Background: Calcium ions (Ca2+) suppress microtubule assembly.
Results: Calmodulin interacts with the Par17 N terminus in a Ca2+-dependent manner, thereby preventing Par17-promoted microtubule assembly.
Conclusion: Ca2+ sensitivity of the tubulin polymerization process is mediated via binding of Ca2+/calmodulin to Par17 and other microtubule-associated proteins.
Significance: Elucidation of this Ca2+-dependent Par17/calmodulin interaction adds to the understanding of Ca2+ influence on microtubule formation.
Keywords: calcium, calmodulin (CaM), microtubule-associated protein (MAP), nuclear magnetic resonance (NMR), protein conformation, protein-protein interaction, tubulin, chemical shift perturbation (CSP) analysis, parvulin
Abstract
Recently we have shown that the peptidyl-prolyl cis/trans isomerase parvulin 17 (Par17) interacts with tubulin in a GTP-dependent manner, thereby promoting the formation of microtubules. Microtubule assembly is regulated by Ca2+-loaded calmodulin (Ca2+/CaM) both in the intact cell and under in vitro conditions via direct interaction with microtubule-associated proteins. Here we provide the first evidence that Ca2+/CaM interacts also with Par17 in a physiologically relevant way, thus preventing Par17-promoted microtubule assembly. In contrast, parvulin 14 (Par14), which lacks only the first 25 N-terminal residues of the Par17 sequence, does not interact with Ca2+/CaM, indicating that this interaction is exclusive for Par17. Pulldown experiments and chemical shift perturbation analysis with 15N-labeled Par17 furthermore confirmed that calmodulin (CaM) interacts in a Ca2+-dependent manner with the Par17 N terminus. The reverse experiment with 15N-labeled Ca2+/CaM demonstrated that the N-terminal Par17 segment binds to both CaM lobes simultaneously, indicating that Ca2+/CaM undergoes a conformational change to form a binding channel between its two lobes, apparently similar to the structure of the CaM-smMLCK796–815 complex. In vitro tubulin polymerization assays furthermore showed that Ca2+/CaM completely suppresses Par17-promoted microtubule assembly. The results imply that Ca2+/CaM binding to the N-terminal segment of Par17 causes steric hindrance of the Par17 active site, thus interfering with the Par17/tubulin interaction. This Ca2+/CaM-mediated control of Par17-assisted microtubule assembly may provide a mechanism that couples Ca2+ signaling with microtubule function.
Introduction
Peptidyl-prolyl cis/trans isomerizations are slow conformational interconversions in the polypeptide backbone that are known to be rate-limiting in many protein folding reactions. As a consequence, peptidyl prolyl cis/trans isomerases (PPIases,8 EC 5.2.1.8) are ubiquitously distributed enzymes, which accelerate the interconversion between prolyl bond isomers of proline-containing polypeptide chains and are, therefore, also referred to as “folding-helper enzymes” (1, 2). The enzyme class of PPIases includes three protein families: parvulins, cyclophilins, and FK506-binding proteins.
Eukaryotic parvulins have been shown to be involved in many cellular processes including cell proliferation and cell cycle progression (3–6). The three-dimensional structure of parvulin-type PPIase domains is highly conserved between the biological kingdoms, suggesting that the main structural features are restricted by evolution (7).
The human parvulin family of PPIases comprises the proteins Pin1, Par14, and Par17 (7, 8). Pin1 shows a high specificity for phosphorylated serine/threonine-proline substrates due to a positively charged surface at the interaction site (3, 9). Besides the PPIase domain, Pin1 possesses an N-terminal WW domain that is involved in protein-protein interactions and targeting to the nucleus (10). This enzyme has been shown to be involved in cell cycle regulation, cancer, and Alzheimer disease (4). Pin1 also restores the ability of phosphorylated Tau protein to bind microtubules and to promote microtubule assembly in vitro (11).
Par14 and Par17 are both encoded by the Par14/Pin4 locus on chromosome Xq13 yet feature different transcription initiation sites. As a consequence, the only difference between their amino acid sequences is a segment of 25 additional residues at the N terminus of Par17 (8, 12). In contrast to Pin1, both Par14 and Par17 show no preference for phosphorylated substrates but specificity for substrates with basic amino acid residues preceding proline (13–16). In the case of Par14, it was demonstrated that this substrate selectivity is related to the presence of a negatively charged surface at the substrate binding site (14, 17).
Par14 is found mainly inside the nucleus and has been associated with cell cycle progression and chromatin remodeling (18). The PPIase domain structure of human Par1436–131 was solved by NMR (17) and x-ray crystallography (16), and the structure of the PPIase domain plus 6 residues belonging to the N-terminal segment, i.e. human Par1430–131, was solved by NMR (14). In all cases the global fold was analogous to the PPIase domain of Pin1 (19), consisting of a four-stranded twisted β-sheet surrounded by several helices. The residues mainly involved in the substrate interaction (i.e. Met-90, Val-91, and Phe-94, which are identical to Met-115, Val-116, and Phe-119 in Par17) are located in the loop connecting the second β-strand with the last helix. Besides the PPIase domain, Par14 possesses a lysine- and glycine-rich segment at the N terminus, which seems to be important for nuclear localization and DNA binding (17, 18), but whose structure could not be resolved.
Although Par17 is also able to bind DNA, it is located mainly in the cytoplasm, where it is found in the mitochondrial matrix or colocalized with microtubules (12, 20). It was, therefore, suggested that the 25 residues composing the N-terminal elongation in Par17 compared with Par14 (i) may be masking the nuclear localization signal of the Par14 N terminus (21) and (ii) could furthermore function as a novel mitochondrial targeting signal (12).
Recently it was demonstrated that Par17 catalyzes tubulin polymerization in a GTP-dependent manner (20). Tubulin heterodimers form long cylindrical structures called microtubules, one of the main components of the cytoskeleton (22). The polymerization process of microtubules is highly regulated, as these dynamic structures change in response to different stimuli. Two tubulin-binding sites were identified in Par17 (20): (i) the amphipathic α-helix defined by the segment Lys-75 to Lys-84, and (ii) the segment Gln-103–Pro-118 located near the putative active site, which includes residues Leu-107 and Met-115 that constitute a part of the substrate binding pocket in the Par14 homolog. Par17-induced microtubule assembly seems to depend on the isomerase activity in the active site of Par17, yet it is 2.5-fold more efficient than Par14. Therefore, the N-terminal elongation of Par17 may represent just one component of a more complex tubulin interaction scenario or it merely accelerates the reaction.
Tubulin polymerization is promoted by proteins called microtubule-associated proteins (MAPs), which interact with the C-terminal tubulin domain and induce microtubule stabilization (23). Microtubule assembly is subsequently regulated by calcium-loaded calmodulin (Ca2+/CaM) via direct binding to MAPs (24–26). Calmodulin (CaM) is a relatively small calcium-binding protein involved in many signaling pathways that regulate a large number of crucial cellular processes (27). The CaM structure consists of two globular lobes connected by a flexible linker (28, 29). Each lobe is able to bind two calcium (Ca2+) ions, which results in a change of the lobe structure (30, 31). Both in the Ca2+-free and the Ca2+-bound state, CaM is able to interact with a plethora of other proteins (9, 32, 33). Of all the CaM complexes reported to date, the structure of CaM bound to the smooth muscle myosin light chain kinase (smMLCK), where the N- and C-terminal lobes of CaM both wrap around a helical segment of smMLCK to form a globular complex (34), turned out to be of particular relevance for the Par17-Ca2+/CaM interaction.
In the present work we investigated the structure and function of the 25-residue elongation at the Par17 N terminus to further elucidate the features responsible for the functional differences to Par14. Based on our results, we could (i) demonstrate that Par17 is able to interact with CaM in a Ca2+-dependent manner, thus preventing microtubule assembly, (ii) identify a Ca2+/CaM-binding site at the 25-residue elongation of the Par17 N terminus, and (iii) propose that the structure of the Par17-Ca2+/CaM complex resembles the one of CaM bound to smMLCK. Hence, a Ca2+-mediated regulation of microtubule assembly via Ca2+/CaM and Par17 may be envisioned.
Experimental Procedures
Peptides and Proteins
Par17, GST-Par14, GST-Par17, and His6-CaM were expressed and purified as reported previously (20, 35, 36). Par172–22 peptide was generated by solid-phase synthesis as previously described (37). Full-length Par17 and the Par172–22 peptide each were produced as the Gln-16/Arg-18 isoform. Bovine brain tubulin was purchased from Cytoskeleton Inc. (Denver). smMLCK797–813 was obtained from Merck Biosciences (Calbiochem).
Database Searches
Amino acid sequence alignments were performed using the program ClustalW (38). Potential CaM-binding sites were identified using the online server “CaM target database.” Helix properties were predicted employing the online server “heliquest” (39).
Protein Microarray
The ProtoArray® Human Protein Microarray v5.0 from Invitrogen was used to identify new binding partners of Par17. This microarray contains 9000 human GST-tagged proteins expressed in Sf9 insect cells and spotted in duplicate on the nitrocellulose-coated glass slide. ProtoArray® was used according to the recommendations of the manufacturer. Briefly, the slides were equilibrated for 15 min at 4 °C and then blocked for 1 h on ice by incubating with 1% BSA and 20 mm reduced glutathione in 50 mm HEPES, pH 7.5, 200 mm NaCl, 0.08% Triton X-100, 25% glycerol, and 1 mm DTT. Next, the buffer was removed, and 1 μm Par17 diluted in PBS buffer containing 1% BSA and 0.1% Tween 20 was spread carefully on the array and capped with a coverslip. After 90 min of incubation on ice, the cover was carefully removed, and the protein arrays were rinsed 5 times for 10 min each in PBS buffer containing 1% BSA and 0.1% Tween 20. After washing, the slides were incubated with rabbit anti-Par14/17 antibody (pab productions, Herbertshausen, Germany; diluted 1:1000) for 1 h at 4 °C. Next, the slides were incubated with an Alexa Fluor 647-labeled goat anti-human IgG (Invitrogen; diluted 1:2000 in sample buffer) for 90 min at 4 °C under slow agitation in the dark.
The protein arrays were washed 5 times with PBS buffer containing 1% BSA and 0.1% Tween 20 and dried by centrifugation at 1000 rpm for 1 min at room temperature. Slides were scanned on a ScanArray 5000 (Packard BioChip Technologies, Billerica, MA) to produce red (Alexa Fluor 647) images. The Genepix Pro 6.0 image analysis software (Axon Laboratories) was employed for data quantification.
GST Pulldown Assay
Equal amounts (20 μg) of GST-Par14, GST-Par17, GST-Par171–25, and GST were bound to glutathione-Sepharose beads (GE Healthcare). Excess protein was removed by extensive washing. Then, glutathione-Sepharose beads were incubated with 20 μg of CaM in 60 μl of incubation buffer (20 mm Tris, pH 7.5, 150 mm NaCl, 5 mm MgCl2, 3% BSA, and either 5 mm EGTA or 0.5 mm CaCl2). After 1 h of incubation at 4 °C, unbound protein was removed by 3 washes with incubation buffer. Interacting proteins were eluted by boiling with sample buffer. Samples were subjected to 17.5% SDS-polyacrylamide gel electrophoresis (SDS-PAGE), and the proteins were visualized by silver staining.
Tubulin Polymerization Assay
The tubulin polymerization assay was performed according to the manufacturer's protocol (tubulin polymerization assay kit; Cytoskeleton Inc., Denver, CO). In brief, bovine brain tubulin (>99% purity) was resuspended in G-PEM buffer (containing 80 mm PIPES, pH 6.9, 2 mm MgCl2, 0.5 mm EGTA, and 1 mm GTP) and 5% glycerol. Polymerization was started by dilution of tubulin to a final concentration of 6.7 μm in 80 μl of G-PEM buffer at a temperature of 37 °C. The polymerization of tubulin was monitored as the increase in absorbance at 340 nm over a period of 60 min using a UV-visible spectrophotometer. To investigate the effect of CaM and Ca2+ on the Par17-promoted microtubule assembly, 1 mm CaCl2 and 30 μm CaM were added to the reaction mixture containing 6.7 μm tubulin and 16 μm concentration of the respective GST-parvulin construct.
Isothermal Titration Calorimetry (ITC)
ITC experiments were performed with a VP-ITC system (Microcal Inc., Northampton, MA). The standard ITC conditions were 10 mm MES, 50 mm KCl, 6 mm CaCl2, pH 6.8, at 20 °C. For a typical titration of Par172–22 peptide with CaM, 50 μm peptide was placed in the reaction cell, whereas a solution of 300 μm CaM was placed in the ITC syringe. For the titration of full-length Par17 with CaM, 10 μm Par17 was placed in the reaction cell, whereas a solution of 100 μm CaM was placed in the ITC syringe. The titration of Par14 was performed in an analogous manner but with three times the Par14 and CaM concentrations for higher sensitivity. In all titrations, CaM was injected into the reaction cell every 5 min in 15 μl aliquots. Data were analyzed using the MicroCal Origin 7.0 software, and the data were fitted with a “one set of sites” model yielding binding enthalpy (H), stoichiometry (n), entropy (S), and association constant (Ka).
PPIase Activity Assay
PPIase activity of Par17 was determined in a coupled assay with the protease α-chymotrypsin. The assay exploits the specificity of α-chymotrypsin for the trans isomer of proline-containing oligopeptides. Thus, α-chymotrypsin only cleaves off the 4-nitroanilide from the substrate succinyl-Ala-Lys-Pro-Phe-4-nitroanilide when the Lys-Pro bond is in the trans conformation. At the applied high concentration of α-chymotrypsin, hydrolysis of the trans peptide occurs immediately in a few seconds. Release of 4-nitroaniline from the remaining cis peptide depends on the rate of cis-to-trans isomerization of the prolyl bond and can be followed by an increase in absorbance at 390 nm. Par17 catalyzed cis/trans isomerization was measured at 10 °C for 6 min in a reaction mixture containing 35 mm HEPES buffer, pH 7.8, 40 μm succinyl-Ala-Lys-Pro-Phe-4-nitroanilide, 2 μm GST-Par17, and 1 mm CaCl2 either with or without 15 μm recombinant human CaM. The reaction was started by the addition of 0.1 mg/ml α-chymotrypsin, and the release of 4-nitroanilide was monitored at 390 nm.
NMR Spectroscopy
NMR data were collected the way previously described (36). Briefly, all NMR spectra were collected in 10 mm MES buffer (50 mm KCl, 6 mm CaCl2, pH 6.8). For resonance assignment purposes, triple-resonance experiments were collected at 18 °C, and in addition titration data were employed to trace signals that showed significant line-broadening with increasing concentration of the binding partner.
1H,15N TROSY experiments for chemical shift perturbation (CSP) analysis were performed on a Bruker DRX 500 spectrometer operating at 500.13 MHz with a 5-mm TXI probe featuring gradient capability. The Par17-CaM complex samples generally contained the 15N-labeled component at 0.5–0.6 mm concentration. All spectra were analyzed with SPARKY 3 (University of California, San Francisco, CA). Chemical shift differences in the amide proton (Δδ1HN) and amide nitrogen (Δδ15N) resonances of the free and complexed protein forms were combined for each residue by using the expression [(Δδ1HN)2 + (Δδ15N/6.5)2]1/2 (40).
Peptide Microarray
Peptide microarrays were produced by JPT Peptide Technologies GmbH (Berlin, Germany). Overlapping peptide scans of Par17 were synthesized by SPOT synthesis and printed on peptide microarrays, resulting in 48 peptides. Each 15-mer peptide was immobilized in three consecutive spots, resulting in 144 spots per subarray. For quality control, peptide microarrays contained three subarrays displaying the features of each subarray in triplicate, thus resulting in 9-fold redundancies of the data. To investigate CaM binding to the microarray, the Par17 peptide microarray was incubated for 30 min at 10 °C with 1 μm CaM in 200 μl of 20 mm Tris, pH 7.5, 150 mm NaCl, 5 mm MgCl2, 3% BSA, 5% glycerol, and either 5 mm EGTA or 0.5 mm CaCl2 followed by 3 washing steps with 10 mm Tris, 150 mm NaCl, 0.1% (v/v) Tween 20, pH 7.5, and additional washes with water. Subsequently, the peptide microarray was incubated with mouse anti-CaM antibody followed by an Alexa Fluor 647 goat anti-mouse IgG. Finally, peptide microarrays were dried using a stream of nitrogen. Peptide microarrays were scanned using an Axon 4200AL microarray scanner at a wavelength of 635 nm (MDS Analytical Technologies, Toronto, Canada) and analyzed using the microarray image analysis software package Axon GenePix Pro 7.1 (MDS Analytical Technologies). The scanner recorded a 16-bit numeric image in tagged-image file format, resulting in a range of signal intensities from 0 to 65,535, which are referred to as relative fluorescence intensities.
Results
Par17 Interacts with Ca2+/CaM
To identify hitherto unknown Par17-binding proteins, which may provide new insights into the biological function of Par17, ProtoArray® chips were employed. These microarrays contained 9000 human proteins expressed as N-terminal GST fusion proteins using a baculovirus expression system. Subsequent analysis revealed an interaction between Par17 and Ca2+/CaM, whereas no other Par17 binding partners could be identified.
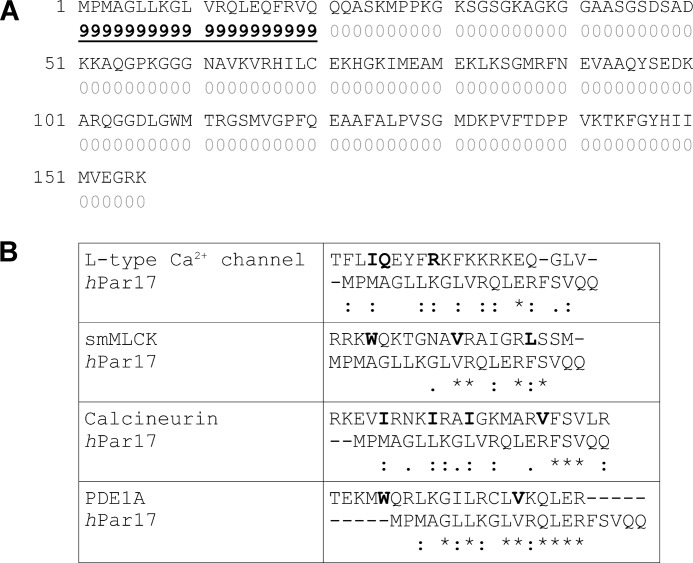
To obtain further evidence for a CaM-binding site in the Par17 sequence, we performed a CaM binding sequence prediction using the online server CaM target database. This program identified a potential CaM-binding site at the N terminus of Par17 (Fig. 1A). Moreover, amino acid sequence alignment of the first 21 N-terminal residues of Par17 with several known CaM binding motifs revealed a high degree of sequence conservation (Fig. 1B), in particular to the CaM-binding site of human phosphodiesterase 1A. But also the CaM-binding sites of smMLCK, L-type Ca2+ channel and calcineurin show notable sequence homologies to the Par17 N terminus. Although the CaM binding sequence of parvulin 17 does not belong to any of the originally described CaM binding motifs, like the IQ motif, 1–14 motif, 1–10 motif, or 1–16 motif, a novel CaM binding motif has been proposed for caldesmon and phosphodiesterases: the so-called 1–12 motif, where the hydrophobic residues are separated by 10 residues and where several basic residues occur within the binding site (41). We propose that such a 1–12 CaM binding motif also exists in the Par17 segment between the bulky hydrophobic residues Leu-6 and Phe-17, which includes several lysine or arginine residues (Lys-8, Arg-12, and Arg-16). Interestingly, these 1–12 motifs have been shown to bind calmodulin primarily in the presence of calcium (42).
FIGURE 1.
A, analysis of potential CaM binding sequences by the online server CaM target database. The figure shows the sequence of human Par17 (hPar17). The scores below the amino acids indicate the probability for the location of a CaM-binding site. A consecutive series of high values suggests the existence of a putative CaM-binding site. B, alignment of CaM binding motifs with the N-terminal sequence of hPar17 (*, identical; :, conserved). Key residues that classify the CaM-binding sites to a particular motif are highlighted in bold letters. The L-type Ca2+ channel displays an IQ motif, and smMLCK comprises a 1-8-14 motif, whereas calcineurin and phosphodiesterase 1A (PDE 1A) encompass a binding sequence of the 1-5-8-14 and the 1-12 class, respectively.
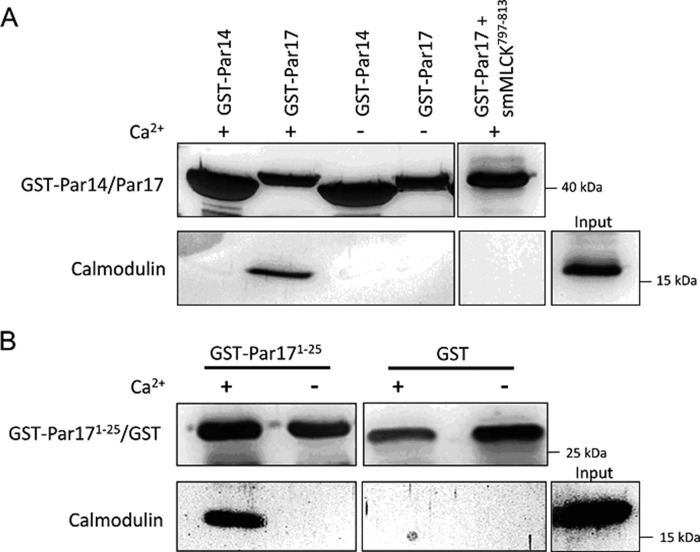
To confirm the interaction of the Par17 N terminus with CaM, we performed a GST fusion protein precipitation assay. To this end GST fusions of both the full-length Par17 protein (GST-Par17) and the Par17 N terminus (GST-Par171–25) were employed. In addition, GST-Par14, which possesses an identical amino acid sequence as Par17 but lacks the 25 N-terminal Par17 residues comprising the potential CaM-binding site, was generated. The precipitation experiments with GST beads demonstrated the association between GST-Par17 and CaM in the presence of Ca2+, whereas no such binding was observed in the absence of Ca2+ (Fig. 2A). When experiments were conducted in the presence of the high affinity CaM-binding peptide smMLCK797–813, the protein-protein interaction was completely abolished, indicating that the smMLCK peptide blocks the Par17-binding site in CaM. GST-Par14, on the other hand, did not interact with CaM either with or without Ca2+ (Fig. 2B), suggesting that the N-terminal 25-residue extension of Par17 contains the binding site for Ca2+/CaM. In fact, the GST fusion of Par171–25 also interacted with CaM in a Ca2+-dependent manner.
FIGURE 2.
Par17 interacts with Ca2+/CaM via its N-terminal extension. For the CaM coprecipitation experiment, glutathione-Sepharose beads loaded with 20 μg of GST fusion proteins or GST alone were blocked with 3% BSA in the incubation buffer. A, Par14- and Par17-bound resins were incubated for 1 h at room temperature with 20 μm CaM in the (i) absence of Ca2+, (ii) presence of Ca2+, and (iii) presence of both Ca2+ and 20 μm smMLCK797–813 peptide. B, GST-Par171–25-loaded as well as GST-loaded GSH beads were incubated with 20 μm CaM in the absence and presence of Ca2+.
Thermodynamic Characterization of the Par17-CaM Interaction Using ITC
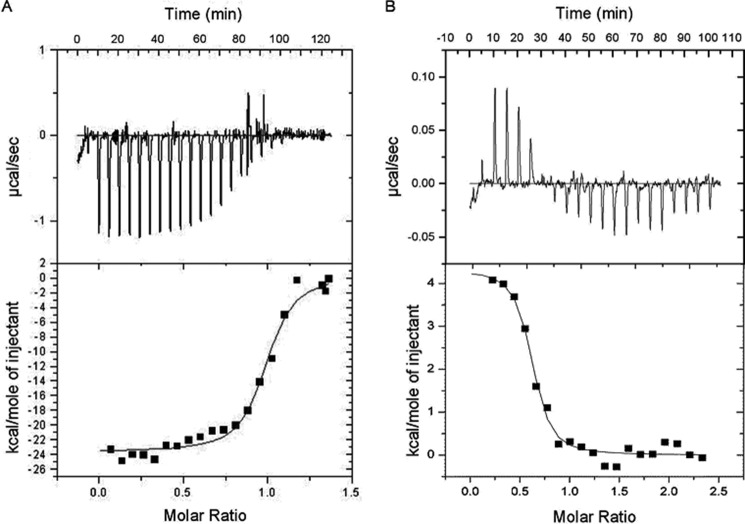
To gain more quantitative information on the Par17-CaM interaction, we examined CaM binding to the N-terminal segment of Par17 (Par172–22) and to full-length Par17 by ITC. Thermodynamic data revealed that Ca2+/CaM and Par172–22 form a one-to-one binding complex (Fig. 3A). This interaction is an enthalpically driven process with a dissociation constant of 149 nm.
FIGURE 3.
ITC data of Ca2+/CaM binding to the Par17 N terminus. The calorimetric trace of heat released upon titration of Ca2+/CaM into a solution containing either the Par172–22 peptide (A) or full-length Par17 (B) is shown at the top. The corresponding heat per mole of injected Ca2+/CaM is shown at the bottom. The binding isotherms were analyzed by applying a single-site binding model using the Microcal Origin software package. The analyses revealed the following thermodynamic parameters for the Par172–22 peptide (ΔH = −23.6 ± 0.4 kcal/mol, ΔS = −49.2 cal/(mol·degree), n = 0.97 ± 0.01, Kd = 149 ± 25 nm) and for the full-length Par17 protein (ΔH = 4.3 ± 0.2 kcal/mol, ΔS = 45.8 cal/(mol·degree), n = 0.59 ± 0.02, Kd = 143 ± 41 nm).
In contrast, the titration of full-length Par17 with Ca2+/CaM showed a different, seemingly more complex curve progression due to an endothermic reaction that is followed by exothermic dilution of CaM (Fig. 3B). The best fit of the ITC data was obtained with a one set of binding sites model that led to a dissociation constant of 143 nm and a number of sites (n) value of 0.59. This value of n between 0.5 and 1 may be interpreted in two ways: (i) a one-to-one binding interaction with some error in the protein concentrations during the assay or (ii) a complex between one Ca2+/CaM and two Par17 molecules. As our NMR titration data, which is reported below, clearly supported a 1:1 rather than a 1:2 binding interaction between Ca2+/CaM and Par17, independent of whether Ca2+/CaM was titrated to Par17 or vice versa (data not shown), the value of n obtained by ITC presumably was affected by experimental error. Moreover, because (i) the dissociation constants of the Ca2+/CaM interaction with either Par172–22 or the full-length Par17 protein are very similar and (ii) Par14 showed no binding interaction with Ca2+/CaM, as the ITC titration of Par14 with Ca2+/CaM is the same as in the control Ca2+/CaM injection into buffer (data not shown), both these results strongly suggest that there is no second high affinity binding site between both proteins.
Interestingly, the ITC data revealed a drastic difference in the thermodynamics when Ca2+/CaM was titrated to either Par172–22 or full-length Par17 protein. The reaction with the free peptide is exothermic, but with the Par17 protein an endothermic reaction is observed, thus indicating that the binding (ΔG = −9 kcal/mol in both reactions) is driven by the enthalpy change (ΔH = −23.6 kcal/mol) when the Par17 N terminus is isolated, whereas the interaction between the full-length proteins is driven by the entropy change (TΔS = −13.4 kcal/mol). This difference in thermodynamic behavior could be explained by the fact that the structure and stability of the isolated Par172–22 peptide is not the same as when the peptide is part of the Par17 protein. As reported below, we have strong evidence for an intramolecular interaction between the N terminus and the PPIase domain in Par17. Hence, the absence of this interaction in the isolated peptide could lead to a different peptide conformation and thus a different kind of CaM binding process.
Ca2+/CaM Interferes with Par17-catalyzed Tubulin Polymerization
Previously, we had demonstrated that Par17 interacts with tubulin in a GTP-dependent manner and thereby promotes the formation of microtubules (20). Par14, which lacks the N-terminal CaM-binding site, was 2.5-fold less efficient in the promotion of microtubule assembly compared with Par17. This prompted us to investigate whether Ca2+/CaM influences Par17-catalyzed tubulin assembly despite the fact that PPIase activity measurements in the presence and absence of Ca2+/CaM revealed no inhibition of the catalytic activity of Par17 by CaM (Fig. 4A), suggesting that the Par17 active site is not directly involved in CaM binding.
FIGURE 4.
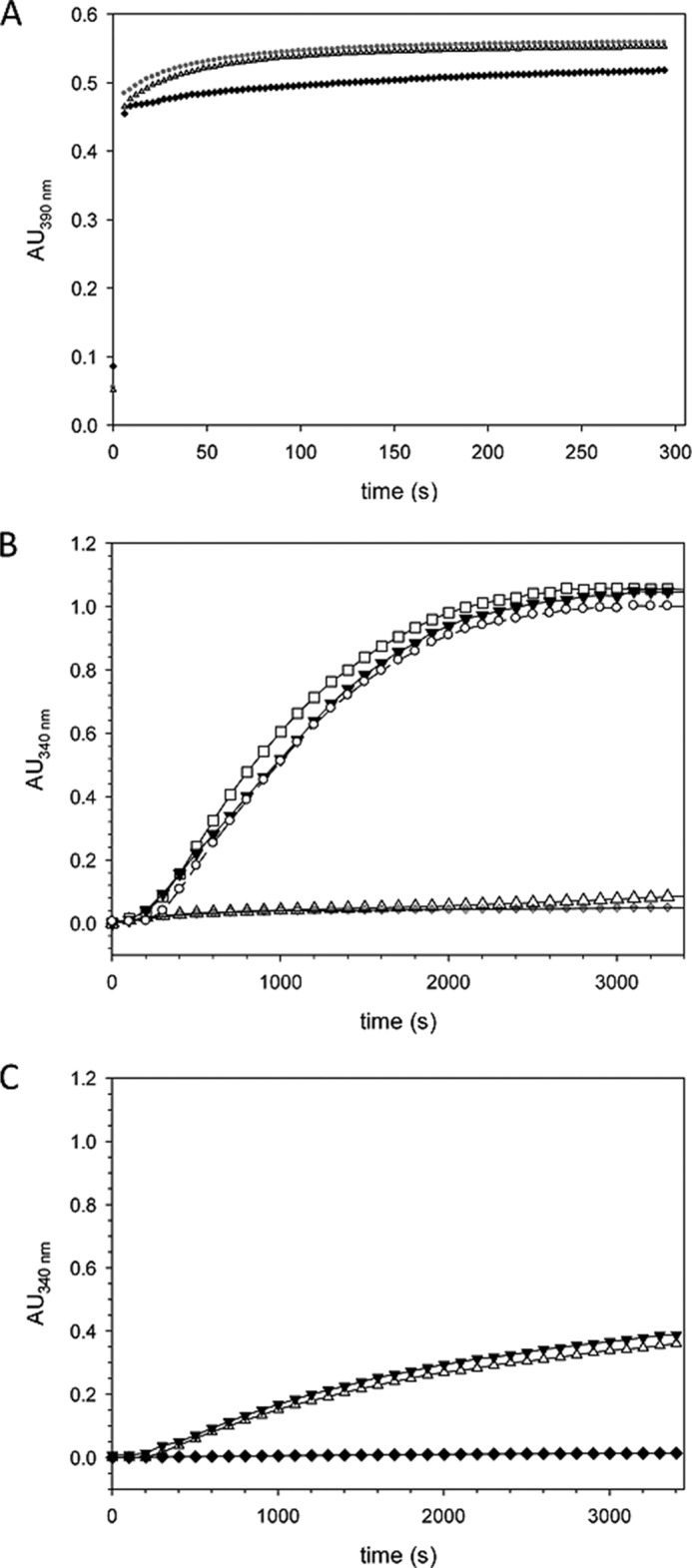
Ca2+/CaM inhibits Par17-catalyzed tubulin assembly in an in vitro tubulin polymerization assay. A, the PPIase activity of Par17 in the presence of Ca2+/CaM was determined using a protease-coupled assay that employs as substrate the oligopeptide succinyl-AKPF-4-nitroanilide, as previously described (20). The PPIase activity of Par17 was measured in a reaction mixture containing 2 μm GST-Par17 and 1 mm CaCl2 either with (▴) or without (●) 15 μm CaM. The uncatalyzed cis-to-trans isomerization in the absence of Par17 is shown as a control (♦). Calculation of the first-order rate constants for the cis/trans isomerization (1.47 ± 0.02 × 10−2 in the presence and 1.5 ± 0.03 × 10−2 in the absence of Ca2+/CaM) indicated that Ca2+/CaM does not affect Par17 PPIase activity (insensitivity of Par17 and CaM to proteolytic digestion by α-chymotrypsin was verified for the duration of the measurements). B, assembly of microtubules was initiated by dilution of ice-cold tubulin to a final concentration of 6.7 μm in 80 μl of G-PEM buffer set to 37 °C. Polymerization was measured in absence of Par17 (♦) and in presence of 16 μm GST-Par17 (▾), 16 μm GST-Par17, and 1 mm Ca2+ (○) and 16 μm GST-Par17 and 30 μm CaM (□) as well as 16 μm GST-Par17, 1 mm Ca2+, and 30 μm CaM (▵). C, tubulin polymerization was monitored in absence of GST-Par14 (♦), in the presence of 16 μm GST-Par14 (▾), and in presence of 16 μm GST-Par14, 1 mm Ca2+, and 30 μm CaM (▵). AU, absorbance units.
Next, on the other hand, an in vitro tubulin polymerization assay was performed in the presence and absence of Ca2+/CaM and Par17. As shown in Fig. 4B, no spontaneous tubulin assembly occurred under assay conditions in the absence of Par17, when the tubulin concentration was kept under the critical concentration that would allow spontaneous tubulin polymerization (43). Subsequent addition of Par17 resulted in a significant increase in absorbance at 340 nm, indicating microtubule formation. This Par17-promoted microtubule assembly was not affected by either Ca2+ or CaM alone. In the presence of Ca2+/CaM, however, no Par17-mediated tubulin assembly was detected. These results thus indicate that Ca2+-dependent binding of CaM to the N terminus of Par17 interferes with the Par17-tubulin interaction and thus Par17-catalyzed microtubule assembly.
To exclude the possibility that Ca2+/CaM prevents tubulin polymerization by direct contact to tubulin, we investigated the effect of Ca2+ and CaM on Par14-promoted microtubule assembly, as Par14 lacks the N-terminal extension of Par17 and, therefore, should not interact with Ca2+/CaM. In fact, Ca2+/CaM had no effect on Par14-promoted tubulin polymerization (Fig. 4C), thus corroborating that parvulin-catalyzed microtubule assembly is blocked primarily as a result of the high affinity binding of Ca2+/CaM to the Par17 N-terminal extension.
NMR-based Comparison between Par14 and Par17 Structural Features
All the above results raised our interest in the characterization of the structural details related to the Par17-Ca2+/CaM complex. In particular, our goal was to see whether CaM binding induces structural changes in the Par17 molecule or vice versa. Therefore, the Par17-Ca2+/CaM interaction was investigated using heteronuclear NMR spectroscopy with isotopically enriched proteins.
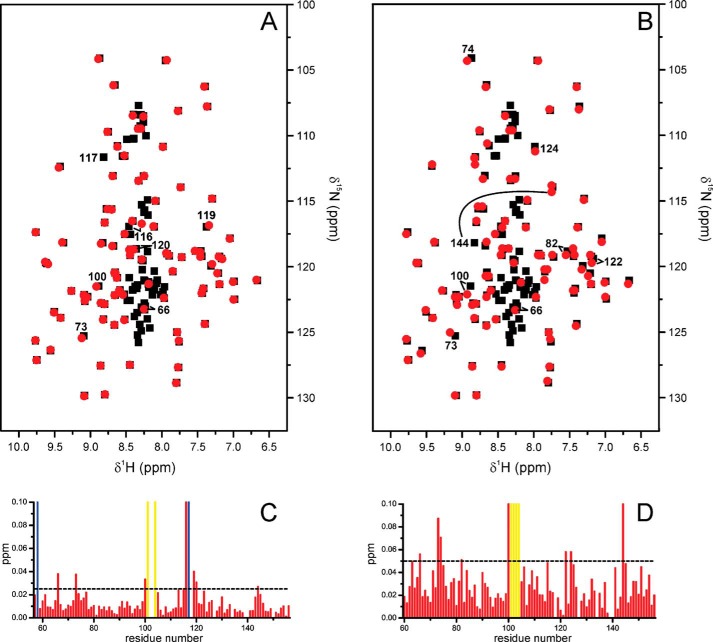
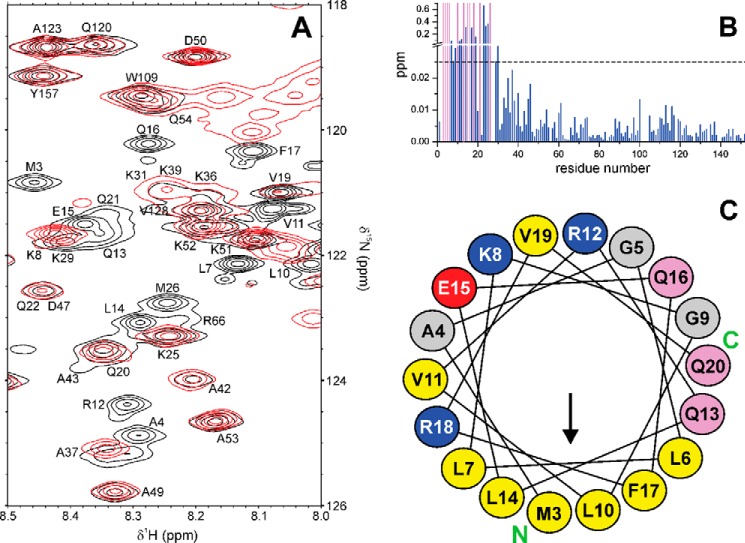
To better understand why Par17 binds Ca2+/CaM, whereas Par14 does not, we first compared the structural elements of both parvulin types to identify any conformational feature(s) that might be responsible for this fundamental biological difference. A chemical shift index analysis of Par17 had previously shown (36) that the 25 additional residues at the Par17 N terminus do not significantly affect the three-dimensional structure of the PPIase domain compared with Par14. Also a direct comparison between the Par14 and Par17 amide resonances, which was focused on the PPIase domain (i.e. Par17 residues Asn-61–Lys-156) due to a lack of assignments in the first 30 residues of Par14 (14, 17), revealed nearly identical chemical shift values (Fig. 5, A and B), thus indicating that both parvulin types share a very similar overall domain fold, as was to be expected based on the identical amino acid sequences.
FIGURE 5.
Comparison of the Par14 and Par17 backbone amide resonances. In the two upper panels, the amide signal positions in the 1H,15N HSQC spectrum of Par17 (Biological Magnetic Resonance Bank (BMRB) accession code 18615; black squares) are overlaid with those of Par14 (red circles) based on the assignment by Terada et al. (14) (A) and Sekerina et al. (Ref. 17; Biological Magnetic Resonance Bank (BMRB) accession code 4768) (B). Shifted peaks are labeled with the respective Par17 residue number and in some cases connected by a line to denote the displacement. Signals belonging to the Par17 N terminus, which corresponds to the segment missing in Par14, are generally found as non-superposed black squares in the 1H resonance range between 8.5 and 8.0 ppm, thus indicating a random-coil structure. In the two lower panels, a CSP analysis for each residue within the structured PPIase domain again compares Par17 with the Par14 assignments of Terada (C) and Sekerina (D). Residues, whose backbone amide resonance assignments are missing in either Par17 or Par14 are indicated by yellow and blue bars, respectively.
The amide chemical shift differences (Δδ) between Par17 and Par14 are thus quite small (Fig. 5, C and D), except for Par17 residues Lys-100, Val-116, and Lys-144 where larger shift differences were observed possibly as a result of misassignments. In the comparison with the Par14 resonance assignment by Terada et al. (14), Δδ values are generally smaller than 0.04 ppm, whereas the comparison with the Par14 assignment by Sekerina et al. (17) resulted in Δδ values that are usually smaller than 0.09 ppm. Interestingly, however, in both resonance comparisons the more pronounced chemical shift differences (i.e. Δδ > 0.025 ppm and Δδ > 0.05 ppm, respectively) occur basically within the same sequence regions. Although some of these effects could also be related to differences in pH and/or temperature between the Par17 and Par14 samples, the rather conspicuous shifts of the amide signals in the Par17 segment Lys-100–Ala-125 are particularly intriguing for several reasons; this segment includes residues that belong to (i) the substrate binding site of Par14 (corresponding to Par17 residues Met-115, Val-116, and Phe-119), (ii) one of the tubulin-binding sites of Par17 (i.e. Gln-103–Pro-118), and (iii) a 310-helical loop (16) that is poorly defined by the solution NMR data. This latter 310-helical loop represents the region with the least well defined resonance assignments not only in Par17 (36) but according to Terada et al. (14) also in Par14, and it apparently exhibits a high degree of backbone flexibility in the structurally homologous archaeal parvulin csPinA (6). Hence, the Par17 segment Lys-100–Ala-125, which overlaps with the tubulin-binding site Gln-103–Pro-118, (i) might be structurally less well defined in solution compared with the crystalline state and (ii) could feature small differences either in conformation or in chemical environment between Par14 and Par17, which may be responsible, at least in part, for the diverging efficiencies in microtubule formation.
Moreover, comparison of the apparently non-structured N termini of Par14 and Par17 (14, 36) revealed a remarkable difference in line-broadening effects. The first 29 residues of Par14 could not be assigned due to extreme broadening of the amide signals (14), indicating a dynamic behavior that is independent of the PPIase domain. The amide signals of Par17 residues Met-1–Gly-60, on the other hand, were all clearly observable and featured intensities comparable to the signals of the PPIase domain (Fig. 6A), suggesting a similar overall tumbling rate rather than a freely mobile, highly dynamic N terminus. Hence, the Par17 N-terminal segment appears to be involved in some kind of intramolecular interaction with its PPIase domain, as observed for example also in case of AtFKBP42 (44). Such an anchoring of the non-structured N-terminal Par17 extension Met-1–Lys-25 at the PPIase domain (i) would explain the above-described chemical shift differences observed in the Par17 segment Lys-100–Ala-125 compared with Par14 and (ii) might confer the unique biological properties that distinguish Par17 from Par14, such as for example the hiding of the nuclear signal (21).
FIGURE 6.
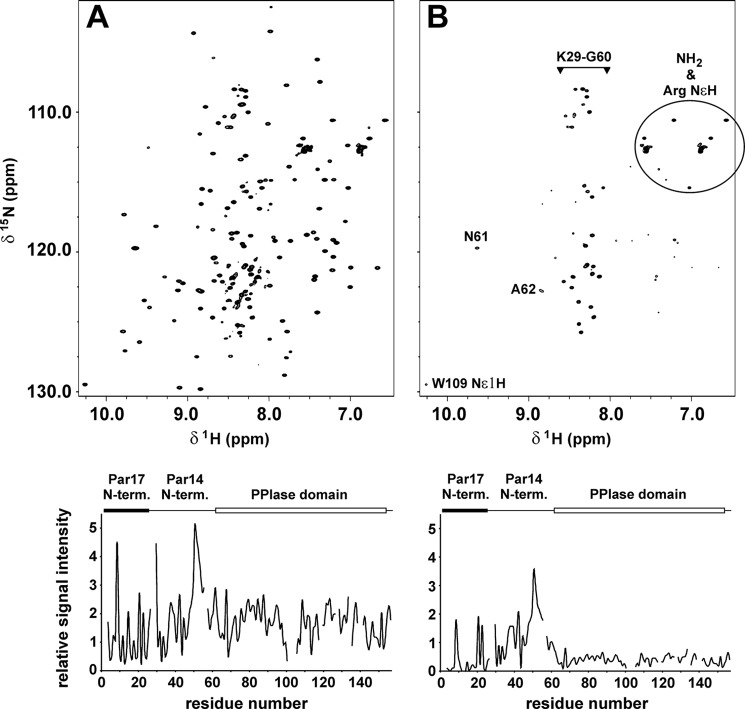
1H,15N HSQC spectra of 0. 9 mm13C/15N-labeled Par17 collected at 18 °C and pH 6.8 in the absence of Ca2+/CaM (A) and in the presence of a 1.4-fold excess of Ca2+/CaM (B). Relative intensities of the Par17 backbone amide signals are shown in the lower panels, where the first 25 residues comprising the N-terminal extension of Par17 as well as the PPIase domain are indicated above the intensity plots by black and white boxes, respectively. In B, the side-chain NH and NH2 signals are marked accordingly; all remaining strong peaks belong to the backbone amide groups in segment Lys-29–Ala-62.
The most likely candidate at the Par17 N terminus for such an intramolecular interaction with the PPIase domain is the hydrophobic ring of Phe-17; however, although several significant changes in chemical shift values between the isolated Par172–22 peptide and the corresponding Par17 N terminus hint at differences in secondary structure (in particular the HN resonances of Lys-8 and Gly-9 as well as Hα of Phe-17 indicate a more extended conformation in the N-terminal segment of full-length Par17), no long range NOE connectivities could be observed for either Phe-17 or any other N-terminal Par17 residue, possibly due to the presumably rather weak nature of this intramolecular interaction.
Ca2+/CaM Binding to 15N-labeled Par17
Although CaM is known to interact with a plethora of different peptides and proteins, certain consensus sequences for CaM recognition exist (45). Moreover, some of the CaM-binding peptides reported to date have the potential to fold into an amphipathic α-helix, displaying hydrophobic residues in conserved positions (46, 47). The presence of such a sequence motif for potential CaM binding had been identified within the first 20 N-terminal residues of Par17 using the CaM target database, as mentioned above, and an amphipathic α-helix was predicted within the same segment by the online server heliquest. As a consequence, we aimed to confirm this CaM-binding site by performing a CSP analysis with 15N-labeled Par17 in the absence and presence of non-labeled CaM. To this end, we employed Ca2+/CaM, as the above-described pulldown experiments had revealed a Ca2+ dependence of CaM binding to Par17.
Interestingly, comparison of 1H,15N HSQC spectra featuring 13C/15N-labeled Par17 in the absence and in presence of Ca2+/CaM revealed a drastic decrease in the Par17 signal intensities upon CaM binding not only for the N-terminal segment of Par17 but also for the PPIase domain (Fig. 6B). With the exception of residues Gly-30 and Ser-32, all backbone amide signals in the sequence comprising Lys-29 to Ala-62 (i.e. the linker region between the N-terminal segment and the PPIase domain of Par17, which corresponds to the N terminus of Par14) remained basically unaffected by the addition of Ca2+/CaM, whereas all other signals showed considerable line-broadening, suggesting the likely existence of not just one but two intermolecular interaction sites.
Because of the strong line-broadening effects in the Par17-Ca2+/CaM complex, we subsequently employed 1H,15N TROSY spectra of 15N-labeled Par17 (Fig. 7A), which clearly showed that the signals corresponding to the 25 N-terminal Par17 residues exhibited significant alterations upon the addition of Ca2+/CaM. Very pronounced chemical shift changes were observed for residues Leu-7, Val-11, Arg-12, Leu-14, Phe-17, Val-19, Ala-23, Ser-24, and Lys-25. In addition, extreme line-broadening effects were detected for a number of residues within the segment spanning from Met-3 to Met-26, whose amide signals disappeared completely (Fig. 7B). Most of the residues displaying very pronounced effects in the presence of CaM are thus identical to those predicted as potential CaM-binding site with propensity to form an amphipathic helix.
FIGURE 7.
Ca2+/CaM binding to 15N-labeled Par17. A, central region of superposed 1H,15N TROSY spectra collected at 25 °C and pH 6.8 with 15N-labeled Par17 (0.6 mm) in the absence of Ca2+/CaM (black) and in presence of a 1.4-fold molar Ca2+/CaM excess (red). B, CSP effects in Par17 upon Ca2+/CaM binding are shown as blue and magenta bars, indicating either signal shifts or extreme line broadening, respectively. C, helical wheel representation of the predicted amphipathic α-helix at the N terminus of Par17. Hydrophobic, polar, acidic, and basic residues are highlighted in yellow, pink, red, and blue, respectively. The arrow indicates the hydrophobic moment of the helix (65); the N- and C-terminal ends of the wheel are marked by green capital letters.
In the absence of Ca2+/CaM, the N-terminal elongation of Par17 exhibits a mostly random-coil conformation in solution, as indicated by the corresponding signal distribution in the 1H,15N HSQC spectra (Fig. 5, A and B) as well as by chemical shift index analysis (36). According to the below-described results, however, it is feasible that the N terminus of Par17 adopts a helical conformation upon Ca2+/CaM binding, with the primary CaM-binding site of Par17 presumably defined mainly by the hydrophobic face of the putative amphipathic α-helix, i.e. residues Met-3, Leu-6, Leu-7, Leu-10, Leu-14, and Phe-17 (Fig. 7C). Unfortunately, however, it was not possible to sequentially assign the Par17 N terminus in the Par17-Ca2+/CaM complex due to the rather extreme line-broadening effects of several signals in the spectra of the bound Par17 form, thus rendering a structure determination or prediction of the N-terminal region impractical.
In addition to the large CSP effects at the N terminus, a relatively high frequency of small CSP effects (∼0.01 ppm) could be identified within the segment Gly-105–Gln-120 (Fig. 7B), which corresponds to the tubulin-binding site that coincides with the Par17 substrate binding pocket. Considering the drastic change in signal intensities in the PPIase domain upon the addition of Ca2+/CaM, these relatively small CSP effects near the active site provide further evidence that a second CaM-binding site may exist in the PPIase domain of Par17, which could result in the formation of a bidentate complex between Par17 and Ca2+/CaM.
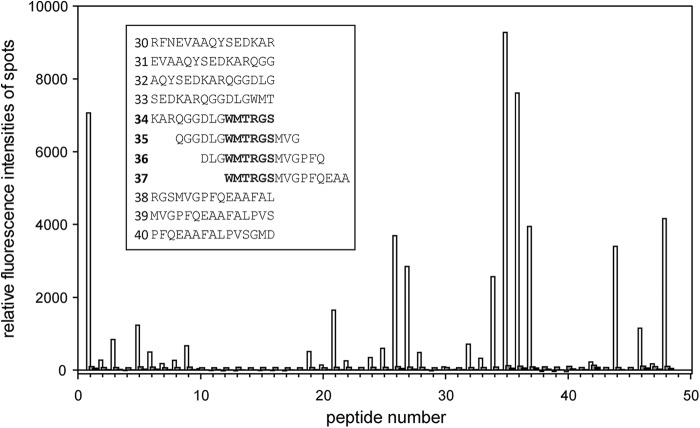
To search for this potential CaM-binding site within the Par17 PPIase domain, a peptide microarray experiment was performed. For this purpose, a series of overlapping 15-mer peptides covering the sequence of Par17 was synthesized and tested for CaM binding. In the presence of Ca2+, CaM bound to the peptides 34–37, which share the core motif 109WMTRGS114, whereas in the absence of Ca2+ no significant CaM binding was observed (Fig. 8). This result further corroborates the likely existence of an additional CaM-binding site near the Par17 active site.
FIGURE 8.
Peptide microarray binding pattern of Ca2+/CaM. To display Ca2+/CaM binding to the microarray of Par17 15-mer peptides, relative fluorescence intensities of spots, which were corrected for the background, are plotted against the peptide number. Incubation of peptide microarrays were performed in the presence of either Ca2+ (white bars) or the Ca2+-chelator EGTA (black bars). As a control, the array was incubated only with anti-mouse-IgG (gray bars). The inset shows the amino acid sequences of peptides 30–40, comprising the dominant binding site for Ca2+/CaM in the Par17 peptide microarray.
Par17 Binding to 15N-Labeled Ca2+/CaM
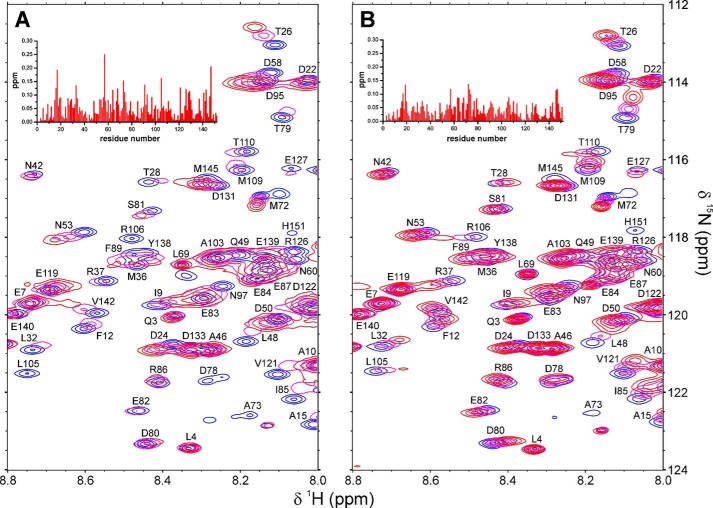
To further characterize the Par17-CaM interaction, we subsequently changed the 15N labeling and collected 1H,15N TROSY spectra of 15N-enriched Ca2+/CaM at various concentrations of non-labeled Par17. Again, both significant chemical shift changes as well as extreme line-broadening effects were observed (Fig. 9A). Subsequent CSP analysis revealed that residues located in both CaM lobes as well as in the flexible interlobe linker were affected by Par17 binding. A very similar CSP pattern was obtained (Fig. 9B) upon binding of merely the corresponding N-terminal Par17 peptide that comprises residues Pro-2–Gln-22 (i.e. Par172–22). Hence the apparently minor effects caused by the potential second binding site again seem to be superseeded by the more drastic effects resulting from binding of the Par17 N-terminal segment.
FIGURE 9.
Par17 binding to 15N-labeled Ca2+/CaM. A, superposed 1H,15N TROSY spectra collected at 25 °C and pH 6.8 with 15N-labeled Ca2+/CaM (0.5 mm) in the absence of Par17 (blue) and in presence of Par17 at a 5-fold (magenta) and 2-fold (red) molar excess of Ca2+/CaM. B, superposed 1H,15N TROSY spectra collected at 25 °C and pH 6.8 with 15N-labeled Ca2+/CaM (0.5 mm) without Par172–22 (blue), with Par172–22 at 1:1 molar ratio (magenta), and with Par172–22 in 3-fold molar excess (red). The CSP effects observed in Ca2+/CaM upon binding of Par17 (at 2:1 molar stoichiometry) and Par172–22 (at 1:3 molar stoichiometry) are shown as insets in panels A and B, respectively.
The CSP effects observed for Ca2+/CaM upon Par17 binding can be the result of either direct intermolecular contacts between both complex partners or indirect effects resulting from conformational changes. Due to the large number of CSP effects in both CaM lobes, we decided to compare our data to the complex of Ca2+/CaM with a 20-residue peptide from chicken smMLCK, where the two CaM lobes clamp around the helical smMLCK796–815 peptide, which is thus located inside a mainly hydrophobic channel (34). Such a binding scenario has been observed also in other CaM complexes (48–50). In all of these cases, binding is driven largely by non-polar interactions, as the channel features a hydrophobic arc that is (i) formed by the hydrophobic patches of both CaM lobes, including numerous methionine residues, and (ii) surrounded by several charged residues, which create the charged outlets of the binding channel. As a consequence, CaM-binding peptides usually are amphipathic α-helices that also display a charge distribution on their molecular surface to determine their relative binding orientation (32).
Using the CaM structure known from the smMLCK complex (34), structural mapping of the most pronounced CSP effects caused by Par17 showed an arrangement where the majority of the affected CaM residues face the bound peptide (Fig. 10A). Additional effects were observed in the CaM interlobe linker, most likely due to changes in its conformation when the CaM molecule clamps around the bound peptide. Taking into consideration the rather large number of hydrophobic residues in the N-terminal Par17 segment (12 of 25 residues), which apparently can form an amphipathic α-helix that includes the aromatic ring of Phe-17 at the hydrophobic face (Fig. 7C), we hence conclude that the Par17 N terminus most likely features a CaM binding scenario that is very similar to the one known from the CaM-smMLCK complex. In fact, a direct comparison between the CaM residues found in close contact (<4 Å) with the smMLCK796–815 peptide according to the x-ray structure (34) and the CaM residues affected by the presence of Par17 according to our CSP data revealed a quite good agreement, in particular with respect to the C-terminal half of the peptides, which (i) is primarily responsible for CaM binding (51), and where (ii) both peptide sequences show a fairly high degree of homology (Fig. 10B).
FIGURE 10.
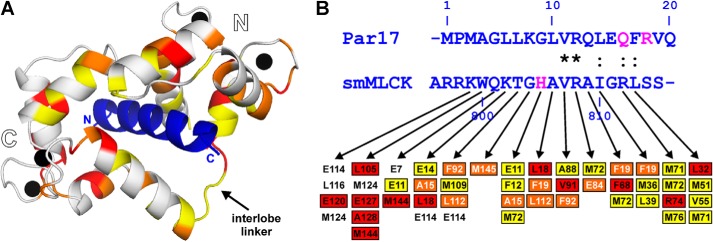
A, structural mapping of the CSP effects observed in Ca2+/CaM upon Par17 binding based on the structure of the complex (PDB ID code 1CDL) between Ca2+/CaM and the smMLCK796–815 peptide from chicken (blue ribbon). Ca2+/CaM residues that showed significant line-broadening effects are highlighted in yellow, whereas residues featuring CSP effects >0.10 ppm or >0.15 ppm are colored in orange and red, respectively. The CaM lobes and interlobe linker as well as the N- and C-terminal ends of the smMLCK796–815 peptide are marked. B, sequence comparison between the predicted CaM-binding site at the N terminus of human Par17 and the smMLCK796–815 peptide. Sequence similarities are found only in the second half of the peptides, as indicated in the row between the sequences (*, identical; :, conserved). Arrows below the smMLCK sequence denote close contacts (<4 Å) to CaM residues according to the x-ray structure (PDB ID code 1CDL). Colored boxes illustrate where these CaM residues or one of their immediate sequential neighbors showed CSP effects in the presence of Par17 (same color code as in panel A). Magenta-colored residues in the peptide sequences indicate positions of naturally occurring amino acid substitutions; (i) due to single-nucleotide polymorphism, a second human Par17 isoform exists featuring the point mutations Q16R and R18S, and (ii) in mammalian smMLCK, H805 is generally replaced by an asparagine.
Discussion
In the present study we provide first evidence that Ca2+/CaM interacts with Par17 in a physiologically relevant way by preventing Par17-promoted microtubule assembly. In contrast, Par14, which is basically identical to Par17 yet lacks the first 25 N-terminal residues of the Par17 sequence, does not interact with Ca2+/CaM at all. Pulldown experiments further revealed that the 25 additional N-terminal residues of Par17, which hold the potential to fold into an amphipathic α-helix, include a high affinity CaM-binding site. Hence, this N-terminal segment confers a unique property to Par17 in comparison with Par14.
Analysis of NMR spectroscopic data (14, 17, 36) has shown that the resonance assignments of the Par17 and Par14 PPIase domains are quite similar, indicating that the 25 additional residues at the N terminus of Par17 do not affect the overall domain fold. Nevertheless, despite the fact that the N-terminal segments of both parvulin types do not seem to display any regular secondary structure elements, the amide signals of the Par17 N terminus were fully observable in contrast to Par14 and exhibited intensities similar to the rest of the protein, thus indicating an overall tumbling rate analogous to the PPIase domain. Moreover, the minor chemical shift differences in the Par17 region Lys-100–Ala-125 compared with the corresponding Par14 residues hint at an intramolecular docking interaction of the Par17 N terminus in the region Lys-100–Ala-125.
Furthermore, the CSP effects observed for the Par17-CaM interaction demonstrated that the N-terminal segment of Par17 binds to both CaM lobes simultaneously, indicating that CaM undergoes a conformational change to form a channel between its two lobes, apparently similar to the structure of the CaM-smMLCK796–815 complex. Moreover, the sequence similarities in the C-terminal half of the peptides smMLCK796–815 and Par171–20 suggest an analogous CaM binding arrangement. Finally, a set of minor CSP effects in the Par17 region Lys-100–Ala-125 upon Ca2+/CaM binding hint at a second CaM-binding site. The peptide microarray experiment has provided additional evidence for the existence of a second, low affinity CaM-binding site in the Par17 region Lys-100–Ala-125, as CaM recognized the core epitope WMTRGS, which encompasses the Par17 residues Trp-109–Ser-114. However, except for the very first peptide in the array, CaM did not bind significantly to the peptides representing the N-terminal high affinity CaM-binding site (Fig. 8), thus indicating (i) that these N-terminal peptides do not adopt an α-helical structure under the conditions of the microarray spots and, moreover, (ii) that the formation of an amphipathic α-helix is crucial for CaM binding.
Interactions of CaM with others PPIases, such as FKBP73 (52) or FKBP38 (37, 53) have been reported before. However, those CaM binding scenarios are markedly different to the one we describe here. The CaM-binding site of FKBP73 is located at its C terminus; the 138 C-terminal residues of FKBP73, including the tetratricopeptide repeat (TPR) domain, are essential for proper CaM binding (52). In the case of FKBP38, two distinct CaM-binding sites exist; (i) a C-terminal motif, which forms a complex with the C-terminal CaM lobe only in the presence of Ca2+, and (ii) a region in the catalytic domain of FKBP38, which binds to the N-terminal CaM lobe in a Ca2+-independent manner (53). Certainly, the presence of a high affinity CaM-binding site in Par17 but not in Par14 could be a key to differentially regulate the function of these two otherwise basically identical proteins.
In a previous study we showed that Par17 interacts with tubulin and thereby promotes the formation of microtubules (20). These microtubule assembly-related properties seem to correlate directly with PPIase activity, because (i) catalytically deficient variants of Par17 were unable to promote microtubule formation and (ii) inhibitors of Par17 activity also prevented parvulin-catalyzed tubulin polymerization. Remarkably, Par14, which comprises an identical PPIase domain, was less efficient in the catalysis of microtubule assembly, indicating an involvement of the 25 additional residues at the N terminus of Par17 in tubulin binding. The N-terminal Par17 segment alone, however, was not sufficient to mediate the tubulin interaction and thus to promote microtubule assembly. Analysis of the tubulin interaction sites revealed that tubulin predominantly interacts with the substrate binding pocket of Par17.
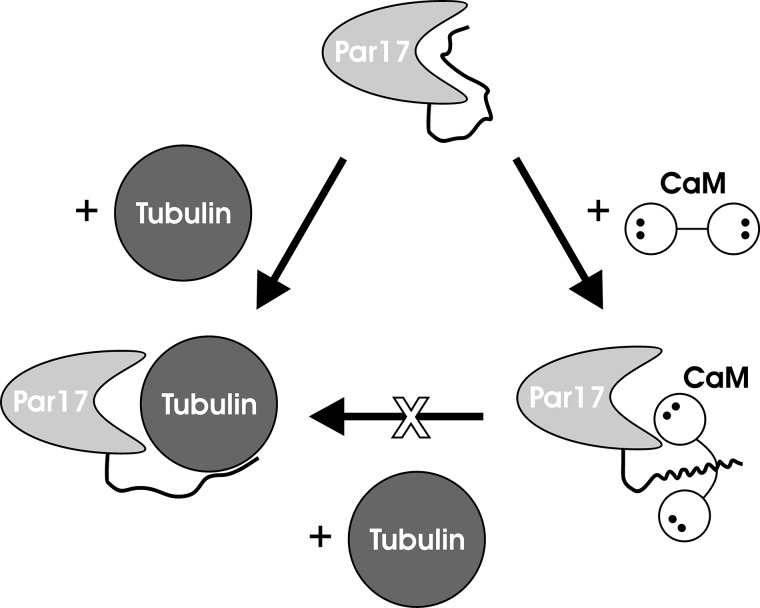
In the present study we were able to show that the addition of Ca2+/CaM completely blocks Par17-catalyzed tubulin polymerization in vitro. The analysis of CaM-binding sites using pulldown and NMR experiments revealed that Ca2+/CaM primarily binds to the N-terminal extension of Par17, which in native parvulin apparently interacts with the active site in the PPIase domain. Par14, which lacks the N-terminal extension, did not interact at all with Ca2+/CaM in pulldown or ITC experiments. These results, therefore, suggest the following interaction scenario, as depicted in Fig. 11. 1) In the native state of Par17, the N-terminal 25-residue extension is located in close proximity to the substrate binding pocket due to a weak intramolecular interaction, thereby controlling the access of substrates to the active site. 2) Tubulin binding to Par17 apparently is favored by the presence of this N-terminal extension, as the tubulin polymerization efficiency is higher in Par17 compared with Par14. 3) Ca2+/CaM interferes with tubulin-Par17 interaction by binding to the N-terminal extension in the Par17 sequence, thereby apparently also causing hindrance of the Par17 active site either sterically or due to the second, low affinity CaM-binding site.
FIGURE 11.
Schematic representation of the Ca2+/CaM-dependent blocking of the Par17-promoted tubulin polymerization. In native Par17 (top), the N-terminal extension interacts with the substrate binding pocket. In the tubulin-Par17 complex (bottom left), tubulin seems to continue to interact with the Par17 N terminus after replacing it at the substrate binding pocket, thus ensuring a higher polymerization efficiency compared with Par14. Ca2+/CaM binds strongly to the Par17 N terminus (bottom right); at the same time, the close proximity of CaM to the substrate binding pocket of the PPIase domain apparently restricts the access to the active site for larger ligands such as tubulin but not for small substrates such as the oligopeptide succinyl-AKPF-4-nitroanilide.
The role of Ca2+/CaM in the regulation of microtubule interactions with MAPs is not unique to Par17. In fact, Ca2+/CaM was already reported to bind MAPs other than Par17 (24, 25, 54), as for example the stable tubulin-only polypeptides (STOPs) (55). Ca2+/CaM binding inhibits the ability of STOPs to cold-stabilize microtubules (56). In this case, Ca2+/CaM and tubulin exhibit overlapping binding sites. However, STOPs merely cold-stabilize microtubules but do not promote microtubule assembly (55), whereas EF-1α, another Ca2+/CaM-interacting MAP, merely promotes microtubule assembly but does not cold-stabilize microtubules (57). Ca2+/CaM binding inhibits EF-1α-mediated tubulin polymerization, similar to the effect observed in case of Par17. MAP2 and the Tau protein represent two further important MAPs that are regulated by Ca2+/CaM binding and also display overlapping binding sites for CaM and tubulin (25, 58). These findings show that Ca2+/CaM-mediated inhibition of tubulin polymerization could be related to direct binding of CaM to different MAPs, thus preventing the interaction of these latter proteins with tubulin. Which MAP is controlled by Ca2+/CaM may depend on both spatial and temporal localization as well as on the impact of the respective MAP on microtubules. For example, Tau protein and MAP2 exist in neuronal cells (59, 60), whereas STOPs, EF-1α, and Par17 are present in many other cell types (8, 61). Moreover, MAPs also exhibit different subcellular localizations; MAP2 is sequestered into dendrites, whereas Tau is localized in the distal portions of axons (62), STOPs preferentially localize to kinetochore microtubules in the spindle (63), and EF-1α immunolocalizes to both mitotic and interphase arrays (64).
Hence, we propose that Par17 also belongs to the group of CaM-regulated MAPs. These MAPs generally share common features, including overlapping binding sites for CaM and tubulin as well as a strict Ca2+ dependence in their CaM interaction. The Ca2+-mediated regulation of microtubule assembly via CaM and a MAP such as Par17 may thus represent another mechanism that serves to intertwine microtubule function with other cellular events.
Acknowledgments
We thank Monika Seidel for excellent technical assistance, Dr. Mike Schutkowski (University Halle-Wittenberg) for helpful suggestions and support regarding the microarrays, Dr. Gunter Fischer (MPI Göttingen) for providing access to the laboratory resources at the Max Planck Research Unit for Enzymology of Protein Folding in Halle (Saale), and Dr. F. Luis González Flecha (Institute of Biochemistry and Biophysics, Buenos Aires) for assistance with ITC data analysis.
The authors declare that they have no conflicts of interest with the contents of this article.
- PPIase
- peptidyl prolyl cis/trans isomerase
- CaM
- calmodulin
- CSP
- chemical shift perturbation
- FKBP
- FK506-binding protein
- ITC
- isothermal titration calorimetry
- MAP
- microtubule-associated protein
- Par
- parvulin
- smMLCK
- smooth muscle myosin light chain kinase
- TROSY
- transverse relaxation optimized spectroscopy
- STOP
- stable tubulin-only polypeptide.
References
- 1. Galat A., Metcalfe S. M. (1995) Peptidylproline cis/trans isomerases. Prog. Biophys. Mol. Biol. 63, 67–118 [DOI] [PubMed] [Google Scholar]
- 2. Fanghänel J., Fischer G. (2004) Insights into the catalytic mechanism of peptidyl prolyl cis/trans isomerases. Front. Biosci. 9, 3453–3478 [DOI] [PubMed] [Google Scholar]
- 3. Yaffe M. B., Schutkowski M., Shen M., Zhou X. Z., Stukenberg P. T., Rahfeld J. U., Xu J., Kuang J., Kirschner M. W., Fischer G., Cantley L. C., Lu K. P. (1997) Sequence-specific and phosphorylation-dependent proline isomerization: a potential mitotic regulatory mechanism. Science 278, 1957–1960 [DOI] [PubMed] [Google Scholar]
- 4. Lu K. P. (2003) Prolyl isomerase Pin1 as a molecular target for cancer diagnostics and therapeutics. Cancer Cell 4, 175–180 [DOI] [PubMed] [Google Scholar]
- 5. Pemberton T. J., Kay J. E. (2005) Identification and comparative analysis of the peptidyl-prolyl cis/trans isomerase repertoires of H. sapiens, D. melanogaster, C. elegans, S. cerevisiae and Sz. pombe. Comp. Funct. Genomics 6, 277–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jaremko Ł., Jaremko M., Elfaki I., Mueller J. W., Ejchart A., Bayer P., Zhukov I. (2011) Structure and dynamics of the first archaeal parvulin reveal a new functionally important loop in parvulin-type prolyl isomerases. J. Biol. Chem. 286, 6554–6565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mueller J. W., Bayer P. (2008) Small family with key contacts: Par14 and Par17 parvulin proteins, relatives of Pin1, now emerge in biomedical research. Perspect. Medicin. Chem. 2, 11–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mueller J. W., Kessler D., Neumann D., Stratmann T., Papatheodorou P., Hartmann-Fatu C., Bayer P. (2006) Characterization of novel elongated Parvulin isoforms that are ubiquitously expressed in human tissues and originate from alternative transcription initiation. BMC Mol. Biol. 7, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jurado L. A., Chockalingam P. S., Jarrett H. W. (1999) Apocalmodulin. Physiol. Rev. 79, 661–682 [DOI] [PubMed] [Google Scholar]
- 10. Lu P. J., Zhou X. Z., Shen M., Lu K. P. (1999) Function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science 283, 1325–1328 [DOI] [PubMed] [Google Scholar]
- 11. Lu P. J., Wulf G., Zhou X. Z., Davies P., Lu K. P. (1999) The prolyl isomerase Pin1 restores the function of Alzheimer-associated phosphorylated tau protein. Nature 399, 784–788 [DOI] [PubMed] [Google Scholar]
- 12. Kessler D., Papatheodorou P., Stratmann T., Dian E. A., Hartmann-Fatu C., Rassow J., Bayer P., Mueller J. W. (2007) The DNA binding parvulin Par17 is targeted to the mitochondrial matrix by a recently evolved prepeptide uniquely present in Hominidae. BMC Biol. 5, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Uchida T., Fujimori F., Tradler T., Fischer G., Rahfeld J. U. (1999) Identification and characterization of a 14 kDa human protein as a novel parvulin-like peptidyl prolyl cis/trans isomerase. FEBS Lett. 446, 278–282 [DOI] [PubMed] [Google Scholar]
- 14. Terada T., Shirouzu M., Fukumori Y., Fujimori F., Ito Y., Kigawa T., Yokoyama S., Uchida T. (2001) Solution structure of the human parvulin-like peptidyl prolyl cis/trans isomerase, hPar14. J. Mol. Biol. 305, 917–926 [DOI] [PubMed] [Google Scholar]
- 15. Zoldák G., Aumüller T., Lücke C., Hritz J., Oostenbrink C., Fischer G., Schmid F. X. (2009) A library of fluorescent peptides for exploring the substrate specificities of prolyl isomerases. Biochemistry 48, 10423–10436 [DOI] [PubMed] [Google Scholar]
- 16. Mueller J. W., Link N. M., Matena A., Hoppstock L., Rüppel A., Bayer P., Blankenfeldt W. (2011) Crystallographic proof for an extended hydrogen-bonding network in small prolyl isomerases. J. Am. Chem. Soc. 133, 20096–20099 [DOI] [PubMed] [Google Scholar]
- 17. Sekerina E., Rahfeld J. U., Müller J., Fanghänel J., Rascher C., Fischer G., Bayer P. (2000) NMR solution structure of hPar14 reveals similarity to the peptidyl prolyl cis/trans isomerase domain of the mitotic regulator hPin1 but indicates a different functionality of the protein. J. Mol. Biol. 301, 1003–1017 [DOI] [PubMed] [Google Scholar]
- 18. Surmacz T. A., Bayer E., Rahfeld J. U., Fischer G., Bayer P. (2002) The N-terminal basic domain of human parvulin hPar14 is responsible for the entry to the nucleus and high-affinity DNA-binding. J. Mol. Biol. 321, 235–247 [DOI] [PubMed] [Google Scholar]
- 19. Ranganathan R., Lu K. P., Hunter T., Noel J. P. (1997) Structural and functional analysis of the mitotic rotamase Pin1 suggests substrate recognition is phosphorylation dependent. Cell 89, 875–886 [DOI] [PubMed] [Google Scholar]
- 20. Thiele A., Krentzlin K., Erdmann F., Rauh D., Hause G., Zerweck J., Kilka S., Pösel S., Fischer G., Schutkowski M., Weiwad M. (2011) Parvulin 17 promotes microtubule assembly by its peptidyl-prolyl cis/trans isomerase activity. J. Mol. Biol. 411, 896–909 [DOI] [PubMed] [Google Scholar]
- 21. Reimer T., Weiwad M., Schierhorn A., Ruecknagel P. K., Rahfeld J. U., Bayer P., Fischer G. (2003) Phosphorylation of the N-terminal domain regulates subcellular localization and DNA binding properties of the peptidyl-prolyl cis/trans isomerase hPar14. J. Mol. Biol. 330, 955–966 [DOI] [PubMed] [Google Scholar]
- 22. Downing K. H., Nogales E. (1998) Tubulin and microtubule structure. Curr. Opin. Cell Biol. 10, 16–22 [DOI] [PubMed] [Google Scholar]
- 23. Maccioni R. B., Cambiazo V. (1995) Role of microtubule-associated proteins in the control of microtubule assembly. Physiol. Rev. 75, 835–864 [DOI] [PubMed] [Google Scholar]
- 24. Lee Y. C., Wolff J. (1984) Calmodulin binds to both microtubule-associated protein 2 and tau proteins. J. Biol. Chem. 259, 1226–1230 [PubMed] [Google Scholar]
- 25. Padilla R., Maccioni R. B., Avila J. (1990) Calmodulin binds to a tubulin binding site of the microtubule-associated protein tau. Mol. Cell. Biochem. 97, 35–41 [DOI] [PubMed] [Google Scholar]
- 26. Lefèvre J., Savarin P., Gans P., Hamon L., Clément M. J., David M. O., Bosc C., Andrieux A., Curmi P. A. (2013) Structural basis for the association of MAP6 protein with microtubules and its regulation by calmodulin. J. Biol. Chem. 288, 24910–24922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cohen P., Klee C. B. (1988) Calmodulin, Elsevier Science Publishing Co., Inc., New York [Google Scholar]
- 28. Babu Y. S., Sack J. S., Greenhough T. J., Bugg C. E., Means A. R., Cook W. J. (1985) Three-dimensional structure of calmodulin. Nature 315, 37–40 [DOI] [PubMed] [Google Scholar]
- 29. Barbato G., Ikura M., Kay L. E., Pastor R. W., Bax A. (1992) Backbone dynamics of calmodulin studied by nitrogen-15 relaxation using inverse detected two-dimensional NMR spectroscopy: the central helix is flexible. Biochemistry 31, 5269–5278 [DOI] [PubMed] [Google Scholar]
- 30. Potter J. D., Strang-Brown P., Walker P. L., Iida S. (1983) Ca2+ binding to calmodulin. Methods Enzymol. 102, 135–143 [DOI] [PubMed] [Google Scholar]
- 31. Gilli R., Lafitte D., Lopez C., Kilhoffer M., Makarov A., Briand C., Haiech J. (1998) Thermodynamic analysis of calcium and magnesium binding to calmodulin. Biochemistry 37, 5450–5456 [DOI] [PubMed] [Google Scholar]
- 32. Vetter S. W., Leclerc E. (2003) Novel aspects of calmodulin target recognition and activation. Eur. J. Biochem. 270, 404–414 [DOI] [PubMed] [Google Scholar]
- 33. Yamniuk A. P., Vogel H. J. (2004) Calmodulin's flexibility allows for promiscuity in its interactions with target proteins and peptides. Mol. Biotechnol. 27, 33–57 [DOI] [PubMed] [Google Scholar]
- 34. Meador W. E., Means A. R., Quiocho F. A. (1992) Target enzyme recognition by calmodulin: 2.4 Å structure of a calmodulin-peptide complex. Science 257, 1251–1255 [DOI] [PubMed] [Google Scholar]
- 35. Edlich F., Weiwad M., Erdmann F., Fanghänel J., Jarczowski F., Rahfeld J. U., Fischer G. (2005) Bcl-2 regulator FKBP38 is activated by Ca2+/calmodulin. EMBO J. 24, 2688–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lin Y. J., Schmidt A., Burgardt N. I., Thiele A., Weiwad M., Lücke C. (2013) 1H, 13C, and 15N resonance assignments of human parvulin 17. Biomol. NMR Assign. 7, 325–329 [DOI] [PubMed] [Google Scholar]
- 37. Maestre-Martínez M., Haupt K., Edlich F., Jahreis G., Jarczowski F., Erdmann F., Fischer G., Lücke C. (2010) New structural aspects of FKBP38 activation. Biol. Chem. 391, 1157–1167 [DOI] [PubMed] [Google Scholar]
- 38. Thompson J. D., Higgins D. G., Gibson T. J. (1994) ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gautier R., Douguet D., Antonny B., Drin G. (2008) HELIQUEST: a web server to screen sequences with specific α-helical properties. Bioinformatics 24, 2101–2102 [DOI] [PubMed] [Google Scholar]
- 40. Mulder F. A., Schipper D., Bott R., Boelens R. (1999) Altered flexibility in the substrate-binding site of related native and engineered high-alkaline Bacillus subtilisins. J. Mol. Biol. 292, 111–123 [DOI] [PubMed] [Google Scholar]
- 41. Zhang R., Liu Z., Qu Y., Xu Y., Yang Q. (2013) Two distinct calmodulin binding sites in the third intracellular loop and carboxyl tail of angiotensin II (AT1A) receptor. PLoS ONE 8, e65266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yap K. L., Kim J., Truong K., Sherman M., Yuan T., Ikura M. (2000) Calmodulin target database. J. Struct. Funct. Genomics 1, 8–14 [DOI] [PubMed] [Google Scholar]
- 43. Barton J. S., Riazi G. H. (1980) Evidence for two growth steps in microtubule polymerization. Biochim. Biophys. Acta 630, 392–401 [DOI] [PubMed] [Google Scholar]
- 44. Burgardt N. I., Linnert M., Weiwad M., Geisler M., Lücke C. (2012) NMR assignments of the FKBP-type PPIase domain of FKBP42 from Arabidopsis thaliana. Biomol. NMR Assign. 6, 185–188 [DOI] [PubMed] [Google Scholar]
- 45. Rhoads A. R., Friedberg F. (1997) Sequence motifs for calmodulin recognition. FASEB J. 11, 331–340 [DOI] [PubMed] [Google Scholar]
- 46. Cox J. A., Comte M., Fitton J. E., DeGrado W. F. (1985) The interaction of calmodulin with amphiphilic peptides. J. Biol. Chem. 260, 2527–2534 [PubMed] [Google Scholar]
- 47. O'Neil K. T., DeGrado W. F. (1990) How calmodulin binds its targets: sequence independent recognition of amphiphilic α-helices. Trends Biochem. Sci. 15, 59–64 [DOI] [PubMed] [Google Scholar]
- 48. Ikura M., Clore G. M., Gronenborn A. M., Zhu G., Klee C. B., Bax A. (1992) Solution structure of a calmodulin-target peptide complex by multidimensional NMR. Science 256, 632–638 [DOI] [PubMed] [Google Scholar]
- 49. Wall M. E., Clarage J. B., Phillips G. N. (1997) Motions of calmodulin characterized using both Bragg and diffuse x-ray scattering. Structure 5, 1599–1612 [DOI] [PubMed] [Google Scholar]
- 50. Kurokawa H., Osawa M., Kurihara H., Katayama N., Tokumitsu H., Swindells M. B., Kainosho M., Ikura M. (2001) Target-induced conformational adaptation of calmodulin revealed by the crystal structure of a complex with nematode Ca2+/calmodulin-dependent kinase kinase peptide. J. Mol. Biol. 312, 59–68 [DOI] [PubMed] [Google Scholar]
- 51. Bagchi I. C., Huang Q. H., Means A. R. (1992) Identification of amino acids essential for calmodulin binding and activation of smooth muscle myosin light chain kinase. J. Biol. Chem. 267, 3024–3029 [PubMed] [Google Scholar]
- 52. Kurek I., Dulberger R., Azem A., Tzvi B. B., Sudhakar D., Christou P., Breiman A. (2002) Deletion of the C-terminal 138 amino acids of the wheat FKBP73 abrogates calmodulin binding, dimerization and male fertility in transgenic rice. Plant Mol. Biol. 48, 369–381 [DOI] [PubMed] [Google Scholar]
- 53. Edlich F., Maestre-Martínez M., Jarczowski F., Weiwad M., Moutty M. C., Malesević M., Jahreis G., Fischer G., Lücke C. (2007) A novel calmodulin-Ca2+ target recognition activates the Bcl-2 regulator FKBP38. J. Biol. Chem. 282, 36496–36504 [DOI] [PubMed] [Google Scholar]
- 54. Sobue K., Fujita M., Muramoto Y., Kakiuchi S. (1981) The calmodulin-binding protein in microtubules is tau factor. FEBS Lett. 132, 137–140 [DOI] [PubMed] [Google Scholar]
- 55. Pirollet F., Derancourt J., Haiech J., Job D., Margolis R. L. (1992) Ca2+-calmodulin regulated effectors of microtubule stability in neuronal tissues. Biochemistry 31, 8849–8855 [DOI] [PubMed] [Google Scholar]
- 56. Job D., Fischer E. H., Margolis R. L. (1981) Rapid disassembly of cold-stable microtubules by calmodulin. Proc. Natl. Acad. Sci. U.S.A. 78, 4679–4682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Durso N. A., Cyr R. J. (1994) A calmodulin-sensitive interaction between microtubules and a higher plant homolog of elongation factor-1α. Plant Cell 6, 893–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kindler S., Schulz B., Goedert M., Garner C. C. (1990) Molecular structure of microtubule-associated protein 2b and 2c from rat brain. J. Biol. Chem. 265, 19679–19684 [PubMed] [Google Scholar]
- 59. Shin R. W., Iwaki T., Kitamoto T., Tateishi J. (1991) Hydrated autoclave pretreatment enhances tau immunoreactivity in formalin-fixed normal and Alzheimer's disease brain tissues. Lab. Invest. 64, 693–702 [PubMed] [Google Scholar]
- 60. Johnson G. V., Jope R. S. (1992) The role of microtubule-associated protein 2 (MAP-2) in neuronal growth, plasticity, and degeneration. J. Neurosci. Res. 33, 505–512 [DOI] [PubMed] [Google Scholar]
- 61. Denarier E., Fourest-Lieuvin A., Bosc C., Pirollet F., Chapel A., Margolis R. L., Job D. (1998) Nonneuronal isoforms of STOP protein are responsible for microtubule cold stability in mammalian fibroblasts. Proc. Natl. Acad. Sci. U.S.A. 95, 6055–6060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tucker R. P. (1990) The roles of microtubule-associated proteins in brain morphogenesis: a review. Brain Res. Brain Res. Rev. 15, 101–120 [DOI] [PubMed] [Google Scholar]
- 63. Margolis R. L., Rauch C. T., Pirollet F., Job D. (1990) Specific association of STOP protein with microtubules in vitro and with stable microtubules in mitotic spindles of cultured cells. EMBO J. 9, 4095–4102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hasezawa S., Kumagai F., Nagata T. (1997) Sites of microtubule reorganization in tobacco BY-2 cells during cell-cycle progression. Protoplasma 198, 202–209 [Google Scholar]
- 65. Eisenberg D., Weiss R. M., Terwilliger T. C. (1982) The helical hydrophobic moment: a measure of the amphiphilicity of a helix. Nature 299, 371–374 [DOI] [PubMed] [Google Scholar]