| WCUH29-M4 (n = 1) | 4 | None | None |
| Parent PVL-1 | 2 | NA | NA |
| PVL-1-M1 (n = 1) | >128 | V59D | None |
| Parent T6466 | 2 | NA | NA |
| T6466-M1 (n = 1) | >128 | V59D | None |
| S. pyogenes | |||
| Parent 201308 | 0.5 | NA | NA |
| 201308-M1 (n = 6)b | >64 | None | M228fsc |
| Parent 202130 | 1 | NA | NA |
| 202130-M1 (n = 1)b | >64 | None | D144Nc |
| 202130-M2 (n = 5) | >64 | None | I121S |
| Parent 202370 | 1 | NA | NA |
| 202370-M1 (n = 1)b | >64 | None | I192fs |
| 202370-M2 (n = 1)b | >64 | None | E131stop |
| 202370-M3 (n = 1)b | >64 | None | Q52stopc |
| Parent 63294 | 0.5 | NA | NA |
| 63294-M1 (n = 2)b | >64 | None | FM insertion at L5 |
| 63294-M2 (n = 1)b | >64 | None | P193fs |
| 63294-M3 (n = 5)d | >64 | None | None |
| S. pneumoniae | |||
| Parent 1629 | 1 | NA | NA |
| 1629-M1 (n = 1) | 16 | A123P | None |
| 1629-M2 (n = 1) | 8 | A123D | None |
| 1629-M3 (n = 1) | 16 | V71A | None |
| 1629-M4 (n = 1) | 32 | V71G | None |
| Parent 403346 | 0.06 | NA | NA |
| 403346-M1 (n = 1) | 1 | V71Ac | None |
| 403346-M2 (n = 1) | 16 | V71Dc | None |
| 403346-M3 (n = 1) | 1 | V71F | None |
| Parent 1307006S | 1 | NA | NA |
| 1307006S-M1 (n = 3) | 32 | V71Fc | None |
| Parent 1302004S | 0.125 | NA | NA |
| 1302004S-M1 (n = 6) | 0.5 | None | None |
| Parent 340643 | 1 | NA | NA |
| 340643-M1 (n = 1) | 32 | A123P | None |
| 340643-M2 (n = 2) | 32 | V71F | None |
| Parent 1314005S | 0.5 | NA | NA |
| 1314005S-M1 (n = 6) | 8-16 | V71Fc | None |
| 1314005S-M2 (n = 18)d | 4-32 | None | None |
| Parent 121175 | 1 | NA | NA |
| 121175-M1 (n = 1) | 4 | None | P113S |
| Parent 339881 | 1 | NA | NA |
| 121175-M1 (n = 1) | 64 | None | G300stop |
| Parent 302305 | 0.5 | NA | NA |
| 302305-M1 (n = 1) | 4 | Q57K | None |
NA, not applicable.
Demonstrated ≥4-fold decrease in tiamulin MIC.
Growth studies were performed.
Unstable resistance phenotype when passaged in the absence of drug.
S. pyogenes.
The FoR to GSK1322322 in four S. pyogenes strains ranged from 1 × 10−6 to <2 × 10−8 at 4× MIC and from 7 × 10−7 to 5 × 10−9 at 10× MIC (Table 2). Susceptibility testing of 37 isolates, representing all parent strains, showed that all mutants were highly resistant to GSK1322322, with MICs of >64 μg/ml, and that ∼60% of them were hypersensitive to tiamulin (Table 2), a characteristic that in S. aureus can differentiate FMT mutants from mutants with target-based mutations, which always retain the parent tiamulin MIC. To identify the mutation responsible for the resistance phenotype, both the pdf and fmt genes from a set of 23 isolates, representing all four S. pyogenes parent strains, were amplified by PCR and sequenced. The six isolates obtained from S. pyogenes 201308, all of which showed hypersensitivity to tiamulin (Table 2), carried identical mutations, i.e., insertion of T at nucleotide 684, which caused a frameshift in the FMT protein (Table 3). Genetic characterization of the six isolates from S. pyogenes 202130 revealed that all carried missense mutations in the fmt gene. The five mutants with wild-type tiamulin MICs had identical FMT I121S substitutions. The D144N substitution, which was found in the remaining isolate, conferred hypersensitivity to tiamulin (Table 2). As all nine mutants selected for characterization from S. pyogenes 202370 were hypersensitive to tiamulin (Table 2), only three were genetically characterized. All of them carried loss-of-function mutations that inactivated FMT, with stop codons at position Q52 or E131 or a frameshift at position I192 (Table 3). Six of the 16 mutants selected from S. pyogenes 63294 for phenotypic characterization were hypersensitive to tiamulin. Three of those six mutants were genetically characterized and carried either a 6-bp insertion resulting in the addition of two amino acid residues (FM) at position 5 or a frameshift at position 193 of the FMT protein (Table 3). Five of the 10 mutants that were not hypersensitive to tiamulin had lost their resistance phenotype when they were replated from glycerol stocks. Genetic characterization of the other five mutants revealed no mutations in either the pdf gene or the fmt gene, but further stability studies performed with three of those isolates showed that their resistance phenotype was lost after three passages in the absence of drug. The instability of their resistance phenotype suggests that the original mutants might have carried unstable mutations in FMT that reverted to the wild type in the absence of drug. A mutant-revertant mixed culture would show high-level resistance to GSK1322322 (as resistance is dominant) and wild-type susceptibility to tiamulin (as hypersensitivity is recessive) and, depending on the mutant/revertant ratio, a mutation in the fmt gene might not be detected by PCR amplification.
All S. pyogenes FMT mutants identified in these studies carried mutations that inactivated the protein, had a small/tiny-colony phenotype on plates, and showed decreases in hemolysin production on blood agar plates, in comparison with their corresponding parent strains. In order to evaluate the fitness costs of the FMT mutations, growth studies were performed with three S. pyogenes FMT mutants that represented the major types of mutations encountered, i.e., missense (D144N, in S. pyogenes 202130-M1), nonsense with addition of a stop codon (Q52stop, in S. pyogenes 202370-M3), or frameshift (M228fs, in S. pyogenes 201308-M1). Although all initial inocula had identical optical density values, S. pyogenes FMT mutant strains were substantially less viable than their corresponding parent strains, showing 4 to 60 times lower bacterial counts (Fig. 1). In fact, no obvious growth was observed for the M228fs and D144N mutant strains by either optical density (Fig. 1A) or viable count (Fig. 1B) methodologies. The Q52stop mutant strain grew slightly (Fig. 1A) but showed a substantial (∼2 log10) decrease in viable bacterial counts with respect to its parent strain (Fig. 1B).
FIG 1.
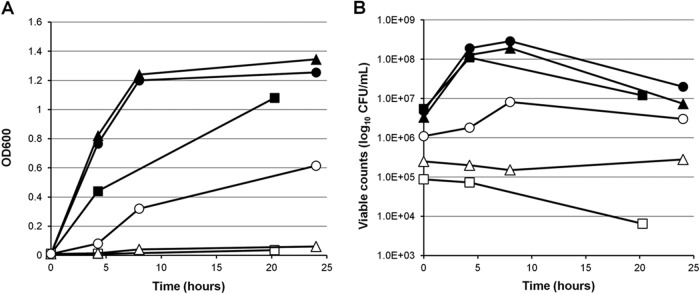
Growth of representative S. pyogenes FMT mutants and their corresponding parent strains, measured as optical density at 600 nm (A) or viable counts (B). ▲, wild-type S. pyogenes 202130; △, S. pyogenes 202130 FMT D144N mutant; ●, wild-type S. pyogenes 202370; ○, S. pyogenes 202370 FMT Q52stop mutant; ■, wild-type S. pyogenes 201308; □, S. pyogenes 201308 FMT M228fs mutant.
These results indicate that, although FMT is not essential for S. pyogenes viability, FMT mutations confer severely impaired growth and are so unstable in some cases that reversion of the mutation is observed after a few passages in the absence of drug. As no resistant mutants with mutations in the PDF protein were identified from any of the strains tested, the target-based FoR was low in S. pyogenes, i.e., <5 × 10−8 (Table 2).
S. pneumoniae.
Resistance studies with S. pneumoniae and PDF inhibitors have always resulted in the isolation of target-based mutants (21, 22). Therefore, it was unexpected when studies performed with GSK1322322 in S. pneumoniae 1314005S yielded a large number of tiny colonies, in addition to other colonies that were bigger in size. Phenotypic and genetic characterization of 11 colonies resulted in the identification of two types of mutants. All six isolates with tiny colonies carried mutations in the fmt gene, which conferred 4- to 128-fold increases in the GSK1322322 MIC and 0- to 32-fold decreases in the tiamulin MIC. The other five isolates, with the mutations V71F (four mutants) and I169T (one mutant) in the PDF protein, showed 4- to 32-fold increases in the GSK1322322 MIC and always had wild-type susceptibility to tiamulin. While the S. aureus fmt gene could always be readily deleted, attempts by us and others (21) to knock out S. pneumoniae fmt were never successful, indicating that the FMT function was essential for this organism. The fact that the S. pneumoniae FMT mutants isolated here showed such substantial growth defects on plates and did not carry mutations that resulted in an inactive protein supports that observation. In order to determine the target-based FoR to GSK1322322 in this species and to confirm the frequency of isolation of FMT mutants, studies were performed at 4 and 10 times the GSK1322322 MIC with four S. pneumoniae strains and at 10 times the MIC with an additional 17 strains. Resistant mutants were not obtained for 10 of the strains tested. Susceptibility to GSK1322322 and tiamulin was analyzed in all or a number of representative isolates from the other 11 strains, in order to focus the genetic characterization on isolates that could carry mutations in PDF, i.e., those with wild-type susceptibility to tiamulin. Frequencies obtained with the different strains are summarized in Table 2. All isolates investigated from the S. pneumoniae 237442 and 395259 strains were hypersensitive to tiamulin (Table 2); therefore, it was assumed that they carried mutations in the fmt gene. Of all the colonies isolated from S. pneumoniae 121175 and 339881, only one in each case showed wild-type susceptibility to tiamulin and thus was genetically characterized (Table 3). Both mutants had amino acid substitutions in the FMT protein (Table 3). All isolates with reduced susceptibility to GSK1322322 that were obtained from S. pneumoniae strains 1629, 403346, 1307006S, and 340643 carried mutations at residue V71 (10 mutants) or A123 (3 mutants) of the PDF protein, which increased the GSK1322322 MIC by 8- to 256-fold (Table 3). Only one mutant of the representative set selected from S. pneumoniae 302305 had a wild-type tiamulin MIC; it carried the Q57K substitution in the PDF protein, which conferred a moderate level of resistance to GSK1322322 (Table 3). All of the isolates selected for phenotypic characterization from S. pneumoniae 1302004S and 1314005S demonstrated wild-type susceptibility to tiamulin (Table 2) and were genetically characterized. All 6 isolates from S. pneumoniae 1302004S maintained their resistance phenotype and did not carry mutations in the pdf, fmt, folD, or glyA genes or their promoter regions (Table 3). Six of the 24 mutants characterized from S. pneumoniae 1314005S carried the V71F substitution in the PDF protein (Table 3). No mutations were identified in the other isolates, but stability studies showed that those isolates had unstable resistance phenotypes, which reverted to the wild type after a few passages in the absence of drug. As speculated for S. pyogenes, it is possible that those mutants originally carried a mutation in the fmt gene but, due to the severe fitness cost of the mutation, wild-type revertants outgrew them very rapidly. Therefore, target-based mutants were identified for only 6 of the 21 S. pneumoniae strains investigated in this study, with frequencies ranging between 3 × 10−8 and 2 × 10−9 (Table 2).
Competitive growth studies were performed with S. pneumoniae mutants carrying different target-based mutations, namely, S. pneumoniae 1307006S-M1 and 1314005S-M14 (V71F), S. pneumoniae 403346-M1 (V71A), S. pneumoniae 403346-M2 (V71D), and S. pneumoniae 1314005S-M5 (I169T), in order to assess the in vitro fitness costs of the mutations. Only the S. pneumoniae 403346-M2 PDF mutant, with the V71D substitution, showed a colony size consistently smaller than that of its parent strain throughout these experiments. As expected, the PDF V71D substitution, which resulted in a 256-fold increase in the GSK1322322 MIC, appeared to involve a significant fitness cost in the mutant strain, compared to the wild-type parent, as shown by a considerable reduction in the mutant/wild-type ratio after three competitive growth cycles (Fig. 2). None of the other mutations, i.e., V71F, V71A, and I169T, which resulted in 16- to 32-fold, 16-fold, and 4-fold increases of the GSK1322322 MIC, respectively, was associated with a significant fitness cost in these studies, with all of the mutants growing similarly to their parent strains (data not shown).
FIG 2.
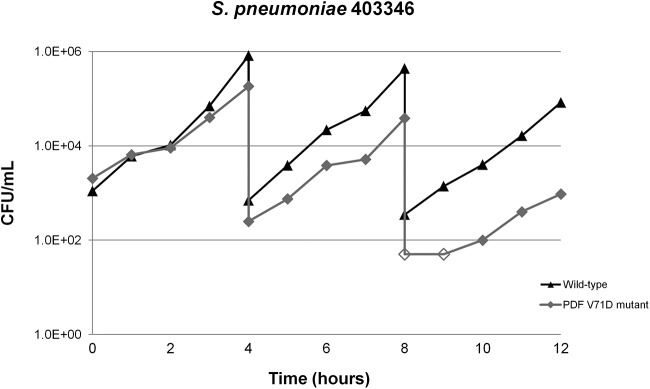
Growth competition studies with S. pneumoniae 403346 and its corresponding V71D mutant. Unfilled symbols, count values below the limit of detection.
Analysis of PDF mutations conferring resistance to GSK1322322.
A mutation in residue V71 (using the S. pneumoniae numbering system) (Fig. 3) was the most common cause of target-based resistance to GSK1322322. The single H. influenzae mutant, 9 of the 11 S. aureus mutants, and 20 of the 25 S. pneumoniae mutants isolated in these studies carried this mutation. This residue is located in motif 1, which is a stretch of highly conserved amino acids involved in catalysis that is present in both Gram-negative and Gram-positive PDF enzymes (27). The V71A substitution was most frequently found in S. aureus, whereas V71F was more common in S. pneumoniae. Those two substitutions, together with V71G, conferred 16- to 32-fold increases in the GSK1322322 MIC and did not seem to translate into obvious in vitro growth fitness costs. In contrast, the V71D substitution, although more unusual, resulted in a decrease in the growth rate and a >64-fold increase in the GSK1322322 MIC. Mutations at amino acid I169 conferred low-level resistance to GSK1322322. This residue is present in all three proteins (Fig. 3) and is in close proximity to motif 3, an area that is also highly conserved across species and has been implicated in active site metal binding and catalysis (28). In contrast, Q57 and A123, whose mutations provide a moderate level of resistance, are located in areas conserved only in Gram-positive PDF proteins (Fig. 3) (27).
FIG 3.

Sequence alignment of PDF proteins from H. influenzae Rd KW20, S. pneumoniae R6, and S. aureus JH9. Residues conserved in all three proteins are bold. Mutated residues are red, and blue and green residues are amino acids that are generally conserved in Gram-negative and Gram-positive PDF proteins, respectively (27).
DISCUSSION
While the eventual appearance of resistance to novel antibacterial agents is a predictable consequence of their clinical use, it is still important to develop drugs with low potential for resistance development. Although many factors affect the frequency at which resistance appears in clinical situations, the determination of the in vitro FoR and the in vivo fitness of the mutants isolated has become an intrinsic part of the early drug discovery process for any new antibacterial agent. The occurrence of resistance depends on the type and number of genes in which mutations can yield a selectable phenotype. In the simplest instances, resistance to an antibacterial agent can arise from mutations that affect its target or its access to or removal from the cell. In the case of PDF inhibitors, mutations that affect formylation of the initiator Met-tRNA, of which those in FMT are the most common, have also been associated with resistance in certain pathogens (12–18).
As reported previously for other PDF inhibitor-strain combinations (12, 13), the overall FoR to GSK1322322 was high in S. aureus, with 87 to 100% of the characterized mutants carrying mutations that inactivated the FMT protein. The fmt mutations were stably maintained in the absence of selective pressure and, as the need for deformylation was bypassed, FMT mutants were highly resistant to PDF inhibitors. In the studies performed with four S. pyogenes strains, all of the isolated mutants had high GSK1322322 MICs and carried loss-of-function mutations that inactivated FMT; unlike the S. aureus FMT mutants, however, they lost substantial viability and had severe difficulty growing, even in complex media. In fact, some of the FMT mutations in S. pyogenes seemed to convey general instability, and wild-type phenotypes were recovered after only a few rounds of replication in the absence of drug. Clearly, for a normal in vitro growth rate, S. pyogenes seems to be more dependent than S. aureus on the initiation of protein synthesis with formyl-methionine. Severe effects on growth have also been observed with Escherichia coli and Salmonella enterica FMT mutants (13, 16, 29).
Mutants with alterations in fmt could be isolated in 28.6% of the S. pneumoniae strains tested (6/21 strains). Those isolates did not carry mutations that completely inactivated the FMT protein, were not always highly resistant to GSK1322322, and showed an unstable phenotype in the absence of selective pressure. These data clearly indicate that FMT plays an essential role in protein synthesis in this organism, as already suggested by the unsuccessful attempts to delete the gene from the chromosome (21). Therefore, although resistance to GSK1322322 can occur through mutations in the FMT protein in S. pyogenes and even S. pneumoniae, the mutations are associated with severe in vitro fitness costs, and it can be anticipated that the mutants would not be able to subsist under more challenging in vivo conditions. In addition, although the loss of FMT function in S. aureus does not seem to have a pronounced fitness cost in vitro, recent studies have demonstrated that FMT mutants are nonhemolytic and cannot cause productive infections in animal models (20). Mutations in the PDF protein, in contrast, could be expected to yield fitter stable mutants.
In general, target-based FoR was low in H. influenzae (≤3 × 10−9) and S. pyogenes (<5 ×10−8) and more moderate in S. aureus (≤2 × 10−8) and S. pneumoniae (≤3 × 10−8). No PDF mutants were isolated in the studies performed with four S. pyogenes strains, and only one mutant, with the PDF substitution I45N (V71 in S. pneumoniae PDF), was identified in the studies carried out with four H. influenzae strains. Resistance studies with H. influenzae N65044 with another PDF inhibitor, LBM415, yielded a larger number of mutants, most of them with mutations in the acrR gene, a repressor that controls expression of the AcrAB-TolC efflux pump, resulting in 8-fold decreases in susceptibility to LBM415 (24). An additional mutant with a chromosomal rearrangement that caused copy number amplification of the pdf gene showed a higher level of resistance (23). Target-based resistance to PDF inhibitors in S. aureus had been described only twice before (19, 20). Studies performed with four methicillin-resistant S. aureus strains yielded only one or no mutants in three of the strains, with higher frequencies observed in the remaining strain. Seven mutants contained identical mutations at position V59 (V71 in S. pneumoniae PDF), and two mutants carried modifications in the promoter region that could be the cause of the low-level resistance observed. Nearly 50% of the S. pneumoniae strains (10/21 strains) tested in the present study did not yield colonies on GSK1322322-containing plates. Mutants with reduced susceptibility to GSK1322322 that were isolated from six of the remaining strains had mutations in the PDF protein, with 20 of the 25 mutants characterized carrying mutations at position V71, three at position A123, and one each at positions Q57 and I169. Mutations at PDF residues similar or identical to those mutations, conferring resistance to different inhibitors in S. pneumoniae, have been described previously (21, 22).
The PDF family of proteins can be divided into two classes. Class I includes PDFs from Gram-negative organisms, whereas class II includes many Gram-positive PDFs, which have three major insertions and a different C terminus, relative to the E. coli sequence (28). Although there is low overall sequence identity between class I and class II PDFs, the two types of enzymes possess similar features in their tertiary structures and share three characteristic areas of highly conserved amino acids, i.e., motif 1 (70-GXGXAAXQ-77), motif 2 (128-EGCLS-132), and motif 3 (173-HEXXH-177) (using the S. pneumoniae numbering system), which form the active site around the metal ion (27, 28). The I169 residue lies immediately upstream of the HEXXH motif that is shared by all PDF proteins and is characteristic of zinc hydrolases. A123 is not conserved among PDF proteins but is located one residue upstream of a corresponding isoleucine in the E. coli enzyme, which has been defined as part of the substrate-binding pocket (13), and five residues upstream of the EGCLS motif, which is involved in binding of the metal ion (28, 30, 31). Q57 is part of a conserved region present only in class II enzymes, and V71 is located within the highly conserved motif 1, which is involved in the structural stability and catalytic mechanism of PDF. It has been shown that PDF inhibitors such as GSK1322322 display strong time-dependent inhibition, which contributes to their antibacterial potency (32). Recent kinetic studies support the hypothesis that the time-dependent nature of PDF inhibition is due to both the binding of the compounds to the active site metal of PDF and the hydrogen-bonding network between the inhibitor and certain key residues of the protein, including V71 (33). Changes in hydrogen bonding between PDF V71 and the inhibitor modulate the rate of binding to the active site metal and play a critical role in maintaining time dependency, and therefore potency, of PDF inhibitors. Substitution of this valine with another hydrophobic amino acid, such as alanine, phenylalanine, or glycine, decreased the antibacterial potency of GSK1322322 by 16- to 32-fold but had no effect on the growth rate of the mutants. In contrast, substitution with a charged amino acid, such as aspartic acid, resulted in not only high-level resistance to GSK1322322 but also considerable fitness costs, confirming the involvement of the residue not only in the interactions of the protein with GSK1322322 but also in the stability and catalytic activity of the protein, as described previously (28).
In summary, the target-based FoR to GSK1322322 is low in H. influenzae and S. pyogenes and low to moderate (depending on the strain) in S. aureus and S. pneumoniae. Further in vitro, in vivo, and clinical studies will need to be performed to determine the clinical relevance of these findings. Of the 35 mutations identified within the PDF protein among the organisms tested, 30 occur at residue V71 and confer moderate to high levels of resistance to GSK1322322, although the mutation V71D (in 3/30 mutants) seems to have a substantial effect on the growth rate as well. It will be interesting to design other PDF inhibitors with hydrogen bonds to alternative residues and to determine whether they maintain time-dependent inhibition while reducing the FoR in certain strains.
ACKNOWLEDGMENT
We thank the GlaxoSmithKline sequencing department for technical assistance.
REFERENCES
- 1.Adams JM. 1968. On the release of the formyl group from nascent protein. J Mol Biol 33:571–589. doi: 10.1016/0022-2836(68)90307-0. [DOI] [PubMed] [Google Scholar]
- 2.Ball LA, Kaesberg P. 1973. Cleavage of the N-terminal formylmethionine residue from a bacteriophage coat protein in vitro. J Mol Biol 79:531–537. doi: 10.1016/0022-2836(73)90404-X. [DOI] [PubMed] [Google Scholar]
- 3.Livingston DM, Leder P. 1969. Deformylation and protein biosynthesis. Biochemistry 8:435–443. doi: 10.1021/bi00829a059. [DOI] [PubMed] [Google Scholar]
- 4.Aubart K, Zalacain M. 2006. Peptide deformylase inhibitors. Prog Med Chem 44:109–143. doi: 10.1016/S0079-6468(05)44403-3. [DOI] [PubMed] [Google Scholar]
- 5.Lofland D, Difuntorum S, Waller A, Clements JM, Weaver MK, Karlowsky JA, Johnson K. 2004. In vitro antibacterial activity of the peptide deformylase inhibitor BB-83698. J Antimicrob Chemother 53:664–668. doi: 10.1093/jac/dkh129. [DOI] [PubMed] [Google Scholar]
- 6.Fritsche TR, Sader HS, Cleeland R, Jones RN. 2005. Comparative antimicrobial characterization of LBM415 (NVP PDF-713), a new peptide deformylase inhibitor of clinical importance. Antimicrob Agents Chemother 49:1468–1476. doi: 10.1128/AAC.49.4.1468-1476.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Dwyer K, Hackel M, Hightower S, Hoban D, Bouchillon S, Qin D, Aubart K, Zalacain M, Butler D. 2013. Comparative analysis of the antibacterial activity of a novel peptide deformylase inhibitor, GSK1322322. Antimicrob Agents Chemother 57:2333–2342. doi: 10.1128/AAC.02566-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naderer OJ, Dumont E, Zhu J, Kurtinecz M, Jones LS. 2013. Single-dose safety, tolerability, and pharmacokinetics of the antibiotic GSK1322322, a novel peptide deformylase inhibitor. Antimicrob Agents Chemother 57:2005–2009. doi: 10.1128/AAC.01779-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naderer OJ, Jones LS, Zhu J, Kurtinecz M, Dumont E. 2013. Safety, tolerability, and pharmacokinetics of oral and intravenous administration of GSK1322322, a peptide deformylase inhibitor. J Clin Pharmacol 53:1168–1176. doi: 10.1002/jcph.150. [DOI] [PubMed] [Google Scholar]
- 10.Naderer OJ, Dumont E, Zhu J, Kurtinecz M, Jones LS. 2013. Safety, tolerability and pharmacokinetics of repeat dosing of the antibiotic GSK1322322, a peptide deformylase inhibitor: a randomized placebo-controlled study. J Antimicrob Chemother 68:1901–1909. doi: 10.1093/jac/dkt097. [DOI] [PubMed] [Google Scholar]
- 11.Corey R, Naderer OJ, O'Riordan WD, Dumont E, Jones LS, Kurtinecz M, Zhu JZ. 2014. Safety, tolerability, and efficacy of GSK1322322 in the treatment of acute bacterial skin and skin structure infections. Antimicrob Agents Chemother 58:6518–6527. doi: 10.1128/AAC.03360-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Margolis PS, Hackbarth CJ, Young DC, Wang W, Chen D, Yuan Z, White R, Trias J. 2000. Peptide deformylase in Staphylococcus aureus: resistance to inhibition is mediated by mutations in the formyltransferase gene. Antimicrob Agents Chemother 44:1825–1831. doi: 10.1128/AAC.44.7.1825-1831.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clements JM, Beckett RP, Brown A, Catlin G, Lobell M, Palan S, Thomas W, Whittaker M, Wood S, Salama S, Baker PJ, Rodgers HF, Barynin V, Rice DW, Hunter MG. 2001. Antibiotic activity and characterization of BB-3497, a novel peptide deformylase inhibitor. Antimicrob Agents Chemother 45:563–570. doi: 10.1128/AAC.45.2.563-570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duroc Y, Giglione C, Meinnel T. 2009. Mutations in three distinct loci cause resistance to peptide deformylase inhibitors in Bacillus subtilis. Antimicrob Agents Chemother 53:1673–1678. doi: 10.1128/AAC.01340-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Newton DT, Creuzenet C, Mangroo D. 1999. Formylation is not essential for initiation of protein synthesis in all eubacteria. J Biol Chem 274:22143–22146. doi: 10.1074/jbc.274.32.22143. [DOI] [PubMed] [Google Scholar]
- 16.Nilsson AI, Zorzet A, Kanth A, Dahlstrom S, Berg OG, Andersson DI. 2006. Reducing the fitness cost of antibiotic resistance by amplification of initiator tRNA genes. Proc Natl Acad Sci U S A 103:6976–6981. doi: 10.1073/pnas.0602171103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Apfel CM, Locher H, Evers S, Takacs B, Hubschwerlen C, Pirson W, Page MG, Keck W. 2001. Peptide deformylase as an antibacterial drug target: target validation and resistance development. Antimicrob Agents Chemother 45:1058–1064. doi: 10.1128/AAC.45.4.1058-1064.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guillon JM, Mechulam Y, Schmitter JM, Blanquet S, Fayat G. 1992. Disruption of the gene for Met-tRNAfMet formyltransferase severely impairs growth of Escherichia coli. J Bacteriol 174:4294–4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watters AA, Jones RN, Leeds JA, Denys G, Sader HS, Fritsche TR. 2006. Antimicrobial activity of a novel peptide deformylase inhibitor, LBM415, tested against respiratory tract and cutaneous infection pathogens: a global surveillance report (2003–2004). J Antimicrob Chemother 57:914–923. doi: 10.1093/jac/dkl093. [DOI] [PubMed] [Google Scholar]
- 20.Lewandowski T, Huang J, Fan F, Rogers S, Gentry D, Holland R, Demarsh P, Aubart K, Zalacain M. 2013. Staphylococcus aureus formyl-methionyl transferase mutants demonstrate reduced virulence factor production and pathogenicity. Antimicrob Agents Chemother 57:2929–2936. doi: 10.1128/AAC.00162-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Margolis P, Hackbarth C, Lopez S, Maniar M, Wang W, Yuan Z, White R, Trias J. 2001. Resistance of Streptococcus pneumoniae to deformylase inhibitors is due to mutations in defB. Antimicrob Agents Chemother 45:2432–2435. doi: 10.1128/AAC.45.9.2432-2435.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kosowska-Shick K, Credito KL, Pankuch GA, DeWasse B, McGhee P, Appelbaum PC. 2007. Multistep resistance selection and postantibiotic-effect studies of the antipneumococcal activity of LBM415 compared to other agents. Antimicrob Agents Chemother 51:770–773. doi: 10.1128/AAC.01150-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dean CR, Narayan S, Richards J, Daigle DM, Esterow S, Leeds JA, Kamp H, Puyang X, Wiedmann B, Mueller D, Voshol H, van Oostrum J, Wall D, Koehn J, Dzink-Fox J, Ryder NS. 2007. Reduced susceptibility of Haemophilus influenzae to the peptide deformylase inhibitor LBM415 can result from target protein overexpression due to amplified chromosomal def gene copy number. Antimicrob Agents Chemother 51:1004–1010. doi: 10.1128/AAC.01103-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dean CR, Narayan S, Daigle DM, Dzink-Fox JL, Puyang X, Bracken KR, Dean KE, Weidmann B, Yuan Z, Jain R, Ryder NS. 2005. Role of the AcrAB-TolC efflux pump in determining susceptibility of Haemophilus influenzae to the novel peptide deformylase inhibitor LBM415. Antimicrob Agents Chemother 49:3129–3135. doi: 10.1128/AAC.49.8.3129-3135.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clinical and Laboratory Standards Institute. 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard— 9th ed Document M07-A9. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 26.Schmitt E, Blanquet S, Mechulam Y. 1996. Structure of crystalline Escherichia coli methionyl-tRNAfMet formyltransferase: comparison with glycinamide ribonucleotide formyltransferase. EMBO J 15:4749–4758. [PMC free article] [PubMed] [Google Scholar]
- 27.Baldwin ET, Harris MS, Yem AW, Wolfe CL, Vosters AF, Curry KA, Murray RW, Bock JH, Marshall VP, Cialdella JI, Merchant MH, Choi G, Deibel MR Jr. 2002. Crystal structure of type II peptide deformylase from Staphylococcus aureus. J Biol Chem 277:31163–31171. doi: 10.1074/jbc.M202750200. [DOI] [PubMed] [Google Scholar]
- 28.Meinnel T, Lazennec C, Villoing S, Blanquet S. 1997. Structure-function relationships within the peptide deformylase family: evidence for a conserved architecture of the active site involving three conserved motifs and a metal ion. J Mol Biol 267:749–761. doi: 10.1006/jmbi.1997.0904. [DOI] [PubMed] [Google Scholar]
- 29.Apfel CM, Evers S, Hubschwerlen C, Pirson W, Page MG, Keck W. 2001. Peptide deformylase as an antibacterial drug target: assays for detection of its inhibition in Escherichia coli cell homogenates and intact cells. Antimicrob Agents Chemother 45:1053–1057. doi: 10.1128/AAC.45.4.1053-1057.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meinnel T, Lazennec C, Blanquet S. 1995. Mapping of the active site zinc ligands of peptide deformylase. J Mol Biol 254:175–183. doi: 10.1006/jmbi.1995.0609. [DOI] [PubMed] [Google Scholar]
- 31.Chan MK, Gong W, Rajagopalan PT, Hao B, Tsai CM, Pei D. 1997. Crystal structure of the Escherichia coli peptide deformylase. Biochemistry 36:13904–13909. doi: 10.1021/bi9711543. [DOI] [PubMed] [Google Scholar]
- 32.Van Aller GS, Nandigama R, Petit CM, DeWolf WE Jr, Quinn CJ, Aubart KM, Zalacain M, Christensen SB, Copeland RA, Lai Z. 2005. Mechanism of time-dependent inhibition of polypeptide deformylase by actinonin. Biochemistry 44:253–260. doi: 10.1021/bi048632b. [DOI] [PubMed] [Google Scholar]
- 33.Totoritis R, Duraiswami C, Taylor AN, Kerrigan JJ, Campobasso N, Smith KJ, Ward P, King BW, Murrayz-Thompson M, Jones AD, Van Aller GS, Aubart KM, Zalacain M, Thrall SH, Meek TD, Schwartz B. 2011. Understanding the origins of time-dependent inhibition by polypeptide deformylase inhibitors. Biochemistry 50:6642–6654. doi: 10.1021/bi200655g. [DOI] [PubMed] [Google Scholar]


