Background: The largest N-glycan in plants is the paucimannosidic N-glycan with Man3XylFuc(GlcNAc)2 structure.
Results: A sophisticated mechanism producing the largest N-glycan in plants is proposed.
Conclusion: Limited addition of the 6-arm GlcNAc to the common N-glycan acceptor (GlcNAcMan3(GlcNAc)2) facilitates formation of the largest N-glycan in plants.
Significance: This sophisticated mechanism expands our knowledge of the energy-efficient N-glycan processing in plants.
Keywords: carbohydrate processing, glycosylation, glycosyltransferase, plant, post-translational modification (PTM)
Abstract
The most abundant N-glycan in plants is the paucimannosidic N-glycan with core β1,2-xylose and α1,3-fucose residues (Man3XylFuc(GlcNAc)2). Here, we report a mechanism in Arabidopsis thaliana that efficiently produces the largest N-glycan in plants. Genetic and biochemical evidence indicates that the addition of the 6-arm β1,2-GlcNAc residue by N-acetylglucosaminyltransferase II (GnTII) is less effective than additions of the core β1,2-xylose and α1,3-fucose residues by XylT, FucTA, and FucTB in Arabidopsis. Furthermore, analysis of gnt2 mutant and 35S:GnTII transgenic plants shows that the addition of the 6-arm non-reducing GlcNAc residue to the common N-glycan acceptor GlcNAcMan3(GlcNAc)2 inhibits additions of the core β1,2-xylose and α1,3-fucose residues. Our findings indicate that plants limit the rate of the addition of the 6-arm GlcNAc residue to the common N-glycan acceptor as a mechanism to facilitate formation of the prevalent N-glycans with Man3XylFuc(GlcNAc)2 and (GlcNAc)2Man3XylFuc(GlcNAc)2 structures.
Introduction
N-Glycosylation is an important co- and post-translational modification affecting the physicochemical properties of proteins in eukaryotic cells (1, 2). N-Glycosylation influences a wide range of biological processes including protein folding (3–5), activity (6), intracellular trafficking (7, 8), secretion (9), and cell-cell communication (10). The early steps in the N-glycosylation pathway, which take place in the ER4 and form the oligomannosidic structure, are highly conserved across all eukaryotic organisms (11). By contrast, the later steps that form complex N-glycans in the Golgi apparatus diverge significantly among different species and kingdoms (12). The major N-glycan (Man3XylFuc(GlcNAc)2) found in plants lacks β1,2-linked N-acetylglucosamine (GlcNAc), galactose (13, 14), and N-acetylneuraminic acid (sialic acid) residues at the non-reducing ends, and thus the complex N-glycans in plants are generally smaller than their mammalian counterparts (Fig. 1A). The predominant N-glycan in plants is paucimannosidic N-glycan with core β1,2-xylose and α1,3-fucose residues (PNGXF) at the β-linked mannose of the trimannosyl core and proximal GlcNAc residues, respectively (13, 14). PNGXF also represents the major class of cross-reactive carbohydrate determinant recognized by immunoglobulin E (IgE) antibodies in the sera of allergic patients (2, 15–17). The allergenic potential of PNGXF is a major limitation to clinical applications of recombinant glycoproteins produced in plants for human use. In addition, some biopharmaceuticals require relatively larger N-glycan structures with galactose and sialic acid residues at non-reducing termini for their efficient delivery and/or extended circulation half-life (18–20). The structure of PNGXF is therefore not compatible with those biopharmaceuticals. These considerations mean that to design plants producing N-glycan structures suitable for biopharmaceuticals it is first necessary to understand the molecular genetic mechanisms that create PNGXF in plants.
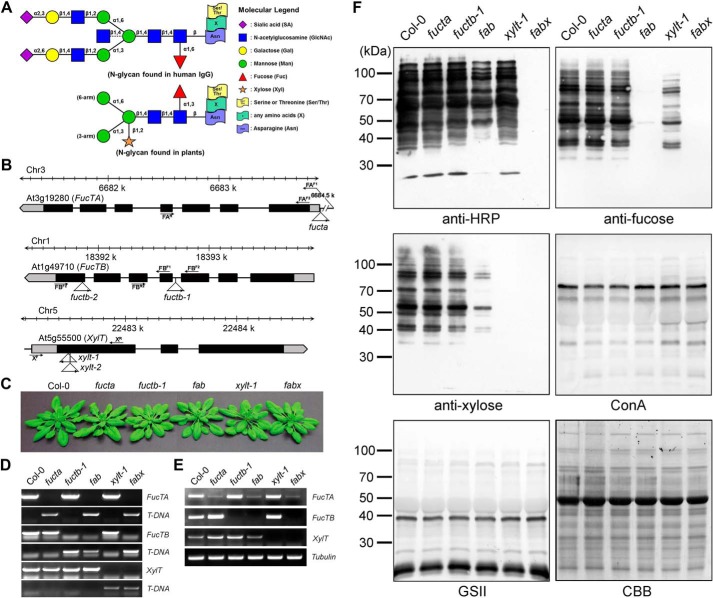
FIGURE 1.
Antibodies show different binding affinities for the proteins extracted from Col-0, fucta, fuctb-1, fab, xylt-1, and fabx. A, schematic representation of the largest N-glycan structure found in human IgG and plants. The largest N-glycan found in human IgG contains a core fucose and is often terminated with a sialic acid. The bisecting arm of the GlcNAc indicated with a dashed line represents around 10% of human IgG glycoforms. PNGXF at the β-linked mannose of the trimannosyl core and proximal GlcNAc residues, respectively, is the predominant N-glycan in plants. B, schematic representation of the T-DNA insertion sites in the FucTA, FucTB, and XylT genes. Boxes represent exons, and black denotes the coding region. T-DNA insertion sites and left border directions are indicated with triangles and arrowheads. PCR primers used in genotyping are indicated by arrows. C, phenotypes of the mutant plants compared with that of Col-0. Plants were grown on soil for 30 days. fucta, fuctb-1, and xylt-1 single mutants were crossed successively to produce fab and fabx. D, PCR-based genotyping of the mutants. Gene-specific primers were designed and used in combination with T-DNA left border primer. E, RT-PCR analysis of the mutants. Levels of transcripts in Col-0 and indicated mutants were determined by RT-PCR with gene-specific primers. Tubulin was used as a control. F, total proteins extracted from 3-week-old seedlings were subjected to immunoblot and lectin blot analyses. The immunoblots were probed with anti-HRP, anti-xylose, and anti-fucose antibodies, and lectin blots were probed with ConA and GSII. Coomassie Brilliant Blue (CBB) staining was used to show equal loading of the proteins.
N-Acetylglucosaminyltransferase I (GnTI) converts N-glycan Man5(GlcNAc)2 structure to GlcNAcMan5(GlcNAc)2 (21, 22). Mutant plants lacking GnTI activity cannot produce hybrid and complex-type N-glycans (23–25). The addition of β1,2-linked GlcNAc to Man5(GlcNAc)2 by GnTI is essential for the action of the subsequent N-glycan-processing enzyme Golgi α-mannosidase II (26–28). The GlcNAcMan3(GlcNAc)2 structure produced by Golgi α-mannosidase II can be used as a common acceptor for β1,2-xylose, α1,3-fucose, and β1,2-GlcNAc residues by the activities of β1,2-xylosyltransferase (XylT), α1,3-fucosyltransferase (FucTA and FucTB in Arabidopsis), and GnTII, respectively (29). It has not yet been elucidated how the addition of each sugar residue affects that of the others and how XylT, FucTA, FucTB, and GnTII share the common acceptor (GlcNAcMan3(GlcNAc)2). β-N-Acetylhexosaminidase 1 (HEXOI) and β-N-acetylhexosaminidase 3 (HEXOIII), residing in different subcellular compartments, are known to be involved in the formation of PNGXF in Arabidopsis thaliana (14). However, why plants retain an energy-inefficient process involving the addition and removal of the non-reducing GlcNAc residues to produce PNGXF is still not fully understood.
We hypothesized that additions of the core β1,2-xylose, α1,3-fucose, and 6-arm β1,2-GlcNAc residues to the common acceptor (GlcNAcMan3(GlcNAc)2) (17, 28, 30–34) are relatively determined by the different activity and/or substrate occupancy of the enzymes of the corresponding genes in plants. To examine this hypothesis, artificial in vivo N-glycosylation conditions were created by genetic combination of Arabidopsis T-DNA insertion mutants lacking XylT, FucTA, FucTB, asparagine-linked glycosylation 3 (ALG3; an ER-resident α1,3-mannosyltransferase), GnTI, and GnTII activities. Structural and quantitative analyses of the N-glycans in the mutants and Col-0 were conducted through immunoblotting, lectin blotting, and mass spectrometry. Here we present evidence that limited addition of the 6-arm β1,2-GlcNAc residue by lower activity and/or substrate occupancy of GnTII is involved in the formation of the largest Man3XylFuc(GlcNAc)2 N-glycan structure in A. thaliana.
Experimental Procedures
Plant Material
A. thaliana ecotype Columbia (Col-0) and mutant plants were grown in a growth chamber at 22 °C under long day conditions (16/8-h light/dark photoperiod, 100–200 μmol m−2 s−1 photon flux density, and 60–70% relative humidity) on soil or on 1× Murashige and Skoog medium (Duchefa), pH 5.8 supplemented with 3% sucrose and 0.25% gellan gum (PhytoTechnology Laboratories). All seeds were cold-treated for 4 days in the dark before incubation at 22 °C. Nicotiana benthamiana plants were grown in a growth chamber at 24 °C with a 16/8-h light/dark photoperiod on soil. Five- to 6-week-old plants were used for agroinfiltration experiments.
Identification of T-DNA Mutants
Seeds of Arabidopsis T-DNA insertion lines fucta (Salk_087481), fuctb-1 (CS468139), xylt-1 (CS875073), alg3 (Salk_064006), and cgl1-T (Salk_073650) were obtained from the Arabidopsis Biological Resource Center, and seed of the gnt2 (Flag_394A11) line was obtained from the Versailles Arabidopsis stock center at the Jean-Pierre Bourgin Institute of the National Institute for Agricultural Research. Homozygous mutants were crossed and allowed to self-pollinate in the F1 generation. Double and triple mutants were analyzed in the F2 generation. Genomic DNA was extracted from leaf tissue using phenol-chloroform. PCR with each combination of specific primers was used to verify the insertion site and homozygosity of the T-DNA. Genotyping primers are listed in Table 1.
TABLE 1.
Primers used in this study
| Primer name | Sequence (5′ to 3′) | Use |
|---|---|---|
| FAF1 | 5′-GAGGAGGCAAAAATTACATGTATATGCTCATCC-3′ | Genotyping |
| FAF2 | 5′-ATGGGTGTTTTCTCCAATCTTCGAGGT-3′ | RT-PCR |
| FAR | 5′-CAGCGACTAGAGATTGGAAGAACTTCTCTGTG-3′ | Genotyping and RT-PCR |
| FBF1 | 5′-TAGTTGACAAGGTTGAAGCTCTTAAGCGA-3′ | Genotyping and RT-PCR |
| FBF2 | 5′-TGTCTCCGGTACAGCCAAAAACTGAGAG-3′ | Genotyping and RT-PCR |
| FBR1 | 5′-TCCAGAGGAATGAAACTAGACGAAGACAACCT-3′ | Genotyping and RT-PCR |
| FBR2 | 5′-AAGCAGCAGGGTTAGCTGCGAGATACTT-3′ | Genotyping and RT-PCR |
| XF | 5′-CACAGAGAGGAATGATGGAATCTTCAGCTT-3′ | Genotyping and RT-PCR |
| XR | 5′-ATTCAACATCTCATCATTCACCAGCCG-3′ | Genotyping and RT-PCR |
| ALG3F | 5′-AGAGGATACATTCAATCTTTGTTCTCCG-3′ | Genotyping and RT-PCR |
| ALG3R1 | 5′-TGCATAATCAAAACTATAATTCGCTGGA-3′ | Genotyping |
| ALG3R2 | 5′-AGAAACGGCAGTCCCACGAGTAT-3′ | RT-PCR |
| CGL1F | 5′-AGGCTGCAGCTAGTCTCATGGA-3′ | Genotyping and RT-PCR |
| CGL1R1 | 5′-GTCATATGCAGCGAACATAGGTCA-3′ | Genotyping |
| CGL1R2 | 5′-TGCATCAGGAATTTCGAATTCC-3′ | RT-PCR |
| GnTIIF1 | 5′-ATGGCAAATCTTTGGAAGAAGCAGA-3′ | RT-PCR |
| GnTIIF2 | 5′-GGTGGATGATGAACACTGTATGGGATGG-3′ | Genotyping |
| GnTIIR | 5′-TCATGGAGATGCACTGCTACTGCTGTAAC-3′ | Genotyping and RT-PCR |
| pDonrF | 5′-TTTATAATGCCAACTTTGTACAAAAAAGCAGGCT-3′ | Localization and BiFC |
| pDonrR | 5′-TCTTATAATGCCAACTTTGTACAAGAAAGCTGGGT-3′ | Localization and BiFC |
| FAF3 | 5′-GTACAAAAAAGCAGGCTTAATGGGTGTTTTCTC-3′ | Localization and BiFC |
| FAR2 | 5′-GTACAAGAAAGCTGGGTAGACAAAGACAACTTCG-3′ | Localization and BiFC |
| FBF3 | 5′-GTACAAAAAAGCAGGCTTAATGGGTGTTTTCTC-3′ | Localization and BiFC |
| FBR3 | 5′-GTACAAGAAAGCTGGGTAGACGAAGACAACCTC-3′ | Localization and BiFC |
| XF2 | 5′-GTACAAAAAAGCAGGCTAGATGAGTAAACGGAATCCG-3′ | Localization and BiFC |
| XR2 | 5′-GTACAAGAAAGCTGGGTAGCAGCCAAGGCTCTT-3′ | Localization and BiFC |
| GnTIIF3 | 5′-GTACAAAAAAGCAGGCTTCATGGCAAATCTTTG-3′ | Localization and BiFC |
| GnTIIR2 | 5′-GTACAAGAAAGCTGGGTATGGAGATGCACTGCTAC-3′ | Localization and BiFC |
| LBSalk | 5′-GCGTGGACCGCTTGCTGCAACT-3′ | Genotyping |
| LBSail | 5′-GCCTTTTCAGAAATGGATAAATAGCCTTGCTTC-3′ | Genotyping |
| LBGabi | 5′-CCCATTTGGACGTGAATGTAGACAC-3′ | Genotyping |
Reverse Transcription-Polymerase Chain Reaction (RT-PCR) Analysis
Total RNA was extracted from leaves with the NucleoSpin RNA Plant kit (Macherey-Nagel) following the manufacturer's instructions. For each sample, 1 μg of purified RNA was used for first strand cDNA synthesis using the RevertAidTM kit (Fermentas) or ReverTraAce-α kit (Toyobo) and a T18 primer according to the manufacturer's instructions. First strand cDNA (1 μl) was used as the template for subsequent PCR. Tubulin primers were used as a control for RNA content. Primers used in this study are presented in Table 1. The thermal profile was 94 °C for 2 min (denaturation); 28 cycles of 94 °C for 15 s (denaturation), 60 °C for 30 s (annealing), and 70 °C for 1 min (extension); and 70 °C for 5 min.
Immunoblot and Lectin Blot Analyses
Plant tissue was ground in liquid nitrogen, resuspended in phosphate-buffered saline (PBS) buffer (pH 7.4, 137 mm NaCl, 10 mm phosphate, 2.7 mm KCl), and cleared by centrifugation (10 min at 15,000 × g). The protein content was determined using a protein assay kit (Bio-Rad) and bovine serum albumin as a standard. Each protein (20 μg) was mixed with SDS-polyacrylamide gel electrophoresis (PAGE) loading buffer, denatured at 95 °C for 5 min, and subjected to 10% SDS-PAGE under reducing conditions. Separated proteins were either stained with Coomassie Brilliant Blue R-250 or transferred to a nitrocellulose membrane (Hybond-ECL, Amersham Biosciences). Blots were blocked in 5% (w/v) nonfat dry milk in Tris-buffered saline (TBS) buffer (pH 7.6, 20 mm Tris-HCl, 137 mm NaCl) for 1 h and incubated in a 1:10,000 dilution of rabbit anti-horseradish peroxidase (Sigma), anti-α1,3-fucose, or anti-β1,2-xylose antibody (Agrisera) in TBS supplemented with 0.1% (v/v) Tween 20. Detection was performed after incubation in a 1:3,000 dilution of a horseradish peroxidase-conjugated goat anti-rabbit antibody (Bio-Rad) in TBS-Tween 20 with Western blotting detection reagents (ECL, Amersham Biosciences). For lectin blot analysis, concanavalin A (ConA; Sigma) and Galanthus nivalis lectin (GNA; EY Laboratories) were used to detect N-glycans with terminal mannose residues, and Griffonia simplicifolia lectin (GSII; EY Laboratories) was used to detect N-glycans with terminal GlcNAc residues.
Bioinformatics
To obtain three-dimensional conformation models, the N-glycan structures of Arabidopsis Col-0, xylt-1, and fuctafuctb-1 (fab) were analyzed with GlyProt (Glycosciences.de of the Deutsches Krebsforschungszentrum Heidelberg).
Preparation of an Isotopically Labeled N-Linked Glycan as an Internal Standard
The internal standard of an isotopically labeled N-linked glycan was synthesized enzymatically by the transglycosylation reaction of endo-β-N-acetylglucosaminidase M (TCI Chemicals) with sialylglycopeptide (Fushimi Pharmaceutical) as an acceptor and 13C6-labeled glucose (ΔM = 6 Da; Cambridge Isotopic Laboratories, Inc.) as a sugar donor. The typical transglycosylation reaction at an analytical level of mass spectrometry was performed with a reaction mixture composed of 20 μg of sialylglycopeptide, 320 μg of 13C6-labeled glucose, and 1 milliunit of endo-β-N-acetylglucosaminidase M in a total volume of 30 μl of 100 mm potassium phosphate buffer, pH 6.0. The released isotopically labeled N-linked glycan was purified from the peptides by a reverse phase cartridge (Sep-Pak C18) with a 5% acetic acid elution solution and dried down for use as an internal standard. The resulting internal standard was used to improve the precision of the relative quantitative analysis for the N-linked glycans from the plants.
N-Linked Glycan Preparation
Plant tissue (3 g) was ground in liquid nitrogen, resuspended in 10 ml of 50 mm Hepes, pH 7.5 buffer containing 20 mm sodium metabisulfite, 5 mm EDTA, 0.1% (w/v) SDS, and 1.7% polyvinylpolypyrrolidone, and insoluble material was eliminated by centrifugation (15 min at 13,000 × g) at 4 °C. Equivalent amounts (200 μg) of protein samples were processed by acetone precipitation three times at 4 °C for 3 h to remove contaminants. Then the samples were digested with 20 μg of trypsin in 100 mm Tris-HCl, pH 8.2 containing 1 mm CaCl2 at 37 °C overnight. The trypsin-digested samples were desalted by a reverse phase cartridge (Sep-Pak C18, Restek) and dried by lyophilization. The dried samples were resuspended in 50 mm sodium citrate buffer, pH 5.0. 100 nmol of glycopeptide in 20–50 μl of sodium citrate buffer without BSA were incubated with 0.2–0.5 milliunit of peptide-N-glycosidase A (Calbiochem) for 18 h at 37 °C. The released N-linked glycans were purified from peptides by reverse phase cartridge (Sep-Pak C18) with a 5% acetic acid elution solution. The samples were dried by lyophilization, and the novel internal standard was added in a constant amount to each sample to improve the precision of quantitative analysis before permethylation. The samples were dried again and permethylated. The permethylated glycans were further cleaned of contaminants by a reverse phase cartridge (Sep-Pak C18) and dried down for the analysis by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS).
Analysis of N-Linked Glycan by Mass Spectrometry
MALDI-TOF MS of the permethylated N-linked glycans was performed in the reflector positive ion mode using α-dihydroxybenzoic acid (20 mg/ml solution in 50% methanol) as a matrix. The spectrum was obtained by using an AB Sciex TOF/TOF 5800 MALDI mass spectrometry system. The peak detection and peak area integration were performed simultaneously and taken as valley to baseline integration by Data ExplorerTM software by Applied Biosystems. The areas of the glycan isotope peaks were summed in all the contributions that each individual isotope makes to that peak. Then all the areas were normalized by the peak areas of the internal standard for quantitative analysis.
Subcellular Localization and Bimolecular Fluorescence Complementation (BiFC) Analyses
cDNAs encoding GnTII, FucTA, FucTB, and XylT were amplified without stop codons by PCR using the primer combinations shown in Table 1 and cloned into the pDONRTM/Zeo vector (Invitrogen) using the BP (attB × attP) recombination reaction to create entry clones. These entry clones in combination with the appropriate destination vectors (pMDC83 for subcellular localization and pVYNE and pVYCE for BiFC) were used to create the final Gateway expression constructs by BP (attL × attR) reactions (Invitrogen). The plasmid expressing the ER-RFP with the signal peptide of the wall-associated kinase 2 at the N terminus of the RFP and the ER retention signal His-Asp-Glu-Leu at its C terminus (35) was used as an ER marker. The plasmid expressing the Man49-mCherry with the N-terminal 49 amino acids of the Glycine max α1,2-mannosidase I (GmManI) at the N terminus of the mCherry (36) was used as a cis-Golgi marker. The plasmid expressing the XylT at the N terminus of the RFP, XylT-RFP (37), was used as a medial Golgi marker. All plasmids were introduced into Agrobacterium tumefaciens strain GV3101 via electroporation. A single colony arising from each transformation was inoculated into 5 ml of Luria-Bertani medium supplemented with 50 μg/ml kanamycin and 25 μg/ml rifampicin and grown to stationary phase (20–24 h) at 28 °C with agitation. Bacterial culture (300 μl) was centrifuged and washed twice with infiltration buffer (10 mm MES, 10 mm MgCl2, and 100 μm acetosyringone). For plant infiltration, resuspended bacteria were diluted to an A600 of 0.8. For experiments requiring coexpression of two different constructs, appropriate volumes of each bacterial suspension were injected through the stomata on the lower epidermal surface of fully expanded leaves using a 1-ml plastic syringe with gentle pressure. Infiltrated plants were returned to the growth chamber. Fluorescent protein expression was analyzed in lower epidermal cells 2–3 days postinfiltration. Small leaf pieces were randomly cut out of the infiltrated area and mounted in water such that the abaxial epidermis was facing upward. Fluorescence imaging was carried out using a model FV1000 confocal laser scanning microscope (Olympus) with excitation at 488 and 543 nm and emission at 510–540 nm for GFP and 587–625 nm for mCherry and RFP.
Prediction of Transmembrane Helices
The length of the transmembrane helices and topology of GmManI, GnTII, XylT, FucTA, and FucTB were predicted using HMMTOP software.
Results
Anti-HRP Antibody Shows Different Binding Affinity for the Proteins Extracted from Col-0, fuctafuctb-1, and xylt-1, Respectively
The transfer of β1,2-xylose and α1,3-fucose residues to N-glycans is dependent on XylT, FucTA, and FucTB activities in A. thaliana (38). We used Arabidopsis T-DNA insertion mutants fuctafuctb-1 and xylt-1 to make mutant plants lacking FucT or/and XylT activities (the fuctafuctb-1 double mutant is referred to hereafter as fab, and the fuctafuctb-1xylt-1 triple mutant is referred to as fabx; Fig. 1, B–E). The mutants did not exhibit significant phenotypic differences as compared with the Col-0 plants under normal growth conditions (Fig. 1, B–E).
To investigate whether the additions of the core β1,2-xylose and α1,3-fucose residues to the common acceptor (GlcNAcMan3(GlcNAc)2) are affected by each other, quantitative immunoblot and lectin blot analyses were carried out using proteins extracted from Col-0, fucta, fuctb-1, fab, xylt-1, and fabx (Fig. 1F). Anti-HRP antibody recognizes epitopes containing N-glycans with the core α1,3-fucose or β1,2-xylose residues (17) and showed strong interactions with proteins extracted from Col-0, fucta, fuctb-1, and xylt-1. However, the same antibody showed significantly less interaction with proteins extracted from fab and no interaction with proteins extracted from fabx (Fig. 1F). The stronger interaction of the polyclonal anti-HRP antibody with proteins from xylt-1 than that from fab indicates that the antibodies recognizing the core α1,3-fucose residue are more prevalent than the antibodies recognizing the β1,2-xylose residue in the polyclonal anti-HRP antibody (Fig. 1F). To examine the amounts of the core β1,2-xylose and α1,3-fucose residues, respectively, in the N-glycans from Col-0, fucta, fuctb-1, fab, xylt-1, and fabx, the proteins extracted from the plants were subjected to immunoblot analyses using antibodies specific for α1,3-fucose (anti-fucose) or β1,2-xylose (anti-xylose) (Fig. 1F). Like anti-HRP antibody, the anti-fucose and anti-xylose antibodies showed strong interactions with proteins extracted from Col-0, fucta, and fuctb-1 but no interactions with proteins from fabx. The anti-fucose and anti-xylose antibodies displayed no interactions with proteins extracted from fab and xylt-1, respectively, confirming that the additions of the core α1,3-fucose and β1,2-xylose residues are specifically inhibited in the fab and xylt-1 mutants, respectively. It is noteworthy that the anti-fucose and anti-xylose antibodies also showed significantly reduced interactions with proteins extracted from xylt-1 and fab, respectively. This finding suggests that the enzymatic transfers of the α1,3-fucose and β1,2-xylose residues to the N-glycan acceptors might occur in a complementary manner.
Proteins extracted from Col-0, fucta, fuctb-1, fab, xylt-1, and fabx were also subjected to lectin blot analyses using ConA, a lectin that recognizes α-linked mannose and terminal glucose, and GSII lectin, which recognizes terminal α- or β-linked GlcNAc (Fig. 1F). ConA showed slightly increased interactions with the proteins extracted from xylt-1 and fabx relative to those of the other plants, whereas GSII did not show significant differences in its interactions with proteins extracted from any of the lines (Fig. 1F).
It has been reported that the reduced immunogenicity observed in the Arabidopsis hgl1 mutant of epitopes containing N-glycans with the core α1,3-fucose or β1,2-xylose residues might be due to their altered accessibility to anti-complex N-glycan antibodies (33). We wondered whether the PNGXF produced in xylt-1 and fab undergoes such a change of conformation that might reduce interactions with the anti-fucose and anti-xylose antibodies. To address this possibility, we used a three-dimensional model algorithm to analyze the conformations of paucimannose N-glycans predicted to be produced in Col-0, xylt-1, and fab. The N-glycans with Man3Fuc(GlcNAc)2, GlcNAcMan3Fuc(GlcNAc)2, Man3Xyl(GlcNAc)2, and GlcNAcMan3Xyl(GlcNAc)2 structures produced in xylt-1 and fab did not show significant differences in the conformations or torsion angles of β1,2-xylose and α1,3-fucose residues compared with the N-glycans with Man3XylFuc(GlcNAc)2 and GlcNAcMan3XylFuc(GlcNAc)2 produced in Col-0 (Fig. 2). These results suggest that the reduced interactions of the anti-fucose and anti-xylose antibodies with the proteins extracted from xylt-1 and fab, respectively, cannot be explained by altered accessibility of the antibodies to the N-glycan epitopes.
FIGURE 2.
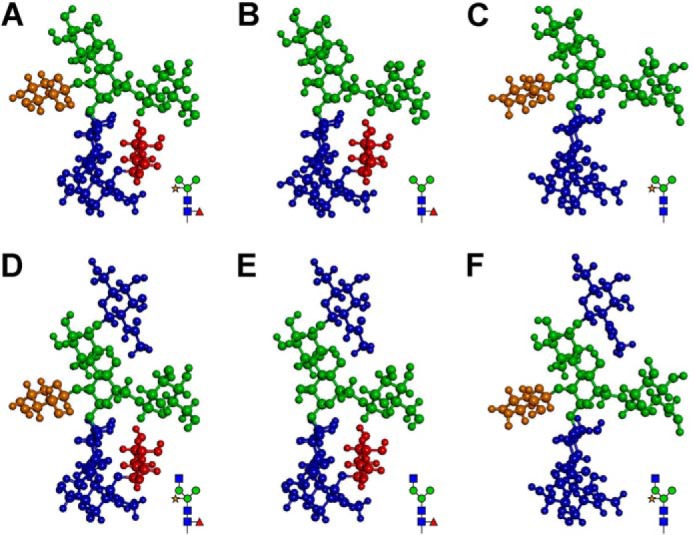
Three-dimensional models do not show significant differences in the conformations of the N-glycans in the mutants and Col-0. A and D, predicted conformations of N-glycans with Man3XylFuc(GlcNAc)2 and GlcNAcMan3XylFuc(GlcNAc)2 structures, respectively, in Col-0. B and E, predicted conformations of N-glycans with Man3Fuc(GlcNAc)2 and GlcNAcMan3Fuc(GlcNAc)2 structures, respectively, in xylt-1. C and F, predicted conformations of N-glycans with Man3Xyl(GlcNAc)2 and GlcNAcMan3Xyl(GlcNAc)2 structures, respectively, in fab. GlcNAc, mannose, xylose, and fucose residues are indicated with dark blue, green, brown, and dark red, respectively.
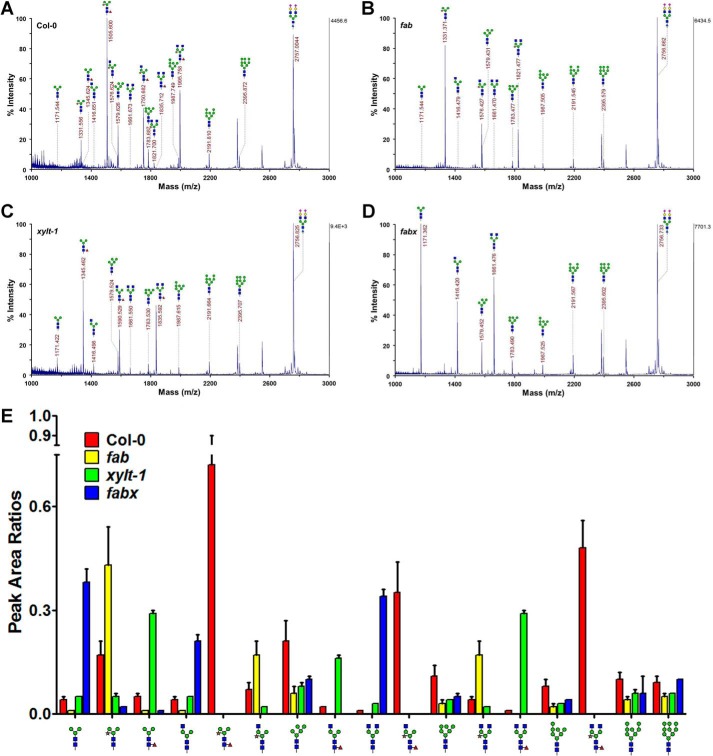
N-Glycans Containing β1,2-Xylose and α1,3-Fucose Residues Are Decreased in fab and xylt-1
To quantitatively examine the presence of the core β1,2-xylose and α1,3-fucose residues in the N-glycans produced in fab and xylt-1, respectively, glycoproteins isolated from Col-0, fab, and xylt-1 were subjected to N-glycan analysis using MALDI-TOF MS. The mass spectra of the total N-glycans from Col-0 plants contained three major peaks assigned to N-glycans with Man3XylFuc(GlcNAc)2 (m/z 1506), (GlcNAc)2Man3XylFuc(GlcNAc)2 (m/z 1996), and GlcNAcMan3XylFuc(GlcNAc)2 (m/z 1751) structures (Fig. 3A). N-Glycans with Man3Xyl(GlcNAc)2 (m/z 1331), GlcNAcMan3Xyl(GlcNAc)2 (m/z 1576), and (GlcNAc)2Man3Xyl(GlcNAc)2 (m/z 1821) structures were the most abundant in fab (Fig. 3B). There was slightly less N-glycan with (GlcNAc)2Man3Xyl(GlcNAc)2 than with GlcNAcMan3Xyl(GlcNAc)2 in fab. This result suggests that the addition of the 6-arm non-reducing β1,2-GlcNAc residue is facilitated by the presence of the core α1,3-fucose residue. In xylt-1, the major peaks were assigned to N-glycans with Man3Fuc(GlcNAc)2 (m/z 1346), (GlcNAc)2Man3Fuc(GlcNAc)2 (m/z 1836), and GlcNAcMan3Fuc(GlcNAc)2 (m/z 1591; Fig. 3C). Similar to Col-0, xylt-1 had more N-glycan with (GlcNAc)2Man3Fuc(GlcNAc)2 than with GlcNAcMan3Fuc(GlcNAc)2; this finding indicates that the addition of the 6-arm non-reducing β1,2-GlcNAc residue is not affected by the presence of the core β1,2-xylose residue.
FIGURE 3.
N-Glycans containing β1,2-xylose and α1,3-fucose residues are decreased in fab and xylt-1. A–D, MALDI-TOF mass spectra of the permethylated total N-glycans extracted from Col-0, fab, xylt-1, and fabx, respectively. The highest peak area from each spectrum was set to 100% for direct comparison of the amounts of N-glycans in each plant. Measured masses are given above or beside the peaks with illustrations of the N-glycan structures. E, relative amounts of N-glycans in Col-0, fab, xylt-1, and fabx. The relative amounts of the N-glycans were compared with the peak area ratios calculated based on that of the internal standard ([M + Na]+ = 2757.3761, theoretical m/z). Illustrations of the N-glycan structures are shown at the bottom of the graph. GlcNAc, mannose, xylose, and fucose residues are represented with dark blue, green, brown, and dark red, respectively. Relative amounts of N-glycans in Col-0, fab, xylt-1, and fabx are shown as red, yellow, green, and blue bars, respectively. Error bars indicate S.D.
The amounts of the N-glycan structures isolated from Col-0, fab, and xylt-1 were compared using peak area ratios calculated based on the amount of artificial N-glycan structure added as an internal control. N-Glycans with the core β1,2-xylose and α1,3-fucose residues were less abundant in fab and xylt-1 than in Col-0 (Fig. 3E). These results are consistent with the immunoblot analysis results, suggesting that the enzymatic transfers of the core β1,2-xylose and α1,3-fucose residues to N-glycan acceptors occur in a complementary manner. It is also noteworthy that the amount of N-glycan with Man3Xyl(GlcNAc)2 (m/z 1331) in fab was higher than that of N-glycan with Man3Fuc(GlcNAc)2 (m/z 1346) in xylt-1 (Fig. 3, B and C). This result is consistent with the previous finding that XylT acts on N-glycan acceptors prior to FucT (34). In the fabx triple mutant, N-glycans with Man3(GlcNAc)2 (m/z 1171), (GlcNAc)2Man3(GlcNAc)2 (m/z 1662), and GlcNAcMan3(GlcNAc)2 (m/z 1416) were the most abundant (Fig. 3D). The amount of N-glycan with (GlcNAc)2Man3(GlcNAc)2 (m/z 1662) in fabx was greater than the amounts of N-glycans with (GlcNAc)2Man3Fuc(GlcNAc)2 (m/z 1836) and (GlcNAc)2Man3Xyl(GlcNAc)2 (m/z 1821) in xylt-1 and fab, respectively; however, it was less than that of N-glycan with (GlcNAc)2Man3XylFuc(GlcNAc)2 (m/z 1996) in Col-0 (Fig. 3E). These results indicate that the addition of the 6-arm non-reducing GlcNAc residue by GnTII may occur using the GlcNAcMan3XylFuc(GlcNAc)2 structure as a preferential acceptor, but the GlcNAcMan3(GlcNAc)2, GlcNAcMan3Fuc(GlcNAc)2, and GlcNAcMan3Xyl(GlcNAc)2 structures can also be utilized as suboptimal acceptors.
N-Glycans Containing β1,2-Xylose and α1,3-Fucose Residues Are Increased in the Mutants with the gnt2 Background
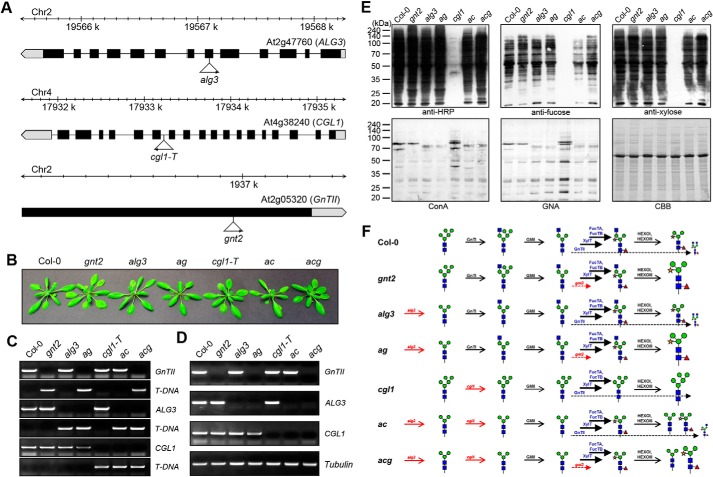
Our results showed that the GlcNAcMan3(GlcNAc)2 N-glycan is used as an acceptor of not only the β1,2-xylose and α1,3-fucose residues but also 6-arm β1,2-GlcNAc by the XylT-, FucT- and GnTII-catalyzed reactions, respectively. We wondered how addition of the 6-arm β1,2-GlcNAc residue to the GlcNAcMan3(GlcNAc)2 N-glycan affected the additions of the core β1,2-xylose and α1,3-fucose residues. To address this question, Col-0, gnt2, alg3, alg3gnt2 (ag), cgl1, alg3cgl1 (ac), and alg3cgl1gnt2 (acg) lines were used to establish artificial in vivo N-glycosylation conditions (Fig. 4, A–D) (29, 39, 40). None of the mutant plants showed significant phenotypic differences as compared with Col-0 under normal growth conditions. To investigate whether the additions of the core β1,2-xylose and α1,3-fucose residues are affected by the absence of the 6-arm β1,2-GlcNAc residue, quantitative immunoblot and lectin blot analyses were carried out using proteins extracted from Col-0 and gnt2 (Fig. 4E). Anti-HRP, anti-fucose, and anti-xylose antibodies showed slightly higher interactions with proteins extracted from gnt2 than with proteins extracted from Col-0 (Fig. 4E). Col-0 and gnt2 proteins were also subjected to lectin blot analyses using ConA and GNA, a lectin recognizing α1,3- and α1,6-linked high mannose structures (Fig. 4E). Both lectins showed slightly reduced interactions with the proteins extracted from gnt2 than with those from Col-0 (Fig. 4E). These results indicate that the additions of the core β1,2-xylose and α1,3-fucose residues to the common acceptor are partially inhibited by the addition of the 6-arm β1,2-GlcNAc residue (Fig. 4F).
FIGURE 4.
Antibodies show different binding affinities for the proteins extracted from Col-0, gnt2, alg3, ag, cgl1, ac, and acg. A, schematic representation of the T-DNA insertion sites in the ALG3, CGL1 (GnTI), and GnTII genes. Boxes represent exons, and black denotes the coding region. T-DNA insertion sites and left border directions are indicated with triangles and arrowheads. B, phenotypes of the mutant plants compared with that of Col-0. Plants were grown on soil for 30 days. gnt2, alg3, and cgl-1T single mutants were used to produce ag, ac, and acg. C, PCR-based genotyping of the mutants. Gene-specific primers were designed and used in combination with T-DNA left border primer. D, RT-PCR analysis of the mutants. Levels of transcripts in Col-0 and the indicated mutants were determined by RT-PCR with gene-specific primers. Tubulin was used as a control. E, immunoblot and lectin blot analyses to assess structure and amount of the N-glycans in Col-0, gnt2, alg3, ag, cgl1, ac, and acg. Total proteins extracted from 3-week-old seedlings were subjected to immunoblot and lectin blot analyses. The immunoblots were probed with anti-HRP, anti-xylose, and anti-fucose antibodies, and lectin blots were probed with ConA and GNA. Coomassie Brilliant Blue (CBB) staining was used to show equal loading of the proteins. F, schematic illustration of the N-glycosylation in the mutant plants. N-Glycan processing in the indicated plants is illustrated with corresponding enzymes and structures of the N-glycans. The additions of the outer α1,3- and α1,6-mannose residues of the core α1,6-mannose of the N-glycan precursor are completely inhibited in alg3, addition of the 3-arm β1,2-GlcNAc residue of the N-glycan is completely inhibited in cgl1, and addition of the 6-arm β1,2-GlcNAc residue is completely inhibited in gnt2. The dominant additions of the core β1,2-xylose and α1,3-fucose residues are indicated with thick solid lines, and limited addition of the 6-arm β1,2-GlcNAc residue is indicated with thin dashed lines. In all mutants with the gnt2 background, the amount of PNGXF has been increased, indicating that the additions of the core β1,2-xylose and α1,3-fucose residues are regulated by the addition of the 6-arm β1,2-GlcNAc residue. Changed amounts of the N-glycans are represented by the enlarged and compressed N-glycan structures.
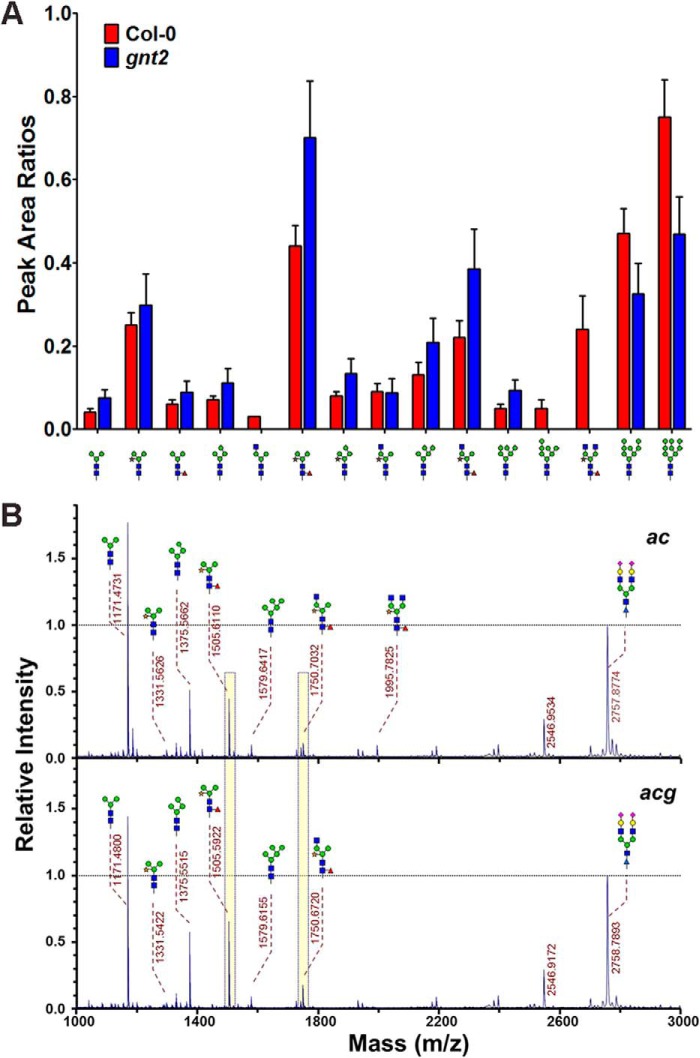
We used MALDI-TOF MS to examine the structures and amounts of N-glycans in Col-0 and gnt2 (Fig. 5A). As expected, the peak assigned to the N-glycan with (GlcNAc)2Man3XylFuc(GlcNAc)2 (m/z 1996) structure was clearly present in Col-0 but not in gnt2 (Fig. 5A). The amounts of N-glycans with the core β1,2-xylose and α1,3-fucose residues were increased, whereas those of high mannose-type N-glycans were decreased in gnt2 compared with in Col-0 (Fig. 5A). These results are consistent with those obtained from the immunoblot and lectin blot analyses. Thus, plants may retain a low GnTII activity and/or substrate occupancy to facilitate efficient production of PNGXF (Fig. 4F).
FIGURE 5.
N-Glycans containing β1,2-xylose and α1,3-fucose residues are increased in gnt2. A, relative amounts of N-glycans in Col-0 (red bars) and gnt2 (blue bars). Relative amounts of the N-glycans were compared with the peak area ratios. Error bars indicate S.D. B, MALDI-TOF mass spectra of the permethylated total N-glycans extracted from ac and acg. An isotopically labeled N-linked glycan was used as an internal standard and set to 1.0 for direct comparison of the amounts of N-glycans. Peak areas of the two major complex N-glycans are highlighted with yellow bars for comparison.
Plants without GnTI activity do not produce hybrid and complex-type N-glycans (23–25). However, when the additions of the outer α1,3- and α1,6-mannose residues to the core α1,6-mannose of the N-glycan precursor (dolichol lipid-linked oligosaccharide) were inhibited by crossing cgl1 with the alg3 loss-of-function mutant to generate the ac double mutant, additions of the β1,2-xylose and α1,3-fucose residues to N-glycans were partially recovered (Figs. 4E and 5B). This result indicates that not only the addition of β1,2-linked GlcNAc to the N-glycan with Man5(GlcNAc)2 structure by GnTI but also the removal of the outer α1,3- and α1,6-mannose residues from the core α1,6-mannose of the GlcNAcMan5(GlcNAc)2 N-glycan by Golgi α-mannosidase II is important for complex N-glycan formation in plants. Based on this finding, the amounts of the N-glycan containing core β1,2-xylose and α1,3-fucose residues were compared under artificial N-glycosylation conditions (in alg3, ac, ag, or acg) with regard to the presence or absence of the 6-arm β1,2-GlcNAc residue in the acceptor (Fig. 4, E and F). Anti-HRP, anti-fucose, and anti-xylose antibodies showed higher interactions with proteins extracted from ag and acg than with proteins extracted from alg3 and ac (Fig. 4E). These results indicate that the amount of the N-glycan containing core β1,2-xylose and α1,3-fucose residues is increased in the mutants with gnt2 background (Fig. 4F). Conversely, ConA and GNA showed a tendency of slightly reduced interactions with the proteins extracted from ag and acg compared with those extracted from alg3 and ac (Fig. 4E). However, anti-HRP, anti-fucose, and anti-xylose antibodies showed no interactions with proteins extracted from cgl1, whereas ConA and GNA showed higher interactions with them (Fig. 4E). These results indicate that the additions of the core β1,2-xylose and α1,3-fucose residues are partially inhibited in the presence of the 6-arm β1,2-GlcNAc residue, whereas they are more facilitated in the mutants with gnt2 background (Fig. 4F).
We next quantified the N-glycans containing core β1,2-xylose and α1,3-fucose residues in ac and acg (Fig. 5B). The peak assigned to the N-glycans with (GlcNAc)2Man3XylFuc(GlcNAc)2 (m/z 1996) was observed in the mass spectrum of the total N-glycans from ac but not from acg (Fig. 5B). N-Glycans with β1,2-xylose and α1,3-fucose residues were more abundant in acg compared with ac (Fig. 5B). These results are also consistent with the immunoblot and lectin blot analyses (Fig. 4E). Taken together, our results indicate that the additions of the core β1,2-xylose and α1,3-fucose residues to the N-glycan acceptor are negatively controlled by the 6-arm β1,2-GlcNAc residue.
Limited Addition of the 6-Arm GlcNAc Is Not Associated with the Order of the Enzymatic Reaction
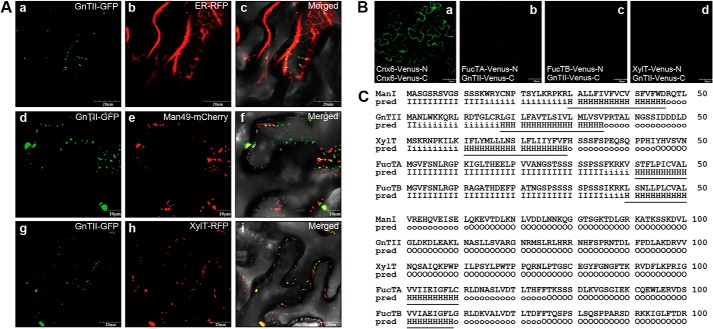
Our results showed that N-glycan with the GlcNAcMan3(GlcNAc)2 structure can be used as a common acceptor of the β1,2-xylose, α1,3-fucose, and 6-arm β1,2-GlcNAc residues; however, the N-glycan with Man3XylFuc(GlcNAc)2 (m/z 1506) structure is the most prevalent in Col-0 plants (Fig. 3A). This result indicates that the additions of the core β1,2-xylose and α1,3-fucose residues to the common acceptor by XylT, FucTA, and FucTB, respectively, are dominant, whereas the addition of the 6-arm non-reducing GlcNAc residue by GnTII is limited. We wondered how the dominant additions of the core β1,2-xylose and α1,3-fucose residues and limited addition of the 6-arm non-reducing GlcNAc residue to the common acceptor are determined in plants. Accordingly, we investigated the spatial arrangement of the GnTII in the Golgi apparatus. Previous studies have shown that GmManI, which acts very early in the Golgi apparatus, is located predominantly in the cis-half of the Golgi and to a low level in the ER, whereas XylT is located mainly in the medial cisternae of the Golgi (36, 37, 41). To examine its distribution within the secretory pathway, GnTII-GFP was coexpressed with ER-RFP, Man49-mCherry, and XylT-RFP in agroinfiltrated leaves of N. benthamiana. The fluorescence images from GnTII-GFP and ER-RFP largely did not overlap with each other, suggesting that the GnTII-GFP is not localized in the ER (Fig. 6A). However, the fluorescence signals from GnTII-GFP considerably overlapped with those of Man49-mCherry and XylT-RFP, suggesting that the GnTII-GFP is positioned in the cis-half of the Golgi cisternae or medial Golgi (Fig. 6A).
FIGURE 6.
GnTII is positioned in the cis-half of the Golgi in a kin-independent manner. A, subcellular localizations of GnTII-GFP, XylT-GFP, FucTA-GFP, and FucTB-GFP are compared with those of ER-RFP (ER marker), Man49-mCherry (cis-Golgi marker), and XylT-RFP (medial-Golgi marker), respectively. Left column, GFP fluorescence obtained with GnTII-GFP (panels a, d, and g); middle column, signals from ER-RFP, Man49-mCherry, and XylT-RFP (panels b, e, and h); right column, overlay of both signals (panels c, f, and i). The indicated fusion constructs were transiently expressed in N. benthamiana epidermal cells and analyzed by confocal laser scanning microscopy. Scale bars, 50 μm. B, BiFC assay. The indicated fusion constructs of Venus-N and Venus-C were transiently expressed in N. benthamiana epidermal cells. Confocal laser scanning microscopy was used for visualization of fluorescence reporting protein-protein interactions. The fluorescence of Cnx6-Venus-N and Cnx6-Venus-C was used as a positive control for the BiFC experiment (panel a). Combinations of the indicated constructs did not produce Venus fluorescence (panels b–d). Scale bars, 10 or 20 μm. C, predicted length of transmembrane helices and topology. The transmembrane helices and topology of GnTII, XylT, FucTA, and FucTB were analyzed at the HMMTOP server. The N-terminal regions predicted (pred) as transmembrane helices are indicated with underlining. I, cytosolic domain; H, transmembrane helix; O, extracellular domain.
Two models have been proposed to explain the retention of the glycosylation enzymes in the secretory pathway of eukaryotic cells. The kin recognition model postulates that the enzymes in the same compartment form aggregates that prevent them from entering the budding vesicles (42). By contrast, the membrane thickness model assumes that the fit between the length of the transmembrane domain and the thickness of the lipid bilayer determines the positioning of the enzymes (43). It has been reported that the length of transmembrane domain acts as a key signal for the spatial arrangement of the Golgi glycosyltransferases in plants (37). We used BiFC to examine whether GnTII, XylT, FucTA, and FucTB interact with each other to be sorted together into the same subset of Golgi cisternae (Fig. 6B). Although coexpression of Cnx6-Venus-N and Cnx6-Venus-C led to reconstitution of fluorescence indicative of interaction, no BiFC signals were detected among different combinations of the GnTII, XylT, FucTA, and FucTB enzymes fused to Venus-N and Venus-C in agroinfiltrated leaves of N. benthamiana (Fig. 6B). This lack of close interaction suggests that GnTII, XylT, FucTA, and FucTB are positioned in the cis-half of the Golgi cisternae or medial Golgi in a different manner from that described in the kin recognition model (42). The length of the transmembrane helices and topology of GmManI, GnTII, XylT, FucTA, and FucTB were analyzed using the HMMTOP algorithm. GmManI, GnTII, XylT, FucTA, and FucTB are predicted to possess 17-, 18-, 19-, 20-, and 20-amino acid transmembrane helices at their N-terminal regions, respectively (Fig. 6C). These results indicate that GnTII with a shorter transmembrane helix may be positioned earlier in the Golgi cisternae than XylT, FucTA, and FucTB. It seems that the limited addition of the 6-arm non-reducing GlcNAc residue by GnTII takes place prior to the dominant additions of the core β1,2-xylose and α1,3-fucose residues to the common acceptor by XylT, FucTA, and FucTB, respectively. Therefore, the limited addition of the 6-arm GlcNAc residue to the common acceptor is not caused by a delayed enzymatic reaction of GnTII compared with those of XylT, FucTA, and FucTB.
Limited Addition of the 6-Arm GlcNAc Is Caused by Regulated GnTII Function
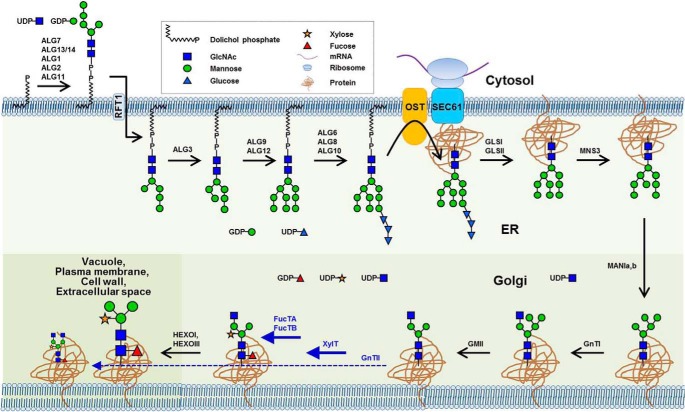
Based on our observations in the gnt2 backgrounds, prior addition of the 6-arm non-reducing GlcNAc residue to the common N-glycan acceptor may inhibit additions of the core β1,2-xylose and α1,3-fucose residues. Thus, addition of the 6-arm non-reducing GlcNAc residue by GnTII might be maintained at a low level to facilitate additions of the core β1,2-xylose and α1,3-fucose residues to the common acceptor by XylT, FucTA, and FucTB, respectively, in plants. Furthermore, the amount of the N-glycan with GlcNAcMan3XylFuc(GlcNAc)2 structure is less than that with (GlcNAc)2Man3XylFuc(GlcNAc)2 structure, suggesting that the GlcNAcMan3XylFuc(GlcNAc)2 is preferentially used to produce PNGXF in plants (Fig. 3A). If the limited addition of the 6-arm non-reducing GlcNAc residue to the common acceptor is caused by a lower activity and/or substrate occupancy of the corresponding enzyme, overexpression of GnTII may lead to an inhibited addition of the core β1,2-xylose and α1,3-fucose residues in plants. To test this prediction of our hypothesis, transgenic Arabidopsis overexpressing GnTII (35S:GnTII) was established and subjected to immunoblot and lectin blot analyses (Fig. 7). Anti-HRP, anti-fucose, and anti-xylose antibodies showed significantly reduced interactions with proteins extracted from 35S:GnTII compared with proteins extracted from Col-0 (Fig. 7). By contrast, ConA and GSII showed increased interactions with proteins extracted from 35S:GnTII relative to proteins extracted from Col-0 (Fig. 7). These results provide evidence that the limited addition of the 6-arm non-reducing GlcNAc residue to the common N-glycan acceptor is caused by lower activity and/or substrate occupancy of GnTII in plants. Taken together, our results indicate that, in addition to the functions of hexosaminidases, regulated GnTII function is also important to facilitate efficient production of PNGXF in plants (Fig. 8).
FIGURE 7.
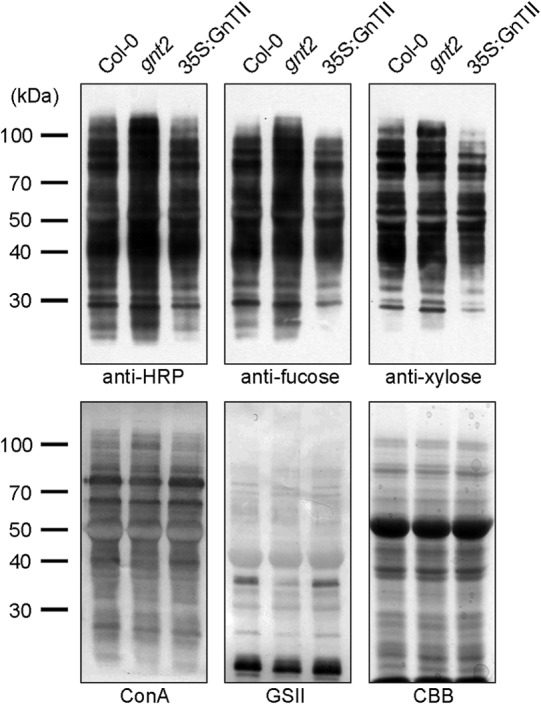
Additions of the core β1,2-xylose and α1,3-fucose are decreased in 35S:GnTII. Total proteins extracted from 3-week-old seedlings were subjected to immunoblot and lectin blot analyses. The immunoblots were probed with anti-HRP, anti-xylose, and anti-fucose antibodies, and lectin blots were probed with ConA and GSII. Coomassie Brilliant Blue (CBB) staining was used to show equal loading of the proteins.
FIGURE 8.
Limited addition of the 6-arm β1,2-GlcNAc is involved in the formation of the largest N-glycan in plants. N-Glycan processing in the ER and Golgi of Arabidopsis is summarized with corresponding enzymes and illustrated structures of the N-glycans (45). The biosynthesis of complex N-glycan initiates with addition of the 3-arm β1,2-GlcNAc residue to the Man5(GlcNAc)2 N-glycan by GnTI at the cis-face of the Golgi complex (23). Subsequently, the outer chain mannose residues are removed by Golgi α-mannosidase II, resulting in the production of the GlcNAcMan3(GlcNAc)2 N-glycan (28, 33). This structure can be used as a common acceptor of the 6-arm β1,2-GlcNAc, core β1,2-xylose, and α1,3-fucose residues by GnTII, XylT, FucTA, and FucTB (17, 28, 30–34) in the medial Golgi compartments. Because the addition of the 6-arm β1,2-GlcNAc residue by GnTII is less efficient than the additions of the core β1,2-xylose and α1,3-fucose residues by XylT, FucTA, and FucTB, the amount of the N-glycan with GlcNAcMan3XylFuc(GlcNAc)2 structure is greater than that with (GlcNAc)2Man3XylFuc(GlcNAc)2 structure in plants. The outer chain GlcNAc residue of the GlcNAcMan3XylFuc(GlcNAc)2 is removed by HEXOI and HEXOIII, resulting in the prevalent PNGXF in plants. The dominant additions of the core β1,2-xylose and α1,3-fucose residues are indicated with thick solid lines, and the limited addition of the 6-arm β1,2-GlcNAc residue is indicated with a thin dashed line. OST, oligosaccharyltransferase.
Discussion
The type and number of glycosylation enzymes and maturation processes of N-glycans through the Golgi apparatus are different according to the species (12). The PNGXF prevalently found in plant glycoproteins acts as an important antigenic determinant in the human body, causing immunological side effects including allergic responses (2, 15–17). Strategies for systematic engineering of the glycosylation pathway need to be established to produce biopharmaceuticals in plants for human therapy. To eliminate allergenic properties of the plant-made biopharmaceuticals, core β1,2-xylose and α1,3-fucose residues of the N-glycan need to be removed. When plant-made biopharmaceuticals with complex N-glycans including α1,6-fucose, β1,4-galactose, and α2,3/6-sialic acid (neuraminic acid) residues are intended to be produced, α1,6-fucosyltransferase, β1,4-galactosyltransferase, α2,6-sialyltransferase, and enzymes involved in the sialic acid synthesis pathway such as UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase, N-acetylneuraminic acid-phosphate synthetase, CMP-N-acetylneuraminic acid synthetase, and CMP-Neu5Ac transporter need to be introduced in plants (32, 44). Strategies to increase addition of the 3- and 6-arm non-reducing GlcNAc residues may also be required to facilitate complex N-glycan production in plants. Furthermore, the possible influence of the glycoengineering on the growth and development of plants needs to be investigated.
A recent study showed that HEXOI and HEXOIII reside in different subcellular compartments and contribute to production of PNGXF in Arabidopsis (14). However, the mass spectra of the total N-glycans from Col-0 plants show that the amount of the N-glycan with (GlcNAc)2Man3XylFuc(GlcNAc)2 is higher than that with GlcNAcMan3XylFuc(GlcNAc)2 (Fig. 3A). If HEXOI and HEXOIII remove the 3- and 6-arm non-reducing β1,2-GlcNAc residues in a nonselective manner and the N-glycan with Man3XylFuc(GlcNAc)2 structure is the end product, the higher amount of the N-glycan with (GlcNAc)2Man3XylFuc(GlcNAc)2 compared with GlcNAcMan3XylFuc(GlcNAc)2 is difficult to interpret. However, if we postulate that HEXOI and HEXOIII preferentially remove the 3-arm non-reducing β1,2-GlcNAc residue over the 6-arm non-reducing β1,2-GlcNAc residue from N-glycans in plants, the GlcNAcMan3XylFuc(GlcNAc)2 structure (with the 3-arm non-reducing β1,2-GlcNAc residue) would be more vulnerable, and the (GlcNAc)2Man3XylFuc(GlcNAc)2 structure would be more resistant to attack by the enzymes. Indeed, when (GlcNAc)2Man3(GlcNAc)2-PA, a pyridylaminated oligosaccharide, is used as a substrate for endogenous β-N-acetylhexosaminidases, significant amounts of unhydrolyzed (GlcNAc)2Man3(GlcNAc)2-PA and GlcNAcMan3(GlcNAc)2-PA are observed even after 16-h incubation (14).
In this study, we have shown that the addition of the 6-arm β1,2-GlcNAc residue is limited compared with the additions of the core β1,2-xylose and α1,3-fucose residues to the common acceptor (GlcNAcMan3(GlcNAc)2). Confocal microscopy and computer-assisted prediction of transmembrane helices suggested that the limited addition of the 6-arm non-reducing GlcNAc residue by GnTII takes place prior to the dominant additions of the core β1,2-xylose and α1,3-fucose residues to the common acceptor by XylT, FucTA, and FucTB, respectively (Fig. 6). Whereas GSII did not show significant differences in interactions with the proteins extracted from fab, xylt-1, and fabx, it showed decreased interaction with the proteins extracted from gnt2 and increased interaction with those from 35S:GnTII. Genetic evidence from fab, xylt-1, fabx, gnt2, and 35S:GnTII indicate that GnTII is limiting in the additions of the sugar residues to the common acceptor (GlcNAcMan3(GlcNAc)2) in plants. The process involving the limited addition of the 6-arm β1,2-GlcNAc residue and preferential use of the GlcNAcMan3XylFuc(GlcNAc)2 as a substrate would be the most cost-effective way to produce PNGXF in an energy-efficient manner in plants. Thus, plants might retain a low GnTII activity to facilitate efficient production of PNGXF. Taken together, our study shows that additions of the core β1,2-xylose and α1,3-fucose residues by XylT, FucTA, and FucTB are dominant, and addition of the 6-arm β1,2-GlcNAc residue by GnTII is limited in plants (Fig. 8). Our results indicate that the prevalent formation of N-glycans with Man3XylFuc(GlcNAc)2 and (GlcNAc)2Man3XylFuc(GlcNAc)2 structures is facilitated not only by hexosaminidase activity but also by the regulated sharing of the common acceptor GlcNAcMan3(GlcNAc)2 in the Golgi of plant cells.
Author Contributions—K. O. L. conceived and coordinated the study and wrote the paper. J. Y. Y., K. S. K., W. I. D. F., R. H., N. K. R., and T. T. designed, performed, and analyzed the experiments shown in Figs. 1–8. H.-K. S., S. P., and J.-M. L. performed and analyzed the experiments shown in Figs. 3 and 5. T. M., J.-M. L., and S. Y. L. provided technical assistance and contributed to the preparation of the figures. All authors reviewed the results and approved the final version of the manuscript.
This work was supported in part by grants from the Next-Generation BioGreen Program (Grant PJ01137901); the Technology Innovation Program funded by the Ministry of Trade, Industry and Energy (Grant 10048311); the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (Grant 2012R1A1A2001074), Republic of Korea; and the Basic Science Research Program and the Priority Research Centers Program through the NRF funded by the Ministry of Education, Science and Technology (Grants NRF 2011-0013961 and NRF 2010-0029634). The authors declare that they have no conflicts of interest with the contents of this article.
- ER
- endoplasmic reticulum
- GnT
- N-acetylglucosaminyltransferase
- PNGXF
- paucimannosidic N-glycan with core β1,2-xylose and α1,3-fucose residues
- XylT
- β1,2-xylosyltransferase
- FucT
- α1,3-fucosyltransferase
- HEXO
- β-N-acetylhexosaminidase
- ALG3
- asparagine-linked glycosylation 3
- ConA
- concanavalin A
- GNA
- G. nivalis lectin
- GSII
- G. simplicifolia lectin
- BiFC
- bimolecular fluorescence complementation
- RFP
- red fluorescent protein
- GmManI
- G. max α1,2-mannosidase I
- fab
- fuctafuctb-1
- fabx
- fuctafuctb-1xylt-1
- ag
- alg3gnt2
- ac
- alg3cgl1
- acg
- alg3cgl1gnt2
- PA
- pyridylamino.
References
- 1. Ruiz-Canada C., Kelleher D. J., Gilmore R. (2009) Cotranslational and posttranslational N-glycosylation of polypeptides by distinct mammalian OST isoforms. Cell 136, 272–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aalberse R. C., Koshte V., Clemens J. G. (1981) Immunoglobulin E antibodies that crossreact with vegetable foods, pollen, and Hymenoptera venom. J. Allergy Clin. Immunol. 68, 356–364 [DOI] [PubMed] [Google Scholar]
- 3. Molinari M., Helenius A. (1999) Glycoproteins form mixed disulphides with oxidoreductases during folding in living cells. Nature 402, 90–93 [DOI] [PubMed] [Google Scholar]
- 4. Sitia R., Braakman I. (2003) Quality control in the endoplasmic reticulum protein factory. Nature 426, 891–894 [DOI] [PubMed] [Google Scholar]
- 5. Spiro R. G. (2004) Role of N-linked polymannose oligosaccharides in targeting glycoproteins for endoplasmic reticulum-associated degradation. Cell. Mol. Life Sci. 61, 1025–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Martina J. A., Daniotti J. L., Maccioni H. J. (2000) GM1 synthase depends on N-glycosylation for enzyme activity and trafficking to the Golgi complex. Neurochem. Res. 25, 725–731 [DOI] [PubMed] [Google Scholar]
- 7. Helenius A., Aebi M. (2001) Intracellular functions of N-linked glycans. Science 291, 2364–2369 [DOI] [PubMed] [Google Scholar]
- 8. Helenius A., Aebi M. (2004) Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 73, 1019–1049 [DOI] [PubMed] [Google Scholar]
- 9. Fiedler K., Simons K. (1995) The role of N-glycans in the secretory pathway. Cell 81, 309–312 [DOI] [PubMed] [Google Scholar]
- 10. Varki A. (1993) Biological roles of oligosaccharides: all of the theories are correct. Glycobiology 3, 97–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schwarz F., Aebi M. (2011) Mechanisms and principles of N-linked protein glycosylation. Curr. Opin. Struct. Biol. 21, 576–582 [DOI] [PubMed] [Google Scholar]
- 12. Wilson I. B. (2002) Glycosylation of proteins in plants and invertebrates. Curr. Opin. Struct. Biol. 12, 569–577 [DOI] [PubMed] [Google Scholar]
- 13. Gutternigg M., Kretschmer-Lubich D., Paschinger K., Rendić D., Hader J., Geier P., Ranftl R., Jantsch V., Lochnit G., Wilson I. B. (2007) Biosynthesis of truncated N-linked oligosaccharides results from non-orthologous hexosaminidase-mediated mechanisms in nematodes, plants, and insects. J. Biol. Chem. 282, 27825–27840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liebminger E., Veit C., Pabst M., Batoux M., Zipfel C., Altmann F., Mach L., Strasser R. (2011) β-N-Acetylhexosaminidases HEXO1 and HEXO3 are responsible for the formation of paucimannosidic N-glycans in Arabidopsis thaliana. J. Biol. Chem. 286, 10793–10802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. van Ree R., Cabanes-Macheteau M., Akkerdaas J., Milazzo J. P., Loutelier-Bourhis C., Rayon C., Villalba M., Koppelman S., Aalberse R., Rodriguez R., Faye L., Lerouge P. (2000) β(1,2)-Xylose and α(1,3)-fucose residues have a strong contribution in IgE binding to plant glycoallergens. J. Biol. Chem. 275, 11451–11458 [DOI] [PubMed] [Google Scholar]
- 16. Manduzio H., Fitchette A. C., Hrabina M., Chabre H., Batard T., Nony E., Faye L., Moingeon P., Gomord V. (2012) Glycoproteins are species-specific markers and major IgE reactants in grass pollens. Plant Biotechnol. J. 10, 184–194 [DOI] [PubMed] [Google Scholar]
- 17. Wilson I. B., Harthill J. E., Mullin N. P., Ashford D. A., Altmann F. (1998) Core α1,3-fucose is a key part of the epitope recognized by antibodies reacting against plant N-linked oligosaccharides and is present in a wide variety of plant extracts. Glycobiology 8, 651–661 [DOI] [PubMed] [Google Scholar]
- 18. Raju T. S., Lang S. E. (2014) Diversity in structure and functions of antibody sialylation in the Fc. Curr. Opin. Biotechnol. 30, 147–152 [DOI] [PubMed] [Google Scholar]
- 19. Ghaderi D., Taylor R. E., Padler-Karavani V., Diaz S., Varki A. (2010) Implications of the presence of N-glycolylneuraminic acid in recombinant therapeutic glycoproteins. Nat. Biotechnol. 28, 863–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Erbayraktar S., Grasso G., Sfacteria A., Xie Q. W., Coleman T., Kreilgaard M., Torup L., Sager T., Erbayraktar Z., Gokmen N., Yilmaz O., Ghezzi P., Villa P., Fratelli M., Casagrande S., Leist M., Helboe L., Gerwein J., Christensen S., Geist M. A., Pedersen L. Ø., Cerami-Hand C., Wuerth J. P., Cerami A., Brines M. (2003) Asialoerythropoietin is a nonerythropoietic cytokine with broad neuroprotective activity in vivo. Proc. Natl. Acad. Sci. U.S.A. 100, 6741–6746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yip B., Chen S. H., Mulder H., Höppener J. W., Schachter H. (1997) Organization of the human β-1,2-N-acetylglucosaminyltransferase I gene (MGAT1), which controls complex and hybrid N-glycan synthesis. Biochem. J. 321, 465–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ioffe E., Stanley P. (1994) Mice lacking N-acetylglucosaminyltransferase I activity die at mid-gestation, revealing an essential role for complex or hybrid N-linked carbohydrates. Proc. Natl. Acad. Sci. U.S.A. 91, 728–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. von Schaewen A., Sturm A., O'Neill J., Chrispeels M. J. (1993) Isolation of a mutant Arabidopsis plant that lacks N-acetyl glucosaminyl transferase I and is unable to synthesize Golgi-modified complex N-linked glycans. Plant Physiol. 102, 1109–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kang J. S., Frank J., Kang C. H., Kajiura H., Vikram M., Ueda A., Kim S., Bahk J. D., Triplett B., Fujiyama K., Lee S. Y., von Schaewen A., Koiwa H. (2008) Salt tolerance of Arabidopsis thaliana requires maturation of N-glycosylated proteins in the Golgi apparatus. Proc. Natl. Acad. Sci. U.S.A. 105, 5933–5938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fanata W. I., Lee K. H., Son B. H., Yoo J. Y., Harmoko R., Ko K. S., Ramasamy N. K., Kim K. H., Oh D. B., Jung H. S., Kim J. Y., Lee S. Y., Lee K. O. (2013) N-Glycan maturation is crucial for cytokinin-mediated development and cellulose synthesis in Oryza sativa. Plant J. 73, 966–979 [DOI] [PubMed] [Google Scholar]
- 26. Altmann F., März L. (1995) Processing of asparagine-linked oligosaccharides in insect cells: evidence for α-mannosidase II. Glycoconj. J. 12, 150–155 [DOI] [PubMed] [Google Scholar]
- 27. Paschinger K., Hackl M., Gutternigg M., Kretschmer-Lubich D., Stemmer U., Jantsch V., Lochnit G., Wilson I. B. (2006) A deletion in the Golgi α-mannosidase II gene of Caenorhabditis elegans results in unexpected non-wild-type N-glycan structures. J. Biol. Chem. 281, 28265–28277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Strasser R., Schoberer J., Jin C., Glössl J., Mach L., Steinkellner H. (2006) Molecular cloning and characterization of Arabidopsis thaliana Golgi α-mannosidase II, a key enzyme in the formation of complex N-glycans in plants. Plant J. 45, 789–803 [DOI] [PubMed] [Google Scholar]
- 29. Strasser R., Steinkellner H., Borén M., Altmann F., Mach L., Glössl J., Mucha J. (1999) Molecular cloning of cDNA encoding N-acetylglucosaminyltransferase II from Arabidopsis thaliana. Glycoconj. J. 16, 787–791 [DOI] [PubMed] [Google Scholar]
- 30. Wilson I. B., Altmann F. (1998) Structural analysis of N-glycans from allergenic grass, ragweed and tree pollens: core α1,3-linked fucose and xylose present in all pollens examined. Glycoconj. J. 15, 1055–1070 [DOI] [PubMed] [Google Scholar]
- 31. Bione H. M., Wilson P. R. (1998) The effect of the mismatch between the core diameter of self-threading dentine pins and the pinhole diameter. Aust. Dent. J. 43, 181–187 [DOI] [PubMed] [Google Scholar]
- 32. Castilho A., Pabst M., Leonard R., Veit C., Altmann F., Mach L., Glössl J., Strasser R., Steinkellner H. (2008) Construction of a functional CMP-sialic acid biosynthesis pathway in Arabidopsis. Plant Physiol. 147, 331–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kaulfürst-Soboll H., Rips S., Koiwa H., Kajiura H., Fujiyama K., von Schaewen A. (2011) Reduced immunogenicity of Arabidopsis hgl1 mutant N-glycans caused by altered accessibility of xylose and core fucose epitopes. J. Biol. Chem. 286, 22955–22964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kajiura H., Okamoto T., Misaki R., Matsuura Y., Fujiyama K. (2012) Arabidopsis β1,2-xylosyltransferase: substrate specificity and participation in the plant-specific N-glycosylation pathway. J. Biosci. Bioeng. 113, 48–54 [DOI] [PubMed] [Google Scholar]
- 35. Gomord V., Denmat L. A., Fitchette-Lainé A. C., Satiat-Jeunemaitre B., Hawes C., Faye L. (1997) The C-terminal HDEL sequence is sufficient for retention of secretory proteins in the endoplasmic reticulum (ER) but promotes vacuolar targeting of proteins that escape the ER. Plant J. 11, 313–325 [DOI] [PubMed] [Google Scholar]
- 36. Nelson B. K., Cai X., Nebenführ A. (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51, 1126–1136 [DOI] [PubMed] [Google Scholar]
- 37. Saint-Jore-Dupas C., Nebenführ A., Boulaflous A., Follet-Gueye M. L., Plasson C., Hawes C., Driouich A., Faye L., Gomord V. (2006) Plant N-glycan processing enzymes employ different targeting mechanisms for their spatial arrangement along the secretory pathway. Plant Cell 18, 3182–3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Strasser R., Altmann F., Mach L., Glössl J., Steinkellner H. (2004) Generation of Arabidopsis thaliana plants with complex N-glycans lacking β1,2-linked xylose and core alpha1,3-linked fucose. FEBS Lett. 561, 132–136 [DOI] [PubMed] [Google Scholar]
- 39. Henquet M., Lehle L., Schreuder M., Rouwendal G., Molthoff J., Helsper J., van der Krol S., Bosch D. (2008) Identification of the gene encoding the α1,3-mannosyltransferase (ALG3) in Arabidopsis and characterization of downstream N-glycan processing. Plant Cell 20, 1652–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Frank J., Kaulfürst-Soboll H., Rips S., Koiwa H., von Schaewen A. (2008) Comparative analyses of Arabidopsis complex glycan1 mutants and genetic interaction with staurosporin and temperature sensitive3a. Plant Physiol. 148, 1354–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pagny S., Bouissonnie F., Sarkar M., Follet-Gueye M. L., Driouich A., Schachter H., Faye L., Gomord V. (2003) Structural requirements for Arabidopsis β1,2-xylosyltransferase activity and targeting to the Golgi. Plant J. 33, 189–203 [DOI] [PubMed] [Google Scholar]
- 42. Nilsson T., Slusarewicz P., Hoe M. H., Warren G. (1993) Kin recognition. A model for the retention of Golgi enzymes. FEBS Lett. 330, 1–4 [DOI] [PubMed] [Google Scholar]
- 43. Bretscher M. S., Munro S. (1993) Cholesterol and the Golgi apparatus. Science 261, 1280–1281 [DOI] [PubMed] [Google Scholar]
- 44. Castilho A., Strasser R., Stadlmann J., Grass J., Jez J., Gattinger P., Kunert R., Quendler H., Pabst M., Leonard R., Altmann F., Steinkellner H. (2010) In planta protein sialylation through overexpression of the respective mammalian pathway. J. Biol. Chem. 285, 15923–15930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yoo J. Y., Ko K. S., Lee S. Y., Lee K. O. (2014) Glycoengineering in plants for the development of N-glycan structures compatible with biopharmaceuticals. Plant Biotechnol. Rep. 8, 357–376 [Google Scholar]