Abstract
In this study, we investigate the amebicidal activities of the pharmaceutical triazole CYP51 inhibitors fluconazole, itraconazole, and voriconazole against Acanthamoeba castellanii and Acanthamoeba polyphaga and assess their potential as therapeutic agents against Acanthamoeba infections in humans. Amebicidal activities of the triazoles were assessed by in vitro minimum inhibition concentration (MIC) determinations using trophozoites of A. castellanii and A. polyphaga. In addition, triazole effectiveness was assessed by ligand binding studies and inhibition of CYP51 activity of purified A. castellanii CYP51 (AcCYP51) that was heterologously expressed in Escherichia coli. Itraconazole and voriconazole bound tightly to AcCYP51 (dissociation constant [Kd] of 10 and 13 nM), whereas fluconazole bound weakly (Kd of 2,137 nM). Both itraconazole and voriconazole were confirmed to be strong inhibitors of AcCYP51 activity (50% inhibitory concentrations [IC50] of 0.23 and 0.39 μM), whereas inhibition by fluconazole was weak (IC50, 30 μM). However, itraconazole was 8- to 16-fold less effective (MIC, 16 mg/liter) at inhibiting A. polyphaga and A. castellanii cell proliferation than voriconazole (MIC, 1 to 2 mg/liter), while fluconazole did not inhibit Acanthamoeba cell division (MIC, >64 mg/liter) in vitro. Voriconazole was an effective inhibitor of trophozoite proliferation for A. castellanii and A. polyphaga; therefore, it should be evaluated in trials versus itraconazole for controlling Acanthamoeba infections.
INTRODUCTION
Acanthamoeba is an opportunistic protist pathogen that is ubiquitously distributed in the environment, being present in rivers, lakes, seawater, air, and soils, as well as in domestic environments, such as heating and air-conditioning systems, swimming pools, aquariums, plants, and jacuzzis (1). Acanthamoeba species are the causative agents of cutaneous lesions, sinus and lung infections, amoebic keratitis of the cornea (AK), and amoebic granulomatous encephalitis (AGE) (1–3). Acanthamoeba has two stages in its life cycle: a motile vegetative trophozoite stage of feeding and reproduction and a dormant cyst stage that occurs when environmental conditions are harsh; this stage confers resistance against extreme temperature, desiccation, UV radiation, and several antiseptic chemicals (1, 4). Therefore, eradication of Acanthamoeba cysts is essential in effective antiacanthamoeba therapies.
Several species of Acanthamoeba (including A. castellanii and A. polyphaga) are known to cause AGE, primarily among immunocompromised patients (1). AGE is a rare infection but is nearly always fatal and involves the protist parasite crossing the blood-brain barrier to infect the brain and central nervous system (CNS), resulting in neuronal and brain damage. Routes of entry include the lower respiratory tract and skin lesions, with infections of the skin and respiratory system lasting several months, followed by infection of the CNS, resulting in death in days to weeks (3, 5–8).
A. castellanii and A. polyphaga are the most common Acanthamoeba subspecies to cause AK in immunocompetent individuals (9), an infection of the cornea that can lead to blindness. Up to 93% of reported AK cases are associated with contact lens usage (10, 11). Infection is usually a result of poor hygiene during storage and handling of contact lenses (12). In non-lens users, AK is associated with trauma and exposure to contaminated water or soil and is prominent among agricultural workers (13). Generally only one eye is affected, with reported cases of bilateral keratitis being rare (1, 14). AK is an emerging disease, with reported cases increasing year upon year, mainly due to increased contact lens usage, better diagnosis, and awareness (13). The number of AK cases in the 1980s and early 1990s was typically one to two cases per million contact lens wearers. By 2000 the incidence of AK had increased to 17 to 70 cases per million contact lens wearers (country dependent) (12), who presently number ca. 120 million worldwide. AK often is difficult to treat, with early detection followed by aggressive treatment being essential for successful outcomes (15). The recommended cysticidal treatment is the use of a biguanide (0.02% polyhexamethylene biguanide or 0.02% chlorhexidine digluconate) in conjunction with a diamidine (0.1% propamidine isethionate or 0.1% hexamidine). If coinfection with bacteria is found, additional antibiotics, such as neomycin or chloramphenicol, also is recommended (3). Combined topical application of 1% miconazole and 1% propamidine, or oral itraconazole together with topically applied 0.1% miconazole and ketoconazole, has stopped the progress of AK (1, 16). Voriconazole also has been used successfully both topically and systemically to treat AK (1, 17, 18). Recent case reports have pointed toward triazoles as an adjunct to biguanide and diamidine therapy, with voriconazole proving effective either applied topically or taken orally (19, 20). However, extended treatment duration was required in order to prevent relapse. Most recently, Acanthamoeba infection has been linked immunologically with multiple sclerosis pathogenesis (21, 22). Therefore, the effective control of Acanthamoeba before infection spreads to the central nervous system may prove beneficial to a far broader range of patients than first thought.
In this study, we cloned a CYP51 homologous gene from Acanthamoeba castellanii into Escherichia coli and expressed and purified the recombinant protein in an active state where its activity was confirmed. In addition, we also characterized the effectiveness of fluconazole, itraconazole, and voriconazole at inhibiting A. castellanii CYP51 activity and in vitro proliferation of Acanthamoeba castellanii and Acanthamoeba polyphaga.
MATERIALS AND METHODS
Construction of the pCWori+:AcCYP51 expression vector.
The Acanthamoeba castellanii CYP51 gene (AcCYP51; UniProtKB accession number L8GJB3) was synthesized by Eurofins MWG Operon (Ebersberg, Germany), incorporating an NdeI restriction site at the 5′ end and a HindIII restriction site at the 3′ end of the gene cloned into the pUC57 plasmid. In addition, the first eight amino acids were changed to MALLLAVF (23), and a six-histidine extension (CATCACCATCACCATCAC) was inserted immediately before the stop codon to facilitate protein purification by Ni2+-nitrilotriacetic acid (NTA) agarose affinity chromatography. The AcCYP51 gene was excised by NdeI/HindIII restriction digestion followed by cloning into the pCWori+ expression vector using Roche T4 DNA ligase. Gene integrity was confirmed by DNA sequencing.
Heterologous expression in E. coli and isolation of recombinant AcCYP51 protein.
The pCWori+:AcCYP51 construct was transformed into competent DH5α E. coli cells, and transformants were selected using 0.1 mg/ml ampicillin. Cells were grown in terrific broth (2.4% yeast extract, 1.2% tryptone, 0.1 M potassium phosphate, pH 7.5, 0.1% glycerol) containing 0.1 mg/ml ampicillin for 7 h at 37°C and 200 rpm. Expression was induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) (1 mM) and 5-aminolevulenic acid (1 mM), followed by incubation at 27°C and 160 rpm for 18 h. Protein isolation was according to the method of Arase et al. (24). The solubilized AcCYP51 protein was purified by Ni2+-NTA agarose affinity chromatography as previously described (25), followed by dialysis against 5 liters of 20 mM potassium phosphate (pH 7.5) and 10% glycerol. Ni2+-NTA agarose-purified AcCYP51 was used for all subsequent spectral and 50% inhibitory concentration (IC50) determinations. Protein purity was assessed by SDS-polyacrylamide gel electrophoresis, followed by staining with Coomassie brilliant blue R-250.
AcCYP51 spectral determinations.
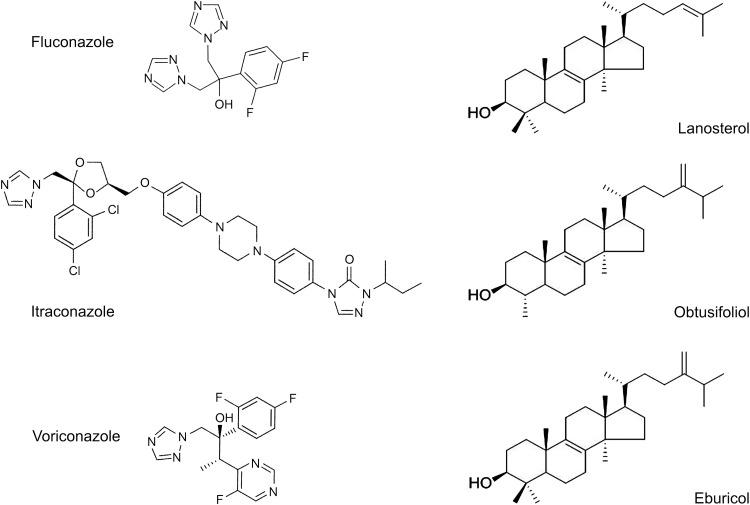
Cytochrome P450 concentration was determined by reduced carbon monoxide difference spectra (26), using an extinction coefficient of 91 mM−1 cm−1 (27), and from the absolute spectrum (600 to 350 nm) (25). Azole ligand binding determinations were performed as previously described (28) using quartz split cuvettes (light path, 4.5 mm) and stock 1, 0.5, 0.2, and 0.1 mg/ml solutions of fluconazole, itraconazole, and voriconazole in dimethyl sulfoxide. Azole antifungals were progressively titrated against 2 μM AcCYP51 in 0.1 M Tris-HCl (pH 8.1) and 25% glycerol at 22°C, with equivalent volumes of dimethyl sulfoxide also being added to the AcCYP51-containing compartment of the reference cuvette. The absorbance difference spectra between 500 and 350 nm were determined after each incremental addition of azole with binding saturation curves constructed from ΔA428-412 against azole concentration. The dissociation constant of the enzyme-azole complex (Kd) for each azole was determined by nonlinear regression (Levenberg-Marquardt algorithm) using a rearrangement of the Morrison equation for tight ligand binding (29, 30). All spectral determinations were performed in triplicate. AcCYP51 was demonstrated to be active by measuring the 14α-demethylation of lanosterol, eburicol, and obtusifoliol using the CYP51 reconstitution assay described below. The chemical structures of the azole antifungals and 14-methylated sterol substrates used in this study are shown in Fig. 1.
FIG 1.
Azole antifungal agents and 14-methylated sterols used in this study. The chemical structures of fluconazole (molecular weight [MW], 306), itraconazole (MW, 706), voriconazole (MW, 349), obtusifoliol (MW, 426), lanosterol (MW, 426), and eburicol (MW, 440) are shown.
CYP51 reconstitution assay system.
IC50 determinations were performed using the CYP51 reconstitution assay system (500-μl final reaction volume) previously described (31), containing 0.6 μM AcCYP51, 1.8 μM Aspergillus fumigatus cytochrome P450 reductase (AfCPR1; UniProtKB accession number Q4WM67), 50 μM obtusifoliol, 50 μM dilaurylphosphatidylcholine, 4% (2-hydroxypropyl)-β-cyclodextrin (HPCD), 0.4 mg/ml isocitrate dehydrogenase, 25 mM trisodium isocitrate, 50 mM NaCl, 5 mM MgCl2, and 40 mM morpholinepropanesulfonic acid (MOPS; pH ∼7.2). Azole antifungal agents were added in 2.5 μl dimethyl sulfoxide, followed by 10 min of incubation at 30°C prior to assay initiation with 4 mM β-NADPH-Na4. Samples then were shaken for 30 min at 30°C. In addition, trial AcCYP51 assays also were performed using 50 μM lanosterol and 50 μM eburicol as substrates. Sterol metabolites were recovered by extraction with ethyl acetate, followed by derivatization with N,O-bis(trimethylsilyl)trifluoroacetamide and tetramethylsilane in pyridine prior to analysis by gas chromatography mass spectrometry (32). IC50 in this study is defined as the inhibitor concentration required causing 50% inhibition of CYP51 activity under the stated assay conditions.
Azole MICs with Acanthamoeba species.
Acanthamoeba polyphaga (ATCC 30461) and Acanthamoeba castellani (ATCC 50491) were kept as continuous cultures in ATCC 712 media at 28°C. Azole susceptibility assays were performed in 96-well microtiter plates using an adaptation of the resazurin cell respiration assay (33, 34). Trophozoites were grown axenically in ATCC 712 media for 48 h at 28°C prior to dilution to 5 × 104 trophozoites/ml with ATCC 712 media. Serial 100× stock azole antifungal agent dilutions were prepared in dimethyl sulfoxide of 6.4, 3.2, 1.6, 0.8, 0.4, 0.2, 0.1, 0.05, 0.025, 0.0125, 0.0625, and 0.03125 mg/ml. These solutions initially were diluted 10-fold using ATCC 712 medium, of which 20 μl was added directly to culture plate wells containing 180 μl trophozoite inoculum (0.9 × 104 cells) to achieve final azole concentrations of 64, 32, 16, 8, 4, 2, 1, 0.5, 0.25, 0.125, 0.0625, and 0.03125 μg/ml. Control wells containing 1% dimethyl sulfoxide and trophozoites also were prepared. The microtiter plates were incubated at 28°C for 48 h, after which 30 μl of 0.02% aqueous resazurin dye was added and the microtiter plates incubated for a further 48 h at 28°C. A color change from purple to pink indicated the presence of respiring cells. MIC determinations were performed in triplicate and scored manually and are defined here as the lowest azole concentration at which cell respiration remained completely inhibited.
Data analysis.
Curve fitting of ligand binding data was performed using the computer program ProFit 6.1.12 (QuantumSoft, Zurich, Switzerland). Spectral determinations were made using quartz semi-micro cuvettes with a Hitachi U-3310 UV-visible spectrophotometer (San Jose, California).
The N-terminal membrane anchor region of AcCYP51 was predicted using the TargetP, version 1.1 (http://www.cbs.dtu.dk/services/TargetP/), software. Subcellular location was predicted using WoLF PSORT (http://www.genscript.com/psort/wolf_psort.html) software. Phylogenetic analyses were performed by comparing the AcCYP51 amino acid sequence (UniProtKB accession number L8GJB3) to selected fungal, plant, and animal CYP51 proteins from the UniProtKB database (http://www.uniprot.org/help/uniprotkb) using BLAST2 (http://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch&PROG_DEF=blastn&BLAST_PROG_DEF=megaBlast&BLAST_SPEC=blast2seq) and ClustalX version 1.81 (http://www.clustal.org/) sequence alignment software with a phylogenetic tree produced from the ClustalX-generated Phylip-dnd file using TreeviewX (https://code.google.com/p/treeviewx/) software. NCBI-BLASTP analysis (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi?CMD=Web&PAGE_TYPE=BlastHome) also was performed using the AcCYP51 sequence as the template.
The fungal CYP51 sequences used were Cryptococcus neoformans var. neoformans (Q5KQ65), Candida albicans (P10613), Malassezia globosa (A8Q3I7), Trichophyton rubrum (F2SHH3), Schizosaccharomyces pombe (Q09736), Ustilago maydis (P49602), Saccharomyces cerevisiae (P10614), Phanerochaete chrysosporium (B6DX27), Botryotinia fuckeliana (Q9P428), Mycosphaerella graminicola (Q5XWE5), Penicillium italicum (Q12664), Aspergillus fumigatus CYP51A (Q4WNT5), Aspergillus fumigatus CYP51B (Q96W81), and Candida glabrata (P50859). The animal CYP51 sequences used were Homo sapiens (Q16850), Danio rerio (Q1JPY5), Xenopus tropicalis (Q28CJ0), Rattus norvegicus (Q64654), and Sus scrofa (O46420). The plant CYP51 sequences used were Cucumis sativus (UPI0002B48861), Solanum lycopersicum (D9J0A9), Oryza sativa (Q2R3E2), Arabidopsis thaliana (Q9SAA9), Solanum chacoense (Q673E9), Populus trichocarpa (B9GMU7), and Sorghum bicolor (P93846). Other CYP51 sequences used were Aphanomyces euteiches (B2ZWE1), Saprolegnia parasitica (BROAD Institute accession number SPRG_09493.2), Galdieria sulfuraria (M2X6V3), Trypanosoma cruzi (Q7Z1V1), Trypanosoma brucei (Q385E8), and Leishmania infantum (A2TEF2).
Chemicals.
All chemicals, unless otherwise stated, were obtained from Sigma Chemical Company (Poole, United Kingdom). Voriconazole was supplied by Discovery Fine Chemicals (Bournemouth, United Kingdom). Growth media, sodium ampicillin, IPTG, and 5-aminolevulenic acid were obtained from Foremedium Ltd. (Hunstanton, United Kingdom). Ni2+-NTA agarose affinity chromatography matrix was obtained from Qiagen (Crawley, United Kingdom).
RESULTS
Analysis of AcCYP51 protein sequence.
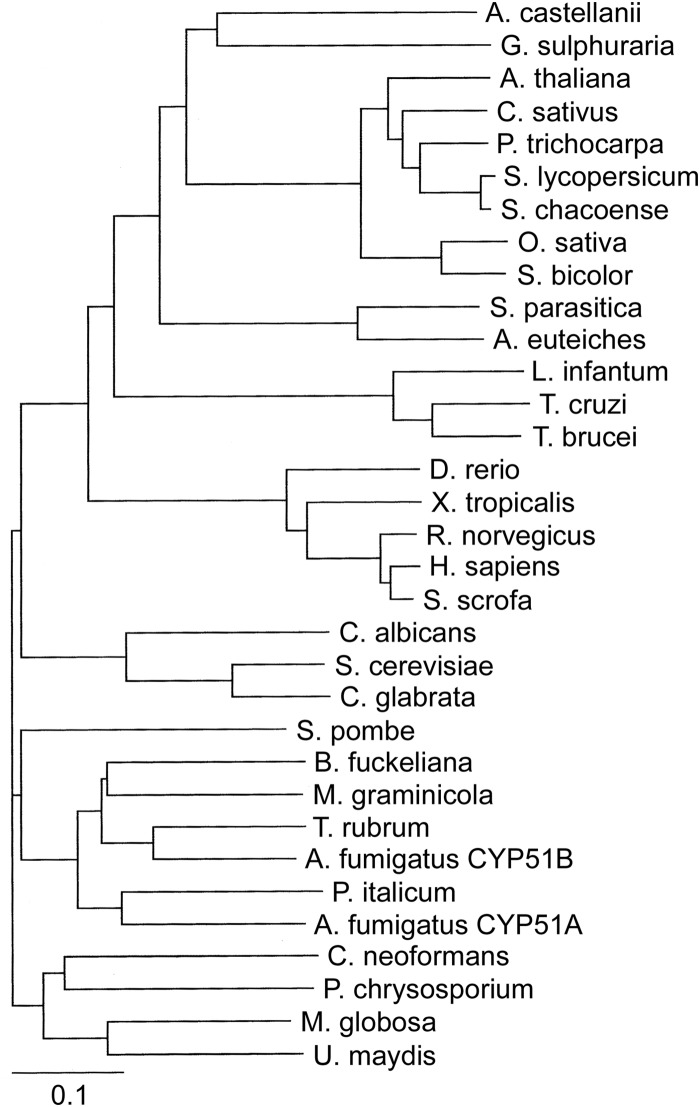
TargetP predicted that the AcCYP51 N-terminal membrane anchor consists of the first 34 amino acid residues, while WoLF PSORT predicted AcCYP51 would be located in the endoplasmic reticulum. This agrees with the observation that eukaryotic CYP51 proteins are associated with the membranes of the endoplasmic reticulum (35). Alignment of AcCYP51 against other eukaryotic CYP51 proteins confirmed that all 23 conserved amino acid residues previously identified for CYP51 proteins (36) were present in AcCYP51. The resultant phylogenetic tree (Fig. 2) showed AcCYP51 shared greatest sequence identity with the thermoacidophilic unicellular red alga G. sulfuraria CYP51 (54%). In addition, AcCYP51 shared 44 to 47% sequence identity with higher plant CYP51 enzymes (47% with cucumber CYP51), 45% sequence identity with the oomycete CYP51 proteins from S. parasitica and A. euteiches, 37 to 39% identity with animal CYP51 proteins (38% identity with human CYP51), 36 to 38% sequence identity with the trypanosomal CYP51 proteins, and 31 to 35% sequence identity with the fungal CYP51 proteins (31% identity with C. albicans CYP51 and 35% identity with A. fumigatus CYP51A). The top 12 NCBI-BLASTP hits against AcCYP51 were cytochrome P450 proteins from Galdieria sulfuraria (sequence identity 54%; http://www.uniprot.org/uniprot/M2X6V3), Guillardia theta CCMP2712 (XP_005826589.1; 53%), Micromonas pusilla CCMP1545 (XP_003061538.1; 50%), Micromonas sp. strain RCC299 (XP_002509219.1; 51%), Ostreococcus lucimarinus CCE9901 (XP_001420297.1; 50%), Bathycoccus prasinos (XP_007509480.1; 50%), Helicosporidium sp. strain ATCC 50920 (KDD76879.1; 50%), Volvox carteri f. nagariensis (XP_002946369.1; 49%), Chlamydomonas reinhardtii (XP_001701788.1; 48%), Geum rivale (CCH26420.1; 46%), Physcomitrella patens (XP_001769413.1; 46%), and Cyanidioschyzon merolae strain 10D (XP_005538940.1; 50%), with cucumber CYP51 (XP_004152160.1) being the 22nd closest match to AcCYP51.
FIG 2.
Phylogenetic tree of CYP51 enzymes. A phylogenetic tree of selected eukaryotic CYP51 proteins, including members from the fungus, plant, and animal kingdoms, in addition to trypanosomal and oomycete CYP51 proteins, was constructed using ClustalX version 1.81 (http://www.clustal.org/) and TreeViewX (https://code.google.com/p/treeviewx/). The individual CYP51 sequences used to construct the phylogenetic tree are detailed in Materials and Methods. Species names have been left nonitalicized for clarity.
Expression and purification of AcCYP51.
The yield of AcCYP51 was ∼50 ± 20 nmol/liter E. coli culture as determined by absolute spectroscopy (25) after purification by Ni2+-NTA agarose chromatography. SDS-polyacrylamide gel electrophoresis confirmed the purity of the Ni2+-NTA agarose-eluted AcCYP51 protein to be greater than 90% when assessed by Coomassie brilliant blue R-250 staining intensity, with an apparent molecular weight of ∼50,000 ± 2,000, which was close to the predicted value of 56,585 and included the six-histidine C-terminal extension.
Spectral properties of recombinant AcCYP51 protein.
The absolute spectrum of AcCYP51 (Fig. 3A) was typical for a ferric cytochrome P450 enzyme predominantly in the low-spin state (25, 37), with α, β, and Soret (γ) spectral bands at 568, 536, and 415 nm, respectively. However, the δ spectral band (∼360 nm) appeared merged with the tail end of the Soret peak. Reduced carbon monoxide difference spectra (Fig. 3B) produced the characteristic red-shifted Soret peak at 448 nm typical of ferrous cytochrome P450 enzymes complexed with CO (26, 27). However, the reduced CO adduct at 448 nm was relatively slow in forming, with a half-life (t1/2) of 3.7 ± 0.2 min. In addition, the presence of the inactive P-420 complex absorbance at 422 nm only slowly diminished with time.
FIG 3.
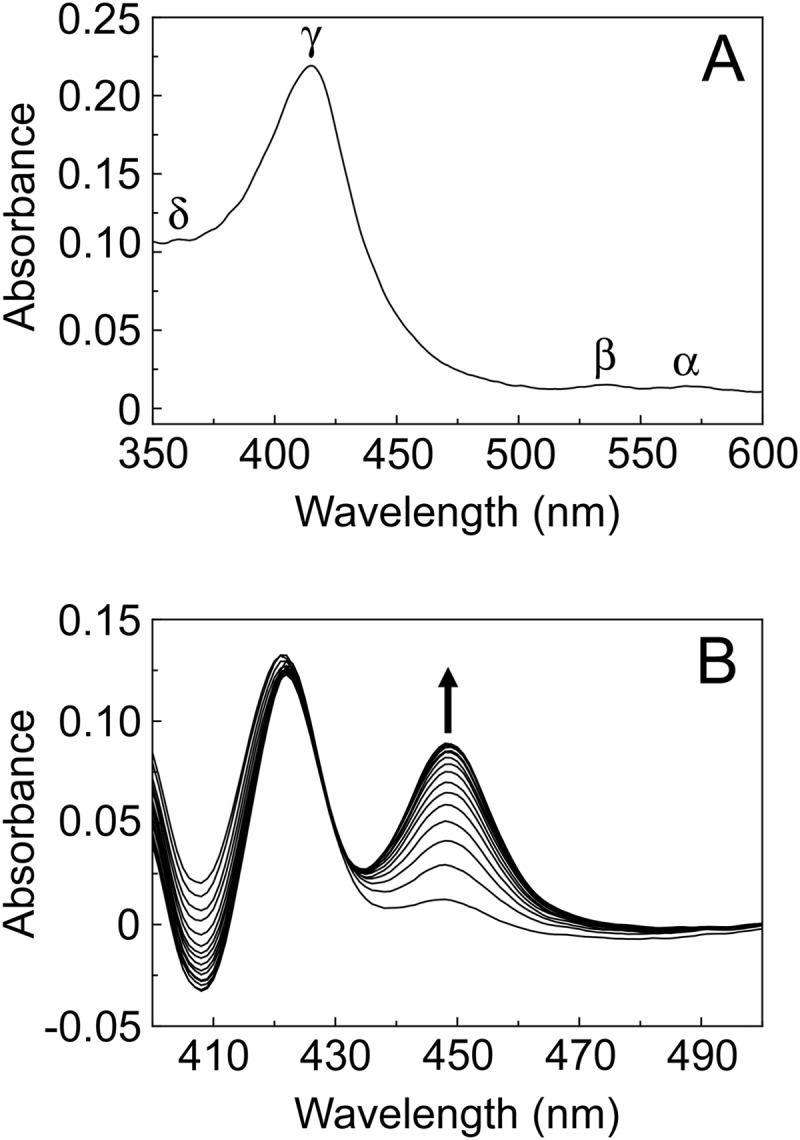
Spectral properties of 4 μM AcCYP51. The absolute oxidized absorption spectrum between 600 and 350 nm (light path, 4.5 mm) (A), in addition to the reduced carbon monoxide difference spectra between 500 and 400 nm at sequential 45-s intervals (light path, 10 mm) (B), is shown.
Type II binding spectra were observed between 2 μM AcCYP51 and fluconazole, itraconazole, and voriconazole (Fig. 4), yielding a peak at ∼428 nm and a trough at ∼412 nm. Type II binding spectra arise from the triazole N-4 nitrogen coordinating as the sixth ligand with the heme iron (38) to form the low-spin CYP51-azole complex, resulting in a red shift of the heme Soret peak. Tight binding normally is observed where the Kd for a ligand is similar to or lower than the concentration of the protein present (39). Azole saturation curves (Fig. 4) confirmed itraconazole and voriconazole bound tightly to AcCYP51 with apparent Kd values of 10.4 ± 1.2 and 13.4 ± 3.8 nM. However, fluconazole binding appeared to be 160- to 205-fold weaker (Kd of 2,137 ± 645 nM). Therefore, the azole ligand binding studies predict that both itraconazole and voriconazole would strongly inhibit AcCYP51 activity while fluconazole would be a weak AcCYP51 inhibitor.
FIG 4.
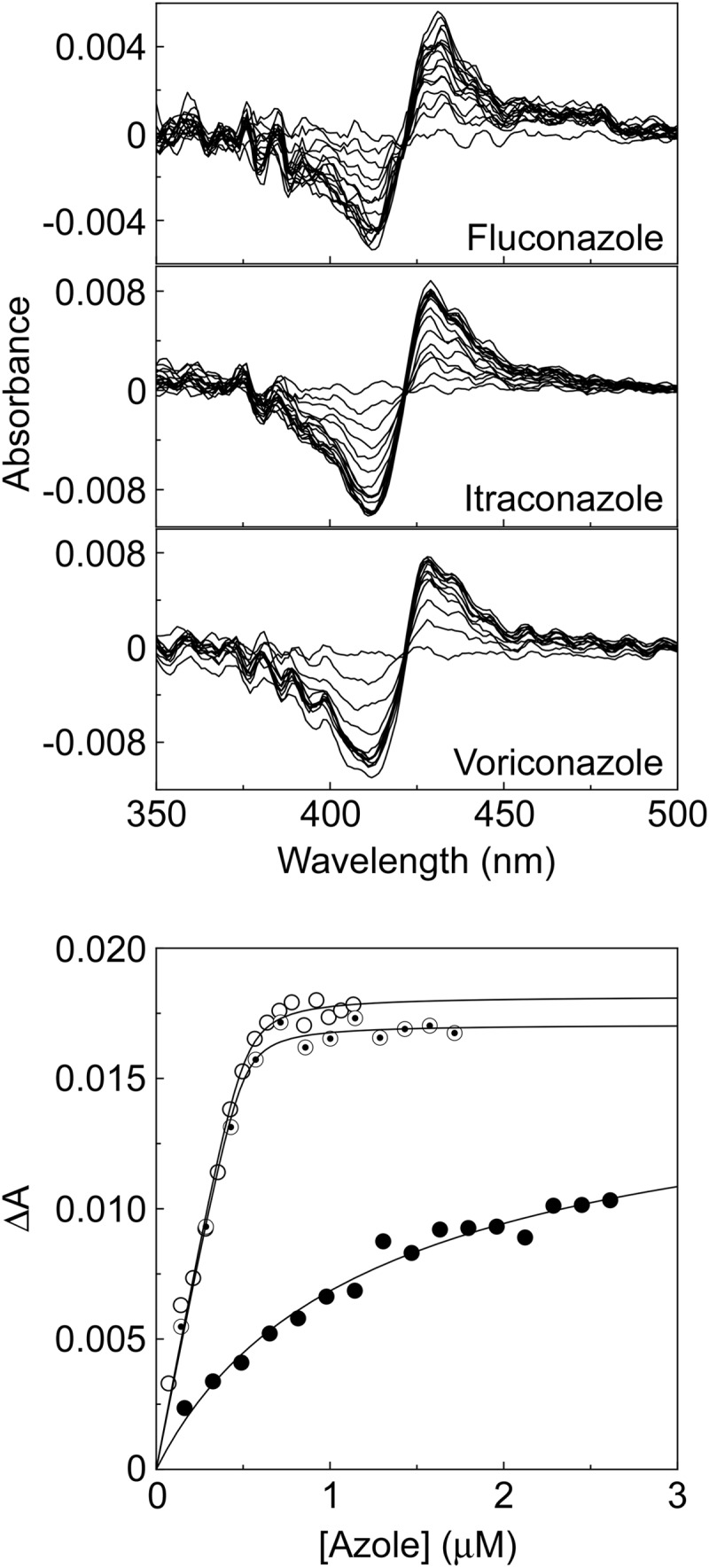
Azole binding to AcCYP51. Type II difference spectra were obtained by progressive titration of 2 μM AcCYP51 with fluconazole, itraconazole, and voriconazole using 4.5-mm-light-path quartz semi-micro cuvettes. Azole saturation curves for AcCYP51 were constructed from the change in absorbance (ΔA428-412) against fluconazole (filled circles), itraconazole (hollow circles), and voriconazole (bullets) concentrations using a rearrangement of the Morrison equation (29, 30) for the tight ligand binding.
Azole inhibition of AcCYP51 sterol 14α-demethylase activity.
Obtusifoliol, lanosterol, and eburicol all were 14α-demethylated in the CYP51 reconstitution assay using purified AcCYP51 protein and AfCPR1 as the redox partner with turnover numbers of 0.474, 0.101, and 0.142/min, respectively. This confirmed the CYP51 enzyme function of the protein and that AcCYP51 had been isolated in a fully functional form. IC50 determinations using 0.6 μM AcCYP51 (Fig. 5) and obtusifoliol as the substrate confirmed that both itraconazole and voriconazole strongly inhibited AcCYP51 activity with IC50s of 0.23 and 0.39 μM. The IC50 for a tight binding azole that cannot be displaced by substrate would be half the CYP51 concentration (∼0.30 μM) observed for itraconazole and voriconazole. In contrast, fluconazole inhibition of AcCYP51 was poor (IC50 of 30 μM), in keeping with the weaker ligand binding observed. These results confirm the predictions made from the azole ligand binding studies and suggest that both itraconazole and voriconazole would be strong inhibitors of Acanthamoeba proliferation.
FIG 5.
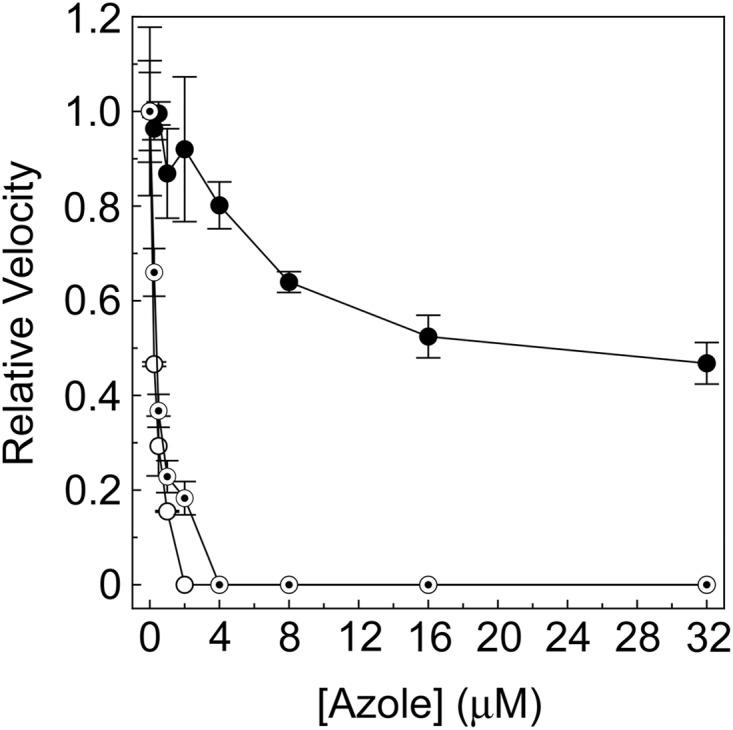
Azole IC50 determinations for AcCYP51. IC50s were determined for fluconazole (filled circles), itraconazole (hollow circles), and voriconazole (bullets). The CYP51 reconstitution assays contained 0.6 μM AcCYP51 and 1.8 μM AfCPR1 using 50 μM obtusifoliol as the substrate. Relative velocities of 1.00 corresponded to actual velocities of 0.474 ± 0.025/min. The mean values of two replicates are shown along with the associated standard deviation bars.
Inhibition of Acanthamoeba proliferation.
MIC studies using A. polyphaga and A. castellani trophozoites (Table 1) confirmed the ineffectiveness of fluconazole at inhibiting cell proliferation of Acanthamoeba with MIC values greater than 64 mg/liter. Surprisingly, itraconazole was a relatively poor inhibitor of Acanthamoeba cell division with MIC values of 16 mg/liter. Only voriconazole effectively inhibited Acanthamoeba proliferation in vitro with MIC values of 1 to 2 mg/liter, confirming voriconazole as an effective anti-amebic agent against trophozoites.
TABLE 1.
MICs for azole antifungal agents against A. polyphaga and A. castellanii trophozoites
| Azole antifungal | MIC (mg/liter) against: |
|
|---|---|---|
| A. polyphaga | A. castellanii | |
| Fluconazole | >64 | >64 |
| Itraconazole | 16 | 16 |
| Voriconazole | 1-2a | 1-2a |
These MICs were between 1 and 2 mg/liter.
DISCUSSION
Phylogenetic analysis of CYP51 amino acid sequences suggested that Acanthamoeba castellanii was more closely related to algae and protists and then plants than higher animals. This was supported by the NCBI-BLASTP results. The relatively low sequence identity (31 to 35%) of AcCYP51 with fungal CYP51 enzymes suggests that existing clinical azole antifungal drugs are not so efficacious against Acanthamoeba infections. The higher AcCYP51 reaction rate observed with obtusifoliol is in keeping with obtusifoliol being the most abundant in vivo CYP51 substrate in Acanthamoeba polyphaga (40), although substrate specificity was not absolute, as has been observed in Trypanosoma brucei CYP51 (41) and plant CYP51 enzymes (42, 43). The substrate specificity of AcCYP51 was broader than that observed with Trypanosoma cruzi CYP51 (44) and Leishmania infantum CYP51 (45).
Azole binding studies suggested fluconazole (Kd of 2,137 nM) would be a poor inhibitor of AcCYP51 activity, while itraconazole and voriconazole (Kd of 10 and 13 nM) would be strong AcCYP51 inhibitors. Itraconazole and voriconazole previously gave similarly low Kd values with Candida albicans CYP51 (46). This was confirmed by azole IC50 determinations using the CYP51 reconstitution assay system (Fig. 5). Previously the affinities of human CYP51 for voriconazole and itraconazole were determined to be 2,290 and 92 to 131 nM, respectively (46). Therefore, the selectivity for AcCYP51 over the human homolog using voriconazole and itraconazole were 170- and 10-fold based on the measured Kd value. In comparison, selectivity for C. albicans CYP51 over the human homolog was 230- and 6-fold for voriconazole and itraconazole (46). This suggested voriconazole would be an effective inhibitor of Acanthamoeba infections in humans due to the high selectivity for the pathogen CYP51 over the host homolog, whereas itraconazole would be less effective. This was borne out during Acanthamoeba MIC determinations where only voriconazole proved effective as an inhibitor of Acanthamoeba proliferation in vitro with a MIC value 8- to 16-fold more potent than that of itraconazole (Table 1). The relatively poor performance of itraconazole against Acanthamoeba may be due in part to poor uptake of the drug into the cells, as itraconazole is a bulky molecule with a long hydrophobic tail compared to the relatively compact voriconazole molecule. MIC values achieved with voriconazole against C. albicans were 24- to 375-fold lower (strain dependent) than those for the Acanthamoeba species used in this study (47), suggesting higher doses of triazoles will be required to be clinically effective against Acanthamoeba infections than those presently used to treat C. albicans infections. However, triazoles have relatively low toxicity and fewer side effects in patients than alternatives such as amphotericin B and can be safely tolerated at higher doses, especially in topical applications. Further investigations are required into the azole uptake preferences of live Acanthamoeba trophozoites to optimize clinical effectiveness.
The widespread occurrence of AK (1–3, 10–14) and the relative difficulty in treating the condition with prolonged use of biguanides and diamidines (15) suggests a role for triazole pharmaceuticals as useful therapeutic adjuncts to conventional AK treatments, especially where secondary yeast/fungal infection is suspected. However, the use of triazole drugs to combat AGE remains limited, because presently there are no recommended treatments as AGE is normally diagnosed postmortem (3), although triazoles, when used in combination with sulfadiazine, pentamidine isethionate, amphotericin B, azithromycin, or rifampin, may be effective if early diagnosis was possible (3). Voriconazole is a strong candidate for the treatment of Acanthamoeba infections in humans, especially AK, as it inhibits the proliferation of trophozoites and benefits from being easily administered either orally as tablets or topically in eye drops. Already some positive case studies and microbiological sensitivity studies have supported this conclusion (1, 16–20), and our study underpins this at the biochemical level. The voriconazole chemical scaffold also could be modified to create a new range of azole therapeutics that have increased effectiveness/potency against Acanthamoeba species, and our work outlines methods by which this can be taken forward.
ACKNOWLEDGMENTS
We are grateful to the Engineering and Physical Sciences Research Council National Mass Spectrometry Service Centre at Swansea University and Marcus Hull for assistance in gas chromatography-mass spectrometry analyses.
This work was supported in part by the European Regional Development Fund/Welsh Government-funded BEACON research program (Swansea University) and the National Science Foundation of the United States grant NSF-MCB-09020212, awarded to W. David Nes (Texas Tech University).
REFERENCES
- 1.Visvesvara GS, Moura H, Schuster FL. 2007. Pathogenic and opportunistic free-living amoebae: Acanthamoeba spp., Balamuthia mandrillaris, Naegleria fowleri and Sappinia diploidea. FEMS Immunol Med Microbiol 50:1–26. doi: 10.1111/j.1574-695X.2007.00232.x. [DOI] [PubMed] [Google Scholar]
- 2.Marciano-Cabral F, Cabral G. 2003. Acanthamoeba spp. as agents of disease in humans. Clin Microbiol Rev 16:273–307. doi: 10.1128/CMR.16.2.273-307.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khan NA. 2006. Acanthamoeba: biology and increasing importance in human health. FEMS Microbiol Rev 30:564–595. doi: 10.1111/j.1574-6976.2006.00023.x. [DOI] [PubMed] [Google Scholar]
- 4.Siddiqui R, Khan NA. 2012. Biology and pathogenesis of Acanthamoeba. Parasit Vectors 5:6. doi: 10.1186/1756-3305-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martinez AJ, Visvesvara GS. 1997. Free-living amphizoic and opportunistic amebas. Brain Pathol 7:583–598. doi: 10.1111/j.1750-3639.1997.tb01076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schuster FL, Visvesvara GS. 2004. Free-living amoebae as opportunistic and non-opportunistic pathogens of humans and animals. Int J Parasitol 34:1001–1027. doi: 10.1016/j.ijpara.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 7.Visvesvara GS, Maguire JH. 2006. Pathogenic and opportunistic free-living amebas Acanthamoeba spp., Balamuthia mandrillaris, Naegleria fowleri, and Sappinia diploidea, p 1001–1027. In Guerrant RL, Walker DH, Weller PF (ed), Tropical infectious diseases, vol 2 Churchill, Livingston, United Kingdom. [Google Scholar]
- 8.Martinez AJ. 1991. Infections of the central nervous system due to Acanthamoeba. Rev Infect Dis 13:S399–S402. doi: 10.1093/clind/13.Supplement_5.S399. [DOI] [PubMed] [Google Scholar]
- 9.John KG, Valerie PJ, Simon K. 2009. Perspective Acanthamoeba keratitis: diagnosis and treatment update 2009. Am J Ophthalmol 1148:487–499. [DOI] [PubMed] [Google Scholar]
- 10.Radford CF, Lehmann OJ, Dart JKG. 1998. Acanthamoeba keratitis: multi-centre survey in England 1992-1996. Br J Ophthalmol 82:1387–1392. doi: 10.1136/bjo.82.12.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stehr-Green JK, Bailey TM, Visvesvara GS. 1989. The epidemiology of Acanthamoeba keratitis in the United States. Am J Ophthalmol 107:331–336. doi: 10.1016/0002-9394(89)90654-5. [DOI] [PubMed] [Google Scholar]
- 12.Lorenzo-Morales J, Martin-Navarro CM, Lopez-Arencibia A, Arnalich-Montiel F, Pinero JE, Valladares B. 2013. Acanthamoeba keratitis: an emerging disease gathering importance worldwide? Trends Parasitol 29:181–187. doi: 10.1016/j.pt.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 13.Radford CF, Minassian DC, Dart JK. 2002. Acanthamoeba keratitis in England and Wales: incidence, outcome and risk factors. Br J Ophthalmol 86:536–542. doi: 10.1136/bjo.86.5.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dart JK, Saw VPJ, Kilvington S. 2009. Acanthamoeba keratitis: diagnosis and treatment update 2009. Am J Ophthalmol 148:487–499. doi: 10.1016/j.ajo.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 15.Perez-Santonja JJ, Kilvington S, Hughes R, Tufail A, Metheson M, Dart JKG. 2003. Persistently culture positive Acanthamoeba keratitis: in vivo resistance and in vitro sensitivity. Ophthalmology 110:1593–1600. doi: 10.1016/S0161-6420(03)00481-0. [DOI] [PubMed] [Google Scholar]
- 16.Amoils SP, Heney C. 1999. Acanthamoeba keratitis with live isolates treated with cryosurgery and fluconazole. Am J Ophthalmol 127:718–720. doi: 10.1016/S0002-9394(98)00426-7. [DOI] [PubMed] [Google Scholar]
- 17.Schuster FL, Guglielmo BJ, Visvesvara GS. 2006. In vitro activity of miltefosine and voriconazole on clinical isolates of free-living amebas: Balamuthia mandrillaris, Acanthamoeba spp., and Naegleria fowleri. J Eukaryot Microbiol 53:121–126. doi: 10.1111/j.1550-7408.2005.00082.x. [DOI] [PubMed] [Google Scholar]
- 18.Arnalich-Montiel F, Martin-Navarro CM, Alio JL, Lopez-Velez R, Martinez-Carretero E, Valladares B, Pinero JE, Lorenzo-Morales J. 2012. Successful monitoring and treatment of intraocular dissemination of Acanthamoeba with voriconazole. J Arch Ophthalmol 130:1474–1475. doi: 10.1001/archophthalmol.2012.2376. [DOI] [PubMed] [Google Scholar]
- 19.Bang S, Edell E, Eghrari AO, Gottsch JD. 2010. Treatment with voriconazole in 3 eyes with resistant Acanthamoeba keratitis. Am J Ophthalmol 149:66–69. doi: 10.1016/j.ajo.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 20.Tu EY, Joslin CE, Shoff ME. 2010. Successful treatment of chronic stromal Acanthamoeba keratitis with oral voriconazole monotherapy. Cornea 29:1066–1068. doi: 10.1097/ICO.0b013e3181cbfa2c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Massilamany C, Marciano-Cabral F, da Rocha-Azevedo B, Jamerson M, Gangaplara A, Steffen D, Zabad R, Illes Z, Sobel RA, Reddy J. 2014. SJL mice infected with Acanthamoeba castellanii develop central nervous system autoimmunity through the generation of cross-reactive T cells for myelin. PLoS One 9:e98506. doi: 10.1371/journal.pone.0098506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Massilamany C, Jamerson M, Madaviputhiya N, Nandakumar R, Marciano-Cabral F, Sejbaek T, Illes Z, Reddy J. 2013. An evidence for a potential linkage between Acanthamoeba infections and multiple sclerosis (P4551). J Immunol 190:197.13. [Google Scholar]
- 23.Barnes HJ, Arlotto MP, Waterman MR. 1991. Expression and enzymatic activity of recombinant cytochrome P450 17α-hydroxylase in Escherichia coli. Proc Natl Acad Sci U S A 88:5597–5601. doi: 10.1073/pnas.88.13.5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arase M, Waterman MR, Kagawa N. 2006. Purification and characterization of bovine steroid 21-hydroxylase (P450c21) efficiently expressed in Escherichia coli. Biochem Biophys Res Commun 344:400–405. doi: 10.1016/j.bbrc.2006.03.067. [DOI] [PubMed] [Google Scholar]
- 25.Bellamine A, Mangla AT, Nes WD, Waterman MR. 1999. Characterization and catalytic properties of the sterol 14α-demethylase from Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 96:8937–8942. doi: 10.1073/pnas.96.16.8937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Estabrook RW, Peterson JA, Baron J, Hildebrandt AG. 1972. The spectrophotometric measurement of turbid suspensions of cytochromes associated with drug metabolism, p 303–350. In Chignell CF. (ed), Methods in pharmacology, vol. 2 Appleton-Century-Crofts, New York, NY. [Google Scholar]
- 27.Omura T, Sato R. 1964. The carbon monoxide-binding pigment of liver microsomes. J Biol Chem 239:2379–2385. [PubMed] [Google Scholar]
- 28.Lamb DC, Kelly DE, Waterman MR, Stromstedt M, Rozman D, Kelly SL. 1999. Characteristics of the heterologously expressed human lanosterol 14α-demethylase (other names: P45014DM, CYP51, P45051) and inhibition of the purified human and Candida albicans CYP51 with azole antifungal agents. Yeast 15:755–763. doi:. [DOI] [PubMed] [Google Scholar]
- 29.Lutz JD, Dixit V, Yeung CK, Dickmann LJ, Zelter A, Thatcher JA, Nelson WL, Isoherranen N. 2009. Expression and functional characterization of cytochrome P450 26A1, a retinoic acid hydroxylase. Biochem Pharmacol 77:258–268. doi: 10.1016/j.bcp.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morrison JF. 1969. Kinetics of the reversible inhibition of enzyme-catalysed reactions by tight-binding inhibitors. Biochim Biophys Acta Enzymol 185:269–286. doi: 10.1016/0005-2744(69)90420-3. [DOI] [PubMed] [Google Scholar]
- 31.Lepesheva GI, Ott RD, Hargrove TY, Kleshchenko YY, Schuster I, Nes WD, Hill GC, Villalta F, Waterman MR. 2007. Sterol 14α-demethylase as a potential target for antitrypanosomal therapy: enzyme inhibition and parasite cell growth. Chem Biol 14:1283–1293. doi: 10.1016/j.chembiol.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parker JE, Warrilow AGS, Cools HJ, Fraaije BA, Lucas JA, Rigdova K, Griffiths WJ, Kelly DE, Kelly SL. 2013. Prothioconazole and prothioconazole-desthio activity against Candida albicans sterol 14α-demethylase (CaCYP51). Appl Environ Microbiol 79:1639–1645. doi: 10.1128/AEM.03246-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sarker SD, Nahar L, Kumarasamy Y. 2007. Microtitre plate-based antibacterial assay incorporating resazurin as an indicator of cell growth, and its application in the in vitro antibacterial screening of phytochemicals. Methods 42:321–324. doi: 10.1016/j.ymeth.2007.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heredero-Bermejo I, Copa-Patino JL, Soliveri J, Gomez R, de la Mata FJ, Perez-Serrano J. 2013. In vitro comparative assessment of different viability assays in Acanthamoeba castellanii and Acanthamoeba polyphaga trophozoites. Parasitol Res 112:4087–4095. doi: 10.1007/s00436-013-3599-5. [DOI] [PubMed] [Google Scholar]
- 35.Lepesheva GI, Waterman MR. 2007. Sterol 14α-demethylase cytochrome P450 (CYP51), a P450 in all biological kingdoms. Biochim Biophys Acta 1770:467–477. doi: 10.1016/j.bbagen.2006.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lepesheva GI, Waterman MR. 2011. Structural basis for conservation in the CYP51 family. Biochim Biophys Acta 1814:88–93. doi: 10.1016/j.bbapap.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jefcoate CR. 1978. Measurement of substrate and inhibitor binding to microsomal cytochrome P-450 by optical-difference spectroscopy, p 258–279. In Fleischer S, Packer L (ed), Methods in enzymology: biomembranes, part C, vol 52 Elsevier Inc., New York, NY. [DOI] [PubMed] [Google Scholar]
- 38.Jefcoate CR, Gaylor JL, Calabrese RL. 1969. Ligand interactions with cytochrome P450. I. Binding of primary amines. Biochemistry 8:3455–3463. [DOI] [PubMed] [Google Scholar]
- 39.Copeland RA. 2005. Evaluation of enzyme inhibitors in drug discovery: a guide for medicinal chemists and pharmacologists, p 178–213. Wiley-Interscience, New York, NY. [PubMed] [Google Scholar]
- 40.Raederstorff D, Rohmer M. 1985. Sterol biosynthesis de novo via cycloartenol by the soil amoeba Acanthamoeba polyphaga. Biochem J 231:609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lepesheva GI, Nes WD, Zhou W, Hill GC, Waterman MR. 2004. CYP51 from Trypanosoma brucei is obtusifoliol-specific. Biochemistry 43:10789–10799. doi: 10.1021/bi048967t. [DOI] [PubMed] [Google Scholar]
- 42.Lamb DC, Kelly DE, Kelly SL. 1998. Molecular diversity of sterol 14α-demethylase substrates in plants, fungi and humans. FEBS Lett 425:263–265. doi: 10.1016/S0014-5793(98)00247-6. [DOI] [PubMed] [Google Scholar]
- 43.Cabello-Hurtado F, Taton M, Forthoffer N, Kahn R, Bak S, Rahier A, Werck-Reichhart D. 1999. Optimized expression and catalytic properties of a wheat obtusifoliol 14α-demethylase (CYP51) expressed in yeast. Eur J Biochem 262:435–446. doi: 10.1046/j.1432-1327.1999.00376.x. [DOI] [PubMed] [Google Scholar]
- 44.Lepesheva GI, Zaitseva NG, Nes WD, Zhou W, Arase M, Liu J, Hill GC, Waterman MR. 2006. CYP51 from Trypanosoma cruzi–a phyla-specific residue in the B′ helix defines substrate preferences of sterol 14α-demethylase. J Biol Chem 281:3577–3585. doi: 10.1074/jbc.M510317200. [DOI] [PubMed] [Google Scholar]
- 45.Hargrove TY, Wawrzak Z, Liu J, Nes WD, Waterman MR, Lepesheva GI. 2011. Substrate preferences and catalytic parameters determined by structural characteristics of sterol 14α-demethylase (CYP51) from Leishmania infantum. J Biol Chem 286:26838–26848. doi: 10.1074/jbc.M111.237099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Warrilow AGS, Parker JE, Kelly DE, Kelly SL. 2013. Azole affinity of sterol 14α-demethylase (CYP51) enzymes from Candida albicans and Homo sapiens. Antimicrob Agents Chemother 57:1352–1360. doi: 10.1128/AAC.02067-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Warrilow AGS, Hull CM, Parker JE, Garvey EP, Hoekstra WJ, Moore WR, Schotzinger RJ, Kelly DE, Kelly SL. 2014. The clinical candidate VT-1161 is a highly potent inhibitor of Candida albicans CYP51 but fails to bind the human enzyme. Antimicrob Agents Chemother 58:7121–7127. doi: 10.1128/AAC.03707-14. [DOI] [PMC free article] [PubMed] [Google Scholar]