Abstract
Heat maps are a commonly used visualization tool for metabolomic data where the relative abundance of ions detected in each sample is represented with color intensity. A limitation of applying heat maps to global metabolomic data, however, is the large number of ions that have to be displayed and the lack of information provided about important metabolomic parameters such as m/z and retention time. Here we address these challenges by introducing the interactive cluster heat map in the data-processing software XCMS Online. XCMS Online (xcmsonline.scripps.edu) is a cloud-based informatic platform designed to process, statistically evaluate, and visualize mass-spectrometry based metabolomic data. An interactive heat map is provided for all data processed by XCMS Online. The heat map is clickable, allowing users to zoom and explore specific metabolite metadata (EICs, Box-and-whisker plots, mass spectra) that are linked to the METLIN metabolite database. The utility of the XCMS interactive heat map is demonstrated on metabolomic data set generated from different anatomical regions of the mouse brain.
Keywords: XCMS Online, Metabolomics, Bioinformatics software, Interactive cluster heat map, Anatomical brain regions, Brain Metabolomics
INTRODUCTION
Untargeted metabolomics measures the levels of thousands of metabolite features in a single analysis, providing a snapshot of metabolism at the systems level (Patti, Tautenhahn, Rinehart, Cho, Shriver, Manchester, Nikolskiy et al. 2012; Patti, Yanes, Siuzdak 2012). Metabolomic experiments generate large data sets and scientists rely greatly on exploratory data analysis including visual pattern recognition when searching for interesting features in the data, like differentially expressed metabolites (Gowda, Ivanisevic, Johnson, Kurczy, Benton, Rinehart, Nguyen et al. 2014; Patti, Tautenhahn, Rinehart, Cho, Shriver, Manchester, Nikolskiy et al. 2012; Xia, Wishart 2011). Several visualization techniques have become common in global metabolomic data analysis such as scores and loadings plots, heat maps, scatter plots, volcano plots and recently designed cloud plots. The cloud plot provides a descriptive visualization of dysregulated metabolite features for quantitative analysis and further structural elucidation. It offers the detailed feature assignment including overlaid extracted ion chromatograms (EICs), Box-Whisker plot, mass spectrum and potential METLIN metabolite database matches (Patti, Tautenhahn, Rinehart, Cho, Shriver, Manchester, Nikolskiy et al. 2012; Tautenhahn, Cho, Uritboonthai, Zhu, Patti, Siuzdak 2012). An interactive cluster heat map is a compelling follow-up to the implementation of the interactive cloud plot, allowing for an added dimension of data visualization to help in sample classification and the description of features that are driving the classification.
Heat maps are one of the most widely used bioinformatic graphic displays (Wilkinson, Friendly 2009). They are especially popular in gene expression analysis and visualization of genomic data sets in general (Eisen, Spellman, Brown, Botstein 1998; Wu, Noble 2004). Similar to genomic experiments, mass spectrometry-based metabolomic experiments present thousands of data points, and while heat map matrices are useful for pattern recognition they are largely limited by their two dimensional representation. The traditional, cluster heat map, with an extensive history of data representation in biological and biomedical publications, is frequently displayed in static form (Wilkinson, Friendly 2009). While color-coded matrix elements and adjacent dendograms indicate functional relationships among variables and samples, traditional heat maps do not offer the opportunity to sort the data on different axes, to filter the data or to focus on specific elements of the map, a difficulty compounded by the large number of represented elements. To overcome this limitation we have developed an “interactive” matrix to display the underlying information, behind the color-coded tiles, about each metabolite feature. The interactive heat map was developed as a tool within XCMS Online interface (Gowda, Ivanisevic, Johnson, Kurczy, Benton, Rinehart, Nguyen et al. 2014), a widely used data processing platform in untargeted metabolomics. The multi-group comparison across three anatomical regions of mouse brain was applied to highlight the illustrative strength of the interactive heat map.
MATERIALS AND METHODS
Metabolome extraction and reconstitution
Animal and tissue preparation protocol is provided in Supplementary Information. The NSG mouse strain was chosen for its abilities to support humanization leading to numerous applications in oncology and infectious disease research. Brain dissection was performed on five specimens from the same genetic strain, males and similar ages. Each subregion of brain tissue was extracted using a MeOH:H2O (4:1, v/v) solvent mixture. An adjusted volume of 1 mL of cold solvent was added per 10 mg tissue, probe sonicated for 5 s, and incubated in liquid nitrogen for 1 min. The samples were then allowed to thaw at room temperature and then probe sonicated for another 5 s. To precipitate proteins, the samples were incubated for 1 h at −20 °C, followed by 15 min centrifugation at 16000 × g and 4 °C. The resulting supernatant was removed and evaporated to dryness in a vacuum concentrator (LABCONCO CentriVap Benchtop). The pellet was reconstituted in water and protein concentrations were measured using Pierce™ BCA Protein Assay Kit (Thermo Scientific, Rockford, IL) as a reference for metabolite reconstitution. The dry extracts were then reconstituted in ACN:H2O (1:1, v/v) normalized by the sample’s protein level, sonicated for 10 min, and centrifuged 15 min at 16000 g and 4 °C to remove insoluble debris. The supernatants were transferred to HPLC vials and stored at −80 °C prior to LC/MS analysis.
LC/MS analysis
Tissue extracts were analyzed on 6550 iFunnel QTOF mass spectrometer (Agilent Technologies) interfaced with 1290 UPLC system (Agilent Technologies). Samples were analyzed using a Luna Aminopropyl, 3 μm, 150 mm × 1.0 mm I.D. HILIC column (Phenomenex). The mobile phase was composed of A = 20 mM ammonium acetate and 40 mM ammonium hydroxide in 95% water and B = 95% acetonitrile. The remaining 5 % were acetonitrile or water, respectively. The linear gradient elution from 100% B (0–5 min) to 100% A (50–55 min) was applied (A = 95% H2O, B = 95% ACN, with appropriate additives). A 10 min re-equilibration time was applied for HILIC, to ensure the column re-equilibration and maintain the reproducibility. The flow rate was 50 μL/min, and the sample injection volume was 5 μL. ESI source conditions were set as follows: dry gas temperature 200 °C and flow 11 L/min, fragmentor 380 V, sheath gas temperature 300 °C and flow 9 L/min, nozzle voltage 500 V, and capillary voltage −2500 V in ESI negative mode. The instrument was set to acquire over the m/z range 50–1000, with the MS acquisition rate of 2 spectra/s.
Data analysis
Data were analyzed by using multi-group method on the web interface for interactive XCMS Online, which is freely available at https://xcmsonline.scripps.edu. It allows users to either upload datasets using a java applet or select pre-uploaded datasets on XCMS Online. Following the upload of raw data files, users can select preset parameters (or customize them) depending on the instrument platform in which the data were acquired. The parameters are displayed in the web browser using the jQuery-UI framework, with each tab organized by category. Users can define parameters for statistical analysis (parametric/non-parametric, paired/unpaired) based on the type of experiment and data. The raw data files are than processed for peak detection, retention-time correction, chromatogram alignment, metabolite feature metadata, statistical evaluation, and putative identification through METLIN standard database matching. Parameter settings for XCMS processing of our demonstration data acquired by HILIC were as follows: centWave for feature detection (Δ m/z = 15 ppm, minimum peak width = 10 sec and maximum peak width = 120 sec); obiwarp settings for retention-time correction (profStep = 1); and parameters for chromatogram alignment, including mzwid = 0.015, minfrac = 0.5 and bw = 5. The relative quantification of metabolite features was based on peak areas. Peak intensity or abundance, expressed in ion counts, refers to peak height and is often used to predict the quality of MS/MS data that can be collected. For comparative analysis across different metabolites in the heat map, peak areas were converted to z-scores. The row Z-score or scaled expression value of each feature was calculated as mean abundance subtracted from the abundance and then divided by the standard deviation across all the samples.
RESULTS AND DISCUSSION
The interactive heat map concept is derived from the recently designed XCMS Online platform which has been developed to deconvolve metabolomic data, simplify data analysis and customize data output. Metabolomic data display has been accomplished through the interactive visualization tools that include cloud plots (two-group and multi-group), PCA scores and loadings plots, and Venn diagrams. The cluster heat map was implemented as an easy-to-use interactive graphic to enable the user to easily explore the data, validate its integrity and provide useful insights about dysregulated metabolite features and sample grouping. The key to our new interactive XCMS Online platform is the integration of univariate and multivariate statistical data processing and metabolite feature assignment. Metabolite identification is facilitated through the link with standard METLIN database (http://metlin.scripps.edu/index.php) (Tautenhahn, Cho, Uritboonthai, Zhu, Patti, Siuzdak 2012; Zhu, Schultz, Wang, Johnson, Yannone, Patti, Siuzdak 2013) providing potential matches and when available, MS/MS spectra and biology relevant information via the link to Human Metabolome Database (HMDB) (Wishart, Knox, Guo, Eisner, Young, Gautam, Hau et al. 2009), LIPID MAPS (Fahy, Sud, Cotter, Subramaniam 2007), and KEGG pathway database (Kanehisa, Goto 2000).
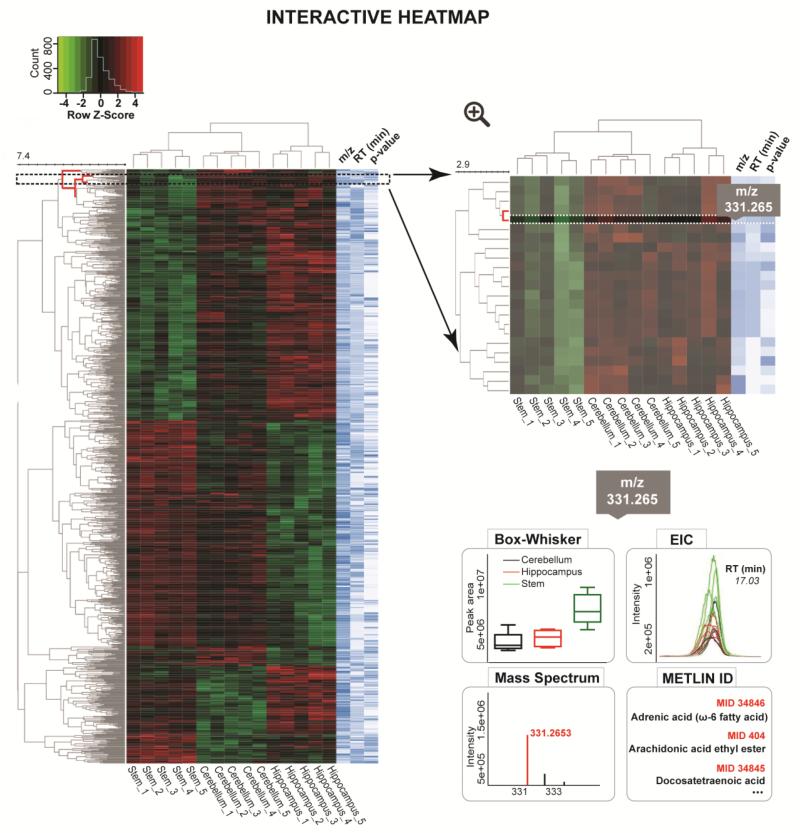
As experiments are processed on XCMS Online, the data matrix comprising the metabolite feature values (peak areas and maximal peak intensities) across samples is collected and stored. When a user selects the interactive heat map visualization tool in XCMS Result Summary menu (Supplementary Figure 1), the web server’s PHP calls a python and R script to load the data file and several JavaScript libraries enable the heat map display and exploration of metadata (Skuta, Bartunek, Svozil 2014). Meta information is made available to users during mouse rollovers or by clicking a link. Depending on the context, data may be dynamically retrieved from a database or file on the server using AJAX technology. The visualization process of large number of metabolite features of interest (top 1000 features ranked by p-value) has been optimized using compression and limiting metadata transfers to maintain a responsive graphical user interface. Only the top 1000 dysregulated features can be explored interactively; however the entire set of dysregulated features can be explored through the Results table and the statistical results can also be exported. Interactive manipulations comprise of modification of display parameters, change of scale, selections of feature tiles and queries related to feature metadata. The display modification for the heat map allows users to sort the table by one of the feature metadata fields, either m/z, RT or p-value (Figure 1, right “heat map” panel in blue-white scale), which is useful for searching underlying patterns that correlate with RT and m/z, such as isotopes and adducts. The key feature introduced during the interactive display is a cursor controlled heat map that provides m/z values, retention times, and p-values by hovering the cursor over each element (Figure 1). For hierarchical clustering analysis (HCA), Euclidean distance is used as a distance measure and the complete linkage is applied as unsupervised clustering method. In the future, the clustering may be upgraded by the addition of different similarity measures (e.g. Correlation, Cosine Correlation, and many more) and clustering algorithms (e.g. single linkage, average linkage, Ward’s method).
Figure 1.
Interactive, sortable heat map with customized metabolomic data visualization. Each row represents a metabolite feature and each column represents a sample. Metabolite features whose levels vary significantly (p < 0.01) across three different brain regions (stem, cerebellum and hippocampus) are projected on the heat map and used for sample clustering. The row Z-score or scaled expression value of each feature is plotted in red-green color scale. The red color of the tile indicates high abundance and green indicates low abundance. When a user scrolls the mouse over the metabolite cluster tree on the left, the selected node is displayed in zoomed-in version. When a feature assignment tile (m/z, retention time or p-value) is selected, its Box-Whisker plot, EIC (Extracted Ion Chromatogram), MS spectrum and METLIN matches appear on the bottom of the main panel.
Once the heat map with associated dendogram has been displayed, the user can zoom into each node of the classification tree to access the more contextual metadata about the metabolite features. The metadata are dynamically loaded after a specific feature is selected. The information includes the variation of peak areas (Box-Whisker plots) and abundances (aligned EICs) across different sample classes, mass spectral data and links to METLIN matches.
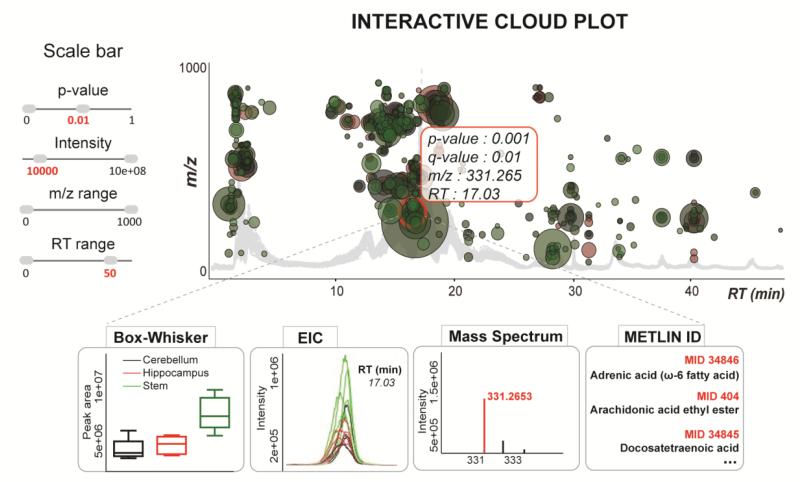
The interactive heat map is an alternative to the cloud plot allowing the user to visualize the large multidimensional untargeted metabolomics results and screen for the significantly altered features by customizing the display. Furthermore, both the interactive heat map and cloud plot allow for zooming to magnify a desired area of the plot, which is very useful for plots with a large number of data points (Gowda, Ivanisevic, Johnson, Kurczy, Benton, Rinehart, Nguyen et al. 2014). On the cloud plot the metabolite features are projected over the aligned total ion chromatograms depending on their retention time (x-axis) and m/z (y-axis) (Figure 2). Each bubble in the plot corresponds to a metabolite feature and the size of the bubble denotes the extent of the fold change (Patti, Tautenhahn, Rinehart, Cho, Shriver, Manchester, Nikolskiy et al. 2012). The heat map mirrors the data table format with the rows representing metabolite features and columns representing the samples, where color gradient denotes the normalized abundance of each metabolite feature across the samples (Deu-Pons, Schroeder, Lopez-Bigas 2014). The complementary value of a cluster heat map in comparison to a cloud plot lies in the ability to identify clusters of samples with similar metabolic patterns as well as groups of discriminating metabolites that drive sample clustering. Since the clustering is an unsupervised method it allows users to see any expected class separation of the samples and the features with a high clustering coefficient (Meunier, Dumas, Piec, Béchet, Hébraud, Hocquette 2006). As an example, we have analyzed global metabolic profiles across three different regions of normal mouse brain: hippocampus, cerebellum and stem (Figure 1). The cluster heat map can be used to visualize the results of two-group as well as multi-group analysis. The untargeted profiling in hydrophilic interaction mode, followed by multi-group comparison enabled the detection of 516 differentially expressed metabolite features (p-value ≤ 0.01, Intensity ≥ 10,000) across three anatomical brain regions. Hierarchical clustering analysis (HCA) confirmed three distinct clusters defined by the samples of hippocampus, cerebellum and stem. The diversity of metabolic patterns across these three regions of brain and their relation to region-specific function should be further investigated by the analysis of statistically discriminative and biochemically related metabolites. For a demonstration, the variation pattern of a metabolite feature with m/z 331.265 is shown across different brain regions by a color pattern on the heat map, by the Box-Whisker plot and the aligned Extracted Ion Chromatograms (Figure 1, zoom right below). This metabolite feature results in 17 matches in METLIN database, using the accurate mass measurement and demonstrates the importance of further MS/MS matching for metabolite identification. The position of this metabolite feature is also indicated on the cloud plot depending on its m/z and retention time following the chromatographic gradient (Figure 2).
Figure 2.
Interactive cloud plot with customized metabolomic data visualization. Metabolite features whose expression level varies significantly (p < 0.01) three different regions of brain (hippocampus, cerebellum and stem) are projected on the cloud plot depending on their retention time (x-axis) and m/z (y-axis). Each metabolite feature is represented by a bubble. Statistical significance (p-value) is represented by the bubble’s color intensity. The size of the bubble denotes feature intensity. When the user scrolls the mouse over a bubble, feature assignments are displayed in a pop-up window (p-value, q-value, m/z, RT). When a bubble is selected by a ‘mouse click’, Box-Whisker plots, the EICs, Mass spectrum, Post-hoc (not shown), and METLIN matches appear on the main panel. Each bubble is linked to the METLIN database to provide putative identifications based on accurate m/z.
Currently little is known about the metabolite distribution across the brain subregions. Brain tissue profiling may be valuable for understanding local metabolic activity and could lead to the observation of metabolic differences across anatomical regions of brain and provide the important insights needed for functional characterization of brain regions and brain metabolism in general (Ivanisevic, Epstein, Kurczy, Benton, Uritboonthai, Fox, Boska et al.). Brain metabolomics has been highlighted, over the last decade, by studies of neurological disorders and enhanced characterization of central nervous system (CNS) metabolome (Dumas, Davidovic 1000; Mandal, Guo, Chaudhary, Liu, Yallou, Dong, Aziat et al. 2012; Nicholson, Holmes, Kinross, Darzi, Takats, Lindon 2012). The potential of untargeted brain metabolomics lies in the comprehensive measurement of small molecules that play an essential role in neurophysiology (for example, neurotransmitters, signaling lipids, and osmolytes) along with regulators of oxidative stress and intermediary and energy currency metabolites (Piomelli, Astarita, Rapaka 2007).
Many additional developments are planned to improve the current implementation of the interactive heat map tool and XCMS Online in general. The essential ones include the increase of raw data upload speed (Rinehart, Johnson, Nguyen, Ivanisevic, Benton, Lloyd, Arkin et al. 2014) biochemical pathway mapping of feature clusters, automated metabolite identification through MS/MS matching against METLIN metabolite database and the exploration of chemical structure similarities within clusters.
CONCLUDING REMARKS
An interactive cluster heat map has been created to improve our ability to explore complex metabolomic data. The metabolomic interactive heat map allows for identification of clusters across data sets and detailed analysis of metabolite features, adding a new dimension to metabolomic data visualization and deconvolution. The incorporation of the interactive heat map into XCMS Online also facilitates rapid data exploration and higher dimensional data displays to provide researchers a novel means of viewing their data to understand biological relationships.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported, in part, by the University of Nebraska Foundation which includes individual donations from Dr. Carol Swarts and Frances and Louie Blumkin and National Institutes of Health grants P01 MH64570, RO1 MH104147, P01 DA028555, R01 NS36126, P01 NS31492, 2R01 NS034239, P01 NS43985, P30 MH062261 and R01 AG043540.
Footnotes
Conflict of interest. The authors declare no Conflict of interests.
Compliance with ethical requirements. NOD scid IL2 receptor gamma chain knockout, NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ, (NSG) mice (The Jackson Laboratories, Bar Harbor, Maine, USA; stock number 005557) were obtained from an established breeding colony and housed under pathogen-free conditions in accordance with ethical guidelines for care of laboratory animals at the National Institutes of Health and the University of Nebraska Medical Center.
REFERENCES
- Deu-Pons J, Schroeder MP, Lopez-Bigas N. jHeatmap: an interactive heatmap viewer for the web. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btu094. doi:10.1093/bioinformatics/btu094. [DOI] [PubMed] [Google Scholar]
- Dumas ME, L Davidovic. Metabolic phenotyping and systems biology approaches to understanding neurological disorders. F1000Prime Rep. 1000;5:5–18. doi: 10.12703/P5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proceedings of the National Academy of Sciences. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahy E, Sud M, Cotter D, Subramaniam S. LIPID MAPS online tools for lipid research. Nucleic Acids Res. 2007;35:21. doi: 10.1093/nar/gkm324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowda H, et al. Interactive XCMS Online: Simplifying Advanced Metabolomic Data Processing and Subsequent Statistical Analyses. Analytical Chemistry. 2014 doi: 10.1021/ac500734c. doi:10.1021/ac500734c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanisevic J, et al. Brain Region Mapping Using Global Metabolomics. Chemistry & Biology. doi: 10.1016/j.chembiol.2014.09.016. doi:10.1016/j.chembiol.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal R, et al. Multi-platform characterization of the human cerebrospinal fluid metabolome: a comprehensive and quantitative update. Genome Med. 2012;4:38. doi: 10.1186/gm337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier B, Dumas E, Piec I, Béchet D, Hébraud M, Hocquette J-F. Assessment of Hierarchical Clustering Methodologies for Proteomic Data Mining. Journal of Proteome Research. 2006;6:358–366. doi: 10.1021/pr060343h. doi:10.1021/pr060343h. [DOI] [PubMed] [Google Scholar]
- Nicholson JK, Holmes E, Kinross JM, Darzi AW, Takats Z, Lindon JC. Metabolic phenotyping in clinical and surgical environments. Nature. 2012;491:384–392. doi: 10.1038/nature11708. [DOI] [PubMed] [Google Scholar]
- Patti GJ, et al. A View from Above: Cloud Plots to Visualize Global Metabolomic Data. Analytical Chemistry. 2012;85:798–804. doi: 10.1021/ac3029745. doi:10.1021/ac3029745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patti GJ, Yanes O, Siuzdak G. Innovation: Metabolomics: the apogee of the omics trilogy. Nat Rev Mol Cell Biol. 2012;13:263–269. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piomelli D, Astarita G, Rapaka R. A neuroscientist’s guide to lipidomics. Nature Reviews Neuroscience. 2007;8:743–54. doi: 10.1038/nrn2233. [DOI] [PubMed] [Google Scholar]
- Rinehart D, et al. Metabolomic data streaming for biology-dependent data acquisition. Nature Biotechnology. 2014;32:524–527. doi: 10.1038/nbt.2927. doi:Doi 10.1038/Nbt.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skuta C, Bartunek P, Svozil D. InCHlib - interactive cluster heatmap for web applications. Journal of Cheminformatics. 2014:6. doi: 10.1186/s13321-014-0044-4. doi:Artn 44 doi 10.1186/S13321-014-0044-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautenhahn R, Cho K, Uritboonthai W, Zhu Z, Patti GJ, Siuzdak G. An accelerated workflow for untargeted metabolomics using the METLIN database. Nat Biotechnol. 2012;30:826–828. doi: 10.1038/nbt.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson L, Friendly M. The History of the Cluster Heat Map. The American Statistician. 2009;63:179–184. doi:10.1198/tas.2009.0033. [Google Scholar]
- Wishart DS, et al. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. 2009;37:25. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Noble WS. Genomic data visualization on the Web. Bioinformatics. 2004;20:1804–1805. doi: 10.1093/bioinformatics/bth154. doi:10.1093/bioinformatics/bth154. [DOI] [PubMed] [Google Scholar]
- Xia J, Wishart DS. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat Protocols. 2011;6:743–760. doi: 10.1038/nprot.2011.319. [DOI] [PubMed] [Google Scholar]
- Zhu Z-J, et al. Liquid chromatography quadrupole time-of-flight mass spectrometry characterization of metabolites guided by the METLIN database. Nat Protocols. 2013;8:451–460. doi: 10.1038/nprot.2013.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.