Background: RWD is a conserved domain in human proteome with unknown function.
Results: We solved the crystal structure of an RWD domain in complex with a Ubc9 homodimer and conducted a biochemical investigation.
Conclusion: The RWD domain binds to a Ubc9 surface that also must interact with E1, E3, and SUMO.
Significance: This study establishes a function for the evolutionary conserved RWD domain.
Keywords: nuclear magnetic resonance (NMR), small ubiquitin-like modifier (SUMO), sumoylation, ubiquitylation (ubiquitination), x-ray crystallography, E2, RWD
Abstract
An RWD domain is a well conserved domain found through bioinformatic analysis of the human proteome sequence; however, its function has been unknown. Ubiquitin-like modifications require the catalysis of three enzymes generally known as E1, E2, and E3. We solved the crystal structure of the E2 for the small ubiquitin-like modifiers (SUMO) in complex with an RWD domain and confirmed the structure using solution NMR analysis. The binding surface of RWD on Ubc9 is located near the N terminus of Ubc9 that is known to be involved in noncovalent binding of the proteins in the conjugation machinery, including a domain of E1, SUMO, and an E3 ligase. NMR data indicate that the RWD domain does not bind to SUMO and E1. The interaction between RWD and Ubc9 has a Kd of 32 ± 4 μm. Consistent with the structure and binding affinity and in contrast to a previous report, the RWD domain and RWDD3 have minimal effects on global SUMOylation. The structural and biochemical information presented here forms the basis for further investigation of the functions of RWD-containing proteins.
Introduction
The RWD domain was identified more than a decade ago as a conserved domain by bioinformatic analysis of the human proteome sequence and was named after the three major families of proteins that contain this domain: RING finger-containing proteins, WD repeat-containing proteins, and DEAD (DEXD)-like helicases (1). The functions of RWD domains and many proteins in the RWD-containing family are not well understood. An RWD domain-containing protein, known as RSUME or RWDD3, was reported to affect post-translational modifications by the small ubiquitin-like modifiers (SUMO).2 It was suggested that RWDD3 promotes the noncovalent interaction between SUMO and Ubc9 (2). However, experimental evidence for this model is lacking.
SUMO modification is one of the more than 10 ubiquitin-like modifications that regulate important cellular functions (3, 4). The enzymes involved in catalyzing ubiquitin and ubiquitin-like modifications share conserved mechanisms (5, 6) that are carried out sequentially by an activating enzyme (E1), conjugation enzyme (E2), and ligase (E3). A single E1 and a single E2 (also known as Ubc9) are responsible for SUMOylation of all SUMO paralogues (7, 8). At least two domains of E1 are involved in binding their corresponding E2s: the ubiquitin fold domain (UFD), which binds to the N-terminal region of E2, and the Cys domain, which binds to a surface near the catalytic Cys of E2 (9, 10). The UFD domain has higher binding affinity for E2 than does the Cys domain and thus is the main contributor to E1-E2 binding affinity (10–13).
In this study, we found that the RWD domain of RWDD3 is the only well structured region in RWDD3 and is responsible for interaction of RWDD3 with Ubc9. The Ubc9 and RWD domain binding interfaces are located near their respective N termini, and the RWD-binding surface on Ubc9 overlaps with the noncovalent SUMO-binding surface on Ubc9. Therefore, it could not enhance Ubc9-SUMO noncovalent interaction, as speculated previously (2). Enzymatic assays revealed that the RWD domain may alleviate the inhibitory effect of Ubc9 on the catalysis of SUMO-activating enzyme at high concentrations of the RWD domain in vitro, but this effect was not observed in cells. In contrast, slight inhibition of SUMOylation was seen in cells, consistent with the mode of interaction between RWD and Ubc9. The moderate cellular effects are consistent with the moderate binding affinity (Kd = 32 ± 4 μm). Taken together, we found that the RWD domain is an E2-interacting module, and the mode of interaction observed here could represent how an RWD domain interacts with an E2 in ubiquitin-like modifications.
Experimental Procedures
Site-directed Mutagenesis and Protein Preparation
Human SUMO E1 (SAE1/SAE2), Ubc9, SUMO-1, SUMO-2, and GST-Sp100 were prepared as previously described (10, 11, 14–16). The DNA fragment encoding RWDD3 was PCR-amplified using primers that generated NdeI and XhoI upstream and downstream restriction sites, respectively. The fragment was then subcloned into the NdeI-XhoI sites of the pET28a vector. The RWD region of RWDD3 was constructed by mutating the residue immediately after the intended C-terminal region to a stop codon. Point mutations were prepared with the QuikChange mutagenesis kit (Stratagene) and confirmed by sequencing. The recombinant DNA plasmid was transformed into Escherichia coli BL21-codon plus (DE3)-RIL for subsequent protein expression.
The unlabeled recombinant proteins were expressed in Luria-Bertani medium as described previously (10, 11, 14–16). For NMR studies, uniformly 15N-labeled RWDD3, RWD, and Ubc9 were expressed in modified M9 medium supplemented with trace minerals and minimal essential medium vitamin solution (Life Technologies Inc.), using 15NH4Cl as the sole nitrogen source as described previously (10, 11, 14). RWDD3 was expressed in inclusion bodies, denatured with 6 m guanidine hydrochloride, and refolded by dialysis against 50 mm Tris buffer (pH 7.5), 500 mm NaCl, and 1 mm DTT prior to purification. The recombinant proteins were purified using nickel-nitriloacetic acid agarose resin (Qiagen) as described previously (10, 11, 14–16). Purity greater than 90% was estimated by Coomassie-stained SDS gel. Proteins were then exchanged to 50 mm Tris buffer (pH 7.5) containing 1 mm DTT or sodium phosphate buffer (pH 7.2) containing 1 mm DTT for conjugation assay and NMR analysis, respectively.
RWDD3/RWD Activity Assays
All SUMO conjugation assays were conducted in a mixture that contained ATP and assay buffer (20 mm HEPES, pH 7.5, 50 mm NaCl, 5 mm MgCl2, and 0.1% Tween). Assay reactions were incubated at 37 °C for the times indicated in the figures and were stopped by addition of SDS loading buffer (with or without DTT). Samples were resolved on 4–12% Bis-Tris NuPAGE SDS-PAGE gels (Invitrogen), and the polypeptide bands were visualized with SimplyBlue SafeStain (Invitrogen). To examine the effect of RWDD3 and RWD on time-dependent formation of SAE2·SUMO and Ubc9·SUMO thioesters, E1 (0.80 μm), Ubc9 (17 μm), and SUMO-1 (40 μm) were incubated in the presence of either full-length RWDD3 (33 μm) or RWD (23 μm). Aliquots of the reactions were withdrawn at 0, 5, 10, 20, and 30 min after initiation of the reaction for SDS-PAGE analysis. To determine the Ubc9 concentration-dependent effect of RWD on Ubc9·SUMO thioester formation, E1 (0.50 μm), SUMO-1 (20 μm), and serial concentrations of Ubc9 (0, 2.5, 5, 10, 15, and 20 μm) were incubated in the presence of RWD (0, 5, and 50 μm). Reactions were terminated after 5 min by adding an equal volume of nonreducing SDS loading buffer. To examine the effect of RWD on the transfer of SUMO from E1 to Ubc9, E1∼SUMO thioester were formed first by incubating E1 (1 μm), and Alexa680 fluorescently labeled SUMO-1 (1 μm). After 15 min, EDTA was added to a final concentration of 30 mm to chelate Mg2+ and subsequently inhibit further ATP-dependent E1 activity. The rate of SUMO transfer from E1 to Ubc9 is fast, and thus the succeeding experiments were conducted in the cold room. All solutions used for the assay were precooled on ice. Ubc9 (0.6 μm) and RWD (0 and 50 μm) were added to the reaction mixtures containing E1·SUMO/EDTA. Aliquots were taken at different time points (0, 15, 30, 45, and 60 s), and reactions were stopped by the addition of equal volumes of nonreducing SDS gel loading buffer containing 4 m urea. The bands were resolved by SDS-PAGE. The fluorescent image of the gel was taken using a LI-COR Odyssey infrared fluorescence scanner.
NMR Sample Preparation and Experiments
All NMR spectra were acquired at 25 °C on a Bruker Avance spectrometer equipped with a cryo-probe and operating at a 600-MHz 1H frequency. The Kd was estimated by titrating an incremental 0.125 molar equivalent of Ubc9 C93A to 15N-RWD WT to a final ratio of 1:2 15N-RWD:Ubc9 followed by recording 1H-15N HSQC. This experiment also provided chemical shift perturbations (CSP) on RWD upon binding Ubc9. Similarly, CSP of Ubc9 was monitored by titrating an incremental 0.125 molar equivalent of RWD to 15N-Ubc9 to a final ratio of 1:2 15N-Ubc9:RWD followed by recording 1H-15N HSQC.
Binding studies of RWD on SUMO, SAE Cys domain and SAE UFD fold were also followed by NMR titration experiments to a final ratio of 1:1.2 of 15N-RWD:SUMO-1, 15N-SAE Cys domain:RWD, and 15N-RWD:SAE UFD fold, respectively. Displacement titration was carried out to determine whether RWD can disrupt the noncovalent interaction of Ubc9 and SUMO. 15N-Ubc9 was titrated with SUMO-1 to a final molar ratio of 1:1, and the 15N-Ubc9-SUMO-1 complex was subsequently titrated with incremental addition of RWD to a final molar ratio of 1:1:1 (15N-Ubc9:SUMO:RWD). All NMR data were processed using NMRPipe and analyzed with the program Sparky.
Sp100 in Vitro Transcription and Translation and SUMO-1 Conjugation Assay
GST-Sp100 (241–360) was labeled with [S35]methionine by in vitro transcription and translation system (TnT coupled wheat germ extract system; Promega), following the manufacturer's instructions. After that, 5 μl of [S35]methionine-labeled GST-Sp100 was mixed with purified E1 (0.1 μm), Ubc9 (17 μm), SUMO-1 (15 μm), RWD, or RWDD3 at the indicated concentrations to perform in vitro SUMO conjugation assay. The reactions were then incubated at 37 °C for 15 min, and the results were verified by SDS-PAGE separation and autoradiography.
In Cell Poly-SUMOylation Assay and Western Blot
pCDNA3.1, pCDNA3-RWD, and pCDNA3-RWDD3 DNAs were transfected into HEK293T cells by Lipofectamine LTX (Life Technologies Inc.) for 48 h. Both RWD and RWDD3 protein have Flag tags at their N termini. The cells were harvested, and the total proteins were first separated by SDS-PAGE, followed by transferring to PVDF membrane. The membrane was first blocked with Odyssey Blocking Buffer (LI-COR) and then blotted by anti-SUMO-2/3 (1:1000; Millipore), HIF1α (1:1000; Cell Signaling Technology), Flag (1:1000; Clontech), and GAPDH (1:1000; Santa Cruz) antibodies. After three washes with 1× PBST buffer (3.2 mm Na2HPO4, 0.5 mm KH2PO4, 1.3 mm KCl, 135 mm NaCl, 0.05% Tween 20, pH 7.4), the fluorophore-conjugated secondary antibodies were then used, and the results were visualized by Odyssey Quantitative Imaging Systems (LI-COR).
Crystallization, Data Collection, and Structure Determination
Initial conditions for the optimal crystal growth were obtained by hanging drop vapor diffusion method at 20 °C using Crystal Screens 1 and 2 from Hampton Research. The complex was prepared by mixing Ubc9 and RWD to a final concentration of 13–18 mg/ml at ∼1:1 stoichiometry. Different volume ratios of protein complex to reservoir solution (1:1, 2:1, and 1:2) were mixed and equilibrated against 500 μl of reservoir solution. Crystals of Ubc9-RWD heterotrimer complex displaying a flat plate morphology were obtained after 4–5 days in the optimized condition of 10% (w/v) PEG8000, 100 mm HEPES/NaOH, pH 8.0, and 8% ethylene glycol at a volume ratio of 1:2 reservoir solution·Ubc9-RWD complex. The crystals were soaked in a cryoprotectant comprised of 20% glycerol in mother liquor prior to data collection. The crystals were frozen in a stream of nitrogen gas at 100 K. Diffraction data were collected at 1.5418 Å on a Rigaku MicroMax-007HF with R-AxisIV++detector (X-ray Crystallography Core Facility of City of Hope). The data were then processed with XDS and scaled with XSCALE (17). The structure of the RWD-Ubc9 heterotrimer was solved by molecular replacement using Molrep (18) with RWD (Protein Data Bank code 2EBK) and Ubc9 (Protein Data Bank code 1U9B) as the search models. The structure was refined in PHENIX (19), with model rebuilding in Coot (20).
Results
The RWD Domain Is the Only Well Structured Region of RWDD3
The human RWDD3 protein (195 amino acid residues), also known as RSUME, has a single RWD domain located at its N terminus (residues 1–115) (2). We acquired and compared the 1H-15N HSQC spectra of both full-length RWDD3 and the RWD domain (residues 1–116) (Fig. 1A). The RWDD3 resonances had much broader line widths, indicating a much larger hydrodynamic radius than the RWD domain. However, the well dispersed resonances of RWDD3 (red) and the RWD domain (blue) overlapped well, indicating that the structure of the RWD domain is not influenced by the C-terminal segment of RWDD3. In addition, most of the RWDD3 resonances that did not overlap with the RWD domain resonances were not well dispersed and corresponded to random coil chemical shifts, suggesting that the RWDD3 region outside of the RWD domain is disordered. The disordered C terminus of RWDD3 likely contributes to its large hydrodynamic radius, resulting in the broad line widths observed for the RWDD3 resonances.
FIGURE 1.
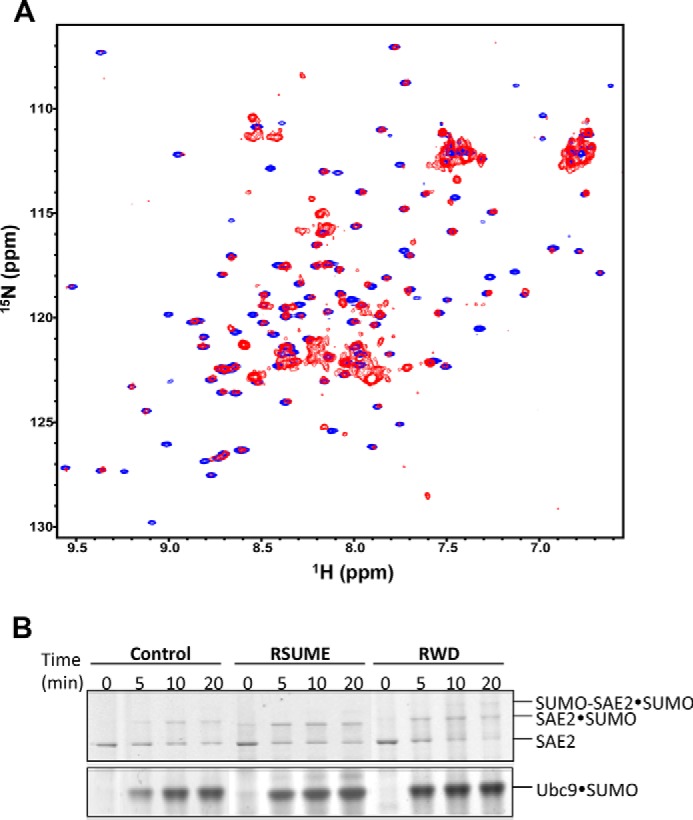
The RWD domain is a well structured region of RWDD3 and confers the same in vitro effects as RWDD3 on SUMOylation. A, superimposed 1H-15N HSQC spectra of full-length RWDD3 (red) and its RWD domain (blue). B, nonreducing SDS-PAGE comparing the enhancement effect of RWDD3 and its RWD domain on formation of the Ubc9·SUMO thioesters, SAE2·SUMO thioester, and SAE2-SUMO isopeptide bonds. Parallel control assays were performed under the same conditions but without RWDD3 or RWD.
Next, we investigated whether the RWD domain is responsible for the reported RWDD3 SUMOylation enhancement effect. To do this, we evaluated the formation rates of the thioester conjugates between SUMO and E2 (Ubc9). Formation of both thioesters is stimulated by RWDD3, as found previously (2). Samples of the conjugation reactions were taken at multiple time points (0, 5, 10, 20, and 30 min) in the presence and absence of RWDD3 or the RWD domain. RWDD3 and the RWD domain similarly stimulated formation of the Ubc9·SUMO thioester conjugate (Fig. 1B). Consistently, they similarly stimulated formation of the SAE2·SUMO thioester and SAE2-SUMO isopeptide (Fig. 1B). Both SAE2 conjugates are dependent on the Ubc9·SUMO thioester. SAE2-SUMO isopeptide conjugate is dependent on the amount of E2·SUMO thioester (21). In addition, the transfer of SUMO from E1 to E2 is reversible, and thus increased Ubc9·SUMO thioester conjugate leads to the increased SAE2·SUMO thioester conjugate. Taken together, the RWD domain is critical for binding Ubc9 and is responsible for the reported in vitro effect on SUMOylationenhancement by the RWDD3 splice variants (22).
The Crystal Structure of Ubc9-RWD Complex and Validation by Solution NMR Studies
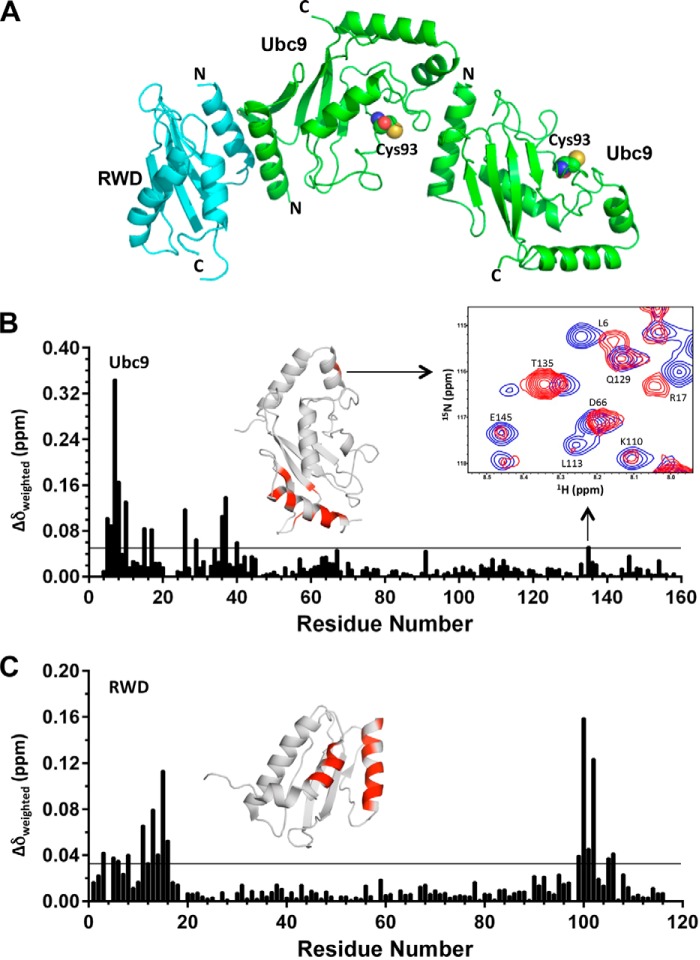
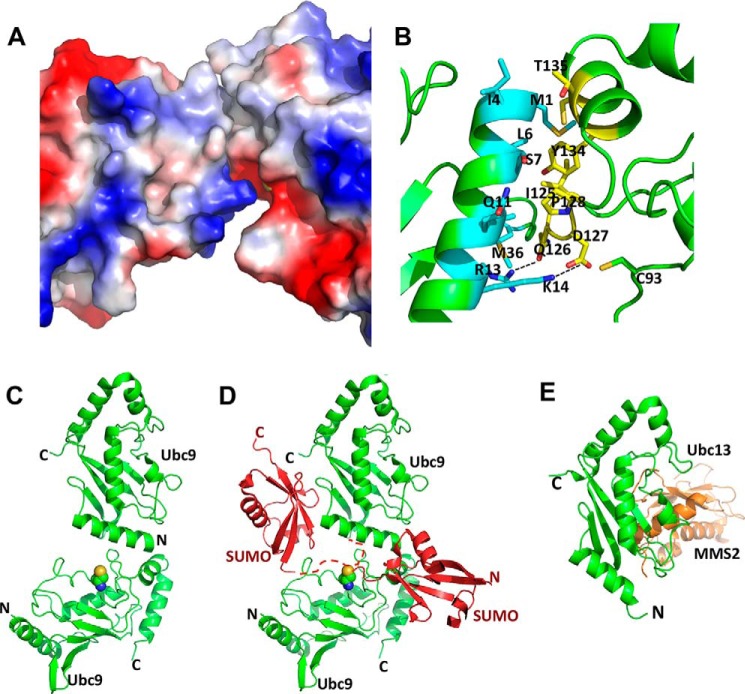
Because the RWD domain had a similar effect on SUMOylation as RWDD3, we used the RWD domain for structural investigations of how it interacts with Ubc9. We obtained crystals of the complex between Ubc9 and the RWD domain of RWDD3 that diffracted to ∼2.7 Å, and the crystal structure was solved by molecular replacement. We observed a heterotrimeric complex with two Ubc9 molecules and one RWD domain in the asymmetric unit (Fig. 2A and Table 1). Through multiple rounds, the structure was built and refined with good overall stereochemical values (Table 1). Based on the model, the N-terminal surface of Ubc9 is independently involved in an interface that forms an asymmetric homodimer with a second Ubc9 and an interface that binds to RWD. The second interface of Ubc9 that participates in the homodimer formation is adjacent to the catalytic Cys-93.
FIGURE 2.
The structure of RWD-Ubc9 complex. A, the overall fold of the crystal structure showing a heterotrimer that contains one RWD and a Ubc9 homodimer (also see Table 1). B and C, plot of CSP upon titration of 15N-labeled Ubc9 with the RWD domain (B) or 15N-labeled RWD domain with Ubc9 (C). The weighted CSP was calculated as Δδweighted = [(ΔδH)2 + (ΔδN/6.7)2]1/2. The horizontal lines (0.05 ppm for Ubc9 and 0.03 ppm for RWD) represent twice the average CSP. The perturbed residues in Ubc9 upon RWD titrations and in RWD upon Ubc9 titrations are indicated in red in ribbon diagrams above their respective CSP versus residue plots.
TABLE 1.
Data collection and processing statistics for the RWD-Ubc9 complex
| Data collection | |
| Space group | P1 21 1 |
| Cell dimensions | |
| a, b, c (Å) | 63.23, 34.86, 114.51 |
| α, β, γ (°) | 90.0, 98.53, 90.0 |
| Resolution (Å) | 33.32-2.77 (2.77-2.70)a |
| Rmeas | 0.112 (1.103) |
| I/σ(I) | 12.8 (1.4) |
| Completeness (%) | 99.5 (100.0) |
| Redundancy | 3.2 (3.3) |
| Refinement | |
| Resolution (Å) | 2.70 |
| No. reflections | 13,981 |
| Rwork/Rfree | 21.6/25.4 |
| No. atoms | |
| Protein | 3310 |
| Water | 56 |
| Root mean square deviation | |
| Bond lengths (Å) | 0.003 |
| Bond angles (°) | 0.705 |
| Ramachandran statistics | |
| Favored | 95.19 |
| Allowed | 3.37 |
| Disallowed | 1.44 |
a Highest resolution shell is shown in parentheses.
The interactions observed in the crystal structure were further validated by NMR CSP. 15N-Labeled Ubc9 was titrated with unlabeled RWD domain, or 15N-labeled RWD domain was titrated with unlabeled Ubc9, and then CSP was monitored using 1H-15N HSQC spectra (Fig. 2, B and C). The weighted CSP upon the complex formation was plotted for each residue of Ubc9 (Fig. 2B) and the RWD domain (Fig. 2C) (excluding Pro and overlapping resonances). Residues 7–10, 26, and 36–37 of Ubc9 exhibited the largest CSPs (Fig. 2B), which are consistent with the RWD-Ubc9 interaction observed in the crystal structure. In addition, CSP on Ubc9 was observed on residues at the homodimer interface near the catalytic Cys-93, i.e. Thr-135. In particular, the resonances of Thr-135 showed increased intensity as well as CSP, despite the fact that complex formation is expected to reduce the overall NMR resonance intensity caused by broadened line widths (Fig. 2B). The broadened resonances in the absence of the RWD domain indicate monomer-oligomer equilibrium, and the sharpened resonance upon addition of the RWD domain suggests its blockage of the N terminus of Ubc9 for Ubc9-Ubc9 dimer formation. For the RWD domain, residues 11–16 and 100–102 showed significant CSP that is consistent with the binding interface observed in the crystal structure (Fig. 2, A and C). Taken together, NMR data verified that the crystal structure exists in solution.
The RWD Domain Binds to a Surface of Ubc9 That Interacts with E1, E3, and SUMO
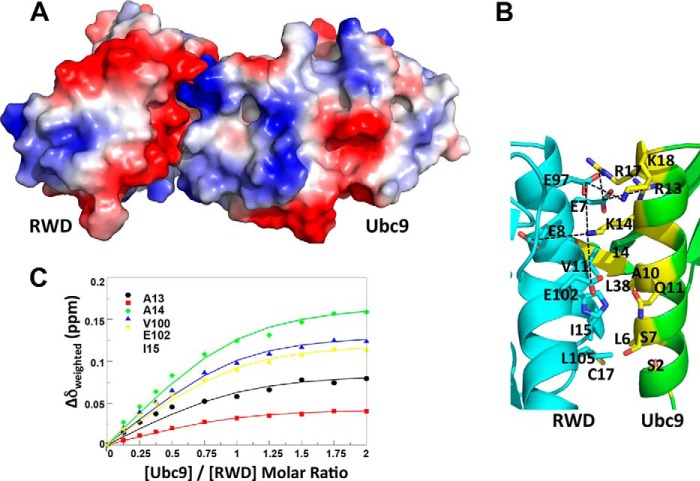
Ubc9 and RWD domain have interfaces that are complementary in both charge and shape (Fig. 3A). The binding interface on Ubc9 has a positive electrostatic potential, whereas the binding interface on RWD has a negative electrostatic potential (Fig. 3A). The interaction is facilitated by charge interactions between Arg-13, Lys-14, Arg-17, and Lys-18 of Ubc9 and Glu-7, Glu-8, Glu-97, and Glu-102 of the RWD domain (Fig. 3B). Hydrophobic and polar residues, such as Leu-6, Ala-10, Thr-35, and Leu-38 of Ubc9 and Val-11, Ile-15, His-101, and Leu-105 of RWD, contribute to the van der Waals contacts (Fig. 3B). The C terminus of the RWD domain faces away from the Ubc9 binding surface (Fig. 3A), and thus the full-length RWDD3, which contains an unstructured C-terminal extension from the RWD domain, likely interacts with Ubc9 in a similar manner as the RWD domain, and the unstructured C-terminal segment is unlikely to affect the RWD-Ubc9 interaction. This is consistent with the similar effects of the RWD and RWDD3 on SUMOylation (Fig. 1B). The dissociation constant (Kd) of the RWD and Ubc9 interaction was estimated as 32 ± 4 μm, using five residues in the RWD domain that displayed the largest chemical shift perturbations (Ala-13, Ala-14, Ile-15, Val-100, and Glu-102) (Fig. 3C).
FIGURE 3.
The binding interface and affinity of the RWD-Ubc9 complex. A, surface representation color-coded by electrostatic potential of the RWD-Ubc9 complex. Red to blue corresponds to negative to positive electrostatic potentials. B, zoomed in view of the RWD-Ubc9 binding interface with key interacting residues shown and labeled. C, estimation of Kd of the complex using CSP of cross-peaks of the indicated residues in 15N-labeled RWD as a function of incremental addition of 0.125 molar equivalent of Ubc9. The Kd of the Ubc9-RWD complex was determined by nonlinear least square fitting of CSP as a function of molar ratios of Ubc9 versus RWD.
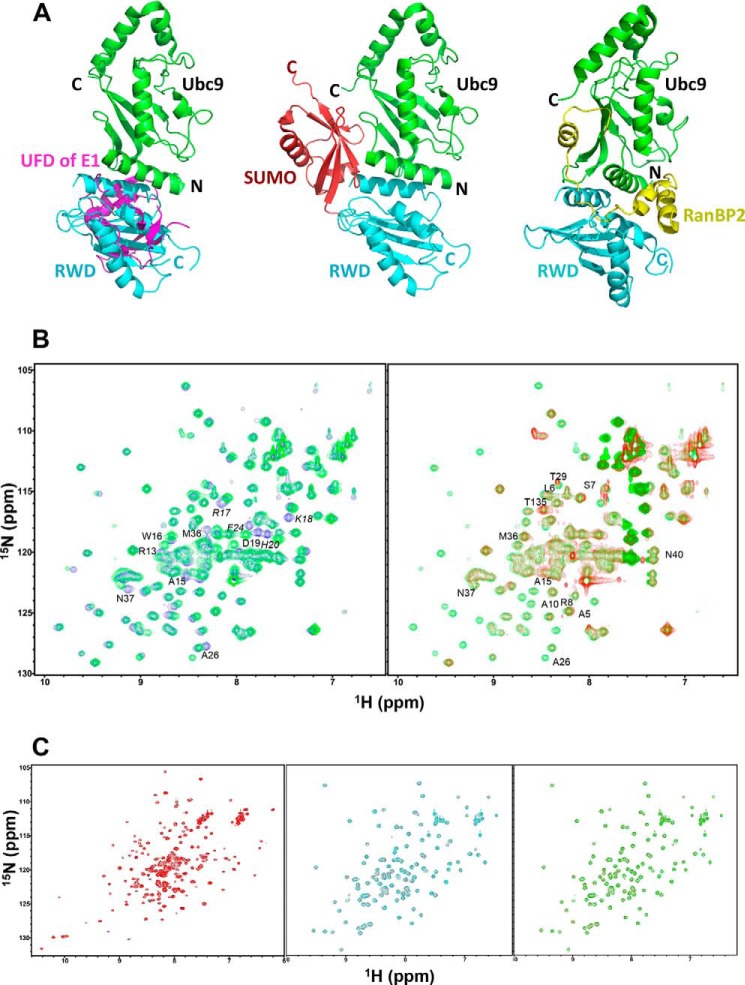
The RWD domain binds to a surface of Ubc9 that is the “hot spot” for interactions with other proteins of the conjugation pathway (E1, SUMO and E3). The RWD-binding surface on Ubc9 overlaps with the surface that binds the UFD of SAE2 (9, 11, 23) (Fig. 4A, left panel). In addition, binding of SUMO to Ubc9 would cause a steric clash with the RWD domain bound to Ubc9 according to available crystal structures (Fig. 4A, middle panel). Consistently, NMR chemical shift perturbation indicated that SUMO and the RWD domain could not simultaneously bind to Ubc9, because addition of the RWD domain to the Ubc9-SUMO complex (Fig. 4B, left panel) did not induce chemical shift changes on Ubc9 residues that are located in the binding interface (Fig. 4B, right panel). No significant chemical shift changes were induced by addition of the RWD domain. The overall resonance intensity reduction upon addition of the RWD domain is due to the increased sample volume and consequently reduced Ubc9 concentration of the sample. Furthermore, the RWD binding surface of Ubc9 overlaps with that of the E3 ligase RanBP2 (Fig, 4A, right panel) (24). The RWD domain does not bind SUMO or E1 (Fig. 4C). The affinity of RWD-Ubc9 interaction (Kd = 32 ± 4 μm; Fig. 3C) is 10–100-fold lower than that of E1-E2, E2-SUMO, and E2-E3 interactions (15, 23). The finding that the RWD domain and SUMO bind to the same region of Ubc9 is inconsistent with the previously proposed model that RWDD3 promotes SUMOylation by promoting noncovalent interaction between Ubc9 and SUMO (2).
FIGURE 4.
The RWD-binding surface on Ubc9 overlaps with the binding surfaces of the UFD of E1, SUMO, and RanBP2 (a SUMO E3 ligase). A, left panel, overlay of Ubc9 in the predicted UFD-Ubc9 complex and the Ubc9-RWD complex. The RWD domain fully overlaps with the UFD domain, indicating their competition for Ubc9. Middle panel, conserved binding site of the ubiquitin-like modifier (SUMO, red) in relation to that of the RWD (cyan) on an E2 (Ubc9, green). Right panel, E3 ligase RanBP2 (yellow) binding site in relation to that of the RWD (cyan) on Ubc9 (green). B, RWD and SUMO-1 do not bind to Ubc9 simultaneously. Left panel, overlay of the 1H-15N HSQC spectra 15N-Ubc9, free (blue) and in complex with 1:1 stoichiometry of unlabeled SUMO-1 (green). Right panel, overlay of the 1H-15N HSQC spectra of the Ubc9-SUMO complex in A (green) and that of the complex with addition of 1:1 stoichiometry of the RWD domain (red). Residues at the protein-protein binding interfaces are indicated. Residues whose resonances disappeared upon complex formation are indicated in italics. C, the RWD domain does not interact with SAE and SUMO. Left panel, overlay of the 1H-15N HSQC spectra of the 15N-labeled Cys domain of SAE, free (black) and in complex with 1.2-fold molar excess of unlabeled RWD domain (red). Middle panel, overlay of the 1H-15N HSQC spectra of the15N-labeled RWD, free (red) and in complex with 1.2 molar excess of unlabeled UFD domain of SAE (cyan). Right panel, overlay of the 1H-15N HSQC spectra of the15N-labeled RWD domain, free (red) and in complex with 1.2 molar excess of unlabeled SUMO-1 (green). No significant chemical shift perturbation was seen upon the formation of all of these complexes.
Effect of the RWD Domain on SUMOylation
We investigated the effect of RWD on enhancing the formation of Ubc9·SUMO-1 thioester conjugates (Fig. 1B), which is consistent with the previous report (2). The structural data have shown that RWD and the UFD domain of E1 bind to the same surface of Ubc9, although the binding affinity of RWD to Ubc9 is 10–100-fold lower than that of UFD binding to Ubc9 (15, 23). Therefore, RWD could reduce the E1-E2 interaction affinity at much higher concentrations than E1. The competition between RWD and E1 for the same surface of Ubc9 suggests that an apparent enhancement of SUMOylation (Fig. 1B) is likely due to the alleviation of the inhibitory effect of E2 on E1 (9).
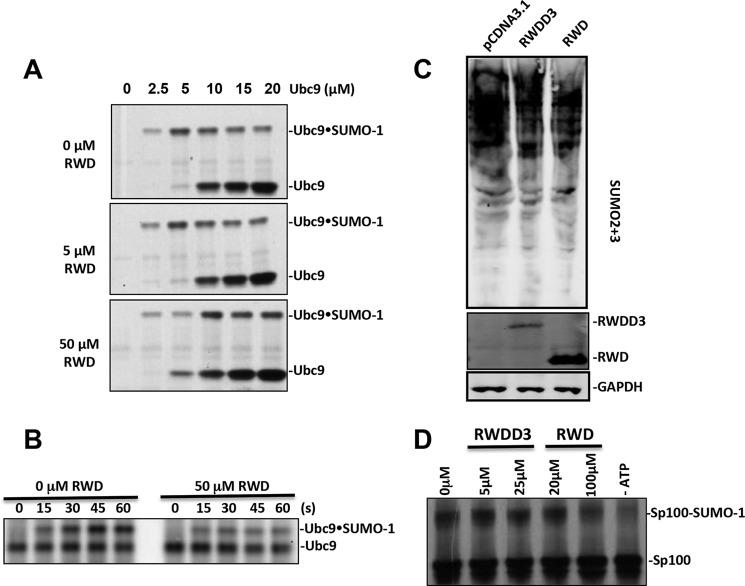
The formation of SUMO·Ubc9 thioester was inhibited by increasing concentrations of Ubc9 caused by the inhibitory effect of the E2 (Fig. 5A) as described previously (9). Under this experimental condition, the SUMO·Ubc9 thioester formation increased with increasing Ubc9 concentrations to 5 μm and then decreased with a further increase in Ubc9 concentrations because of the inhibition of E2 on E1 catalysis (9). The addition of 5 μm of the RWD domain had no effect on this assay, consistent with its low affinity for Ubc9. However, at 50 μm of the RWD domain, the formation of SUMO·Ubc9 thioester is increased by increasing concentrations of Ubc9 to 10 μm. At 5 μm Ubc9 concentration, 50 μm RWD domain had an inhibitory effect on the formation of SUMO·Ubc9 thioester, consistent with the reduced the efficiency of transferring SUMO from E1 to E2 (9). However, at 10 μm or higher Ubc9 concentrations, the RWD domain enhanced the efficiency of SUMO·Ubc9 thioester formation. These data suggest that the RWD domain alleviates the inhibitory effect of Ubc9 on SUMO E1 catalysis, indicating reduced binding affinity of the E2 for the E1 in the presence of 50 μm RWD domain. Experiments that directly measure the transfer of SUMO from E1 to E2 confirmed that the 50 μm RWD domain had an inhibitory effect on the transfer of SUMO from E1 to E2, consistent with inhibited E1-E2 interaction (Fig. 5B).
FIGURE 5.
The effect of RWDD3 and its RWD domain in SUMOylation. A, the effect of various concentrations of the RWD domain on the formation of Ubc9·SUMO thioester conjugate at the indicated Ubc9 concentrations. B, the transfer of SUMO from SAE2 to Ubc9 at the indicated time after the addition of Ubc9 in the presence and absence of 50 μm RWD. C, HEK293T cells were transfected with pCDNA3-RWD, pCDNA3-RWDD3, and pCDNA3.1, respectively. After 48 h of DNA transfection, poly-SUMOylation (SUMO-2 and -3), GAPDH, Flag tag RWDD3, and its RWD domain expressions were detected by Western blot. GAPDH expression was used as the loading control. D, RWDD3 and the RWD domain inhibited SUMO conjugation of the protein Sp100 using 35S-labeled Sp100 obtained from in vitro transcription and translation and detected by autoradiography.
We also investigated the effect of the RWD domain and RWDD3 in cells. An equal amount of RWD or RWDD3 expression plasmids was transfected into 293T cells, and the empty vector pCDNA3.1 was transfected as a control. Expression of RWDD3 appeared to be less than that of its RWD domain. In contrast to the previous report (2), we could not observe an enhancement effect of either RWDD3 or its RWD domain on global SUMOylation levels (Fig. 5C). In fact, some inhibition of global SUMOylation was indicated by the cellular assays. This is consistent with the inhibitory effect of RWDD3 and RWD domain in vitro SUMOylation assays. Consistent with this finding, we observed inhibition of overall SUMOylation at when the concentrations of Ubc9 did not reach inhibitory concentrations (Fig. 5D).
The Ubc9 Homodimer
The observation of a Ubc9 homodimer in the crystal structure (Fig. 2A) is consistent with a previous NMR study that indicated that Ubc9 is not a pure monomer in solution (25). The estimated rotational correlation time τm of Ubc9 at 0.7 mm was 14.02 ± 0.06 ns, which is larger than expected, given the size and shape of Ubc9. At a lower concentration (0.3 mm), the estimated rotational correlation time τm was reduced to 12.03 ± 0.13 ns. The concentration-dependent reduction of τm indicated that Ubc9 undergoes oligomerization in solution. Additionally, the systematically increased order parameter (S2) is also indicative of self-association. The dissociation constant, estimated from the rotational correlation times at two concentrations, based on hydrodynamic principle (26), is ∼1.2 mm (25). The low affinity of the Ubc9 homodimer is consistent with the interface of the Ubc9 homodimer, which buries 405 Å2 of surface area.
The homodimer of Ubc9 involves interfaces that have complementary electrostatic potential (Fig. 6A) and is facilitated by charge interactions between Lys-14 of one Ubc9 molecule and Asp-127 of the other Ubc9 molecule (Fig. 6B). The side chain oxygen of Gln-126 and the guanidinium group of Arg-13 appear to form a hydrogen bond. In addition, there are hydrophobic contacts between Ile-125, Pro-128, Tyr-134, Thr-135, and Cys-138 of one Ubc9 monomer and Met-1, Ile-4, Leu-6, Ser-7, Ala-10, Gln-11, Met-36, and Leu-38 of the other Ubc9 molecule. The binding interface on one Ubc9 molecule has a positive electrostatic potential because of Arg-13 and Lys-14, whereas the binding interface on the other Ubc9 molecule has a negative electrostatic potential because of residue Asp-127.
FIGURE 6.
The Ubc9 homodimer interfaces. A, the Ubc9 homodimer interface is shown with the surfaces color-coded according to electrostatic potentials; red to blue corresponds to negative to positive electrostatic potentials. B, zoomed in view of the Ubc9 homodimer interface with key interacting residues shown and labeled. One Ubc9 molecule is shown in cyan, and the other is in yellow. C, ribbon diagram of the Ubc9 homodimer. D, a model of how Ubc9 homodimer could stimulate SUMO chain formation. The structure of noncovalent Ubc9-SUMO complex is superimposed onto the top Ubc9 molecule of the Ubc9 homodimer, resulting in the position of the SUMO molecule on the upper left side. The dashed red line represents the flexible N-terminal segment of SUMO that contains the SUMOylation site but did not have electron density in x-ray diffraction. Ubc9 is shown in green, and SUMO is shown in red. Side chain atoms of the catalytic Cys-93 of the bottom Ubc9 are shown as spheres. A hypothetical SUMO molecule that forms a thioester conjugate with the bottom Ubc9 shown on the lower right side. E, ribbon diagram of Ubc13/MMS2 complex in which Ubc13 is in a similar orientation as the top Ubc9 molecule in C.
The Ubc9 homodimer observed in this study could fill the missing gap in the molecular mechanism of poly-SUMO chain formation. It was shown previously that the noncovalent interaction between Ubc9 and SUMO was important for poly-SUMO chain formation, but for this interaction to lead to poly-SUMOylation, Ubc9 needs to form a homodimer (27). The Ubc9 homodimer observed in this study provides a logical mechanism for the poly-SUMO chain formation. The crystal structure of the SUMO-Ubc9 complex can be superimposed on to the crystal structure of Ubc9 homodimer observed in this study by superimposing Ubc9 in the SUMO-Ubc9 complex to the top Ubc9 molecule in the asymmetric homodimer (Fig. 6D). There is no steric clash between SUMO and the bottom Ubc9 molecule (Fig. 6D). The SUMOylation consensus sequence in both SUMO-2 and -3 is VKTE at the flexible N-terminal region, where Lys is the poly-SUMO linkage site. A SUMO-2 or SUMO-3 molecule that is noncovalently bound to the top Ubc9 molecule is positioned with its SUMOylation consensus sequence poised to interact with the substrate binding surface near the catalytic Cys-93 of the bottom Ubc9 molecule (28) (Fig. 6D, dashed line). Simultaneous interactions of one SUMO-2 or -3 with two Ubc9 molecules in a homodimer should further stabilize the ternary complex. The bottom Ubc9 molecule is modeled with a thioester-conjugated SUMO molecule (lower right side) that could easily form an isopeptide bond with the SUMO molecule that is noncovalently bonded to the top Ubc9 molecule (upper left side). Although the Ubc9-SUMO complex was solved with SUMO-1 (27), Ubc9 interaction with SUMO-1 and that with SUMO-2 or -3 are nearly identical (15).
The homodimer of Ubc9 is formed through the two protein-protein interaction hot spots on Ubc9: the N-terminal region that interacts with UFD domain of E1, E2, SUMO, and E3 (Fig. 4A) and the region near the catalytic Cys that interacts with the Cys domain of E1 and substrates (10, 28). The low affinity of the homodimer and overlapping interfaces with different members of the SUMOylation machinery make it challenging to specifically investigate the effects of the Ubc9 homodimer on poly-SUMO formation. Our finding will help to formulate future studies on the functions of the Ubc9 homodimer.
Discussion
We have shown for the first time how an RWD domain interacts with an E2 in ubiquitin-like modifications at the atomic level. Sequence alignment of the RWD domains indicates that the residues at the Ubc9 binding surface of RWDD3 are not well conserved among the RWD members, and thus, it is unlikely that the other RWD domains can bind Ubc9 (29). However, it is possible that some of these RWD domains bind other E2s in a manner similar to that described in this study. The RWD-binding surface on E2 does not appear to overlap with the RING-domain binding surface on an E2. Therefore, it is possible that an RWD domain in a RING-containing E3 ligase may participate in binding E2 by providing an additional binding site for E2. On the other hand, an RWD-containing protein could inhibit ubiquitin-like modifications by inhibiting E1-E2 interaction based on the same binding surface of the RWD domain and E1 on E2. The later scenario is consistent with the effects of RWDD3 and its RWD domain in cells (Fig. 5).
Although the structure of RWD resembles the fold of an E2, the Ubc9-RWD binding mode differs from that of the only other known complex between an E2 and an E2 homologue, the Ubc13 and MMS2 complex (Fig. 6E) (30). Ubc9 binds to RWD by using a surface located near the N terminus, whereas MMS2 binds to Ubc13 on its main β-sheet. In addition, the mode of the observed Ubc9 homodimer (Fig. 6C) is different from that of Ubc13-MMS2 complex (30) (Fig. 6E). The top Ubc9 molecule is shown with a similar orientation as Ubc13 (Fig. 6, C and E). These results indicate that the mode of complex formation by between E2s and/or E2 homologues is not conserved.
The RWD domain binds to a surface of Ubc9 that is a protein-protein interaction hot spot involved in binding all of the key proteins in the conjugation machinery, including SUMO, E1, and E3 (Fig. 4A) (9, 11, 12, 24, 27). Future structural studies would provide an understanding of how such protein-protein interaction hot spots interact with various proteins that lack structural similarity. The information provided in this study form the basis for these future investigations.
Acknowledgments
We thank Ning Zheng at University of Washington for help with crystallography and crystal analysis and for helpful comments on the manuscript. The NMR and x-Ray core facilities are supported in part by the City of Hope Comprehensive Cancer Center. We are grateful to Drs. Sumio Sugano and Shigeyuki Yokoyama at RIKEN Yokohama Institute in Japan for providing the expression plasmids of the RWD domains of human RWDD1, RWDD2, RWDD3, C21orf6, and Ring finger protein 25, as well as the expression plasmid of the extended conformation of the RWD domain of human Ring finger protein 25.
This work was supported, in whole or in part, by National Institutes of Health Grants GM102538 and GM086171. The authors declare that they have no conflicts of interest with the contents of this article.
- SUMO
- small ubiquitin-like modifier
- UFD
- ubiquitin fold domain
- HSQC
- heteronuclear single quantum coherence
- CSP
- chemical shift perturbation.
References
- 1. Doerks T., Copley R. R., Schultz J., Ponting C. P., Bork P. (2002) Systematic identification of novel protein domain families associated with nuclear functions. Genome Res. 12, 47–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Carbia-Nagashima A., Gerez J., Perez-Castro C., Paez-Pereda M., Silberstein S., Stalla G. K., Holsboer F., Arzt E. (2007) RSUME, a small RWD-containing protein, enhances SUMO conjugation and stabilizes HIF-1alpha during hypoxia. Cell 131, 309–323 [DOI] [PubMed] [Google Scholar]
- 3. Yeh E. T. (2009) SUMOylation and De-SUMOylation: wrestling with life's processes. J. Biol. Chem. 284, 8223–8227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ulrich H. D. (2009) The SUMO system: an overview. Methods Mol. Biol. 497, 3–16 [DOI] [PubMed] [Google Scholar]
- 5. Chen Y. (2007) The enzymes in ubiquitin-like post-translational modifications. Biosci. Trends 1, 16–25 [PMC free article] [PubMed] [Google Scholar]
- 6. Dye B. T., Schulman B. A. (2007) Structural mechanisms underlying posttranslational modification by ubiquitin-like proteins. Annu. Rev. Biophys. Biomol. Struct. 36, 131–150 [DOI] [PubMed] [Google Scholar]
- 7. Yasugi T., Howley P. M. (1996) Identification of the structural and functional human homolog of the yeast ubiquitin conjugating enzyme UBC9. Nucleic Acids Res. 24, 2005–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dohmen R. J., Stappen R., McGrath J. P., Forrová H., Kolarov J., Goffeau A., Varshavsky A. (1995) An essential yeast gene encoding a homolog of ubiquitin-activating enzyme. J. Biol. Chem. 270, 18099–18109 [DOI] [PubMed] [Google Scholar]
- 9. Wang J., Cai S., Chen Y. (2010) Mechanism of E1-E2 interaction for the inhibition of Ubl adenylation. J. Biol. Chem. 285, 33457–33462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang J., Hu W., Cai S., Lee B., Song J., Chen Y. (2007) The intrinsic affinity between E2 and the Cys domain of E1 in ubiquitin-like modifications. Mol. Cell 27, 228–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang J., Lee B., Cai S., Fukui L., Hu W., Chen Y. (2009) Conformational transition associated with E1-E2 interaction in small ubiquitin-like modifications. J. Biol. Chem. 284, 20340–20348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huang D. T., Hunt H. W., Zhuang M., Ohi M. D., Holton J. M., Schulman B. A. (2007) Basis for a ubiquitin-like protein thioester switch toggling E1-E2 affinity. Nature 445, 394–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Walden H., Podgorski M. S., Huang D. T., Miller D. W., Howard R. J., Minor D. L., Jr., Holton J. M., Schulman B. A. (2003) The structure of the APPBP1-UBA3-NEDD8-ATP complex reveals the basis for selective ubiquitin-like protein activation by an E1. Mol. Cell 12, 1427–1437 [DOI] [PubMed] [Google Scholar]
- 14. Liu Q., Jin C., Liao X., Shen Z., Chen D. J., Chen Y. (1999) The binding interface between an E2 (UBC9) and a ubiquitin homologue (UBL1). J. Biol. Chem. 274, 16979–16987 [DOI] [PubMed] [Google Scholar]
- 15. Tatham M. H., Kim S., Yu B., Jaffray E., Song J., Zheng J., Rodriguez M. S., Hay R. T., Chen Y. (2003) Role of an N-terminal site of Ubc9 in SUMO-1, -2, and -3 binding and conjugation. Biochemistry 42, 9959–9969 [DOI] [PubMed] [Google Scholar]
- 16. Tatham M. H., Kim S., Jaffray E., Song J., Chen Y., Hay R. T. (2005) Unique binding interactions among Ubc9, SUMO and RanBP2 reveal a mechanism for SUMO paralog selection. Nat. Struct. Mol. Biol. 12, 67–74 [DOI] [PubMed] [Google Scholar]
- 17. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Afonine P. V., Grosse-Kunstleve R. W., Echols N., Headd J. J., Moriarty N. W., Mustyakimov M., Terwilliger T. C., Urzhumtsev A., Zwart P. H., Adams P. D. (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 21. Truong K., Lee T. D., Chen Y. (2012) Small ubiquitin-like modifier (SUMO) modification of E1 Cys domain inhibits E1 Cys domain enzymatic activity. J. Biol. Chem. 287, 15154–15163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gerez J., Fuertes M., Tedesco L., Silberstein S., Sevlever G., Paez-Pereda M., Holsboer F., Turjanski A. G., Arzt E. (2013) In silico structural and functional characterization of the RSUME splice variants. PLoS One 8, e57795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huang D. T., Paydar A., Zhuang M., Waddell M. B., Holton J. M., Schulman B. A. (2005) Structural basis for recruitment of Ubc12 by an E2 binding domain in NEDD8's E1. Mol. Cell 17, 341–350 [DOI] [PubMed] [Google Scholar]
- 24. Reverter D., Lima C. D. (2005) Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature 435, 687–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu Q., Yuan Y. C., Shen B., Chen D. J., Chen Y. (1999) Conformational flexibility of a ubiquitin conjugation enzyme (E2). Biochemistry 38, 1415–1425 [DOI] [PubMed] [Google Scholar]
- 26. Garciade la Torre J. G., Bloomfield V. A. (1981) Hydrodynamic properties of complex, rigid, biological macromolecules: theory and applications. Q. Rev. Biophys. 14, 81–139 [DOI] [PubMed] [Google Scholar]
- 27. Knipscheer P., van Dijk W. J., Olsen J. V., Mann M., Sixma T. K. (2007) Noncovalent interaction between Ubc9 and SUMO promotes SUMO chain formation. EMBO J. 26, 2797–2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lin D., Tatham M. H., Yu B., Kim S., Hay R. T., Chen Y. (2002) Identification of a substrate recognition site on Ubc9. J. Biol. Chem. 277, 21740–21748 [DOI] [PubMed] [Google Scholar]
- 29. Nameki N., Yoneyama M., Koshiba S., Tochio N., Inoue M., Seki E., Matsuda T., Tomo Y., Harada T., Saito K., Kobayashi N., Yabuki T., Aoki M., Nunokawa E., Matsuda N., Sakagami N., Terada T., Shirouzu M., Yoshida M., Hirota H., Osanai T., Tanaka A., Arakawa T., Carninci P., Kawai J., Hayashizaki Y., Kinoshita K., Güntert P., Kigawa T., Yokoyama S. (2004) Solution structure of the RWD domain of the mouse GCN2 protein. Protein Sci. 13, 2089–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. VanDemark A. P., Hofmann R. M., Tsui C., Pickart C. M., Wolberger C. (2001) Molecular insights into polyubiquitin chain assembly: crystal structure of the Mms2/Ubc13 heterodimer. Cell 105, 711–720 [DOI] [PubMed] [Google Scholar]