Background: The interaction between the C terminus of claudin proteins and the ZO-1 PDZ1 domain regulates tight junction assembly.
Results: Solved structures of PDZ1 in complex with claudin-1 and claudin-2 and determined binding affinities.
Conclusion: Phosphorylation state of the tyrosine at position-6 regulates claudin/ZO-1 interaction.
Significance: Revealed how post-translational modifications could affect the claudin/ZO-1 interaction and thereby tight junction barrier properties.
Keywords: phosphorylation, phosphotyrosine, scaffold protein, structural biology, tight junction, ZO-1, claudin
Abstract
The molecular seal between epithelial cells, called the tight junction (TJ), is built by several membrane proteins, with claudins playing the most prominent role. The scaffold proteins of the zonula occludens family are required for the correct localization of claudins and hence formation of the TJ. The intracellular C terminus of claudins binds to the N-terminal PDZ domain of zonula occludens proteins (PDZ1). Of the 23 identified human claudin proteins, nine possess a tyrosine at the −6 position. Here we show that the claudin affinity for PDZ1 is dependent on the presence or absence of this tyrosine and that the affinity is reduced if the tyrosine is modified by phosphorylation. The PDZ1 β2-β3 loop undergoes a significant conformational change to accommodate this tyrosine. Cell culture experiments support a regulatory role for this tyrosine. Plasticity has been recognized as a critical property of TJs that allow cell remodeling and migration. Our work provides a molecular framework for how TJ plasticity may be regulated.
Introduction
Tight junctions create selective paracellular barriers between epithelial cells limiting movement of material through the intracellular space (1). The cell-cell junction seal is formed by members of the claudin family. Claudins have four transmembrane domains; it is the extracellular loops connecting the transmembrane domains that form the paracellular barrier. The C-terminal cytoplasmic sequences of claudin bind to the PDZ domains of several distinct scaffolding proteins. The most fully characterized interaction is with PDZ domains of the zonula occludens proteins ZO-1,3 ZO-2, and ZO-3 (2). This is a critical interaction, as without it claudins do not localize to build a tight junction (3). Although less well characterized, claudins also interact with MUPP1 (multi-PDZ domain-containing protein-1) (4) and Pals1-associated tight junction protein (PATJ)) (5) via PDZ-mediated interactions. Although most of the 23 human claudins end in a similar Tyr-Val canonical PDZ binding motif (see Fig. 1), their more upstream residues are both shared and distinct (6). The mechanisms that regulate claudin function remain poorly defined but of considerable interest. Potentially their localization at the barrier, turnover, trafficking, and selective barrier properties might be influenced by the specificity and affinity of binding to cytoplasmic scaffolding proteins, such as ZO-1. Furthermore, it seems reasonable to speculate that the affinity of claudins for the PDZ domains of the scaffolding proteins is dynamically regulated to allow remodeling during cell-cell movements. This led us in the current study to determine the structural determinants of specificity and affinity for claudin binding to their PDZ target (PDZ1) in ZO-1.
FIGURE 1.

Sequence alignment of the 10-most C-terminal residues in human claudins. Humans have 23 annotated claudin (CLDN) genes; CLDN13 is missing in humans; CLD25 has been previously referred to as CLDN21. Of these 23 human CLDN genes, 15 contain a -YV at the C terminus (highlighted in yellow). The tyrosine at position −6 (9 of 23) is highlighted in green.
Because of the prevalence of PDZ domains and their role in myriad cellular events, there has been considerable interest in understanding the structural basis for this type of protein-protein interaction and whether these interactions are regulated (7). Within the human genome alone there are >150 PDZ domain-containing proteins, many with multiple PDZ domains (8). The claudin-binder ZO-1, the focus of this work, contains three tandem PDZ domains in its N terminus, and it is the first PDZ domain (PDZ1) that binds claudins. The second (PDZ2) binds connexins, which are gap junction proteins, showcasing the diverse role of ZO-1 in cellular junctions. Interestingly, PDZ2 is also required for ZO-1 homo- and heterodimerization with other ZO proteins via a unique domain-swapping mechanism (9, 10). This dimerization is critical for normal tight junction barrier formation (11). Finally, the third ZO-1 PDZ domain (PDZ3) binds the tight junction protein JAM-A (12). Thus, ZO-1 can assemble multiprotein complexes with multiple junction proteins via its three PDZ domains. Here we examine in detail the interaction between claudin-1 and -2 to PDZ1 of ZO-1.
The first studies of claudin binding to ZO-1, which were based on mutational deletions, suggested a requirement for the C-terminal residue, position 0 (P0), and the residue at position −2 (P−2) of the ligand for binding to its cognate PDZ-domain (2). The accepted nomenclature used to refer to residues binding to PDZ domains is as follows. The C-terminal residue is referred to as the P0 residue; subsequent residues toward the N terminus are termed P−1, P−2, P−3, etc. More recent studies reveal a role for residues at positions P−1, P−3, P−4, and P−5 in defining specificity (13). In the “general model” the claudin's P0 hydrophobic side chain inserts into a hydrophobic pocket, and the C-terminal carboxylate coordinates with a largely conserved GLGF motif within the PDZ domain (7). The preceding several residues build an antiparallel strand to the β2 strand of the PDZ domain. In contrast to specificity, not much is known about what determines the interaction affinity. Most reported affinities are relatively low and in the micromolar range (14). However, the implication of evidence in several studies is that phosphorylation of claudins near their PDZ binding motifs alters affinity for ZO-1. For example, the cytoplasmic tails of many claudins are documented to be phosphorylated at multiple positions including serines, threonines, and tyrosines (15). Changes in phosphorylation have been correlated with changes in barrier properties, trafficking, and turnover, cited in Van Itallie et al. (6), although no direct evidence has yet been provided that phosphorylation alters affinity of the PDZ domain-target interaction. Similar correlative findings have been reported after introducing phosphomimicking mutations (i.e. mutating the Ser/Thr/Tyr residue to Asp or Glu) although the mechanisms underlying these changes are not defined (16, 17). We were particularly interested in testing a role for the tyrosine at the P−6, which we had previously shown could be phosphorylated in vivo (6) and because this position has a tyrosine residue in 9 of the 23 human claudins (Fig. 1).
To better understand the specificity and affinity for the interaction of different claudins with ZO-1, we defined the structural basis for the interaction of ZO-1 PDZ1 with claudin-1, which lacks Tyr−6, and claudin-2, which contains Tyr−6. Surprisingly, there are distinct and significant differences between the way claudin-1 binds PDZ1 and the way claudin-2 binds PDZ1. Contacts of claudin-2 with the PDZ1 domain include residues more distal from the C terminus than does claudin-1. Specifically, the tyrosine residue present in claudin-2 at P−6 intercalates between the β2-β3 loop of the core PDZ1 domain. This observation suggested that due to the more extensive interaction, claudin-2 would bind more tightly than claudin-1 to PDZ1. Isothermal thermal calorimetry and NMR experiments verified this prediction. The functional relevance of the tyrosine residue was probed by introducing the phosphomimicking mutation Y224E (mimicking not the structure of a phosphorylated tyrosine, but charge), which revealed that the mutation at this site interrupts in vitro binding to PDZ1 and results in loss of the claudin-2 barrier properties in cultured MDCK cells. The modulation of claudin-2 binding to PDZ1 by phosphorylation state of Tyr-224 suggests a potential mechanism for physiologic regulation of affinity. These results can be applied to a better understanding of the regulation of claudin function at the tight junction and potentially be generalized to PDZ-ligand interactions outside the tight junction.
Experimental Procedures
Protein Expression and Purification
For the ZO-1 PDZ1 domain, a DNA fragment encoding for residues 18–110 of human ZO-1 was cloned into the NdeI/BamHI sites of a pET14b expression vector that includes the tobacco etch virus cleavage site. In addition, the QuikChange® site-directed mutagenesis kit from Agilent Technologies was used to fuse 10-residue extensions to the C terminus of PDZ1 to produce PDZ1-Cld1 (extension, -GGGSSGKDYV), PDZ1-Cld2 (extension, -GGGYSLTGYV), and PDZ1-Cld2Y224E (extension, -GGGESLTGYV). Human claudin tails were cloned into the NdeI/BamHI sites of a modified pET14b expression vector that includes a SUMO tag. 13-Residue extensions were thus inserted to generate the following constructs: His-SUMO-Cld1 (extension, -YPKPAPSSGKDYV), His-SUMO-Cld2 (extension, -KSEFNSYSLTGYV), and His-SUMO-Cld2Y224E (extension, -KSEFNSESLTGYV). All recombinant proteins were expressed in Escherichia coli (BL21 C41). Cells were cultured at 37 °C in 2YT medium then induced by 0.1 mm isopropyl β-d-1-thiogalactopyranoside at 0.6 absorbance (600 nm) and left to grow overnight at 22 °C. Cells were then harvested by centrifugation at 5000 rpm for 30 min. Pellets were resuspended and washed with 100 mm KCl, 50 mm Tris-HCl, pH 7.5. Lysis was accomplished by sonication in a buffer composed of 25 mm Tris-HCl, pH 7.5, 100 mm KCl, 10% glycerol, 1% Triton X-100, and 1 mm PMSF. After centrifugation at 30,000 rpm for 1 h, the supernatant was loaded onto a 5-ml His-trap nickel affinity column (GE Healthcare), and the column was washed with 300 ml of buffer A composed of 25 mm Tris-HCl, pH 7.5, and 100 mm KCl supplemented with 25 mm imidazole. Two supplemental wash steps were performed by flowing 100 ml of buffer A supplemented with (i) 50 mm then (ii) 75 mm imidazole. The bound protein was then eluted in buffer A supplemented with 500 mm imidazole. Fractions containing the protein were pooled and concentrated. All ZO-1 PDZ1 constructs were cleaved using tobacco etch virus protease, and the cut protein was injected onto a S-200 gel filtration column (GE Healthcare) equilibrated with 25 mm Tris-HCl, pH 7.5, 500 mm NaCl, pH 7.5. The purity of the protein preparations was confirmed using SDS-PAGE and detected through Coomassie staining. Proteins were concentrated to ∼20 mg/ml (ZO1-PDZ1) and ∼12 mg/ml (ZO1-PDZ1-Clds) and stored at −80 °C until used for crystallization experiments. After elution of the His-trap column, all His-SUMO-Clds constructs were concentrated and stored at −80 °C until used for calorimetry experiments.
Peptide Synthesis
The acetylated heptapeptides corresponding to the cytoplasmic tail of human claudin-1 (sp_O95832), claudin-2 (sp_P57739), claudin-2 with a phosphorylated tyrosine at position −6, and claudin-2Y224E (the mutated claudin-2 at position −6) were prepared by Biomatik Corp. (Cambridge, MA). The amino acid composition and purity (>98%) of peptides were verified by mass spectroscopy and HPLC analysis by the manufacturer.
Isothermal Titration Calorimetry (ITC)
ITC experiments were performed on a VP-ITC instrument (MicroCal). All ITC experiments involved direct titration of the His-SUMO ligand or acetylated peptide ligand into protein solution. PDZ1 (12.5 μm) was thus titrated with 5–10-μl injections of His-SUMO-Claudin tails (330–1600 μm) or ac-Cld2 (450 μm). All proteins were dialyzed against the same buffer of 25 mm Tris-HCl, pH 7.5, 500 mm NaCl, and an aliquot of the latter dialysis buffer was used for dilutions and blank experiments. Experiments were performed at 25 °C; 5–10-μl injections were performed at 200-s intervals, stirring speed used was 307 rpm, and reference power was 10 μcal/s. The first injection peak was systematically discarded because of the backlash effect of the syringe plunger. Heats of dilution were measured in blank titrations by injecting the ligands into buffer and subtracted from the observed raw heat values. Control experiments were performed and consisted of injecting PDZ1 into buffer and His-SUMO into PDZ1 solution, but these titrations did not produce a significant signal. Data-fitting was based on a one-site binding model using the commercial package provided (Origin) to obtain the values of Kd, ΔH, and ΔS. Each titration was recorded at least in duplicate.
NMR Experiments
NMR experiments were performed on a Bruker 900 MHz spectrometer equipped with a cryogenic triple resonance probe. Experimental conditions were 200 μm peptide, ±20 μm protein in 100 mm PO4, pH 7.2, in 90% 1H2O, 10% 2H2O at 25 °C in 3-mm NMR tubes. The WaterLOGSY experiments were performed as described previously (18). The data were collected with a sweep width of 14,370 Hz and a relaxation delay of 2 s. Water was selectively saturated using a 2-ms square-shaped pulse with a mixing time of 2 s. All data were processed by NMRPipe (19) with referencing to the water peak at 4.773 ppm and using a 4-Hz line-broadening window.
Crystallization
Crystals of the PDZ1-Cld1 and PDZ1-Cld2Y224E proteins were grown at 20 °C using hanging drop vapor diffusion against a reservoir containing 20–40% PEG 2000 monomethyl ether, 0.1 m sodium acetate (NaAc), pH 4.6, 0.2 m ammonium sulfate, whereas PDZ1-Cld2 protein crystals where grown at 12 °C against a reservoir containing 2.0 m sodium formate, 0.1 m NaAc, pH 4.6. Drops of 2.0 μl were set up at a 1:1 ratio of reservoir-to-protein solution at a concentration of 7–10 mg/ml. Before data collection, crystals were transferred into a solution containing 80% of the mother liquor, 20% glycerol for cryoprotection.
Data Collection and Structure Solution Determination
Diffraction data for all constructs were collected on the Life Sciences Collaborative Access Team (LS-CAT) ID-G using a MARCCD 300 detector at a temperature of 100 K and a wavelength of 0.9785 Å. Data were processed and scaled with XDS and XSCALE (20). Structures were determined by molecular replacement with MOLREP (21) using the human ZO1-PDZ1 structure (PDB entry 2H3M; Ref. 22) as a search model. Refinement was conducted using REFMAC5 (23) and model building using Coot (24). The unliganded ZO-1 PDZ1 domain structure was obtained from crystals that contained the PDZ1-Cld2Y224E fusion protein. Because we did not observe electron density for the residues corresponding to the modified claudin-2 tail sequence, we designate this as the unliganded ZO-1 PDZ1 structure. This 1.9 Å resolution structure is very similar to the 2.9 Å resolution structure previously reported (root mean square deviation (r.m.s.d.) of 0.22 Å on 82 atoms with PDB entry 2H3M (22). Non-crystallographic symmetry restraints were not used during refinement. Structural figures were prepared using the PyMol Molecular Graphics System, Version 1.6.0 Schrödinger, LLC.
Cell Biology and Physiology Studies
MDCK II Tet-off cells, cells expressing wild type mouse claudin-2 (25), and (−) PDZ binding motif (26) have been previously described. A claudin-2 Y224E mutant was generated using the Agilent QuikChange II site-directed mutagenesis kit with the following sense primer (antisense primers are the reverse complement): 5′-CAAAGCCAAGAGTGAGTTCAACTCAGAAAGCCTGACTGGGT-3′. Stable cell lines were generated as described previously in Tet-off MDCK II cells. pTRE claudin-2 Y224E vectors were co-transfected with pTKhyg, and clones were selected by culturing cells in 200 μg/ml hygromycin. Two clonal cell lines were selected and characterized. For all localization and barrier assays cells were grown on Transwell filters (Corning) for 5 days. Measurement of dilution potential and transepithelial electrical resistance (TER) was carried out as previously described (25). Standard SDS-PAGE immunoblotting and immunofluorescence were carried out as described previously (26, 27). Claudin and tubulin antibodies were purchased from Life Technologies; IR-coupled secondary antibodies were from Rockland (Gilbertsville, PA).
Results and Discussion
Crystal Structures of the ZO-1 PDZ1 Domain with and without Claudin Ligands
For the purpose of studying the structure of the human ZO-1 PDZ1 domain in complex with the C terminus of human claudin-1 and -2, we attempted both co-crystallization and soaking methods using heptapeptides corresponding to the respective claudin sequences. However, we did not observe electron density at the PDZ1 binding region, presumably due to steric limitations caused by crystal packing. Therefore, we took advantage of a previously described strategy in which the claudin C-terminal residues are fused at the C terminus of the PDZ domain via a tri-glycine linker (22, 28, 29), such that the residues corresponding to the claudin would bind to a PDZ1 domain of a neighboring molecule. Using this approach we determined the crystal structures of the ZO-1 PDZ1 domain in complex with the 7-most C-terminal residues of claudin-1 (-SSGKDYVCOOH, referred to as PDZ1-Cld1) and claudin-2 (-YSLTGYVCOOH, PDZ1-Cld2). See Table 1 for detailed data collection and refinement statistics. In addition, because we had previously determined by mass spectrometry that the claudin-2 P−6 tyrosine could be phosphorylated in vivo (6), we crystallized a construct containing PDZ1 followed by a claudin-2 sequence mutated at P−6 with a phosphomimicking mutation: -ESLTGYVCOOH (PDZ1-Cld2Y224E). However, when we solved this structure at 1.9 Å resolution, we could not observe electron density for the claudin-2-related residues and hence regard this structure as a high resolution unliganded PDZ1 structure. All comparisons to claudin-1- and -2-bound PDZ1 complexes are done relative to this high resolution structure.
TABLE 1.
Data collection and refinement statistics
a.u., asymmetric unit.
| PDZ1 | PDZ1-Cld1 | PDZ1-Cld2 | |
|---|---|---|---|
| PDB codes | 4OEO | 4OEP | 4YYX |
| Data collection statistics | |||
| Resolutiona (Å) | 30−1.89 (2.01−1.89) | 30−2.35 (2.49−2.35) | 30−1.79 (1.90−1.79) |
| Number of reflections | |||
| Observed | 59,341 (9,444) | 95,219 (14,861) | 162,977 (13,612) |
| Unique | 24,585 (3,900) | 13,306 (2,056) | 22,067 (3,348) |
| Completeness (%) | 95.1 (94.2) | 99.6 (99.2) | 99.2 (95.1) |
| Rsym (%) | 2.8 (44.8) | 5.8 (65.1) | 7.7 (59.2) |
| Average I/σ(I) | 17.5 (2.7) | 29.3 (3.4) | 16.8 (2.3) |
| Redundancy | 2.4 (2.4) | 7.1 (7.2) | 7.4 (4.1) |
| Space group | C2 | P41212 | P212121 |
| Unit cell (Å): a, b, c | 104.9, 60.4, 62.2 | 106.2, 106.2, 53.9 | 41.3, 72.2, 76.9 |
| α, β, γ | 90.0, 124.2, 90.0 | 90, 90, 90 | 90, 90, 90 |
| Refinement statistics | |||
| Rcryst (%) | 19.5 (34.3) | 21.4 (35.1) | 19.8 (35.7) |
| Rfree (%) | 24.3 (39.3) | 25.6 (32.2) | 25.3 (42.9) |
| Resolution range (Å) | 51.42–1.90 | 75.12–2.35 | 52.64–1.79 |
| Protein molecules per a.u. | 3 | 2 | 2 |
| Number of atoms | |||
| Protein | 2,037 | 1,543 | 1,597 |
| Water molecules | 96 | 43 | 187 |
| SO4− | 10 | ||
| Acetate | 12 | 8 | 16 |
| Formate | 6 | ||
| PEG | 63 | 32 | |
| r.m.s.d. from ideal | |||
| Bond length (Å) | 0.012 | 0.012 | 0.013 |
| Bond angles (°) | 1.558 | 1.688 | 1.561 |
| Average B-factors (Å2) | |||
| Protein | 36.6 | 47.7 | 13.9 |
| Water molecules | 38.9 | 49.6 | 30.7 |
| SO4− | 38.3 | ||
| Acetate | 47.2 | 54.5 | 32.6 |
| Formate | 30.0 | ||
| PEG | 44.7 | 49.4 | |
| Ramachandran plot (%) | |||
| Most favored regions | 88.9 | 90.0 | 89.9 |
| Additionally allowed regions | 10.3 | 8.8 | 10.1 |
| Generously allowed | 0.9 | 1.2 | 0.0 |
| Disallowed regions | 0.0 | 0.0 | 0.0 |
a The last shell is in parentheses.
Only the Four-most C-terminal Residues of Claudin-1 Make Contact with the ZO-1 PDZ1 Domain
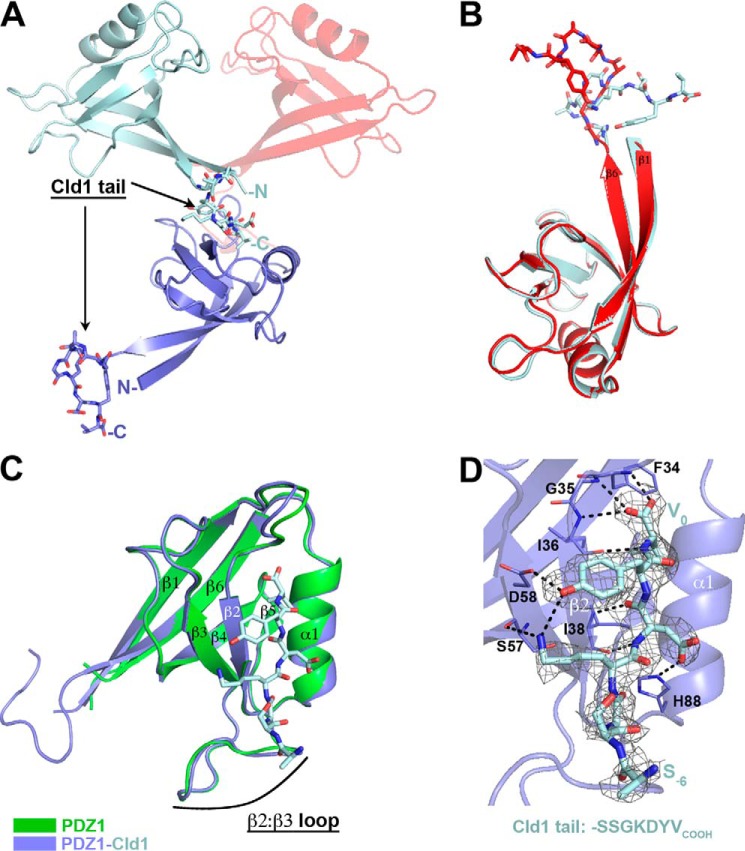
We determined the crystal structure of the ZO-1 PDZ1 domain in complex with the C-terminal residues of human claudin-1 (-SSGKDYV) at a resolution of 2.35 Å (Table 1). PDZ1-Cld1 crystallized in the P41212 space group with two molecules in the asymmetric unit (depicted in light blue and red, Fig. 2A). The residues corresponding to the claudin-1 C-terminal sequence from one protomer (e.g. light blue, Fig. 2A) bind to the PDZ1 domain related by crystallographic symmetry (purple, Fig. 2A). The PDZ1 domains of the two independent molecules in the asymmetric unit are essentially identical (r.m.s.d. of 0.29 Å on 86 atoms) as is the conformation of the claudin-1 residues. However, the precise relative orientation of PDZ1 to claudin-1 differs between the molecules in the asymmetric unit (Fig. 2B), made possible by the three glycine residues that link the PDZ1 domain to the claudin-1 sequence. The observed flexibility of the linker suggests that no artificial constraints (e.g. linker, crystal contacts) determine the precise conformation of the claudin-1 residues.
FIGURE 2.
Claudin-1 (Cld1) uses its 4-C-terminal residues to bind to the ZO-1 PDZ1 domain, which does not undergo any significant conformational change upon Cld1 binding. A, the C-terminal sequence of Cld1 was fused to C terminus of PDZ1 (see “Experimental Procedures”). The two PDZ1-Cld1 molecules in the asymmetric unit are represented in light blue and red. The Cld1 tail from one molecule (light blue) binds at the PDZ binding site of a crystallographically related molecule (purple). B, the two PDZ1-CLD1 molecules in the asymmetric unit are overlaid showing the different orientations of the Cld1 peptides. C, overlay of unliganded PDZ1 (green) on the PDZ1-Cld1 complex reveals no conformational change in PDZ1 upon ligand binding. D, detailed view of the PDZ1-Cld1 interaction. Hydrogen bond interactions are denoted with a dashed line. An Fo − Fc omit map (gray mesh) contoured at 2.0 σ around Cld1 peptide is also shown. Cld1 peptide was removed from the model that then underwent several rounds of refinement to eliminate model bias.
Superimposition of the unliganded and claudin-1-bound PDZ1 structures reveals that binding of claudin-1 does not generate a conformational change within the PDZ1 domain (r.m.s.d. of 0.33 Å on 85 atoms; Fig. 2C). PDZ domains usually recognize sequences of about three-five residues in length with a few examples extending to six residues (30). Here we show that claudin-1 interacts with PDZ1 through its terminal four amino acids, from Val0 to Lys−3 (Fig. 2D and supplemental Fig. S1). As expected, Val0 makes extensive interactions with PDZ1: its carboxyl group forms polar contacts with the main chain atoms of Phe-34 and Gly-35, its side chain orients toward a hydrophobic area, and its NH atom makes a hydrogen bond with the carbonyl of Ile-36. The preceding residue, Tyr−1, is at hydrogen bond distance to Asp-58 of PDZ1, but otherwise lacks extensive hydrophobic interactions. The side chain of Tyr−1 also interacts with the side chain of Lys−3. Farther away from the claudin-1 C terminus, Asp−2 makes main chain hydrogen bond contacts with Ile-38 and through its side chain builds a polar contact with the side chain of His-88. Finally, the side chain of Lys−3 makes a hydrogen bond with the main chain of Ser-57 as well as the aforementioned interaction with Tyr−1. Most relevant to our understanding of the interaction between claudin-1 and the PDZ1 domain, the claudin-1 residues beyond Lys−3 are not observed to make any direct interaction with the PDZ1 domain.
Seven C-terminal Residues of Claudin-2 Make Contact with the ZO-1 PDZ1 Domain
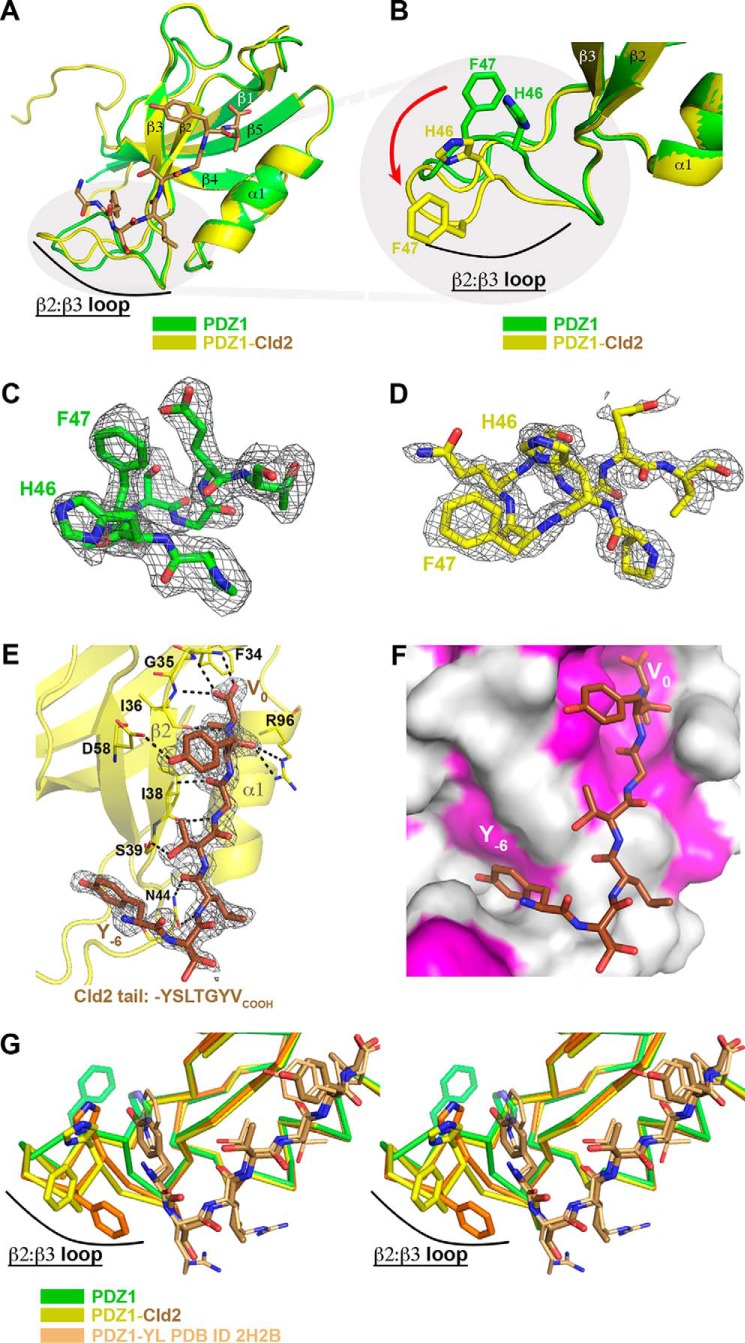
Next we determined the structure of the PDZ1 domain in complex with human claudin-2 (-YSLTGYV) at a resolution of 1.79 Å (Table 1). PDZ1-Cld2 crystallized in the P212121 space group and displays 2 molecules in the asymmetric unit. As observed for PDZ1-Cld1, the C-terminal claudin-2 residues bind to an adjacent crystallographically related PDZ1 domain. Unliganded PDZ1 and PDZ1-Cld2 were superimposed (r.m.s.d. of 0.38 Å on 77 atoms; Fig. 3A). Notably different from the case observed with claudin-1 binding to PDZ1, the binding of claudin-2 involves a main chain conformational change within the β2:β3 loop of the PDZ1 domain (Fig. 3B, red arrow). Unbiased electron density for the β2:β3 loop in the apo- and claudin-2-bound state is shown in Fig. 3, C and D, respectively (see also supplemental Fig. S2). Indeed, Tyr−6 intercalates in the β2:β3 loop (Fig. 3, E and F), which requires a major conformational change of residues His-46 and Phe-47 of PDZ1 to avoid a steric clash (Figs. 3B). Residues Val0 and Tyr−1, common to both claudin-1 and -2, interact with PDZ1 in a similar manner. However, the main chain of Tyr−1 in claudin-2 makes an extra interaction with the side chain of Arg-96 from PDZ1 (Fig. 3E). This interaction is made possible by the presence of a glycine residue at P−2 of claudin-2; any larger claudin residue at that position (it is an Asp in claudin-1) would sterically block the extension of the PDZ1 Arg-96 side chain, the conformation required for interaction with Tyr−1. The aforementioned Gly−2 interacts via its main chain NH and carbonyl atoms with the carbonyl and NH atoms of Ile-38, respectively. The preceding residue in claudin-2, Thr−3, employs its side chain to hydrogen bond with the Ser-39 side chain, whereas the Leu−4 main chain NH and carbonyl atoms make polar contacts with Asn-44 (Fig. 3E). Finally, although Ser−5 faces away from the PDZ1 domain, Tyr−6 inserts in the β2:β3 loop where it is stabilized by hydrophobic interactions (Fig. 3F). A previous study using phage display to identify high affinity peptide ligands of the ZO-1 PDZ1 domain revealed that the best binders have a hydrophobic residue (tryptophan or tyrosine) at P−6 that intercalates in the β2:β3 loop (22), a feature we also observe with the physiologically relevant claudin-2 sequence (Fig. 3G). To summarize the structural results, the PDZ1-Cldn2 complex structure reveals extended interactions to the PDZ1 domain that include the interaction of Tyr−6 with the β2:β3 loop. This is in contrast to the interaction with claudin-1, which is limited to residues between P−2 and P0.
FIGURE 3.
Claudin-2 (Cld2) uses its seven C-terminal residues to bind to the ZO-1 PDZ1 domain, which as a result undergoes a significant conformational change in its β2-β3 loop. A, overlay of unliganded PDZ1 (green) on the PDZ1-Cld2 complex (PDZ1 in yellow, Cld2 residues in brown). Note the conformational change of the β2-β3 loop. B, zoom on the β2-β3 loop. In addition to a change in main chain atoms of the entire loop, most dramatic is the change in position of the His-46–Phe-47 side chains. The red arrow demonstrates the conformational change. C, Fo − Fc omit map (gray mesh) contoured at 2.0 σ around unliganded PDZ1 β2-β3 loop. D, Fo − Fc omit map (gray mesh) contoured at 2.0 σ around PDZ1-Cld2 β2-β3 loop. E, detailed view of the PDZ1-Cld2 interaction. Hydrogen bond interactions are denoted with a dashed line. Fo − Fc omit map (gray mesh) contoured at 2.0 σ around Cld2 peptide is also shown. Cld2 peptide was removed from the model that then underwent several rounds of refinement to eliminate model bias. F, surface representation of PDZ1 with hydrophobic surfaces colored in magenta. Note how Tyr−6 buries its side chain into the area previously occupied by the β2-β3 loop. G, stereoview of the overlay of unliganded PDZ1 (green), Cld2-bound PDZ1 (yellow for PDZ1, brown for Cld2), and PDZ1 in complex with a tight binding peptide identified via phage display (orange). The P−6 tryptophan residue present in the phage display sequence binds in a similar manner to the Cld2 Tyr−6. As seen for PDZ1-Cld2, the β2-β3 loop adjusts its position to accommodate the tryptophan side chain. This suggests that high affinity binders utilize a P−6 residue (Tyr or Trp) that displaces the β2-β3 loop and buries its side chain in the ensuing cavity.
Calorimetric Measurements
The more extended interaction between claudin-2 and PDZ1 relative to that formed by claudin-1 suggest that the binding affinity between PDZ1 and claudin-2 would be greater than that between PDZ1 and claudin-1. To test this prediction we used ITC to determine the affinities of claudin peptides to ZO-1 PDZ1 (Table 2, Fig. 4). To provide the high concentrations of peptide required for titration, we generated and expressed in E. coli constructs in which the claudin-1, -2, and -2Y224E C-terminal sequences are fused to a His-SUMO tag (see “Experimental Procedures” for details). Titrations of claudin peptides (in the context of fusions to His-SUMO) into PDZ1 as well as the experimental fits are given in Fig. 4. Satisfactory titrations to saturation could be performed only for the claudin-2 peptide (Fig. 4A) with a measured Kd of ∼13.5 μm (Table 2), which is consistent with affinity measured by Zhang et al. (13 μm; Ref. 14) using a different method. The same binding experiment was conducted using an N-terminal-acetylated peptide corresponding to the last seven residues of claudin-2 (acCld2: -acYSLTGYV), and equivalent values were obtained (Table 2, Fig. 4B), thus indicating that the His-SUMO tag is not contributing or otherwise affecting the interaction with PDZ1. In contrast to these claudin-2 binding studies, we were not able to reach saturation upon titration of His-SUMO-Cld1 peptide into PDZ1 under the experimental conditions (Fig. 4C), indicating a very low binding affinity. Interestingly, the same lack of binding as observed with claudin-1 was obtained with claudin-2 when tyrosine at P−6 is mutated into the phosphomimetic amino acid (Glu) (Fig. 4D), indicating that phosphorylation at this position would considerably lower the affinity to PDZ1.
TABLE 2.
Calorimetric analysis of PDZ1/claudin peptides interactions from ITC measurements
| Peptide | Na | Kd | ΔH | ΤΔS |
|---|---|---|---|---|
| μm | kcal/mol | kcal/mol | ||
| His-SUMO-Cld1 | NDb | ND | ND | ND |
| His-SUMO-Cld2 | 1.1 ± 0.1 | 13.5 ± 1.3 | −6808 ± 681.6 | −166.9 |
| His-SUMO-Cld2-Y224E | ND | ND | ND | ND |
| ac-Cld2 | 1.0 ± 0.1 | 16.6 ± 1.3 | −6265 ± 853.9 | 256.3 |
a Stoichiometry.
b ND, not detectable under the experimental conditions.
FIGURE 4.
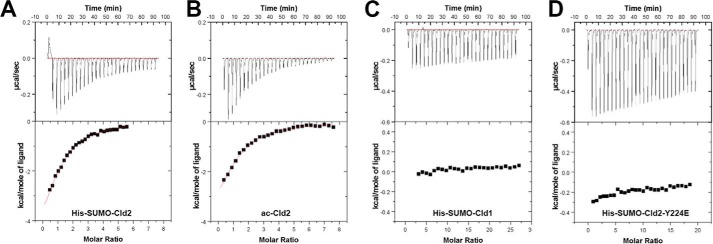
ITC measurements for PDZ1/claudin peptides interactions. Shown are titration data of His-SUMO-Cld2 peptide (A), acetylated Cld2 peptide (B), His-SUMO-Cld1 peptide (C), and His-SUMO-Cld2-Y224E peptide (D) injected into PDZ1 and the integrated heat measurements for the titration corrected from the reference (see “Experimental Procedures”). The measured thermodynamic parameters are reported in Table 2.
NMR Measurements
We have previously shown that WaterLOGSY NMR is useful for the detection of peptide interactions with larger proteins (12). We thus performed WaterLOGSY experiments to detect claudin peptide binding to the ZO1 PDZ1 domain by monitoring the acetyl group present in each peptide. In the WaterLOGSY experiment, the presence of positive resonances occurs when a small molecule such as a peptide binds to a larger biomolecule (18). As shown in Fig. 5A, we observed binding of claudin-2 to PDZ1. On the other hand, we observed little or weak binding of claudin-1 to PDZ1 (Fig. 5B). When we exchanged the Ser−6 residue in claudin-1 by a tyrosine we could detect binding to PDZ1 (Fig. 5C). Conversely, when we mutated the Tyr−6 residue in claudin-2 to a glutamic acid (generating the phosphomimic) or when using a phosphotyrosine-containing peptide the previously strong binding signal was largely diminished (Fig. 5, D and E, respectively). Hence, the NMR studies corroborate the calorimetry results that claudin-2 binds more tightly than claudin-1 to the ZO-1 PDZ1 domain. Additionally, the NMR data shows that the unphosphorylated claudin-2 peptide binds more tightly than its phosphorylated version or its Y224E phosphomimic version. Furthermore, it shows that adding a tyrosine to a claudin that naturally lacks this type of residue at position P−6 can enhance binding. Note that the lack of observed binding of wild type claudin-1 to PDZ1 in this NMR experiment and in the ITC experiment does not mean that it does not bind in cells; it only means that this interaction in isolation and under the experimental conditions is not detectable, so we can only make conclusions as to relative affinities. Because claudins are oligomeric and ZO-1 is at least a homodimer, avidity would greatly increase the affinity in the natural membrane proximity where the interaction between claudins and ZO proteins occurs.
FIGURE 5.
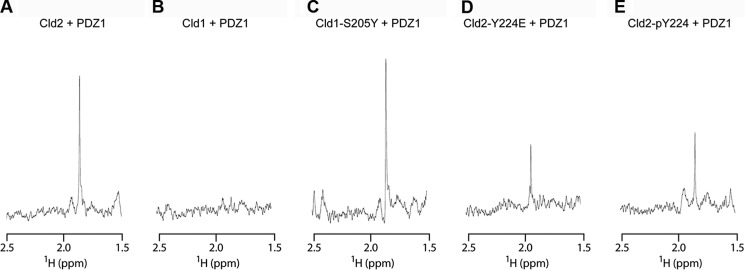
Detection of claudin peptide binding to PDZ1 by NMR. PDZ1 was added to the peptide solution, and WaterLOGSY was used to detect binding. Shown is the acetyl resonance used as a reporter. In this type of experiment the relative height of the signal reflects the degree of binding. Note the strong signal for Cld2 (i.e. strong binding) (A) and lack of signal for Cld1 (i.e. weak binding) (B). C, Cld1 binding could be detected when Ser-205 at P−6 is replaced by a tyrosine. D, the phosphomimetic Cld2-Y224E peptide shows decreased binding relative to Cld2. E, likewise, the phosphotyrosine-containing peptide shows decreased binding relative to the unphosphorylated form.
A Phosphorylation-mimicking Y224E Mutation of Claudin-2 Results in Loss of Its Physiologic Barrier Properties and Decreased Claudin-2 at the Junction
Our structural and biochemical results demonstrate that interaction of Tyr-224 (i.e. Tyr−6) of claudin-2 with the β2:β3 loop of the PDZ1 domain of ZO-1 is critical for creating high affinity binding. We previously showed using mass spectrometry that Tyr-224 can be phosphorylated in MDCK-cultured epithelial cells, although at steady state in confluent monolayers the level is very low (6). Taken together with the structural studies described above leads to speculation that the phosphorylation state at this position might be a physiologic mechanism for modulating the affinity for ZO-1, with possible functional consequences. Claudin-2 normally creates pores through the tight junction with low TER and high Na+ permeability (25, 31). Hence, disruption of claudin-2 function is predicted to result in increased TER and decreased Na+ permeability. To test the possibility that phosphorylation of Tyr-224 alters claudin-2 function, we generated clonal monolayers of MDCK II cells expressing the phosphorylation-mimicking Y224E mutant of claudin-2 under an inducible promoter and determined the effect on barrier properties and cellular localization before and after induction. For comparison, we also determined the effects of expressing previously described wild type claudin-2 (25) and a form of claudin-2 in which the terminal three residues were removed (−3) making it incapable of binding PDZ1 (6).
Expression of all three forms of claudin-2 could be induced in MDCK monolayers as determined by immunoblotting (Fig. 6A). When additional wild type claudin-2 is induced, as previously shown (25), there is no change from the normally low monolayer resistance and high Na+ permeability (Fig. 6B, left bars). In contrast, when the Y224E mutant is induced there is a highly significant doubling in the electrical resistance (Fig. 6B, middle bars) and a decrease in the dilution potential both accounted for a specific decrease in Na+ permeability (Fig. 6C, middle bars). The same electrophysiological phenotype is observed when the (−3) mutant is expressed (Fig. 6, B and C, right bars). This is the predicted outcome if there is less functional claudin-2 in the tight junction.
FIGURE 6.
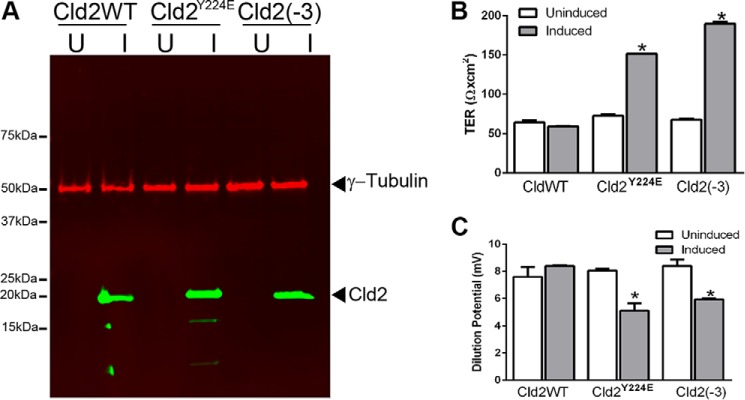
The tyrosine phosphomimic mutant Y224E affects the TER and dilution potential in MDCK monolayers relative to the wild type. A, inducible expression of claudin-2 (Cld2), the Y224E phosphomimic (Cld2Y224E) and a three-residue C-terminal truncation variant (Cld2(−3)). Immunoblot analysis demonstrates inducible expression of the three claudin forms in representative clonal monolayers (U = uninduced; I = induced). B, induction of the wild type claudin-2 results in no change in TER, whereas expressing either the Y224E or (−3) mutants result in a significant increase in TER. C, induction of the wild type claudin-2 results in no change in dilution potential, whereas expressing either the Y224E or (−3) mutants results in a significant decrease. Data represent the means ± S.E. of determinations from duplicate Snapwells for wild type claudins and mutants. Significance was calculated using Student's t test.
A decreased level of claudin-2 at the tight junction was confirmed by confocal immunofluorescent imaging (Fig. 7). ZO-1 was used to define the location of the tight junction in z-axis confocal images. The localization of ZO-1 (green) and endogenous claudin-2 (red) normally coincides resulting in a yellow merged localization (Fig. 7, top row). When additional wild type claudin-2 was induced the continuous junctional colocalization (yellow) intensifies. There is also a small accumulation of claudin-2 in intracellular vesicles (Fig. 7, second row). In contrast, when expression of the Y224E mutant is induced there is less claudin-2 at the junction, and it is less continuous. The merged junctional signal becomes greener, indicating that the ratio of claudin-2 to ZO-1 at the junction has decreased (Fig. 7, bottom row). There is also a visible increase in the relative amount of claudin-2 in vesicles. The antibody used recognizes both the wild type and Y224E forms of claudin-2. Removal of wild type claudin-2 from the junction was also observed after inducing expression of claudin-2 lacking its C-terminal three residues (data not shown).
FIGURE 7.
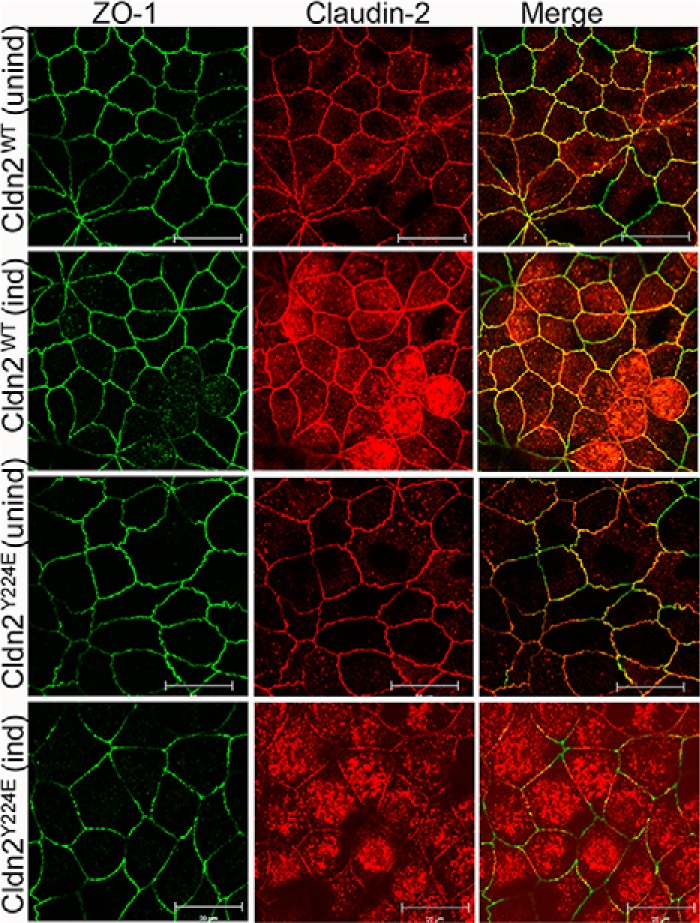
The tyrosine phosphomimic mutant Y224E fails to localize to tight junctions and reduces the wild type claudin-2 at the junction. Shown is confocal immunofluorescence localization ZO-1 (green), claudin-2 (red), and merged imaged (yellow) before and after induction of Cldn2WT and Cldn2Y224E. ZO-1 and endogenous and induced Cldn2WT are well colocalized at the junction (top two rows). In contrast, induction of Cldn2Y224E (bottom row) resulting in less continuous claudin-2 staining and a shift from yellow to green in the merged image is shown.
Taken together we interpret these data as indicating that when the affinity of claudin-2 for PDZ1 is lowered by the phosphorylation-mimicking mutation, Cld2Y224E, or when the C-terminal three residues of the PDZ binding motif are removed Cld2(−3), its physiologic influence as a pore is diminished, possibly through a change in trafficking that results in accumulation in vesicles. We previously showed that claudin-2 forms dimers in MDCK cells (32). We speculate that the mutant forms of claudin-2 bind wild type endogenous claudin-2 resulting in its coincident defective targeting. These results are consistent with a model that has the phosphorylation state of Tyr−6 acting as a modulator of claudin-2 function, specifically, by influencing its affinity (high, non-phosphorylated; low, phosphorylated) to the ZO-1 PDZ1 domain.
Role of the β2-β3 Loop of PDZ Domains in Ligand Binding Affinity
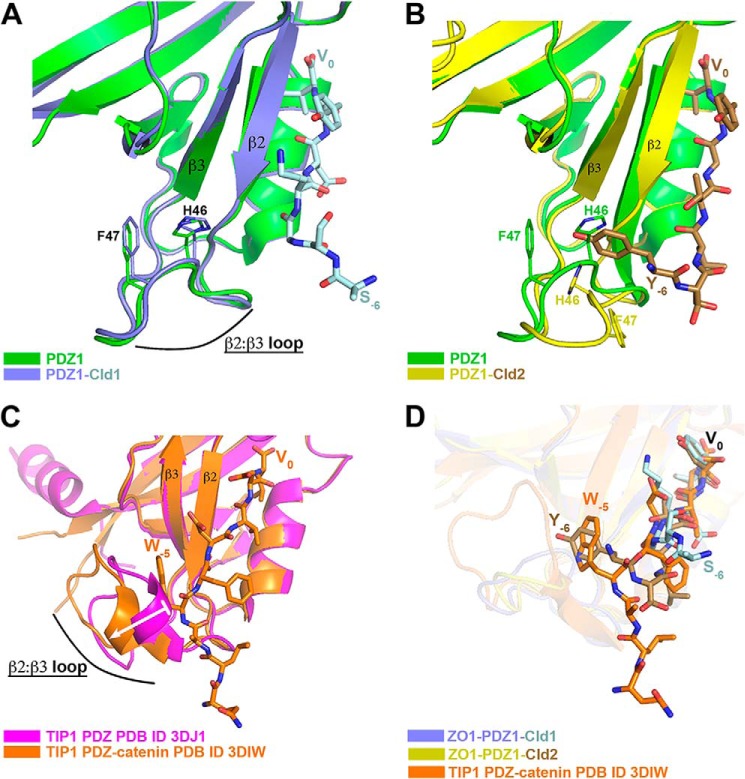
The canonical binding of C-terminal protein sequences by PDZ domains involves a binding site relatively distant from the PDZ domain β2-β3 loop. Therefore, it is not surprising that for most PDZ domains, binding of their particular protein target does not impact the conformation of this loop. Indeed, this is also the case we observe when claudin-1 binds PDZ1; the β2-β3 loop is unperturbed relative to its unliganded state (Fig. 8A). Surprisingly, this is not the case upon the binding of claudin-2 by this same PDZ domain. In this case, a significant conformation change of the β2-β3 loop occurs, which is presumably induced by Tyr−6 that buries itself in the space previously filled by this loop (Fig. 8B). This is not the first example for a change in the β2-β3 loop conformation upon ligand binding. Recently it was shown that the Tax-interacting protein-1 (TIP1) undergoes a conformational change in its β2- β3 loop upon binding of ligand β-catenin (Fig. 8C) (33). In the case of β-catenin binding to TIP1, it is its tryptophan residue at P−5 that displaces the β2-β3 loop of the TIP1 PDZ domain. Notably, the P−5 tryptophan residue occupies a very similar position as the P−6 tyrosine residue of claudin-2 when bound to the ZO-1 PDZ1 domain (Fig. 8D). An additional commonality to our result with claudin-2 showed that mutation of the tryptophan (in the case to an alanine) reduces the binding affinity by 100-fold (33). A similar change in the PDZ β2-β3 loop conformation between the unbound and its ligand-bound state was observed with the PDZ of SHANK1 (34). Hence, it appears that the intrinsic flexibility of the β2-β3 loop present in certain PDZ binders (such as TIP1, SHANK1, and ZO-1 PDZ1) is exploited for attaining increased binding affinity. However, unlike TIP1 and SHANK1, where the ligands (β-catenin and ARHGEF7, respectively) insert a tryptophan residue, claudin-2 binding to PDZ1 utilizes a tyrosine residue. This is an important distinction, as Tyr−6 can be phosphorylated, and this posttranslational modification can be used to modulate the affinity and hence the function of claudin-2.
FIGURE 8.
Role of PDZ domain β2-β3 loop flexibility in ligand affinity. A, overlay of the unliganded ZO-1 PDZ1 domain (green) and Cld2-bound PDZ1 (PDZ1 in purple, Cld1 in light blue). The β2-β3 loop does not change conformation nor participates in ligand binding when Cld1 binds PDZ1. B, in contrast, the β2-β3 loop does change conformation and does participate in ligand binding when Cld2 binds PDZ1 (PDZ1 in yellow, Cld1 in brown). Cld2 inserts Tyr−6 into the space previously occupied by the β2-β3 loop. C, a similar change in the β2-β3 loop PDZ conformation (represented by a white arrow) was observed for TIP1 (unliganded shown in magenta) when bound to β-catenin (complex in orange). D, focus on Cld1/Cld2/β-catenin binding to the PDZ domain (PDZ domains shown as transparent objects). Cld2 makes a similar binding mode as β-catenin, where Tyr−6 or Trp−5, respectively, insert into the space vacated by the β2-β3 loop.
The above analysis shows that ligand affinity to PDZ is increased by the presence of a large hydrophobic residue (Trp or Tyr) at P−5 or P−6. But is the ability to modulate this affinity by changing the phosphorylation state of Tyr−6 unique to claudin-2 or a general principal exploited by claudins and possibly other PDZ binders? While an open question, we note that a very recent study of claudin-3, which also has a Tyr−6 (Tyr-214), shows that mutation of that tyrosine to an aspartic acid (i.e. a phosphomimetic) perturbs claudin-3 trafficking to junctions and thereby decreases junctional dynamics (17). In another study, increased phosphorylation of the claudin-3 Tyr-214 was detected in epidermal growth factor receptor- and c-MET-positive colon cancer (35). The disruption of barrier function is often observed in cancers, and our data suggest that the detected observed phosphorylation of claudin-3 Tyr-214 would diminish its affinity to the ZO-1 PDZ domain.
Looked upon from a different perspective, it was shown that ZO-3 is unable to restore claudin assembly into tight junction strands in cells lacking ZO-1 and ZO-2 (3). Sequence analysis indicates that there is a notable difference in the β2-β3 loop of ZO-3 relative to the other ZO proteins; specifically, there is a 5-residue deletion and loss of the consecutive His-Phe sequence that we identified as playing an important role in Tyr−6 binding (Fig. 9). While requiring confirmation by additional experiments, our data suggest that the inability of ZO-3 to compensate for the lack of ZO-1 and ZO-2 stems from the inability of its PDZ1 domain from forming the high affinity interaction with Tyr−6-containing claudins.
FIGURE 9.

Sequence alignment of PDZ1 from human ZO-1, 2, and -3 shows large conservation except for the β2-β3 loop region in ZO-3. β2-β3 loop residues in ZO-1 are highlighted in green. The His-Phe residues within the β2-β3 loop, in which side chains dramatically change in position are highlighted in gray. The residues missing in ZO-3 are highlighted in red. Stars denote conserved, colons highly homologous, and a period moderately homologous residues.
In summary, our studies, presenting the first crystal structures of claudins binding to the ZO-1 PDZ1 domain, reveal divergent binding modes between claudin-1 and -2. The molecular explanation for the higher PDZ1 affinity of claudin-2 is the presence of Tyr−6, which forms interactions not present in the case of claudin-1 binding. This interaction is made possible by the intrinsic flexibility of the β2-β3 loop. Moreover, we show that phosphorylation of this residue can be exploited to reduce the affinity to PDZ1. Hence, we suggest that this is one possible mechanism used by the large subset of claudins that possesses a Tyr−6 for regulating their localization at the tight junction and perhaps a more general mechanism exploited by PDZ binders for controlling their binding affinity.
Supplementary Material
Acknowledgments
We thank Dr. Gerd Prehna of the University of Illinois at Chicago Center for Structural Biology for assistance with ITC experiments. We also thank the staff at Life Sciences Collaborative Access Team for assistance in data collection.
Author Contributions—J. N. performed all structural and ITC experiments, interpreted data, and helped to write the manuscript, A. A. performed the NMR experiments, M. C. designed and analyzed the NMR experiments, C. M. V. and J. M. A. performed the cell biology experiments and helped write the manuscript, A. S. F. consulted on experimental design, data analysis, and helped to write the manuscript, and A. L. designed the structural and biophysical experiments, analyzed data, and helped write the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants DK61397 (to A. S. F. and A. L.) and AI101676 (to M. C.). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Fig. S1 and S2.
The atomic coordinates and structure factors (codes 4OEO, 4OEP, and 4YYX) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- ZO
- zonula occludens
- MDCK
- Madin-Darby canine kidney cell
- ITC
- isothermal titration calorimetry
- r.m.s.d.
- root mean square deviation
- TER
- transepithelial electrical resistance
- TIP1
- Tax-interacting protein-1
- Cld
- Claudin.
References
- 1. Anderson J. M., Van Itallie C. M. (2009) Physiology and function of the tight junction. Cold Spring Harb. Perspect. Biol. 1, a002584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Itoh M., Furuse M., Morita K., Kubota K., Saitou M., Tsukita S. (1999) Direct binding of three tight junction-associated MAGUKs, ZO-1, ZO-2, and ZO-3, with the COOH termini of claudins. J. Cell Biol. 147, 1351–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Umeda K., Ikenouchi J., Katahira-Tayama S., Furuse K., Sasaki H., Nakayama M., Matsui T., Tsukita S., Furuse M., Tsukita S. (2006) ZO-1 and ZO-2 independently determine where claudins are polymerized in tight-junction strand formation. Cell 126, 741–754 [DOI] [PubMed] [Google Scholar]
- 4. Jeansonne B., Lu Q., Goodenough D. A., Chen Y. H. (2003) Claudin-8 interacts with multi-PDZ domain protein 1 (MUPP1) and reduces paracellular conductance in epithelial cells. Cell. Mol. Biol. 49, 13–21 [PubMed] [Google Scholar]
- 5. Poliak S., Matlis S., Ullmer C., Scherer S. S., Peles E. (2002) Distinct claudins and associated PDZ proteins form different autotypic tight junctions in myelinating Schwann cells. J. Cell Biol. 159, 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Van Itallie C. M., Tietgens A. J., LoGrande K., Aponte A., Gucek M., Anderson J. M. (2012) Phosphorylation of claudin-2 on serine 208 promotes membrane retention and reduces trafficking to lysosomes. J. Cell Sci. 125, 4902–4912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Luck K., Charbonnier S., Travé G. (2012) The emerging contribution of sequence context to the specificity of protein interactions mediated by PDZ domains. FEBS Lett. 586, 2648–2661 [DOI] [PubMed] [Google Scholar]
- 8. Wang C. K., Pan L., Chen J., Zhang M. (2010) Extensions of PDZ domains as important structural and functional elements. Protein Cell 1, 737–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fanning A. S., Lye M. F., Anderson J. M., Lavie A. (2007) Domain swapping within PDZ2 is responsible for dimerization of ZO proteins. J. Biol. Chem. 282, 37710–37716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen J., Pan L., Wei Z., Zhao Y., Zhang M. (2008) Domain-swapped dimerization of ZO-1 PDZ2 generates specific and regulatory connexin43-binding sites. EMBO J. 27, 2113–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rodgers L. S., Beam M. T., Anderson J. M., Fanning A. S. (2013) Epithelial barrier assembly requires coordinated activity of multiple domains of the tight junction protein ZO-1. J. Cell Sci. 126, 1565–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nomme J., Fanning A. S., Caffrey M., Lye M. F., Anderson J. M., Lavie A. (2011) The Src homology 3 domain is required for junctional adhesion molecule binding to the third PDZ domain of the scaffolding protein ZO-1. J. Biol. Chem. 286, 43352–43360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ye F., Zhang M. (2013) Structures and target recognition modes of PDZ domains: recurring themes and emerging pictures. Biochem. J. 455, 1–14 [DOI] [PubMed] [Google Scholar]
- 14. Zhang Y., Yeh S., Appleton B. A., Held H. A., Kausalya P. J., Phua D. C., Wong W. L., Lasky L. A., Wiesmann C., Hunziker W., Sidhu S. S. (2006) Convergent and divergent ligand specificity among PDZ domains of the LAP and zonula occludens (ZO) families. J. Biol. Chem. 281, 22299–22311 [DOI] [PubMed] [Google Scholar]
- 15. Findley M. K., Koval M. (2009) Regulation and roles for claudin-family tight junction proteins. IUBMB Life 61, 431–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ikari A., Kinjo K., Atomi K., Sasaki Y., Yamazaki Y., Sugatani J. (2010) Extracellular Mg2+ regulates the tight junctional localization of claudin-16 mediated by ERK-dependent phosphorylation. Biochim. Biophys. Acta 1798, 415–421 [DOI] [PubMed] [Google Scholar]
- 17. Twiss F., Oldenkamp M., Hiemstra A., Zhou H., Matheron L., Mohammed S., de Rooij J. (2013) HGF signaling regulates Claudin-3 dynamics through its C-terminal tyrosine residues. Tissue Barriers 1, e27425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Antanasijevic A., Cheng H., Wardrop D. J., Rong L., Caffrey M. (2013) Inhibition of influenza H7 hemagglutinin-mediated entry. PloS One 8, e76363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 20. Kabsch W. (2010) Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vagin A., Teplyakov A. (1997) MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 22. Appleton B. A., Zhang Y., Wu P., Yin J. P., Hunziker W., Skelton N. J., Sidhu S. S., Wiesmann C. (2006) Comparative structural analysis of the Erbin PDZ domain and the first PDZ domain of ZO-1. Insights into determinants of PDZ domain specificity. J. Biol. Chem. 281, 22312–22320 [DOI] [PubMed] [Google Scholar]
- 23. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 24. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 25. Colegio O. R., Van Itallie C., Rahner C., Anderson J. M. (2003) Claudin extracellular domains determine paracellular charge selectivity and resistance but not tight junction fibril architecture. Am. J. Physiol. Cell Physiol. 284, C1346–C1354 [DOI] [PubMed] [Google Scholar]
- 26. Van Itallie C. M., Holmes J., Bridges A., Anderson J. M. (2009) Claudin-2-dependent changes in noncharged solute flux are mediated by the extracellular domains and require attachment to the PDZ-scaffold. Ann. N.Y. Acad. Sci. 1165, 82–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Van Itallie C. M., Fanning A. S., Bridges A., Anderson J. M. (2009) ZO-1 stabilizes the tight junction solute barrier through coupling to the perijunctional cytoskeleton. Mol. Biol. Cell 20, 3930–3940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Karthikeyan S., Leung T., Ladias J. A. (2002) Structural determinants of the Na+/H+ exchanger regulatory factor interaction with the β2 adrenergic and platelet-derived growth factor receptors. J. Biol. Chem. 277, 18973–18978 [DOI] [PubMed] [Google Scholar]
- 29. Birrane G., Chung J., Ladias J. A. (2003) Novel mode of ligand recognition by the Erbin PDZ domain. J. Biol. Chem. 278, 1399–1402 [DOI] [PubMed] [Google Scholar]
- 30. Niethammer M., Valtschanoff J. G., Kapoor T. M., Allison D. W., Weinberg R. J., Craig A. M., Sheng M. (1998) CRIPT, a novel postsynaptic protein that binds to the third PDZ domain of PSD-95/SAP90. Neuron 20, 693–707 [DOI] [PubMed] [Google Scholar]
- 31. Amasheh S., Meiri N., Gitter A. H., Schöneberg T., Mankertz J., Schulzke J. D., Fromm M. (2002) Claudin-2 expression induces cation-selective channels in tight junctions of epithelial cells. J. Cell Sci. 115, 4969–4976 [DOI] [PubMed] [Google Scholar]
- 32. Van Itallie C. M., Mitic L. L., Anderson J. M. (2011) Claudin-2 forms homodimers and is a component of a high molecular weight protein complex. J. Biol. Chem. 286, 3442–3450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang J., Yan X., Shi C., Yang X., Guo Y., Tian C., Long J., Shen Y. (2008) Structural basis of β-catenin recognition by Tax-interacting protein-1. J. Mol. Biol. 384, 255–263 [DOI] [PubMed] [Google Scholar]
- 34. Im Y. J., Kang G. B., Lee J. H., Park K. R., Song H. E., Kim E., Song W. K., Park D., Eom S. H. (2010) Structural basis for asymmetric association of the beta PIX coiled coil and shank PDZ. J. Mol. Biol. 397, 457–466 [DOI] [PubMed] [Google Scholar]
- 35. Guo A., Villén J., Kornhauser J., Lee K. A., Stokes M. P., Rikova K., Possemato A., Nardone J., Innocenti G., Wetzel R., Wang Y., MacNeill J., Mitchell J., Gygi S. P., Rush J., Polakiewicz R. D., Comb M. J. (2008) Signaling networks assembled by oncogenic epidermal growth factor receptor and c-Met. Proc. Natl. Acad. Sci. U.S.A. 105, 692–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.