Abstract
Peptidoglycan (PG) is unique to bacteria, and thus, the enzymes responsible for its biosynthesis are promising antibacterial drug targets. The membrane-embedded enzymes in PG remain significant challenges in studying their mechanisms due to the fact that preparations of suitable enzymatic substrates require time-consuming biological transformations or chemical synthesis. Lipid I (prenyl diphosphoryl-MurNAc-pentapeptide) is an important PG biosynthesis intermediate to study the central enzymes, translocase I (MraY/MurX) and MurG. Lipid I isolated from nature contains the C50-or C55-prenyl unit that shows extremely poor water-solubility that renders studies of translocase I and MurG enzymes difficult. We have studied biological transformation of water soluble lipid I fluorescent probes using bacterial membrane fractions and purified MraY enzymes. In our investigation of the minimum structural requirements of the prenyl phosphates in MraY-catalyzed lipid I synthesis, we found that (2Z,6E)-farnesyl phosphate (C15-phosphate) can be recognized by E. coli MraY to generate the water-soluble lipid I fluorescent probes in high-yield. Under the optimized conditions, the same reaction was performed by using the purified MraY from Hydrogenivirga spp. to afford the lipid I analog with high-yield in a short reaction time.
Keywords: Translocase I (MraY/MurX), MurG, Lipid I, Lipid II, Chemoenzymatic synthesis
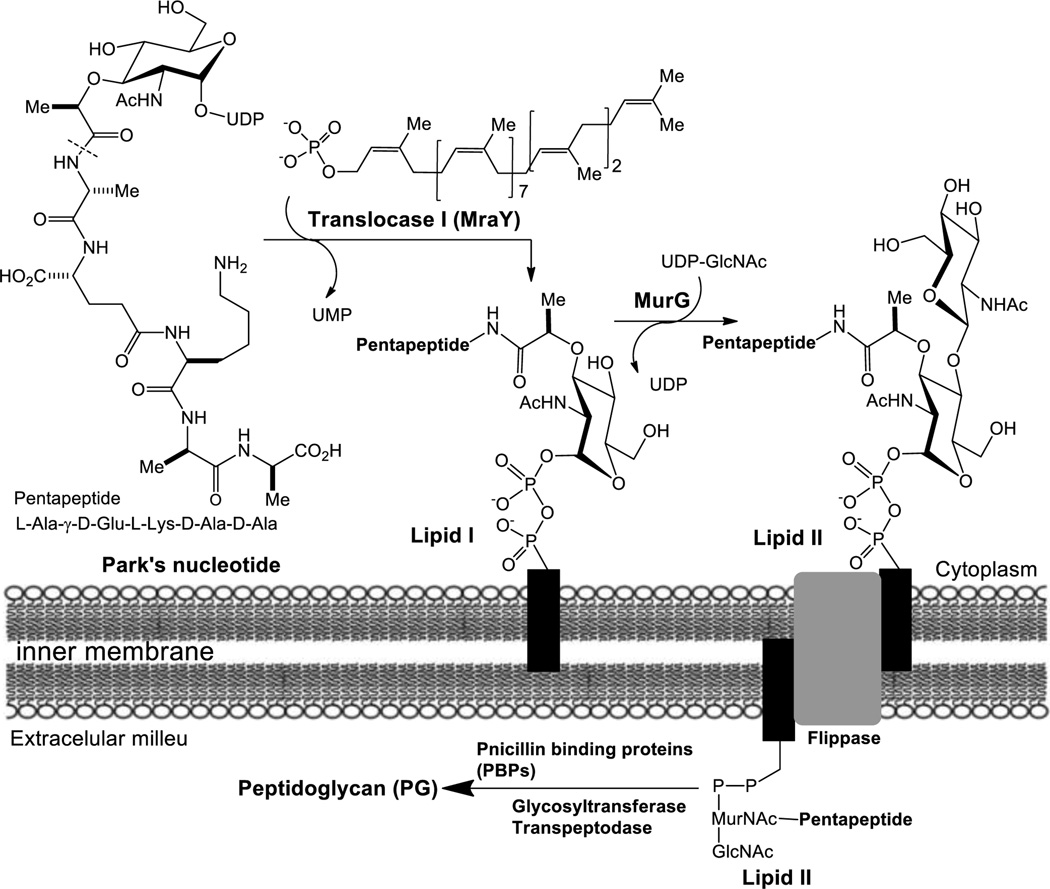
Since peptidoglycan (PG) is an essential bacterial cell-wall polymer, the machinery for PG biosynthesis provides a unique and selective target for antibiotic action.1–3 To date, only a few enzymes in PG biosynthesis such as the transpeptidase of penicillin binding proteins (PBPs) have been studied extensively. Thus, the machinery for PG synthesis is still considered to be a source of unexploited drug targets.4,5 We have been studying bacterial phosphotransferases and glycosyltransferases aiming at developing new antibacterial agents for multi-drug resistant (MDR) pathogens.5–13 Especially, the membrane-associated proteins translocase I (MraY/MurX) and MurG, which catalyze the transformations from Park’s nucleotide (UDP-MurNAc-pentapeptide) to lipid I (MurNAc(pentapeptide)-pyrophosphoryl prenol) and from lipid I to lipid II (GlcNAc-MurNAc(pentapeptide)-pyrophosphoryl prenol), are our research interests (Fig. 1). Syntheses of lipid I and lipid II have been achieved by total chemical synthetic or chemoenzymatic approaches; however, these require sophisticated synthetic techniques and skills in purifying amphiphilic molecules.6,14–19 In addition, the cost of acquisition of enough decaprenyl phosphate is very high. Recently, we reported biosynthesis of a water-soluble lipid I-neryl (C10) analog 7 from Park’s nucleotide-N[ε]-dansylthiourea 1 (Fig. 2 and 3) and neryl phosphate using M.
Figure 1.
Biosynthesis of lipid II from Park’s nucleotide in E. coli.
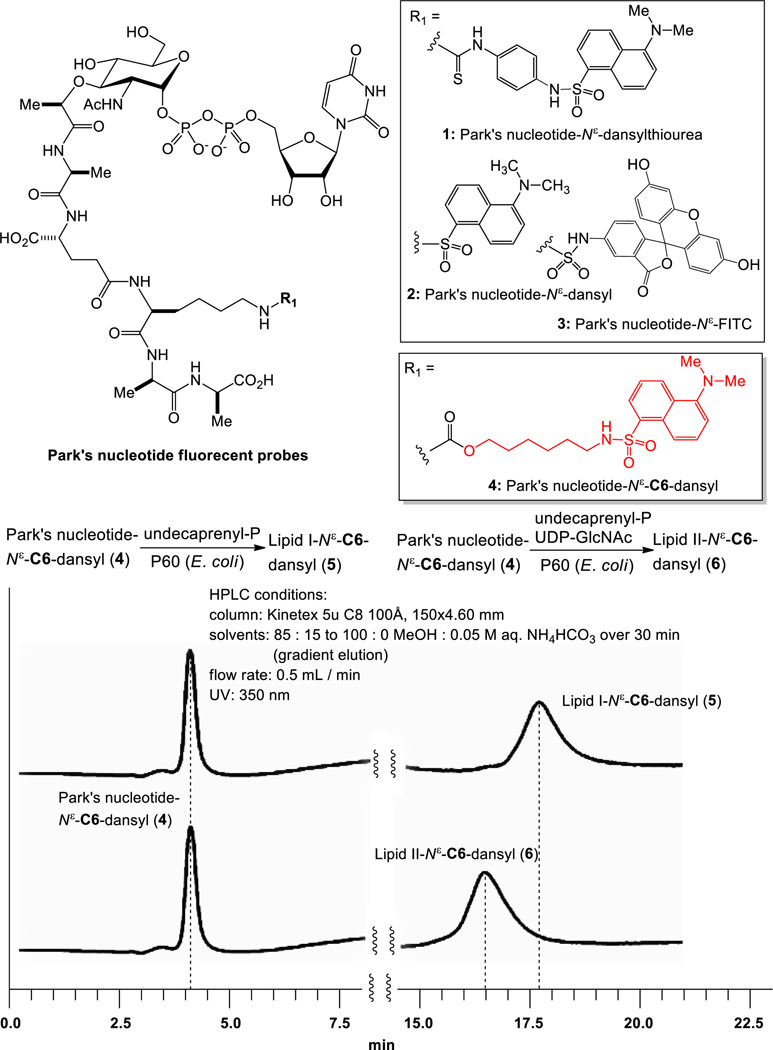
Figure 2.
Identification of a fluorescent probe for MraY/MurG-catalyzed biotransformations.
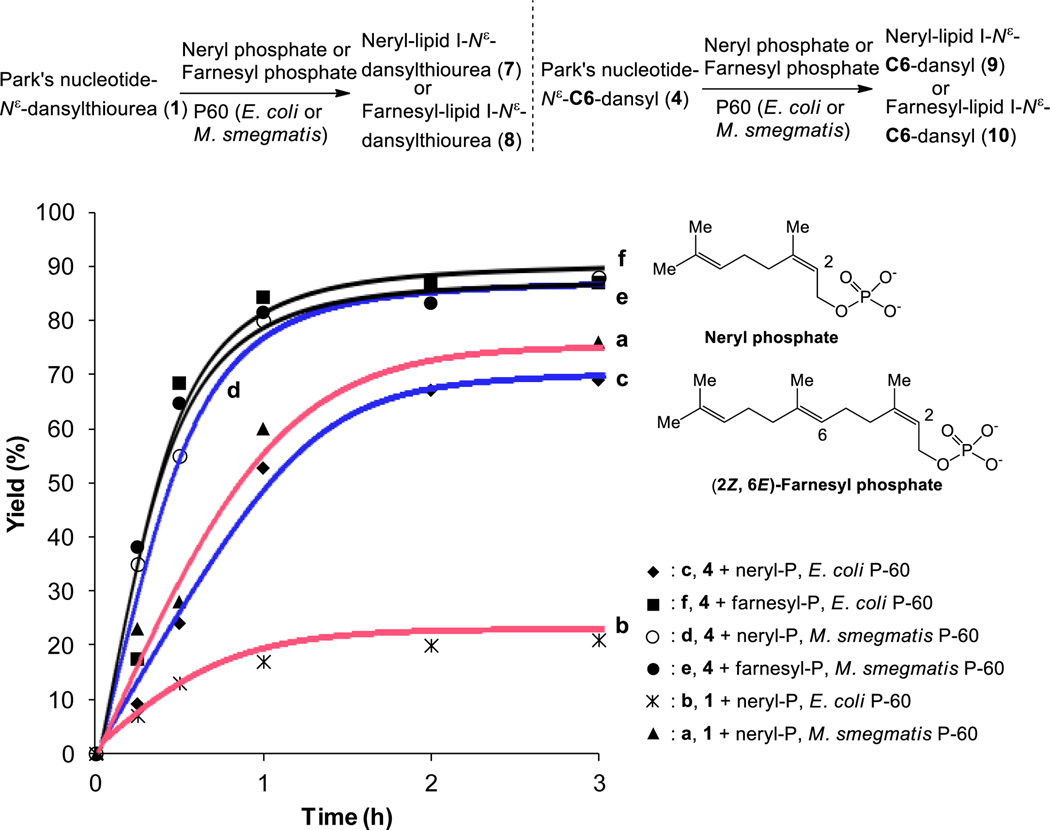
Figure 3.
Biosyntheses of lipid I analogs via P-60 membrane fractions.
smegmatis P-60 (MraY-containing membrane fraction).5 A water-soluble lipid I generated in the reaction could be quantitated conveniently via reverse-phase HPLC without tedious extraction procedures. These synthetic protocols could be applied to a development of robust MraY/MurX assay for identifying novel antibacterial agents. Investigation of versatile MraY/MurG enzyme substrate mimics that can efficiently be transformed to lipid I and lipid II fluorescent derivatives enzymatically requires further structural studies of Park’s nucleotide and lipid I. In the present work, we report a new Park’s nucleotide fluorescent probe 4 that can be recognized by MraY and MurG from a wide range of bacteria and the efficient biosynthesis of a water soluble lipid I fluorescent probe 10 with E. coli MraY and a purified MraY.
We realized that MurX- and MraY-containing membrane fractions (P-60)20 obtained from M. tuberculosis, M. smegmatis, S. aureus, and E. coli could recognize Park’s nucleotide fluorescent probes (1–3, Fig. 2) and they could be converted to the corresponding lipid I analogs in 15–25% yields when 2–10 equivalents of undecaprenyl phosphate were utilized under the optimized conditions.21 On the other hand, Park’s nucleotide fluorescent probes (1–3) were not effective in biosynthesis of lipid II analogs using P-60 in the presence of UDP-GlcNAc; no lipid II derivative was detected in HPLC analyses. These results indicate that the binding affinity of lipid I, whose lysine residue was modified directly with commercially available fluorophores, with MurG is markedly decreased. On the other hand, Park’s nucleotide-N[ε]-C6-dansyl 4 was converted to the corresponding lipid I and lipid II in 65–70% yield using E. coli P-60 in the absence or presence of UDP-GlcNAc (Fig. 2). It is interesting to note that a nitrobenzoxadiazole (NBD) linked N-glycolyl Park’s nucleotide was applied to the biosynthesis of lipid II, but the desired lipid II derivative was isolated in a trace amount when E. coli membrane fraction was used.22 Thus, we could identify a linker, 6-aminohexan-1-ol that can effectively conjugate the fluorophore with Park’s nucleotides without loss of the binding affinities with MraY and MurG. Because Park’s nucleotide-N[ε]-C6-dansyl 4 could be converted to undecaprenyl lipid II via P-60 derived from M. tuberculosis, M. smegmatis, S. aureus, and E. coli, it was concluded that 4 is a versatile Park’s nucleotide probe to study functions and mechanisms of MraY/MurX and MurG. In order to confirm the efficiency of 4 in biosynthesis of lipid I-fluorescent probes, a series of experiments were performed. In biosynthesis of neryl-lipid I analog with Park’s nucleotide-N[ε]-dansylthiourea 1, the reaction with M. smegmatis P-60 furnished the neryl-lipid I analog in >50% yield within 2h, whereas, the same reaction with E. coli P-60 did not provide the desired product with satisfactory yield (<20%) (a vs. b in Fig. 3). Park’s nucleotide-N[ε]-C6-dansyl 4 has a significant advantage in biosyntheses of lipid I analogs; the corresponding neryl-lipid I derivative 9 could be synthesized in over 60% and 80% yield in 3h using E. coli and M. smegmatis P-60, respectively (c vs. d in Fig. 3). Upon further investigation of minimum structure requirement of the prenyl phosphate in the MraY-catalyzed lipid I analog synthesis with 4, it was found that (2Z,6E)-farnesyl phosphate (C15-phosphate) is a better mimic for prenyl phosphate than neryl phosphate (C10-phosphate); in the product/time course experiments, no obvious difference in reaction rate was observed between E. coli and M. smegmatis P-60 catalyzed lipid I analog syntheses (e vs. f in Fig. 3).
Comparison of kinetic parameters of E. coli P-60-catalyzed lipid I-neryl and -farnesyl syntheses with 4 clearly supported the observed reaction rates (c vs. f in Fig. 3); E. coli MraY has over 5 times higher Km value for neryl phosphate than that for farnesyl phosphate (5.66 × 103 µM for neryl-P and 9.80 × 102 µM for farnesyl-P). The Vmax for farnesyl-lipid I-N[ε]-C6-dansyl 10 synthesis by E. coli MraY was determined to be 7.96 × 10−7 µM/sec through the Michaelis-Menten plot, whereas 1.23 × 10−6 µM/sec for neryl-lipid I-N[ε]-C6-dansyl 9.23 Significant difference in MraY-catalyzed farnesyl-lipid I analog synthesis compared to the neryl-lipid I version can be attributed to the fact that more hydrophobic characteristics of the farnesyl group increases the affinity with the hydrophobic catalytic site of MraY. However, it is worth mentioning that farnesyl phosphate applied in these experiments has 2Z,6E-configuration, albeit, the natural substrates has 2Z,6Z-isoprenyl units (Fig. 1).
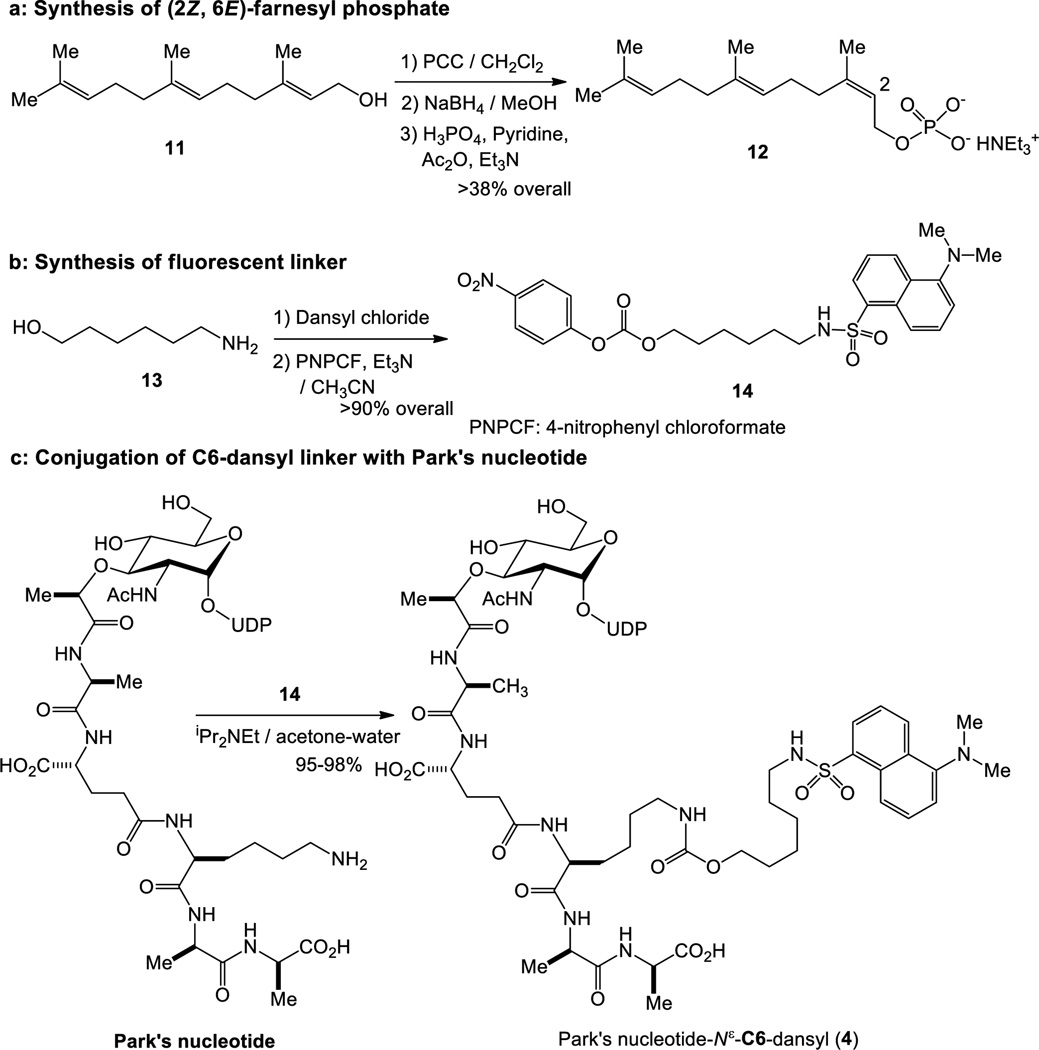
(2Z,6E)-Farnesyl phosphate was synthesized from farnesol (11) in three steps in >38% overall yield,24 thus, unlike undecaprenyl phosphate, a sufficient amount of the water-soluble counterpart are readily available. Park’s nucleotides can be relatively easily synthesized via total chemical synthesis or enzymatic processes using crude MurA-F from UDP-GlcNAc.6,9 As illustrated in Scheme 1, the fluorescent linker 14 was synthesized in two steps in >90% overall yield;25 14 could be utilized for conjugation with Park’s nucleotides without purification. Conjugation between Park’s nucleotide and 14 was accomplished with iPr2NEt in water-containing solvents (see, supporting information).26 Park’s nucleotide-N[ε]-C6-dansyl 4 could be stored in DMSO or in pH 8 tris-buffer at −20 °C for over three months without detectable degradation. Thus, practical and convenient enzymatic substrates were identified to study MraY and MurG enzymes.
Scheme 1.
Syntheses of MraY enzyme substrates for biotransformation from 4 to 10.
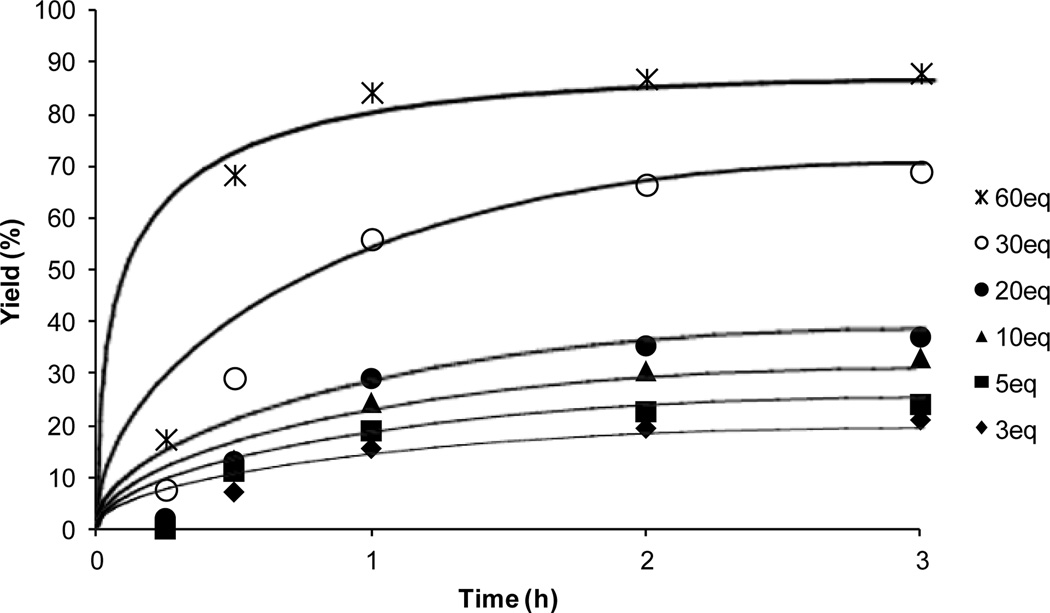
The MraY-catalyzed transformation of lipid I from Park’s nucleotide is believed to be a reversible process.27 Therefore, it is difficult to achieve a complete conversion with natural enzyme substrates. On the contrary, under the optimized conditions synthesis of farnesyl-lipid I 10 with Park’s nucleotide-N[ε]-C6-dansyl 4 and E. coli P-60 could be achieved in >80% yield within 1h when 60 equivalents of farnesyl phosphate 12 was used (Fig. 4). Even using 3 equivalents of 12, a useful level of conversion was achieved for studying functions of MraY.
Figure 4.
Effect of concentrations of (2Z,6E)-farnesyl phosphate in biosynthesis of farnesyl-lipid I-N[ε]-C6-dansyl 10.
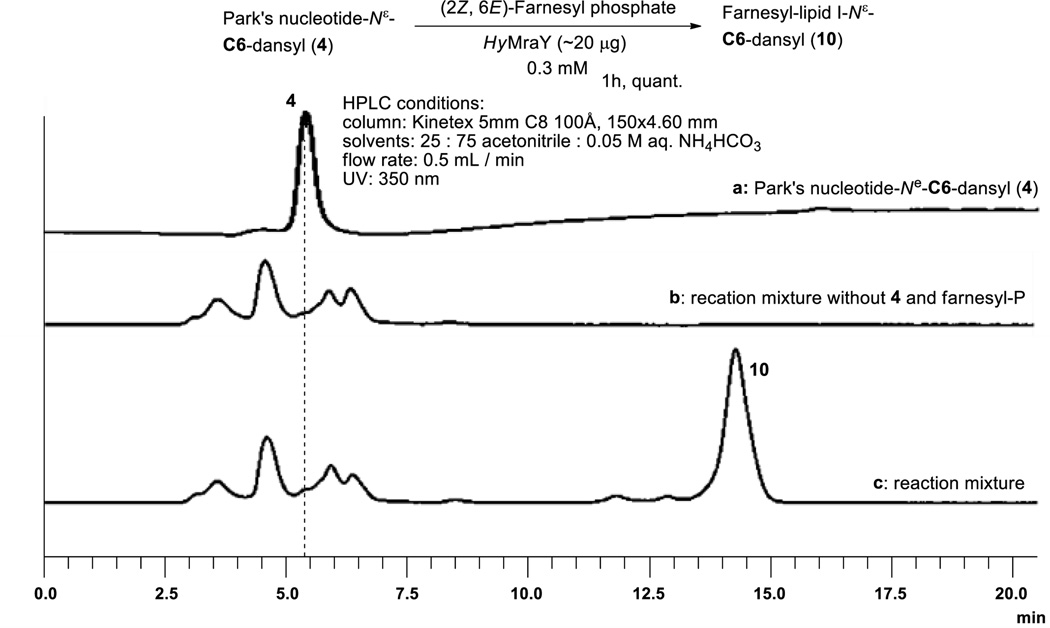
For structural and functional studies, recombinant MraY enzymes from thermophilic eubacteria, such as Aquifex spp. or Hydrogenobacter spp. have been expressed and purified by several research groups.28 In our attempt at development of a rapid and practical preparation of lipid I fluorescent probes, the purified MraY (40.1 kDa) from Hydrogenivirga spp. 128-5-R1-1 was applied for the synthesis of farnesyl-lipid I-N[ε]-C6-dansyl 10.29 When HyMraY was applied to the transformation from Park’s nucleotide-N[ε]-dansylthiourea 1 to neryl-lipid I-N[ε]-dansylthiourea 7 with neryl phosphate (60 equivalents), denaturing of the purified MraY was observed during long reaction time required for neryl-lipid I synthesis; the reaction was terminated in 40–60% conversion within 1h, whereas, the same reaction with M. smegmatis P-60 furnished the desired product 7 in near quantitative yield after 12h (in 0.075 mM solution for Park’s nucleotide).5 Using membrane fractions (e.g. P-60) are convenient approach for pilot scale syntheses and especially for biological assays. However, the reactions using P-60 are impractical when one aims to synthesize over-milligram quantities of substrates due to a requirement of large volume of P-60 in solvents and a phase-transfer catalyst (detergent). In contrast, HyMraY-catalyzed lipid I synthesis with Park’s nucleotide-N[ε]-C6-dansyl 4 and farnesyl phosphate 12 (30 equivalents) could be accomplished within 1h to afford 10 in quantitative yield. Notably, this reaction could readily be scaled-up for the synthesis of a few milligram of 10 with HyMraY (20 µg) in 0.3 mM concentrations (for Park’s nucleotide) (Fig. 5). The reaction mixture was lyophilized and filtered through a membrane filter to afford the crude product, which was purified by reverse-phase HPLC (solvent: CH3CN:0.05 M aq. NH4HCO3 = 25 : 75), yielding 10 in 80–90% yield.30 All physical data including the retention time (in HPLC) of farnesyl-lipid I-N[ε]-C6-dansyl 10 synthesized in Fig. 5 were agreed with 10 synthesized via a total chemical synthesis.31
Figure 5.
HyMraY-catalyzed transformation from 4 to 10.
In summary, we have demonstrated efficient biosynthesis of a water-soluble lipid I fluorescent probe with a convenient membrane fraction (P-60) prepared from E. coli. In previous works, biosynthesis of lipid I analogs with Park’s nucleotide-N[ε]-dansylthiourea 1 were demonstrated via P-60 from Mycobacterium spp; the affinity of 1 with the other MraY enzymes are weaker than that from Mycobacterium spp. A new probe, Park’s nucleotide-N[ε]-C6-dansyl 4 exhibited sufficient affinity with MraY enzymes of a wide range of bacterial species.32 (2Z,6E)-Farnesyl phosphate 12 was identified as a water soluble counterpart of undecaprenyl phosphate for MraY enzymes. The combination of the MraY enzyme substrate mimics, 4 and 12 enabled the syntheses of lipid I analogs with a purified MraY at high concentrations; as demonstrated in Fig. 5 a few milligram quantities of farnesyl-lipid I-N[ε]-C6-dansyl 10 could be synthesized with HyMraY (~20 µg) within 1h. We have accomplished the synthesis of over 300 mg quantities of Park’s nucleotide.9 Therefore, lipid I and lipid II fluorescent probes can be synthesized chemoenzymatically according to the procedures illustrated in Scheme 1 and Fig. 3. Lipid I-N[ε]-C6-dansyl 5 was recognized by MurG to form the corresponding lipid II 6. A water-soluble lipid I-N[ε]-C6-fluorecent probe will be a valuable asset to study functions of MurG and the other enzymes associated with MurG.33–35 New lipid I fluorescent probes and their kinetic parameters for MurG will be disclosed elsewhere.
Supplementary Material
Acknowledgments
The National Institutes of Health is greatly acknowledged for financial support of this work (AI084411 to MK and DP1GM105385 to WMC). We also thank University of Tennessee for generous financial support. NMR data were obtained on instruments supported by the NIH Shared Instrumentation Grant. The following reagent was obtained through BEI Resources, NIAID, NIH: Mycobacterium tuberculosis, Strain H37Rv and Gamma-Irradiated Mycobacterium tuberculosis, NR-14819.
Footnotes
Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.tetlet.2015.
References and notes
- 1.Van Heijenoort J. Natural Pro. Reports. 2001;18:503–519. doi: 10.1039/a804532a. [DOI] [PubMed] [Google Scholar]
- 2.Higashi Y, Strominger JL, Sweeley CC. J. Biol. Chem. 1970;245:3697–3702. [PubMed] [Google Scholar]
- 3.Parr TR, Saier MH. Res. Microbiol. 1992;143:443–447. doi: 10.1016/0923-2508(92)90089-7. [DOI] [PubMed] [Google Scholar]
- 4.Wright GD. Science. 2007;315:1373–1374. doi: 10.1126/science.1140374. [DOI] [PubMed] [Google Scholar]
- 5.Siricilla S, Mitachi K, Skorupinska-Tudek K, Swiezewska E, Kurosu M. Anal. Biochem. 2014;461:36–45. doi: 10.1016/j.ab.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kurosu M, Mahapatra S, Narayanasamy P, Crick DC. Tetrahedron Lett. 2007;48:799–803. [Google Scholar]
- 7.Kurosu M, Li K, Crick DC. Org. Lett. 2009;11:2393–2397. doi: 10.1021/ol900458w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurosu M, Narayanasamy P, Crick DC. Heterocycles. 2007;72:339–352. [Google Scholar]
- 9.Li K, Kurosu M. Heterocycles. 2008;76:455–469. [Google Scholar]
- 10.Aleiwi BA, Schneider CM, Kurosu M. J. Org. Chem. 2012;77:3859–3867. doi: 10.1021/jo300205b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Siricilla S, Aleiwi BA, Kurosu M. Chem. Eur. J. 2013;19:13847–13858. doi: 10.1002/chem.201302389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siricilla S, Mitachi K, Wan B, Franzblau SG, Kurosu M. J. Antibiot. 2014;58:6413–6423. doi: 10.1038/ja.2014.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanaka S, Clemons WM. Mol. Microbiol. 2012;85:975–985. doi: 10.1111/j.1365-2958.2012.08153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hitchcock SA, Eid CN, Aikins JA, Zia-Ebrahimi M, Blaszack LC. J. Am. Chem. Soc. 1998;120:1916–1917. [Google Scholar]
- 15.Van Nieuwenhze MS, Mauldin SC, Zia-Ebrahimi M, Aikins JA, Blaszczak LC. J. Am. Chem. Soc. 2001;123:6983–6988. doi: 10.1021/ja016082o. [DOI] [PubMed] [Google Scholar]
- 16.Schwartz B, Markwalder JA, Wang Y. J. Am. Chem. Soc. 2001;123:11638–11643. doi: 10.1021/ja0166848. [DOI] [PubMed] [Google Scholar]
- 17.Saha SL, Van Nieuwenhze MS, Hornback WJ, Aikins JA, Blaszczak LC. Org. Lett. 2001;3:3575–3577. doi: 10.1021/ol016692t. [DOI] [PubMed] [Google Scholar]
- 18.Lebar MD, Lupoli TJ, Tsukamoto H, May JM, Walker S, Kahne D. J. Am. Chem. Soc. 2013;135:4632–4635. doi: 10.1021/ja312510m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shih HW, Chen KT, Cheng TJR, Wong CH, Cheng WC. Org. Lett. 2011;13:4600–4003. doi: 10.1021/ol201806d. [DOI] [PubMed] [Google Scholar]
- 20.Rezwan M, Laneelle MA, Sander P, Daffe M. J. Microbiol. Methods. 2007;68:32–39. doi: 10.1016/j.mimet.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 21.Typical procedure using P-60 membrane fraction: Tris-buffer (pH = 8.0; 50 mM, 38.7 µL), Triton X 100 (0.5%; 11.2 µL), MgCl2 (0.5 M; 10 µL), KCl (2 M, 10 µL), prenyl phosphate (20mM, 2–60 equivalents) and Park’s nucleotide (2mM; 3.8 µL) were placed in a 1.5 mL Eppendorf tube. To a reaction mixture, P-60 membrane fractions (1 mg/µL, 15 µL) was added. The reaction mixture was incubated for 1–15h at room temperature and quenched with CHCl3 (200 µL). Two phases were centrifuged at 25,000g for 10 min. The upper aqueous phase was collected. 10 µL of the aqueous phase was injected into the reverse-phase HPLC (CH3CN : 0.05 M NH4HCO3 = 25:75, UV: 350 nm, flow rate: 0.5 mL/min, Column: Kinetex 5u C8 100Å, 150 × 4.60mm) and the area of the peak for the lipid I derivative was quantified to obtain the product yield.
- 22.Chen KT, Kuan YC, Fu WC, Liang PH, Cheng TJR, Wong CH, Cheng WC. Chem. Euro. J. 2013;19:834–838. doi: 10.1002/chem.201203251. [DOI] [PubMed] [Google Scholar]
- 23.Ma Y, Münch D, Schneider T, Sahl HG, Bouhss A, Ghoshdastider U, Wang J, Dötsch V, Wang X, Bernhard F. J. Biol. Chem. 2011;286:38844–38853. doi: 10.1074/jbc.M111.301085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Data for (2Z,6E)-farnesyl phosphate 12: 1H NMR (400 MHz, Deuterium Oxide) δ 5.40 (t, J = 7.3 Hz, 1H), 5.20 – 5.12 (m, 2H), 4.35 (t, J = 7.0 Hz, 2H), 2.15 – 2.00 (m, 8H), 1.73 (s, 3H), 1.66 (s, 3H), 1.59 (s, 6H); LRMS (EI) calcd for C15H28O4P [M + H]+: 303.17, found: 303.19.
- 25.Data for the linker 14: 1H NMR (400 MHz, Chloroform-d) δ 8.61 (d, J = 8.5 Hz, 1H), 8.33 (d, J = 7.4 Hz, 1H), 8.30-8.24 (m, 1H), 8.27 (d, J = 9.2 Hz, 2H), 7.57 (q, J = 7.9 Hz, 2H), 7.37 (d, J = 9.2 Hz, 2H), 7.25 (brs, 1H), 4.74 (t, J = 6.2 Hz, 1H), 4.18 (t, J = 6.6 Hz, 2H), 2.95 (s, 6H), 2.97 – 2.85 (m, 2H), 1.65 – 1.56 (m, 2H), 1.47 – 1.38 (m, 2H), 1.29 – 1.21 (m, 4H); LRMS (EI) calcd for C25H30N3O7S ([M + H]+): 516.18, found: 516.26.
- 26.Data for Park’s nucleotide-Nε-C6-dansyl (4): 1H NMR (400 MHz, Deuterium Oxide) δ 8.46 (d, J = 8.6 Hz, 1H), 8.27 (d, J = 8.7 Hz, 1H), 8.23 (d, J = 7.3 Hz, 1H), 7.89 (d, J = 8.1 Hz, 1H), 7.66 (q, J = 7.4 Hz, 2H), 7.38 (d, J = 7.6 Hz, 1H), 5.91 (d, J = 3.7 Hz, 1H), 5.89 (d, J = 7.9 Hz, 1H), 5.41 (dd, J = 7.3, 3.2 Hz, 1H), 4.32 – 4.26 (m, 2H), 4.25 – 4.02 (m, 10H), 3.90 (dt, J = 9.7, 3.0 Hz, 1H), 3.82 – 3.70 (m, 3H), 3.66 – 3.54 (m, 3H), 3.04 (t, J = 6.3 Hz, 2H), 2.92 (t, J = 6.5 Hz, 2H), 2.84 (s, 6H), 2.26 – 2.19 (m, 2H), 2.13 – 2.02 (m, 1H), 1.95 (s, 3H), 1.87 – 1.78 (m, 1H), 1.74 – 1.62 (m, 2H), 1.47 – 1.32 (m, 4H), 1.37 (d, J = 7.2 Hz, 3H), 1.34 (d, J = 6.7 Hz, 3H), 1.29 (d, J = 7.7 Hz, 3H), 1.27 (d, J = 7.6 Hz, 3H), 1.13 – 1.06 (m, 2H), 1.04 – 0.97 (m, 2H), 0.85 – 0.68 (m, 4H); LRMS (EI) calcd for C59H90N11O30P2S ([M + H]+): 1526.51, found: 1526.40.
- 27.Stachyra T, Dini C, Ferrari P, Bouhss A, van Heijenoort J, Mengin-Lecreulx D, Blanot JD, Biton D, Le Beller D. Antimicrob. Agents Chemother. 2004;48:897–902. doi: 10.1128/AAC.48.3.897-902.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chung BC, Zhao J, Gillespie RA, Kwon D-Y, Guan Z, Hong J, Zhou P, Lee S-Y. Science. 2013;341:1012–1016. doi: 10.1126/science.1236501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MraY, a pET22b vector containing an N-terminal 6×Histagged HyMraY was used to transform Nico(pLemo) cells for expression.
- 30.Synthesis of lipid I analog with HyMraY: Tris-buffer (pH = 8.0; 50mM, 1.7 µL), Triton X 100 (22%; 0.2 µL), MgCl2 (2.5 M; 2 µL), KCl (4 M, 5 µL), (2Z,6E)-farnesyl phosphate (20 mM, 11.3 µL) and Park’s nucleotide-Nε-C6-dansyl (2mM; 3.8 µL) were place in a 1.5 mL Eppendorf tube. To a stirred reaction mixture, HyMraY (~0.1 mg/mL, 6 µL) was added (total volume of reaction mixture: 30 µL). The reaction mixture was incubated for 1h at room temperature. The reaction mixture was lyophilized and filtered through a membrane filter. The crude product was purified by reverse-phase HPLC (CH3CN : 0.05 M ammonium bicarbonate = 25:75). This reaction was scale-up by using Park’s nucleotide-Nε-C6-dansyl (1.5 mg) and (2Z,6E)-farnesyl phosphate (9 mg) in total volume of 1.5 mL.
- 31.Mitachi K, Mohan P, Siricilla S, Kurosu M. Chem. Eur. J. 2014;20:4554–4558. doi: 10.1002/chem.201400307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.The FITC conjugate, Park’s nucleotide-Nε-C6-FITC was equally effective in the syntheses of lipid I and lipid II analogs.
- 33.Pasquina LW, Santa, Maria JPS, Walker S. Curr. Opin. Microbiol. 2013;16:531–537. doi: 10.1016/j.mib.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Helm JS, Hu Y, Chen L, Gross B, Walker S. J. Am. Chem. Soc. 2003;125:11168–11169. doi: 10.1021/ja036494s. [DOI] [PubMed] [Google Scholar]
- 35.Kang C-M, Nyayapathy S, Lee J-Y, Suh JW, Husson RN. Microbiology. 2008;154:725–735. doi: 10.1099/mic.0.2007/014076-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.