Background: The molecular mechanisms of Plasmodium invasion in mosquito midguts are not well understood.
Results: The mosquito midgut peritrophic matrix protein FREP1 binds Plasmodia. Blocking parasite-FREP1 interactions or ablating FREP1 expression reduced P. falciparum infection in mosquitoes.
Conclusion: FREP1 functions as a critical host factor that mediates Plasmodium invasion in mosquito midguts.
Significance: Targeting FREP1 may inhibit Plasmodium transmission to mosquitoes and the spread of malaria.
Keywords: fibrinogen, host-pathogen interaction, insect, invasion, malaria, vaccine
Abstract
Malaria transmission depends on sexual stage Plasmodium parasites successfully invading Anopheline mosquito midguts following a blood meal. However, the molecular mechanisms of Plasmodium invasion of mosquito midguts have not been fully elucidated. Previously, we showed that genetic polymorphisms in the fibrinogen-related protein 1 (FREP1) gene are significantly associated with Plasmodium falciparum infection in Anopheles gambiae, and FREP1 is important for Plasmodium berghei infection of mosquitoes. Here we identify that the FREP1 protein is secreted from the mosquito midgut epithelium and integrated as tetramers into the peritrophic matrix, a chitinous matrix formed inside the midgut lumen after a blood meal feeding. Moreover, we show that the FREP1 can directly bind Plasmodia sexual stage gametocytes and ookinetes. Notably, ablating FREP1 expression or targeting FREP1 with antibodies significantly decreases P. falciparum infection in mosquito midguts. Our data support that the mosquito-expressed FREP1 mediates mosquito midgut invasion by multiple species of Plasmodium parasites via anchoring ookinetes to the peritrophic matrix and enabling parasites to penetrate the peritrophic matrix and the epithelium. Thus, targeting FREP1 can limit malaria transmission.
Introduction
Malaria remains a global public health crisis, and Anopheline mosquitoes transmit malaria parasites, of which Plasmodium falciparum is the most dangerous (1). Female mosquitoes need to feed on blood for egg production (2). Feeding on Plasmodium-infected blood can result in the ingestion of male and female haploid gametocytes that fuse to form diploid ookinetes, a process that initiates Plasmodium infection of the mosquito vector. Ookinetes start invading mosquito midgut epithelial cells between 12 and 24 h after a blood meal feeding (3). Un-fused gametocytes and ookinetes located near the periphery of the blood bolus in the mosquito midgut are susceptible to attacks by diverse digestive proteases and bacteria (4–6), whereas gametocytes and ookinetes inside the blood bolus are protected by blood. However, mature ookinetes must cross and exit the blood bolus to initiate invasion of the midgut epithelium. Blood feeding regulates mosquito gene expression (7, 8) and stimulates the formation of the peritrophic matrix (PM)3 within the midgut (9). The newly formed PM completely surrounds the ingested blood, separating the blood bolus from secretory midgut epithelial cells, providing a second physical barrier that limits the infection by pathogens co-ingested with the blood meal (10). The PM is composed of 3–13% chitin microfibrils and is embedded with many known (3) and unknown proteins (11). Notably, when the ookinetes are mature 12 h after the blood meal (9), the PM also becomes visible in the midgut lumen. To infect mosquitoes, the motile ookinetes must sequentially attach to and penetrate the PM and the midgut epithelium (12). At present, the detailed molecular mechanisms involved in ookinete attachment to and penetration of the PM and the subsequent midgut invasion are unclear.
We recently identified a mosquito gene, fibrinogen-related protein 1 (FREP1), that is implicated in Plasmodium infection in mosquitoes (13). Specific genetic polymorphisms in FREP1 are significantly associated with P. falciparum infection intensity levels in wild Anopheles gambiae populations from Kenya. The FREP1 is a member of the fibrinogen-related protein family (FREPs or FBNs) that contains a highly conserved C-terminal interacting fibrinogen-like (FBN) domain. In vertebrates, fibrinogen molecules usually associate as hexamers and are comprised of two sets of disulfide-bridged α, β, and γ chains that participate as a principal component of both cellular and fluid coagulation (14). In invertebrates, FREPs/FBNs are common pattern recognition receptors (15, 16) responsible mainly for initiating innate immune responses (17). For instance, tachylectin proteins in the horseshoe crab regulate host defense by recognizing bacterial lipopolysaccharides (18). Previous work examining the role and function of FREP/FBN family members in Anopheles mosquitoes has shown that two family members, FBN9 and FBN30, appear to restrict Plasmodium infection of midgut epithelial cells. Silencing the expression of either FBN9 or FBN30 in mosquitoes increased Plasmodium infection (13, 19). Here, we report the role and function of a third FREP/FBN family member, FREP1, during P. falciparum infection of Anopheles mosquitoes. Our genetic and biochemical assays reveal that FREP1 functions as a critical molecular anchor in the PM that facilitates Plasmodium invasion and infection of mosquito midguts. In contrast to FBN9 and FBN30 that inhibit Plasmodium infection, our results show that FREP1 is an important host factor that promotes infection of mosquito midguts by the major human pathogen, P. falciparum. Collectively, our data reveal new insight into Plasmodium-Anopheles interactions and identify FREP1 as a promising transmission-blocking target.
Experimental Procedures
Rearing An. gambiae Mosquitoes
An. gambiae G3 strain was maintained at 27 °C, 80% humidity with a 12-h day/night cycle. Larvae were reared on ground KOI fish food supplements (∼0.1 mg/larvae per day). Adult mosquitoes were maintained with 8% sucrose and fed on ketamine/xylazine-anesthetized mice for egg production.
Generating Anti-FREP1 Polyclonal Antibody
FREP1 was cloned using PCR with primers shown in Table 1 from an An. gambiae mosquito cDNA library. The PCR product and pQE30 plasmid were digested with restriction enzymes XmaI and HindIII. Products were ligated into a His6 expression plasmid and transformed into Escherichia coli JM109. The sequence-verified construct was subsequently transformed into E. coli M15 strain. One mm isopropyl 1-thio-β-d-galactopyranoside was used to induce gene expression in E. coli M15 transformants. Cells were lysed in buffer B (8 m urea, 100 mm NaH2PO4, 10 mm Tris·Cl, pH 8.0). Recombinant FREP1 was purified by a Ni-NTA column using a standard protocol (20). Four ml of E. coli lysate was collected from 200 ml of culture, and the amount of recombinant FREP1 in E. coli lysate was about 4.0 mg according to bands on the SDS-PAGE comparing to standard markers. In the end, about 0.85 mg of purified FREP1 was obtained with a recovery rate of 21%. SDS-PAGE and Coomassie Brilliant Blue R-250 staining confirmed the purity of recombinant FREP1. Purified recombinant FREP1 was then used as an antigen to generate customized polyclonal antibody against FREP1 in rabbits (Thermo Fisher Scientific, Rockford, IL). Anti-FREP1 antibody was purified from antiserum by protein A-agarose affinity chromatography and suspended in PBS.
TABLE 1.
PCR primers
The italic and underlined sequences denote restriction recognition sites. The underlined sequences are T7 promoter. The bold sequence is Kozak consensus sequences. The primers were synthesized through Integrated DNA Technologies Inc.
| Purpose | Primer name | Primer sequence |
|---|---|---|
| In vitro synthesis of FREP1 dsRNA | Forward | 5′-TAATACGACTCACTATAGGAGCTCGAGGTGAAGCAGAG-3′ |
| Reverse | 5′-TAATACGACTCACTATAGGTTCTCCAGCCGGTTGTGT-3′ | |
| Verify FREP1 mRNA expression | Forward | 5′-ACAGGGCAAGTTCGAGAAGA-3′ |
| Reverse | 5′-AAGTCAACCGTACCGTCCTG-3′ | |
| AgS7 gene | Forward | 5′-GGCGATCATCATCTACGTGC-3′ |
| Reverse | 5′-GTAGCTGCTGCAAACTTCGG-3′ | |
| In vitro synthesis of GFP dsRNA | Forward | 5′-TAATACGACTCACTATAGGCAAGTTTGAAGGTGATACCC-3′ |
| Reverse | 5′-TAATACGACTCACTATAGGCTTTTCGTTGGGATCTTTCG-3′ | |
| Clone FREP1 into pQE30 | Forward | 5′-ACCCGGGCACTGCCCTGAACGGTGCAG-3′ |
| Reverse | 5′-GGCAAGCTTCGCGAACGTCGGCACAGTC-3′ | |
| Clone FREP1 into pIB/V5-His | Forward | 5′-TCAAAGCTTCACCATGGTGAATTCATTCGTGTCG-3′ |
| Reverse | 5′-ACTCTAGATTACGCGAACGTCGGCACAGTCGTG-3′ | |
| Reverse with His tag | ACTCTAGAGCGAACGTCGGCACAGTCGTG |
Expressing Recombinant FREP1 in Insect Cells
The complete FREP1 coding sequence was PCR amplified using primers shown in Table 1 from an adult An. gambiae cDNA library. Products were cloned into plasmid pIB/V5-His (Life Technologies) to generate pIB-FREP1 (encoding FREP1) and pIB-FREP1-His (encoding FREP1 with a His6 tag) expression constructs. Constructs were amplified in E. coli DH5α and then purified with endotoxin-free plasmid preparation kits (Sigma). Cabbage looper ovarian cell-derived High Five cells (21) were used to express recombinant FREP1 according to the manufacturer's instructions (22). In brief, endotoxin-free plasmids were mixed with Cellfectin® Reagent (1 μl of Cellfectin/μg of plasmids, Invitrogen) in 5–6 ml of Express Five® SFM medium (Invitrogen). Cells were cultured in 25-cm2 cell culture flasks (Greiner Bio-One, Monroe, NC) for 48 h at 27 °C. Medium and cells were separated by centrifugation at 300 × g for 5 min. The proteins in the medium were concentrated using Amicon® ULTRA-4 Centrifugal Filter Devices (Milipore, Billerica, MA) by centrifugation at 5,000 × g for 10 min. The His6-tagged FREP1 was purified using a Ni-NTA column using a standard protocol (20). In total, 0.3 mg of insect cell-expressed recombinant FREP1 was purified from 50 ml of culture medium with an initial amount of ∼1.0 mg of FREP1 in culture supernatant (estimated based on SDS-PAGE). The yield was ∼30%.
Gel Filtration Chromatography to Determine the FREP1 Size
Using standard protocols (23), 0.03–1 mg of purified High Five-expressed recombinant FREP1 in 0.1 ml of PBS with or without 0.1% Triton X-100 was subjected to fast protein liquid chromatography gel filtration (Bio-Rad). Sephadex G-200 columns (30 cm in length, 1.0 cm in diameter, 5 to 600 kDa resolution) were used with flow rates regulated to 0.2 ml/min. The void volume (V0) of this column is about 8.0 ml, and the total elution volume (Vt) is about 24.0 ml. Fractions of ∼0.1 ml were collected. Absorbance (A280) was monitored constantly, and ELISA was used to detect recombinant FREP1 in each fraction. Briefly, 50 μl of sample from each fraction was coated per well onto an immunoGrade microplate (Brand, Wertheim, Germany) and incubated overnight at 4 °C. Wells were blocked for 1.5 h with 100 μl of 1.0% BSA in PBS, followed by sequential incubation for 1 h with 100 μl of anti-FREP1 antibody (diluted 5,000-fold in PBS-0.2% BSA to a concentration of 0.1 μg/ml) and then 1 h in 100 μl of alkaline phosphatase-conjugated anti-rabbit IgG (Sigma, 1:20,000 dilution with PBS, 0.2% BSA). Wells were washed three times with PBST (0.2% Tween 20 in PBS) between incubations. Wells were developed with 100 μl of p-nitrophenyl phosphate solution (Sigma). When colors in wells were visible, the optical density absorbance at 405 nm was measured. A set of molecular weight standards was used to establish standard curves of molecular masses for the gel filtration columns (aprotinin (6.5 kDa), ribonuclease A (13.7 kDa), carbonic anhydrase (29 kDa), canalbumin (75 kDa), and ferritin (440 kDa)). The elution (retention) volumes were measured to be 21.05, 18.34, 16.91, 13.65, and 9.38 ml, respectively. The linear regression equation of standard curve based on these standards was y = −6.32x + 44.86, where x is log10 transformed molecular mass in daltons and y is elution volume in milliliters. The correlation coefficient between x and y was 0.99.
Immunohistochemical (IHC) Assay to Determine Protein Distribution in Mosquito Tissues
Midguts from 3–5-day-old naive and blood-fed female mosquitoes were dissected in PBS supplemented with protease inhibitors (Thermo Scientific, Rockford, IL). Tissues were embedded in optimal cutting temperature compound and immediately frozen in liquid nitrogen. Frozen midguts were sectioned (8–10 μm), mounted on super frost plus slides (positively charged), air dried for 30 min at room temperature, fixed with 4% paraformaldehyde in PBS for 20 min, and stored at −20 °C until use. Prior to staining, sections were re-hydrated in Tris-buffered saline (50 mm Tris, 150 mm NaCl, pH 7.6) with 0.05% Tween 20 (TBST) for 10 min. Sections were blocked with 5% dry milk in TBST for 30 min, and then incubated for 2 h with 2.5 μg/ml of purified rabbit anti-FREP1 antibodies diluted in blocking solution. Control sections were incubated with blocking solution containing preimmune rabbit antibodies. Samples were washed 3 times for 5 min with TBST and then incubated for 30 min with alkaline phosphatase-conjugated goat anti-rabbit secondary antibody (Sigma) diluted 1:20,000 in blocking solution. Slides were then washed 3 times in TBST and developed with BCIP/NBT Chromogenic solution (Sigma) for 10–20 min. Sections were examined under a microscope at ×4 and 40 magnification.
Quantifying the Pixel Intensity of IHC Staining and Western Blot Images
Photoshop (version CC 2014) software was used to measure the “gray values” of an area of interest. The pixel intensity was obtained by subtracting the gray value from a constant value of 255. For IHC staining images of mosquito midguts, three different rectangles (0.01 square inches each, chosen randomly) within PM were selected with the “Rectangular Marquee Tool.” After clicking “Record Measurements,” the mean gray value of the region of interest was obtained. The average of three measured gray values (mean) from three selected areas was used to represent a value of an experimental image. The same procedure was conducted outside of the PM to establish gray values for background signals. The pixel intensity value of an area was obtained by subtracting the measured gray values of the area within PM from the background gray values. The measurements from three slices were used to calculate the mean ± S.D. The same procedure was conducted to obtain the pixel intensity of control groups. To measure the average pixel intensity on a Western blot, we conducted the same procedure as described above except selecting target bands from experimental groups and control groups, and the areas from experimental groups were the same as the areas from control groups. The pixel intensity of the background was calculated from regions adjacent to the bands. Three replicates of the Western blot that were conducted exactly the same were used to calculate the mean pixel intensity and standard deviations.
Quantification of pixel intensity (gray values) was independently validated by both measuring pixel intensity as a function of exposure time and by using other software (such as Microsoft PowerPoint) to change the brightness and/or contrast of an image. Calculations were consistent across multiple software applications.
Preparation of P. falciparum Gametocytes and Ookinetes
P. falciparum parasites (NF54 strain from MR4) were propagated in O+ fresh human blood (4% red blood cells (RBC), 0.25–0.5% parasitemia). Cultures were maintained in 6-well plates (Corning Inc., Costar) with 5.0 ml of complete RPMI 1640 medium supplemented with 10% heat-inactivated human AB-type serum (Interstate blood bank, Memphis, TN) and 12.5 μg/ml of hypoxanthine. The plates were maintained under 37 °C in a candle jar (24) and the medium was replaced daily until days 15–17. The parasitemia or gametocytemia was checked every other day by Giemsa staining of thin blood smears. For ookinete enrichment, P. falciparum cultures harboring stage V gametocytes were diluted 10-fold in complete RPMI 1640 without sodium bicarbonate and incubated at room temperature for 24 h as described (25). P. falciparum culturing and infection experiments were conducted in the biosafety level 2 (BSL2) laboratory at the University of Oklahoma.
Indirect Immunofluorescence Assays (IFA) to Examine the Binding between Parasites and Insect Cell-expressed Recombinant FREP1
Standard IFA was performed as described previously (26). In brief, P. falciparum cultures were deposited on coverslips (Fisher Scientific). Cells and parasites were immediately fixed in 4% paraformaldehyde in PBS at room temperature for 30 min, which preserved intact (non-permeabilized) cell membranes. Cells were then sequentially incubated with 100 mm glycine in PBS for 20 min, 0.2% bovine serum albumin (BSA) in PBS for 90 min, High Five cell-expressed FREP1 (10 μg/ml) in 0.2% BSA-PBS for 2 h, enhancer (Alexa Fluor® 594 goat anti-mouse SFX kit, Invitrogen) for 30 min, 5 μg/ml of anti-FREP1 antibody in PBS containing 0.2% BSA for 1 h, and 2 μg/ml of secondary antibody (Alexa Fluor® 594 goat anti-rabbit antibody, in PBS containing 0.2% BSA, Invitrogen) for 30 min in the dark. Between each incubation, the cells were washed 3 times for 3 min in PBS containing 0.2% BSA. Coverslips were rinsed in distilled water for 20 s and mounted on glass slides using 50 μl of Vectashield mounting media (Vector Laboratories, Burlingame, CA). Cell staining was examined using a Nikon Eclipse Ti-S fluorescence microscope. The fluoresence (pixel) intensity was measured as described above. The fluoresence intensity of a target region of an image was calculated by substracting the mean background fluoresence intensity (gray values) from the mean target fluoresence intensity (gray values). The fluoresence intensity values from three independent experiments or three controls were measured to calculate the mean ± S.D.
Binding Assay between FREP1 and Midgut PM by ELISA
Three to 5-day-old mosquitoes were fed on anesthetized mice and then maintained with 8% sugar. About 18 h after the blood meal, the engorged mosquitoes were dissected in 1× PBS supplemented with protease inhibitors (PBSI) (Pierce Protease Inhibitor Mini Tablets, EDTA-free, Thermo Scientific, Rockford, IL). The blood bolus in each midgut was removed manually. The dissected midguts were washed three times with PBSI. Each replicate contained at least 10 midguts. The same number of unfed (control) mosquito midguts was examined in parallel. Insect cell-expressed recombinant FREP1 (0.75 μg in 100 μl of PBSI) was incubated with experimental and control midguts for 1.5 h. An equivalent amount (0.75 μg in 100 μl of PBSI) of BSA was used as an additional control. The midguts were then washed three times with PBSI, and shredded with a micro-pestle in PBSI containing 0.5% Tween 20 in 1.5-ml tubes. The insoluble materials were removed by centrifugation at 8,000 × g for 2 min. Supernatants (100 μl) were added into immunoGrade microplates and the plates were incubated overnight at 4 °C. ELISA was used to quantify the relative amount of FREP1 in each reaction.
Ablating FREP1 Expression by dsRNA to Verify Its Function on Susceptibility to P. falciparum Infection in An. gambiae Mosquitoes
FREP1 was cloned from an An. gambiae mosquito cDNA library as described in our previous work (13). Briefly, nested PCR using the primers listed in Table 1 was used to generate a DNA template for synthesis of double-stranded RNA (dsRNA) using the in vitro transcription system T7 Megascript (Amibon, TX). The non-mosquito, negative control sequence was amplified from the Aequoria green fluorescent protein (GFP) gene using the primers listed in Table 1. dsRNA was synthesized from these gene fragments using the in vitro transcription system T7 Megascript (Amibon, TX). The FREP1 targeting construct spans a unique sequence from nucleotide 791 to 1,268, corresponding to amino acids 263 to 423 of FREP1. Blast analyses confirmed that no other genes in An. gambiae share significant DNA sequence identity with the targeted sequence of FREP1. The dsRNA was purified with a Qiagen RNA purification kit. Approximately 207 ng of dsRNA in a 69-nl solution was injected into the hemocoel of each cold-anesthetized, 1-day-old An. gambiae G3 female mosquitoes. Approximately 100 mosquitoes per treatment were used for the RNAi knockdown experiments. Thirty-six hours after dsRNA injection, the treated mosquitoes were fed with P. falciparum-infected blood containing 0.2% gametocytes in standard membrane feeding assays (27). Seven days post-infection and feeding, treated mosquitoes were dissected in PBS and oocysts numbers were counted using light microscopy after staining with 0.1% mercury dibromofluorescein disodium salt in PBS. In the negative control groups, ∼100 mosquitoes injected with dsRNA targeting GFP were exposed to the same parasite cultures, and were processed identically to the experimental groups. The transcript knockdown efficiency was confirmed by the quantitative RT-PCR in five treated mosquitoes that were taken randomly 12 h after infection. In parallel, reductions in FREP1 in mosquito midguts were confirmed using IHC assays as described above, except replacing secondary reagents with Alexa Fluor® 594-conjugated goat anti-rabbit antibody. Fluorescent staining intensity was quantified as described above.
Antibody Blocking Assays of P. falciparum Infection in An. gambiae Mosquitoes
P. falciparum-infected blood cultures containing mature stage V gametocytes were diluted with the fresh O+ type human blood to get the 0.2% final concentration of stage V gametocytes. An equal volume of heat-inactivated (65 °C for 15 min) AB-type human serum was added. Identical volumes of PBS (1/10 volume of blood) containing different concentrations of rabbit polyclonal anti-FREP1 antibody (5, 4, 2, 1, and 0.5 mg/ml) were added to the gametocyte cultures. Artificial membrane feeding was conducted using 3-day-old female naive mosquitoes. After feeding for 15 min, engorged mosquitoes were separated and maintained with 8% sugar in a BSL2 insectary (28 °C, 12-h light/dark cycle, 80% humidity) at the University of Oklahoma. Seven days after infection, midguts were dissected, stained with 0.1% mercurochrome, and examined using light microscopy to count the number of oocysts. Equivalent amounts of purified preimmune rabbit antibody (Thermo Scientific) were used as controls.
Results
Generating Anti-FREP1 Polyclonal Antibody
To begin to understand the basic biochemical characteristics of FREP1, we first examined its functional domains. According to our previous genome annotation (28), full-length FREP1 comprises 738 amino acids, including a 22-amino acid signal peptide at the N terminus, three coiled-coil regions, and a conserved ∼200-amino acid FBN domain at the C terminus (Fig. 1A). All six cysteine amino acid residues are within the C-terminal FBN domain.
FIGURE 1.
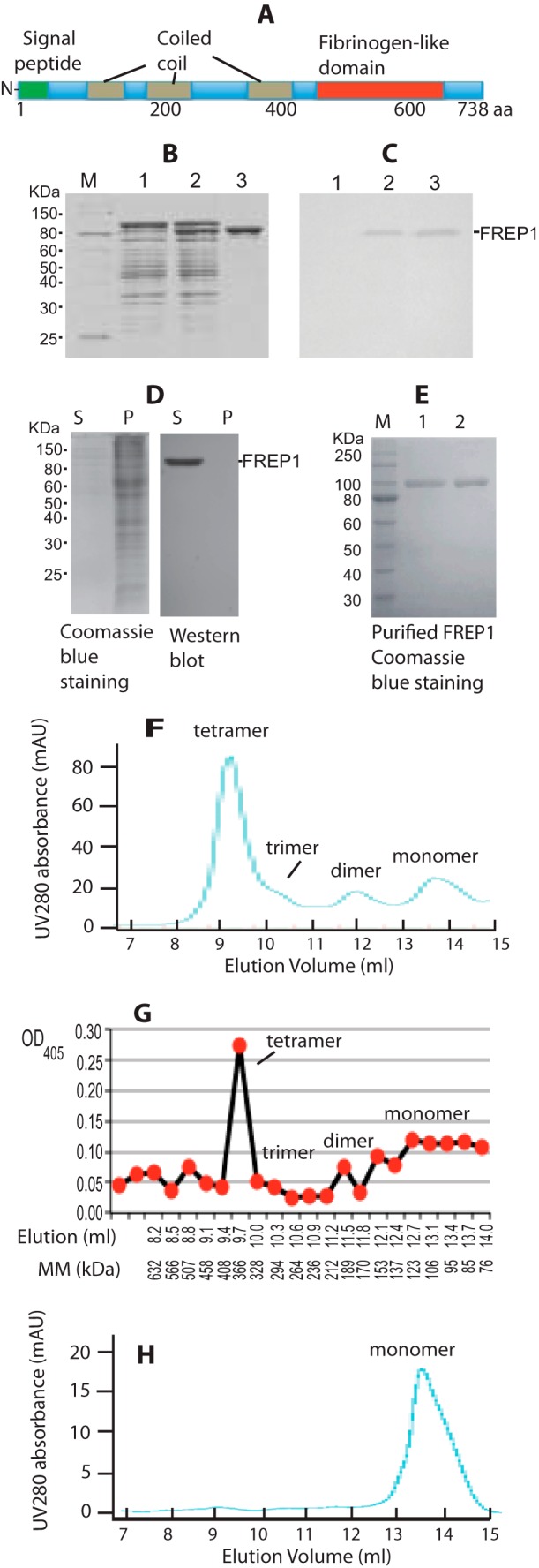
FREP1 is secreted from insect cells and forms tetramers. A, FREP1 has an N-terminal signal peptide, three coiled coils, and a C-terminal FBN domain. B, the FREP1 was cloned into an isopropyl 1-thio-β-d-galactopyranoside regulatable expression vector and transformed into E. coli M15 cells. Lysates from transformants were fractionated on 12% SDS-PAGE gels, which were subsequently stained with Coomassie Brilliant Blue. The amount of total protein loaded in lanes 1–3 was ∼10, 15, and 4 μg, respectively. Lane 1, non-induced; lane 2, isopropyl 1-thio-β-d-galactopyranoside-induced; lane 3, Ni-NTA column purified recombinant FREP1. C, Western blotting using purified anti-FREP1 antibody specifically detects recombinant FREP1. Gels were loaded as in B, proteins were transferred to membranes and probed with 0.2 μg/ml of anti-FREP1 antibody. D, SDS-PAGE (left) and Western blotting (right) showing that the recombinant FREP1 is secreted from High Five cells into the culture medium. S and P represent supernatant and cell pellet and correspond to 1 and 20 μg of protein loaded in gels, respectively. Only one band was detected by anti-FREP1 antibody (0.2 μg/ml). E, the purified recombinant FREP1 expressed in High Five insect cells was subjected to reducing (lane 1, 1.9 μg of protein) and non-reducing (lane 2, 1.9 μg of protein) 12% SDS-PAGE and stained with Coomassie Brilliant Blue. F, the UV280 absorbance profile of the purified insect cell-expressed recombinant FREP1 in PBS using Sephadex G-200 gel filtration chromatography. G, ELISA analysis using anti-FREP1 antibody confirmed that the FREP1 peaks match those identified using gel filtration. The molecular mass (MM) were calculated based on the equation of [(10)]((44.86 − y)/6.32), as described under “Experimental Procedures,” where y is the elution volume in ml. H, the UV280 absorbance profile of the purified FREP1 in PBS containing 0.1% Triton X-100 using Sephadex G-200 gel filtration chromatography. Data in panels B–H represent more than 3 independent experiments.
To generate anti-FREP1 antibodies, the FREP1 coding sequence (excluding the signal peptide) was cloned and expressed in the E. coli M15 strain and the recombinant FREP1 was purified by a Ni-NTA column. The results indicated that the purity of E. coli-expressed recombinant FREP1 is >95% (Fig. 1B, lane 3). Rabbits were immunized with the purified FREP1 to generate anti-FREP1 antiserum and anti-FREP1 antibodies were purified from serum using protein A/G-agarose affinity chromatography. Western blot analysis showed that the purified polyclonal rabbit anti-FREP1 antibody specifically recognizes recombinant FREP1 (Fig. 1C). All subsequent experiments were performed using the purified anti-FREP1 antibodies.
Recombinant FREP1 Is Secreted from Insect Cells and Forms Tetramers
We next examined cellular FREP1 expression patterns in vitro. Full-length FREP1 was expressed in High Five insect cells as described under “Experimental Procedures.” We found that FREP1 was exclusively detected in the cell culture supernatants and absent from the cell pellets (Fig. 1D), supporting that FREP1 is a secreted protein. Notably, Western blotting using the anti-FREP1 antibody identified only a single band.
Next we determined whether in vitro-expressed FREP1 forms distinct quaternary structures. When supernatants were subjected to SDS-PAGE, the purified insect cell-expressed recombinant FREP1 exhibited identical molecular mass under both reducing (with 2-mercaptoethanol) and non-reducing conditions (Fig. 1E), indicating that insect cell-expressed recombinant FREP1 exists as either monomers or multimers that associate via non-covalent bonds. The observed molecular mass of insect-expressed recombinant FREP1 (∼95 kDa, Fig. 1E) is greater than the predicted molecular mass (83.5 kDa), suggesting post-translational modification of secreted FREP1. Gel filtration chromatography was then utilized to separate recombinant FREP1 and protein complexes, and ELISA was subsequently used to quantify amounts of recombinant FREP1 in each fraction. The profile of UV absorbance at 280 nm from FPLC (Fig. 1F) matched the ELISA result (Fig. 1G). Based on the gel-filtration standard curves, the dominant recombinant FREP1 peak eluted between 328 and 408 kDa (Fig. 1, F and G). Given that the observed molecular mass of recombinant FREP1 is ∼95 kDa, these data support that the majority of the secreted, insect cell-expressed FREP1 exists as tetramers (∼380 kDa). In addition to monomers and tetramers, FREP1 dimers and trimers were also apparent (Fig. 1F). Coiled-coil proteins tend to be elongated and yield aberrant molecular masses when subjected to gel filtration, particularly when the calibration standards are globular proteins. For these reasons, we repeated our gel filtration studies and included a non-ionic detergent to confirm the quaternary structure of the FREP1. We found that addition of 0.1% Triton X-100 to FREP1 eliminated the higher-ordered structures (Fig. 1H). To exclude the possibility that the His6 tag contributed to the formation of FREP1 quaternary structures, we additionally examined highly concentrated, non-tagged recombinant FREP1 in gel-filtration studies. These analyses were consistent with Fig. 1G (data not shown). Together, these data show that FREP1 is a secreted mosquito protein that likely forms tetramers through non-covalent hydrophobic interactions.
Endogenous FREP1 Localizes to the Peritrophic Matrix in Mosquito Midguts after Blood Feeding
Published microarray-based gene expression data (29, 30) support that FREP1 is expressed higher in mosquito midguts, compared with other tissues, and FREP1 is up-regulated by blood meals. Thus, we next generated polyclonal anti-FREP1 antibodies and used Western blotting and IHC assays to determined the localization of endogenous FREP1 in mosquito midguts.
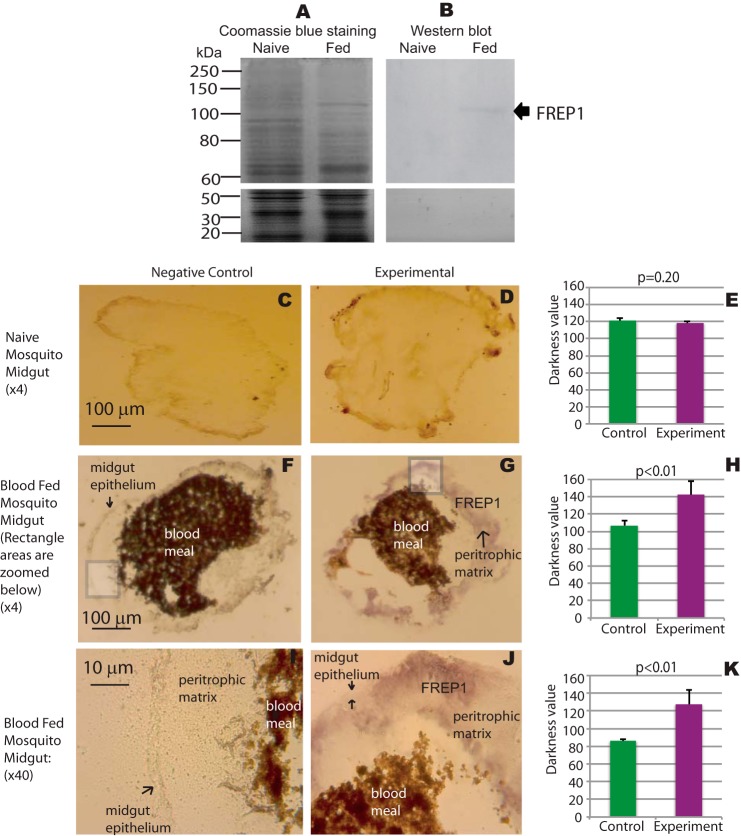
To determine whether anti-FREP1 specifically recognizes endogenous FREP1 in mosquitoes, we dissected midguts from unfed (naive) and blood-fed mosquitoes 18 h after a blood meal. Midgut lysates were fractionated on SDS-PAGE revealing that the protein composition of blood-fed mosquito midguts was distinct from that of naive mosquitoes (Fig. 2A), which is consistent with reports of blood feeding regulation of mosquito gene expression (13). These data are also consistent with microarray transcriptome data showing that blood up-regulates FREP1 expression. Notably, Western blotting revealed only a single band in the blood-fed mosquito midgut lysate, and no band was detected in naive mosquito midgut lysate (Fig. 2B). The molecular mass of endogenous FREP1 (∼100 kDa) is slightly larger than the High Five cell-expressed FREP1 (∼95 kDa), but much greater than E. coli-expressed FREP1 (∼83.5 kDa), supporting that FREP1 in mosquitoes and High Five cells undergo post-translational modifications (Fig. 1D). Together, these data confirm the specificity of the rabbit polyclonal anti-FREP1 antibody for endogenous FREP1 expressed in mosquito midguts following a blood meal feeding.
FIGURE 2.
FREP1 localizes to the mosquito midgut peritrophic matrix. A, midgut proteins from unfed (naive) and blood-fed mosquitoes were fractionated on 10% SDS-PAGE and stained with Coomassie Brilliant Blue. Approximately 10 μg of total protein, extracted from 2 to 3 midguts, was loaded per lane. B, gels were loaded as in A, and proteins were transferred to membranes and probed with 0.2 μg/ml of anti-FREP1 antibody. Western blotting data show that anti-FREP1 antibody specifically recognizes FREP1 in blood-fed mosquito midguts. C, F, and I, negative control staining of naive and blood-fed An. gambiae midgut sections using purified preimmune rabbit antibody. D, G, and J, experimental staining of naive and blood-fed An. gambiae midgut sections using anti-FREP1 rabbit antibody. FREP1 (purple signal, panels G and J) is only detected in blood-fed sections probed with anti-FREP1 antibody; I and J are ×40 magnifications of the boxed areas shown in panels F and G, respectively. Locations of the midgut epithelium, the peritrophic matrix, the FREP1, and the blood bolus are annotated on the images. Summary data (mean ± S.D.) in E, H, and K show the statistical difference of pixel intensity values between negative controls and experimental groups from three independent experiments.
IHC assays were next used to localize FREP1 in mosquito midguts. Comparisons of unfed (naive) midgut tissue sections stained with preimmune antibody (Fig. 2C) versus anti-FREP1 antibody (Fig. 2D) revealed no significant differences (Fig. 2E). However, 12-h post-blood meal, the FREP1 signal (purple color) in sections stained with anti-FREP1 (Fig. 2G) were significantly more intense (p < 0.01, Fig. 2H) compared with sections stained with preimmune antibodies (Fig. 2F). These data further demonstrate that endogenous FREP1 expression is induced in mosquito midguts following a blood meal feeding. These data are also consistent with the Western blotting data (Fig. 2B) and published microarray-based expression data showing up-regulation of FREP1 mRNA expression in mosquitoes 3 h after a blood meal feeding (13). Higher resolution analyses (rectangles in Fig. 2, F and G, were magnified ×40 and displayed in Fig. 2, I and J, respectively) revealed that the majority of FREP1 was localized in the mosquito PM that resides within the midgut lumen. Consistent staining patterns and FREP1 localization were observed in more than 3 independent experiments and the statistical analyses of pixel intensity values between control and experimental groups confirmed that endogenous FREP1 was localized to the mosquito midgut PM (Fig. 2, H and K). Together, our Western blotting and IHC studies consistently show that FREP1 is up-regulated after blood feeding and that the endogenous FREP1 is secreted into mosquito midgut lumen and associated with the PM.
To verify FREP1 association with the PM, we analyzed the interactions between mosquito midguts and FREP1. The insect cell-expressed recombinant FREP1 was incubated with midguts from naive and blood-fed mosquitoes. The ELISAs were then used to quantify the relative amount of FREP1 that bound midgut preparations. We found that significantly more FREP1 was bound in blood-fed mosquito midguts compared with unfed mosquito midguts (p < 0.001, Fig. 3). Substituting recombinant FREP1 with BSA eliminated the binding signals. The pattern was consistent in three experiments. Collectively, these results support that FREP1 localizes to and specifically interacts with the mosquito midgut PM.
FIGURE 3.
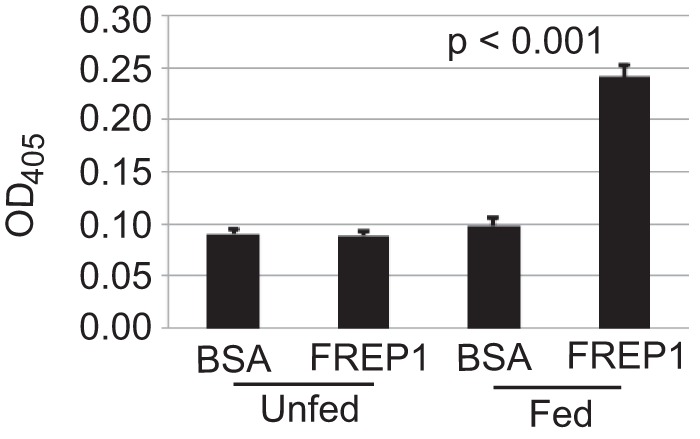
Blood-fed mosquito midguts bound significantly (p < 0.001) more High Five cell-expressed recombinant FREP1 compared with unfed (naive) mosquito midguts. Purified recombinant FREP1 or irrelevant protein (BSA) was incubated with midguts and the midgut-bound FREP1 was quantified by ELISA. Data (mean ± S.D.) are from three independent experiments.
FREP1 Binds P. falciparum
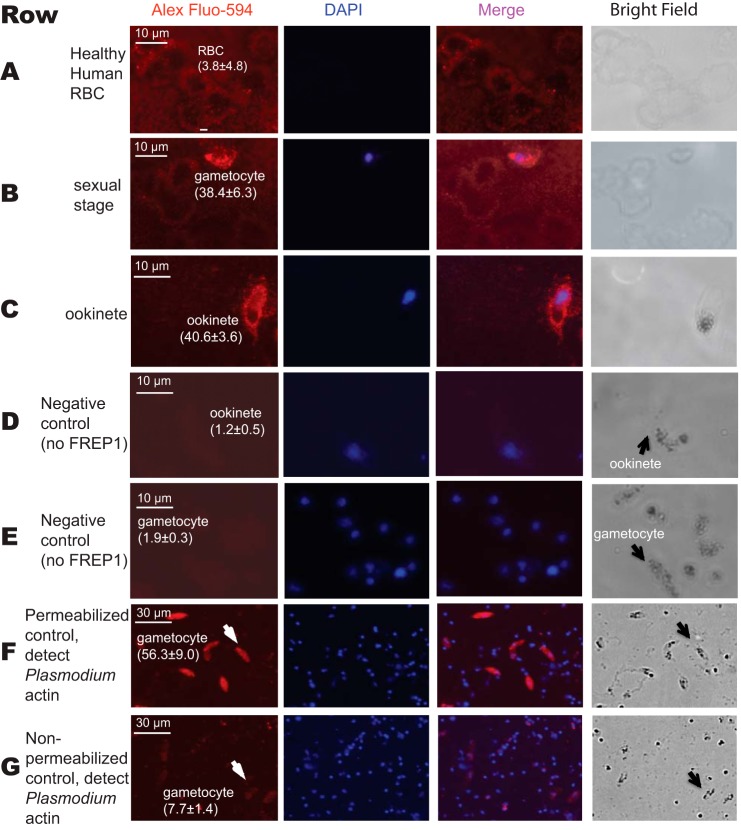
We next sought to determine whether FREP1 could interact with the sexual stage P. falciparum parasites. We fixed non-permeabilized P. falciparum gametocytes and ookinetes on coverslips and then probed the cells with insect cell-expressed recombinant FREP1. Anti-FREP1 antibody and fluorescence-conjugated secondary antibodies were used sequentially to determine whether FREP1 bound to gametocytes and ookinetes. In our fluorescence assays, the bound FREP1 appeared red and cell nuclei stained with 4,6-diamidino-2-phenylindole (DAPI) appeared purple. Our results consistently showed that the fluorescence intensity of P. falciparum gametocytes (Fig. 4, row B) and ookinetes (Fig. 4, row C) were significantly (p < 0.01) higher than those of healthy (non-infected) human RBC (Fig. 4, row A), supporting that the recombinant FREP1 can specifically bind both sexual stage (gametocytes) and mosquito midgut stage (ookinetes) P. falciparum. Of note, non-infected RBC do not contain nuclei and are therefore not stained by DAPI. To confirm that addition of FREP1 is necessary to generate the signals in the IFA assays, we performed the same experiments omitting FREP1 and using only secondary reagents (Fig. 4, rows D and E). The absence of the detectable fluorescence signal further supports that FREP1 binds to the sexual stage of P. falciparum parasites or mosquito invasion stage ookinetes. Finally, to confirm that our non-permeabilized fixation approaches maintain cell membrane integrity, we stained parasite preparations with antibodies against Plasmodium actin (31). Cells containing gametocytes were fixed separately with either 4% paraformaldehyde or 100% methanol on dry ice for 5 min. The results showed that the anti-actin antibody could only stain cells fixed in methanol (Fig. 4F), whereas the fluorescence intensity of gametocytes fixed with 4% paraformaldehyde was similar to the background (Fig. 4G). Together, these data support that both recombinant FREP1 and anti-FREP1 reagents were reacting extracellularly. Thus, FREP1 binds molecules on the surface of the gametocytes and ookinetes.
FIGURE 4.
Interaction between the recombinant FREP1 and P. falciparum demonstrated by immunofluorescence assays. Images in row A depict healthy, non-infected human RBC. Images in rows B and E depict sexual stage gametocytes. Images in the rows C and D are diploid ookinetes. Images in the rows D and E were obtained by conducting the same procedures as rows A–C except no recombinant FREP1 was added. Image F depicts the anti-actin antibody staining of gametocytes that were fixed with 100% methanol. Image G shows that anti-actin antibody does not stain malaria actin within gametocytes that were fixed with 4% paraformaldehyde. Of note, numerous DAPI-positive spots in rows F and G represent extracellular merozoite forms of P. falciparum, which become abundantly released after 10 days of culture. The first and second columns depict FREP1 and nuclear staining, respectively. Merging the first and second columns generated the third column. The last column shows cells under bright field. The numbers within parentheses after the cells are the average fluorescence signal intensity values (± S.D.) for three replicates.
Ablating FREP1 Expression Reduced P. falciparum Infection
To determine whether FREP1 directly regulates P. falciparum infection of Anopheles mosquito vectors, we used a standard dsRNA-mediated gene-silencing assay to knockdown FREP1 mRNA in live mosquitoes, and analyzed the impact of this silencing on P. falciparum infection of mosquito midguts. One-day-old adult female An. gambiae were injected with FREP1 dsRNA and subsequently fed on cultured P. falciparum gametocytes 36 h post-dsRNA injection. Because blood meal feeding increases FREP1 expression, we analyzed the efficacy of FREP1 mRNA knockdown 12 h post-blood meal/infection. Quantitative RT-PCR data show that FREP1 dsRNA injection reduced (∼20-fold) FREP1 mRNA to nearly undetectable levels 12 h post-blood meal feeding (Fig. 5A, lane 2). As a control, we injected mosquitoes in parallel with dsRNA corresponding to an irrelevant target, GFP (Fig. 5A, lane 1). Furthermore, the AgS7 gene, which is constitutively expressed in mosquitoes, was used as a control in the quantitative RT-PCR assays (Fig. 5A, lanes 3 and 4). We also compared both total protein and FREP1 protein expression levels in control and experimental mosquito midguts using Western blotting and IHC assays. The total protein concentration and composition in FREP1 dsRNA-treated mosquito midguts (Fig. 5B, lane 1) was similar to that of control midguts (Fig. 5B, lane 2). However, Western blotting results from five mosquitoes showed that the FREP1 protein was reduced >3-fold in FREP1 dsRNA-treated mosquito midguts, compared with GFP dsRNA-treated mosquito midguts (compare Fig. 5C, lanes 1 and 2).
FIGURE 5.
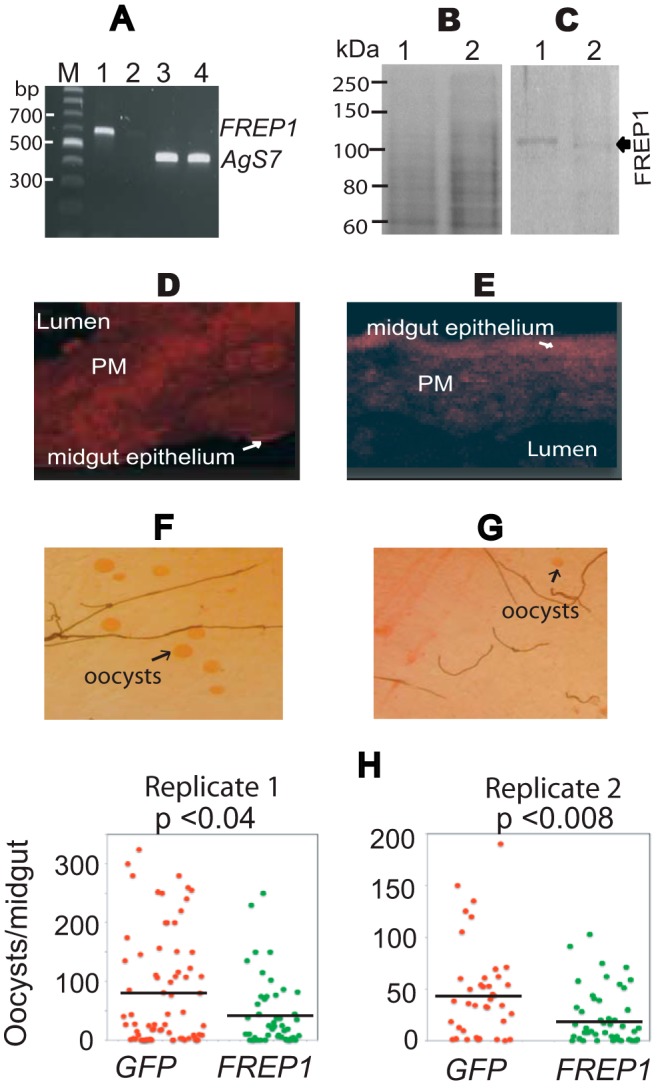
Impact of dsRNA-mediated knockdown of FREP1 on P. falciparum infection in mosquitoes. A, quantitative RT-PCR detection of FREP1 mRNA in dsRNA-treated experimental and control mosquitoes. Lane order: M, DNA ladder; 1, FREP1 expression in GFP dsRNA-treated mosquitoes; 2, FREP1 expression in FREP1 dsRNA-treated mosquitoes; 3, Ags7 expression in GFP dsRNA-treated mosquitoes; 4, Ags7 expression in FREP1 dsRNA-treated mosquitoes. All primer sequences are shown in Table 1. B, GFP dsRNA-treated (lane 1) and FREP1 dsRNA-treated (lane 2) mosquito midgut proteins fractionated on 10% SDS-PAGE and stained with the Coomassie Brilliant Blue. Approximately 10 μg of total protein, extracted from 2 to 3 midguts, was loaded per lane. C, gels were loaded as in B, and proteins were transferred to membranes and probed with 0.2 μg/ml of anti-FREP1 antibody. GFP dsRNA-treated (control, lane 1) and FREP1 dsRNA-treated mosquito (lane 2) midgut proteins are shown. The average pixel intensity values of the FREP1 signal in panel C are 15.93 and 4.24 for the control and experimental group, respectively. D and E, immunohistochemical analysis of midgut tissue sections from GFP dsRNA-treated (D) and FREP1 dsRNA-treated (E) mosquitoes. Sections were probed with anti-FREP1 followed by Alexa Fluor® 594 goat anti-rabbit antibody. The fluoresence intensities of control sections (D) were 17.93 ± 1.77, whereas the fluoresence intensities of FREP1 dsRNA-treated experimental sections (E) were 6.32 ± 0.11. The mean fluoresence intensities and standard deviations were obtained from three sections. F and G, enumeration of oocysts (round red spots) in GFP dsRNA-treated (F) and FREP1 dsRNA-treated (G) mosquito midguts. H, summary data and statistical analyses of the number of oocysts in mosquitoes treated with the dsRNA targeting GFP or FREP1, respectively. y axis represents the number of oocysts per midgut and x axis indicates the treatment groups. The black bars represent the mean oocysts per midgut. The p values between two groups were calculated using Wilcoxon-Mann-Whitney test. Data are representative of two independent experiments.
IHC assays were also used to confirm FREP1 reductions in FREP1 dsRNA-treated mosquitoes. Midgut sections from experimental and control mosquitoes were sequentially probed with the anti-FREP1 rabbit antibodies and fluorescence-conjugated secondary antibodies. The results show that the fluorescence intensity of FREP1 staining in FREP1 dsRNA-treated mosquito midgut PM was 6.32 ± 0.11 (Fig. 5E), significantly (p < 0.01) weaker than the fluorescence intensity of control sections, which was 17.93 ± 1.77 (Fig. 5D). Together, Western blotting and IHC assays confirm the specific and efficacious knockdown of FREP1 by dsRNA treatment.
Given these results, we next examined whether knockdown of FREP1 impacted P. falciparum midgut infection. Seven days post-blood meal/infection, we compared the number of oocysts in FREP1 dsRNA-treated and control GFP dsRNA-treated mosquitoes. The data show that significantly (p < 0.04) fewer oocysts developed in FREP1-depleted An. gambiae midguts (Fig. 5G), compared with the GFP dsRNA-treated mosquitoes (Fig. 5F). Consistent results were observed in two independent dsRNA-mediated gene expression-silencing experiments (Fig. 5H). Together, these data show that FREP1 facilitates P. falciparum infection of the An. gambiae midgut and the expressed FREP1 in the mosquito midgut PM serves as an agonist for P. falciparum invasion.
Anti-FREP1 Antibody Blocks P. falciparum Parasite Infection in Mosquitoes
Our data suggest that FREP1 likely exerts its function through direct interaction with both Plasmodium parasites and the mosquito PM. Thus, we hypothesized that interfering with these interactions would inhibit parasite infection of the mosquito midgut. To test this hypothesis, rabbit anti-FREP1 polyclonal antibody was mixed with P. falciparum gametocyte cultures (0.2% gametocytes, 0.5 mg/ml of antibody) prior to their use in mosquito membrane feeding-based infection assays. An equivalent amount of purified preimmune rabbit antibody was used as a negative control. Strikingly, co-ingestion of gametocytes mixed with anti-FREP1 antibody significantly (p < 10−7) reduced the number of P. falciparum oocysts per midgut by 25- and 60-fold, compared with control antibody-treated cultures in two replicate experiments (Fig. 6A). The infection prevalence also decreased from 85 and 82% to 35 and 16%, respectively (Fig. 6A). To examine dose-dependent effects of anti-FREP1 antibody on blocking P. falciparum infection, we tested a dilution series (0.4, 0.2, 0.1, and 0.05 mg/ml) of antibodies. The results (Fig. 6B) show that as the concentration of anti-FREP1 antibody decreased, the number of oocysts in mosquito midguts increased. Notably, antibody concentrations as low as 0.1 mg/ml, which is 5 times lower than the antibody concentration in the original undiluted rabbit serum, still mediated significant (p < 0.003) reductions in Plasmodium parasite infection in mosquitoes. Thus, vaccination with FREP1 can potentially elicit physiologically relevant titers of anti-FREP1 antibodies capable of exerting transmission-blocking activity in blood feeding mosquitoes.
FIGURE 6.
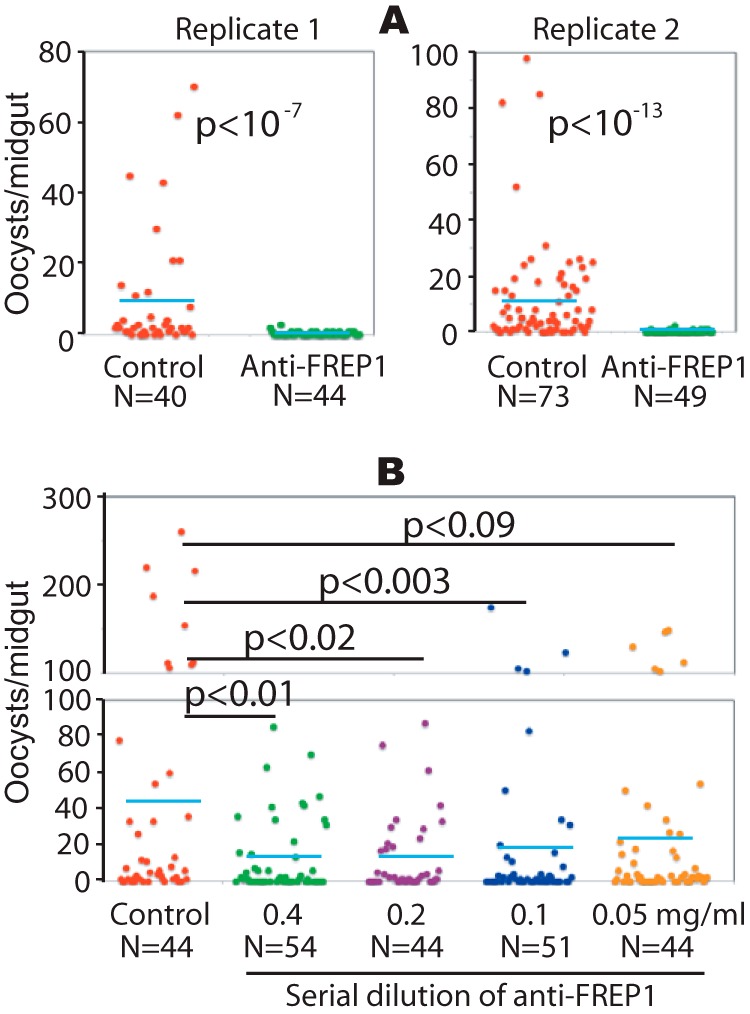
The anti-FREP1 antibody blocks P. falciparum parasite invasion in mosquitoes. Three-day-old female An. gambiae were fed with P. falciparum-infected blood containing 0.2% stage V gametocytes and 0.5 mg/ml of purified anti-FREP1 antibodies (A) or a series of dilutions of anti-FREP1 antibodies (0.4, 0.2, 0.1, and 0.05 mg/ml) (B). Midguts from engorged (fed) mosquitoes were dissected 7 days post-infection and the numbers of oocysts in mosquito midguts were counted microscopically. N represents the number of mosquitoes and the blue bars represent the mean number of oocysts per midgut in each group. Control in panels A and B represent mosquitoes fed on infected blood treated with purified preimmune rabbit antibodies (0.5 and 0.4 mg/ml in A and B, respectively). The p values between experimental and the control groups were calculated using Wilcoxon-Mann-Whitney tests. Data represent two independent experiments.
To exclude the possibility that the rabbit anti-FREP1 antibody affected parasite viability before feeding, we examined P. falciparum viability following addition of purified anti-FREP1 antibody. Anti-FREP1 was added into cultures at 0.4 mg/ml and an equivalent amount of the purified preimmune rabbit antibody was used as a control. Twenty-four hours later, the number of P. falciparum-infected cells was counted. In three independent experiments we observed equivalent parasitemias (in percentage) in non-treated cultures and cultures containing anti-FREP1 or preimmune antibody (6.2 ± 0.53, 7.2 ± 0.81, and 6.4 ± 1.32, respectively). No statistical differences in viable parasite numbers were observed among the three groups (p = 0.20). Therefore, anti-FREP1 antibodies do not impact parasite viability outside of the mosquito host. Collectively, these data support that anti-FREP1 antibodies can inhibit Plasmodium infection in mosquitoes via disruption of FREP1-parasite interactions within the PM of mosquito midguts.
Discussion
We previously reported that specific polymorphisms within FREP1 were strongly associated with reduced P. falciparum infection intensity in wild Anopheles mosquitoes (13). Our current work aimed to investigate the molecular mechanisms and functions of FREP1 during infection of mosquitoes by the human malaria pathogen P. falciparum. First, we demonstrated that FREP1 is expressed in the mosquito midgut and exerts its effects on Plasmodium infection in the PM. It is well known that PM formation occurs ubiquitously in the midguts of hematophagous insects and serves as an important physical barrier to resist or prevent invasion by pathogens in blood (11). Previous studies showed that FREP1 expression is up-regulated by a blood meal (29). Here we show that FREP1 is also up-regulated and the protein product is secreted into the mosquito midgut lumen and associated with the PM after a blood meal. Ablating FREP1 expression by dsRNA reduced FREP1 levels in blood-fed mosquito midguts. It is reported that FBN domains in FREPs can recognize N-acetylglucosamine of chitin (32, 33), other carbohydrates and their derivatives (17, 33), and our data confirmed that FREP1 binds PM. We speculate that the FREP1 is a structural and functional protein in the PM and interacts with chitin and other carbohydrates via the FBN domain of FREP1 (Fig. 7). This interaction keeps FREP1 intimately associated with the PM following blood meal feeding.
FIGURE 7.
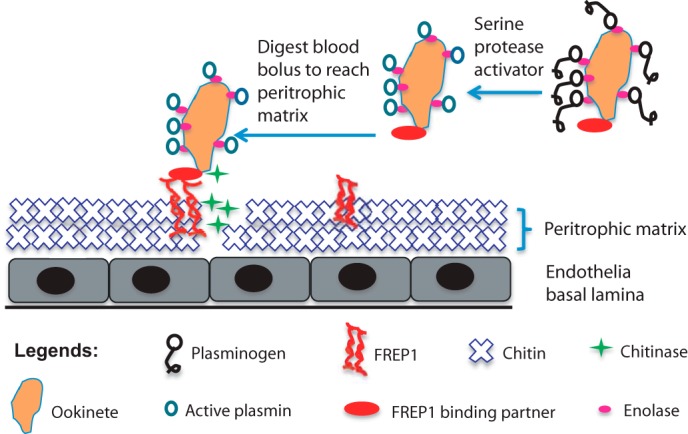
Working molecular model for FREP1-mediated Plasmodium parasite invasion in Anopheline mosquitoes. After a blood meal, FREP1 is secreted into mosquito midgut lumen and associated with the newly formed PM. PM-associated tetrameric FREP1 binds a putative FREP1-binding partner(s) expressed on the surface of Plasmodium parasites, thereby anchoring the parasite within the PM. Stoichiometry of these interactions is under investigation. Continuous secretion of chitinase by anchored ookinetes digests chitin in PM, which results in disruption of PM. Dissolution of the PM enables ookinetes to eventually escape and overcome the PM barrier and subsequently invade the midgut epithelium.
Second, our data support the tetrameric quaternary structure of FREP1. Invertebrate FREP/FBN family members tend to multimerize to exert their physiological functions (34). For example, the An. gambiae mosquito FBN9 protein forms dimers that interact with Gram-positive and Gram-negative bacteria (19). Moreover, the functional forms of horseshoe crab TL-5A and TL-5B proteins form propeller-like structures with each blade corresponding to a disulfide-linked dimer (35). Similarly, our results show that FREP1 forms tetramers through hydrophobic forces, rather than disulfide bonds, because non-reducing SDS-PAGE revealed FREP1 monomers and gel filtration showed that the dominant species of FREP1 were founds as tetramers. Moreover, non-ionic detergent disassociated these oligomers. Consistent with this, coiled-coil motifs in proteins have been reported to mediate homodimer complexes (36) and tetramer complex formation (37). Therefore, interactions between the coiled-coil domains of FREP1 likely mediate FREP1 tetramer formation.
Third, IFA assays showed that FREP1 interacts with the sexual stage (gametocytes) and mosquito midgut invasion stage (ookinetes) P. falciparum parasites. The putative parasite-expressed FREP1-binding partners are expected to localize on the cell surface of gametocytes and ookinetes, because a non-permeabilization approach was used in our IFA assays to detect the interaction between FREP1 and parasites. Since FREPs were originally proposed to function as pattern recognition molecules (19), the C-terminal FBN domain within FREP1 is likely responsible for mediating interactions between FREP1 and Plasmodium parasites. Studies to fine-map these interactions are currently underway.
Finally, we examined the in vivo function of FREP1 during P. falciparum infection of mosquitoes. Previously, we reported that ablation of FREP1 expression in An. gambiae mosquitoes resulted in a significant reduction of the number of Plasmodium berghei oocysts, a rodent malaria parasite (13). Here, we extend these data and investigate the influence of FREP1 on P. falciparum infection of mosquitoes using RNA interference. Importantly, our data show that FREP1 is also critical for P. falciparum infection. Knockdown of FREP1 expression significantly reduced P. falciparum infection intensity and prevalence in An. gambiae. Moreover, anti-FREP1 antibody blocked P. falciparum invasion, strongly supporting that the interaction between parasites and FREP1 mediates Plasmodium invasion. Collectively, FREP1 regulates infection by both P. berghei and P. falciparum, supporting that FREP1 functions as a conserved host factor that facilitates Plasmodium invasion of the mosquito midgut. Thus, targeting FREP1 and its product may limit the infectivity of multiple Plasmodium species in mosquitoes.
Based on our experimental data, we propose a molecular model for FREP1 activity during Plasmodium invasion (Fig. 7). FREP1 is up-regulated and expressed in midguts after a blood meal and our new data support that FREP1 is secreted into the mosquito midgut lumen. In the lumen, the FREP1 may form tetramers through interactions between coiled-coil regions of FREP1 monomers. Our data suggest that FREP1 tetramers are an integral component of the mosquito PM. Thus, FREP1 is likely to be a structural component of the PM, and the tetramerization of FREP1 may perhaps increase the binding affinity between FREP1, the PM, and Plasmodium parasites. We speculate that the interactions between ookinetes and FREP1 may facilitate localization and positioning of ookinetes within the PM, which may further potentiate the enzymatic activities of the chitinase secreted by ookinetes (38). These interactions and the activity of chitinase, coupled with the digestive activity of plasmin on the ookinete surface (39), may result in a disruption of the PM structure, ultimately facilitating parasite invasion. After ookinetes cross the PM to invade midgut epithelial cells other proteins in mosquito hemolymph may perhaps interact with parasites and impact infection intensity (40, 41).
By extension, our model predicts that blocking FREP1 activity or interaction with Plasmodium parasites will significantly reduce the capacity for mosquitoes to transmit malaria. Consistent with this prediction, our published study (13) and new experimental data show that Plasmodium infection in mosquitoes is markedly reduced following genetic ablation of FREP1 expression or targeting FREP1 by antibodies. It is worth noting that if the concentration of mature gametocytes in a blood meal is low (i.e. capable of generating less than 100 oocysts per mosquito midgut), blocking FREP1 by RNAi or antibodies rendered the majority of mosquitoes wholly resistant to P. falciparum (Fig. 6A) and P. berghei infection (13). On the other hand, when the density of gametocytes in blood is high, the infection intensity is still significantly lower in mosquitoes treated with dsRNA (Fig. 5D) or anti-FREP1 antibodies, compared with control groups (Fig. 6B). Importantly, physiological parasite densities observed in wild P. falciparum-infected An. gambiae is usually very low, e.g. resulting in less than 10 oocysts per midgut (13). Thus, FREP1 remains a critical component of pathways for parasite invasion at physiological parasite densities and targeting FREP1 represents a potentially viable strategy to reduce Plasmodium transmission.
Ookinetes have to overcome and exit the blood bolus barrier before reaching PM. Previous reports showed that mammalian plasminogen can be captured by enolase on the surface of ookinetes (42). Plasminogen is an essential serine protease precursor in vertebrate blood. After activation, plasminogen becomes plasmin that can cleave fibrin or fibronectin to free ookinetes in the blood bolus, thereby facilitating their migration to the midgut PM. After traversing the PM, many ookinetes will manage to invade and cross the mosquito midgut epithelium through multiple pathways (43). Parasite invasion-induced apoptosis of midgut epithelial cells will further determine survival and/or successful ookinete infection (44). Therefore, interactions between the PM and Plasmodium parasites (i.e. the “interactome”) mediated by FREP1 is important for parasite invasion, and targeting this interaction may prove effective for limiting malaria transmission. It is worth emphasizing that proteins in the PM are readily accessible by antibodies co-ingested after a blood meal (2). Thus, FREP1 may serve as an excellent potential antigen for inclusion in experimental malaria transmission blocking vaccines. Consistent with this notion, and as we demonstrated in this report, anti-FREP1 antibodies significantly reduced the mosquito vectorial load of malaria parasites. These antibody protection assays further support our proposed molecular model of FREP1 activity during Plasmodium invasion.
In summary, here we describe a novel molecular mechanism and function of the mosquito FREP1 during Plasmodium infection of the mosquito midgut. We propose that FREP1 acts as an anchor to facilitate ookinete penetration of the PM in mosquito midguts. Consistent with the concept that the PM protects mosquitoes from pathogen infection or invasion, pathogens like Plasmodium have also evolved mechanisms to overcome these physical barriers by exploiting the specific constituents of the PM. Understanding the mechanisms of Plasmodium parasite invasion of mosquito midguts will identify new opportunities and targets for malaria control.
Acknowledgments
We thank Dr. Abhai Tripathi, The Johns Hopkins Malaria Research Institute Parasitology and Insectary core facilities, for providing consult on culturing P. falciparum, and Professor Barbara Safiejko-Mrocka, Department of Biology of University of Oklahoma, for assistance with IHC assays. We also appreciate the anonymous reviewers for their constructive comments.
This work was supported, in whole or in part, by National Institutes of Health Grants 1R21AI115178-01A1 and GM103447 and Oklahoma Center for the Advancement of Science and Technology Grant HR13-055. The authors declare that they have no conflicts of interest with the contents of this article.
- PM
- peritrophic matrix
- Ni-NTA
- nickel-nitrilotriacetic acid
- FBN
- fibrinogen-like
- IHC
- immunohistochemical
- IFA
- immunofluorescence assay
- PBSI
- PBS supplemented with protease inhibitors.
References
- 1. Christophides G. K., Crisanti A. (2013) Vector and vector-borne disease research: need for coherence, vision and strategic planning. Pathog. Glob. Health 107, 385–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhao B., Kokoza V. A., Saha T. T., Wang S., Roy S., Raikhel A. S. (2014) Regulation of the gut-specific carboxypeptidase: a study using the binary Gal4/UAS system in the mosquito Aedes aegypti. Insect Biochem. Mol. Biol. 54, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dinglasan R. R., Devenport M., Florens L., Johnson J. R., McHugh C. A., Donnelly-Doman M., Carucci D. J., Yates J. R., 3rd, Jacobs-Lorena M. (2009) The Anopheles gambiae adult midgut peritrophic matrix proteome. Insect Biochem. Mol. Biol. 39, 125–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Abraham E. G., Jacobs-Lorena M. (2004) Mosquito midgut barriers to malaria parasite development. Insect Biochem. Mol. Biol. 34, 667–671 [DOI] [PubMed] [Google Scholar]
- 5. Pumpuni C. B., Demaio J., Kent M., Davis J. R., Beier J. C. (1996) Bacterial population dynamics in three anopheline species: the impact on Plasmodium sporogonic development. Am. J. Trop. Med. Hyg. 54, 214–218 [DOI] [PubMed] [Google Scholar]
- 6. Gonzalez-Ceron L., Santillan F., Rodriguez M. H., Mendez D., Hernandez-Avila J. E. (2003) Bacteria in midguts of field-collected Anopheles albimanus block Plasmodium vivax sporogonic development. J. Med. Entomol. 40, 371–374 [DOI] [PubMed] [Google Scholar]
- 7. Mead E. A., Li M., Tu Z., Zhu J. (2012) Translational regulation of Anopheles gambiae mRNAs in the midgut during Plasmodium falciparum infection. BMC Genomics 13, 366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Christophides G. K., Vlachou D., Kafatos F. C. (2004) Comparative and functional genomics of the innate immune system in the malaria vector Anopheles gambiae. Immunol. Rev. 198, 127–148 [DOI] [PubMed] [Google Scholar]
- 9. Billingsley P. F. (1990) The midgut ultrastructure of hematophagous insects. Annu. Rev. Entomol. 35, 219–248 [Google Scholar]
- 10. Shao L., Devenport M., Jacobs-Lorena M. (2001) The peritrophic matrix of hematophagous insects. Arch. Insect Biochem. Physiol. 47, 119–125 [DOI] [PubMed] [Google Scholar]
- 11. Toprak U., Baldwin D., Erlandson M., Gillott C., Hegedus D. D. (2010) Insect intestinal mucins and serine proteases associated with the peritrophic matrix from feeding, starved and moulting Mamestra configurata larvae. Insect Mol. Biol. 19, 163–175 [DOI] [PubMed] [Google Scholar]
- 12. Sinden R. E., Billingsley P. F. (2001) Plasmodium invasion of mosquito cells: hawk or dove? Trends Parasitol. 17, 209–212 [DOI] [PubMed] [Google Scholar]
- 13. Li J., Wang X., Zhang G., Githure J. I., Yan G., James A. A. (2013) Genome-block expression-assisted association studies discover malaria resistance genes in Anopheles gambiae. Proc. Natl. Acad. Sci. U.S.A. 110, 20675–20680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mosesson M. W. (2005) Fibrinogen and fibrin structure and functions. J. Thromb. Haemost. 3, 1894–1904 [DOI] [PubMed] [Google Scholar]
- 15. Zhang H., Wang L., Song L., Song X., Wang B., Mu C., Zhang Y. (2009) A fibrinogen-related protein from bay scallop Argopecten irradians involved in innate immunity as pattern recognition receptor. Fish Shellfish Immunol. 26, 56–64 [DOI] [PubMed] [Google Scholar]
- 16. Fan C., Zhang S., Li L., Chao Y. (2008) Fibrinogen-related protein from amphioxus Branchiostoma belcheri is a multivalent pattern recognition receptor with a bacteriolytic activity. Mol. Immunol. 45, 3338–3346 [DOI] [PubMed] [Google Scholar]
- 17. Wang X., Zhao Q., Christensen B. M. (2005) Identification and characterization of the fibrinogen-like domain of fibrinogen-related proteins in the mosquito, Anopheles gambiae, and the fruitfly, Drosophila melanogaster, genomes. BMC Genomics 6, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kawabata S., Iwanaga S. (1999) Role of lectins in the innate immunity of horseshoe crab. Dev. Comp. Immunol. 23, 391–400 [DOI] [PubMed] [Google Scholar]
- 19. Dong Y., Dimopoulos G. (2009) Anopheles fibrinogen-related proteins provide expanded pattern recognition capacity against bacteria and malaria parasites. J. Biol. Chem. 284, 9835–9844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. QIAGEN (2003) The QIAexpressionist: A handbood for high-level expression and purification of 6XHis-tagged proteins, pp. 1–128, QIAGEN, Hilden, Germany [Google Scholar]
- 21. Wickham T. J., Davis T., Granados R. R., Shuler M. L., Wood H. A. (1992) Screening of insect cell lines for the production of recombinant proteins and infectious virus in the baculovirus expression system. Biotechnol. Prog. 8, 391–396 [DOI] [PubMed] [Google Scholar]
- 22. Invitrogen (2008) InsectSelect BSD system: for stable expression of heterologous proteins in lepidopteran insect cell lines using pIB/V5-His, pp. 1–31, Invitrogen, Grand Island, NY [Google Scholar]
- 23. Duong-Ly K. C., Gabelli S. B. (2014) Gel filtration chromatography (size exclusion chromatography) of proteins. Methods Enzymol. 541, 105–114 [DOI] [PubMed] [Google Scholar]
- 24. Jensen J. B., Trager W. (1977) Plasmodium falciparum in culture: use of outdated erthrocytes and description of the candle jar method. J. Parasitol. 63, 883–886 [PubMed] [Google Scholar]
- 25. Beetsma A. L., van de Wiel T. J., Sauerwein R. W., Eling W. M. (1998) Plasmodium berghei ANKA: purification of large numbers of infectious gametocytes. Exp. Parasitol. 88, 69–72 [DOI] [PubMed] [Google Scholar]
- 26. Korochkina S., Barreau C., Pradel G., Jeffery E., Li J., Natarajan R., Shabanowitz J., Hunt D., Frevert U., Vernick K. D. (2006) A mosquito-specific protein family includes candidate receptors for malaria sporozoite invasion of salivary glands. Cell. Microbiol. 8, 163–175 [DOI] [PubMed] [Google Scholar]
- 27. Goodman A. L., Blagborough A. M., Biswas S., Wu Y., Hill A. V., Sinden R. E., Draper S. J. (2011) A viral vectored prime-boost immunization regime targeting the malaria Pfs25 antigen induces transmission-blocking activity. PloS One 6, e29428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li J., Ribeiro J. M., Yan G. (2010) Allelic gene structure variations in Anopheles gambiae mosquitoes. PLoS One 5, e10699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Marinotti O., Calvo E., Nguyen Q. K., Dissanayake S., Ribeiro J. M., James A. A. (2006) Genome-wide analysis of gene expression in adult Anopheles gambiae. Insect Mol. Biol. 15, 1–12 [DOI] [PubMed] [Google Scholar]
- 30. Baker D. A., Nolan T., Fischer B., Pinder A., Crisanti A., Russell S. (2011) A comprehensive gene expression atlas of sex- and tissue-specificity in the malaria vector, Anopheles gambiae. BMC Genomics 12, 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Angrisano F., Riglar D. T., Sturm A., Volz J. C., Delves M. J., Zuccala E. S., Turnbull L., Dekiwadia C., Olshina M. A., Marapana D. S., Wong W., Mollard V., Bradin C. H., Tonkin C. J., Gunning P. W., Ralph S. A., Whitchurch C. B., Sinden R. E., Cowman A. F., McFadden G. I., Baum J. (2012) Spatial localisation of actin filaments across developmental stages of the malaria parasite. PLoS One 7, e32188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sugimoto R., Yae Y., Akaiwa M., Kitajima S., Shibata Y., Sato H., Hirata J., Okochi K., Izuhara K., Hamasaki N. (1998) Cloning and characterization of the Hakata antigen, a member of the ficolin/opsonin p35 lectin family. J. Biol. Chem. 273, 20721–20727 [DOI] [PubMed] [Google Scholar]
- 33. Garlatti V., Belloy N., Martin L., Lacroix M., Matsushita M., Endo Y., Fujita T., Fontecilla-Camps J. C., Arlaud G. J., Thielens N. M., Gaboriaud C. (2007) Structural insights into the innate immune recognition specificities of L- and H-ficolins. EMBO J. 26, 623–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hanington P. C., Zhang S. M. (2011) The primary role of fibrinogen-related proteins in invertebrates is defense, not coagulation. J. Innate Immun. 3, 17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gokudan S., Muta T., Tsuda R., Koori K., Kawahara T., Seki N., Mizunoe Y., Wai S. N., Iwanaga S., Kawabata S. (1999) Horseshoe crab acetyl group-recognizing lectins involved in innate immunity are structurally related to fibrinogen. Proc. Natl. Acad. Sci. U.S.A. 96, 10086–10091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Qiu J. J., Chu H., Lu X., Jiang X., Dong S. (2011) The reduced and altered activities of PAX5 are linked to the protein-protein interaction motif (coiled-coil domain) of the PAX5-PML fusion protein in t(9;15)-associated acute lymphocytic leukemia. Oncogene 30, 967–977 [DOI] [PubMed] [Google Scholar]
- 37. Syguda A., Bauer M., Benscheid U., Ostler N., Naschberger E., Ince S., Stürzl M., Herrmann C. (2012) Tetramerization of human guanylate-binding protein 1 is mediated by coiled-coil formation of the C-terminal α-helices. FEBS J. 279, 2544–2554 [DOI] [PubMed] [Google Scholar]
- 38. Huber M., Cabib E., Miller L. H. (1991) Malaria parasite chitinase and penetration of the mosquito peritrophic membrane. Proc. Natl. Acad. Sci. U.S.A. 88, 2807–2810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ploplis V. A., Castellino F. J. (2000) Nonfibrinolytic functions of plasminogen. Methods 21, 103–110 [DOI] [PubMed] [Google Scholar]
- 40. Povelones M., Bhagavatula L., Yassine H., Tan L. A., Upton L. M., Osta M. A., Christophides G. K. (2013) The CLIP-domain serine protease homolog SPCLIP1 regulates complement recruitment to microbial surfaces in the malaria mosquito Anopheles gambiae. PLoS Pathog. 9, e1003623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Povelones M., Upton L. M., Sala K. A., Christophides G. K. (2011) Structure-function analysis of the Anopheles gambiae LRIM1/APL1C complex and its interaction with complement C3-like protein TEP1. PLoS Pathog. 7, e1002023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ghosh A. K., Coppens I., Gårdsvoll H., Ploug M., Jacobs-Lorena M. (2011) Plasmodium ookinetes coopt mammalian plasminogen to invade the mosquito midgut. Proc. Natl. Acad. Sci. U.S.A. 108, 17153–17158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vega-Rodríguez J., Ghosh A. K., Kanzok S. M., Dinglasan R. R., Wang S., Bongio N. J., Kalume D. E., Miura K., Long C. A., Pandey A., Jacobs-Lorena M. (2014) Multiple pathways for Plasmodium ookinete invasion of the mosquito midgut. Proc. Natl. Acad. Sci. U.S.A. 111, E492–E500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Han Y. S., Thompson J., Kafatos F. C., Barillas-Mury C. (2000) Molecular interactions between Anopheles stephensi midgut cells and Plasmodium berghei: the time bomb theory of ookinete invasion of mosquitoes. EMBO J. 19, 6030–6040 [DOI] [PMC free article] [PubMed] [Google Scholar]