Background: Single molecules of the kinesin-5 Cin8 were previously demonstrated to be minus-end-directed under high-ionic-strength conditions.
Results: Under high-ionic-strength conditions, Cin8 lacking the tail domain is bidirectional.
Conclusion: The tail domain is one of the factors that regulate Cin8 directionality.
Significance: An important structural element was identified that regulates the directionality of kinesin-5 motors.
Keywords: kinesin, microtubule, mitosis, mitotic spindle, molecular motor, kinesin-5, single-molecule biophysics
Abstract
The bipolar kinesin-5 motors are one of the major players that govern mitotic spindle dynamics. Their bipolar structure enables them to cross-link and slide apart antiparallel microtubules (MTs) emanating from the opposing spindle poles. The budding yeast kinesin-5 Cin8 was shown to switch from fast minus-end- to slow plus-end-directed motility upon binding between antiparallel MTs. This unexpected finding revealed a new dimension of cellular control of transport, the mechanism of which is unknown. Here we have examined the role of the C-terminal tail domain of Cin8 in regulating directionality. We first constructed a stable dimeric Cin8/kinesin-1 chimera (Cin8Kin), consisting of head and neck linker of Cin8 fused to the stalk of kinesin-1. As a single dimeric motor, Cin8Kin switched frequently between plus and minus directionality along single MTs, demonstrating that the Cin8 head domains are inherently bidirectional, but control over directionality was lost. We next examined the activity of a tetrameric Cin8 lacking only the tail domains (Cin8Δtail). In contrast to wild-type Cin8, the motility of single molecules of Cin8Δtail in high ionic strength was slow and bidirectional, with almost no directionality switches. Cin8Δtail showed only a weak ability to cross-link MTs in vitro. In vivo, Cin8Δtail exhibited bias toward the plus-end of the MTs and was unable to support viability of cells as the sole kinesin-5 motor. We conclude that the tail of Cin8 is not necessary for bidirectional processive motion, but is controlling the switch between plus- and minus-end-directed motility.
Introduction
Chromosome segregation during mitosis is mediated by the dynamic microtubule-based mitotic spindle. One of the major factors that govern the mitotic spindle dynamics is the function of kinesin-5 motor proteins (1–5), which share homotetrameric structure, with two catalytic domains located at the opposite ends of a coiled-coil stalk (6, 7). This bipolar structure enables kinesin-5 motors to cross-link and slide apart antiparallel microtubules (MTs)4 (8, 9), emanating from the opposing spindle poles, in yeast spindle pole bodies (SPBs) (10–12). Kinesin-5 motors are believed to thus provide the necessary SPB-separating force to perform essential functions in spindle assembly and maintenance of the bipolar spindle structure (10, 11) and anaphase B spindle elongation (1–4, 13).
Sliding apart of antiparallel MTs by the kinesin-5 motors can only be accomplished by the simultaneous plus-end-directed motility of both pairs of catalytic domains in the homotetrameric complex. Plus-end-directed motility was demonstrated in vitro for a number of kinesin-5 motors in MT surface gliding assays (8), in single-molecule fluorescence motility assays (14, 15), and in MT sliding assays (9, 16). Surprisingly, single molecules of Saccharomyces cerevisiae kinesin-5 Cin8 were recently shown to move in the minus-end direction of the MTs, in high-ionic-strength conditions (17–19). Although the mechanism of this minus-end-directed motility is not yet understood, it is consistent with the suggested role for Cin8 in clustering the kinetochores (20, 21) during S. cerevisiae mitosis (17). Cin8 was furthermore shown to switch from fast minus-end-directed motility to slow plus-end-directed motility when bound between antiparallel MTs (17, 18) and when the ionic strength was lowered (17, 19). Furthermore, the unusually large, ∼100-amino acid long loop-8 of Cin8 was shown to be involved in the regulation of its directionality (17). It has remained unclear which other structural elements of Cin8 are involved in regulating the directionality switching, whether processive minus-end-directed motility of Cin8 can be produced by its catalytic head domains alone, or whether it needs interaction with domains outside of the heads.
Previous studies have shown that the tail domains of kinesin motors regulate various aspects of their activity. For example, the tail domain of dimeric kinesin-1 has been shown to inhibit its motility (22) by cross-linking the two catalytic domains (23). In addition, it has been reported that the Xenopus laevis kinesin-5 Eg5 contains a non-motor MT binding site in its tail (24). Finally, it has been shown that the tail domains of the Drosophila melanogaster kinesin-5 Klp61F are located close enough to the motor domains to interact (25). A curious feature that was found for Klp61F is that the central BASS domain of the tetrameric stalk is constructed such that the two pairs of catalytic heads are rotated by 90° to each other (26). Binding between antiparallel MTs could thus cause twist in the stalk that might change the tail-head interaction and serve as a switch mechanism (27). Such an allosteric communication mechanism between the two ends of the tetramer would actually be necessary to switch the motor on, as in the case of Eg5, or to switch directionality, as in the case of Cin8, when the motor is bound between two MTs.
Here we tested the hypothesis that the tail domain of Cin8 regulates its motor functions. For this purpose, we examined the activity of a stable dimeric chimera of Cin8, missing stalk and tail and fused to stalks of Drosophila kinesin-1, and of a tetrameric “tailless” Cin8 variant in which only the tail was deleted (Cin8Δtail). Both mutants were bidirectional in high-ionic-strength conditions, indicating that the catalytic domains of Cin8 are sufficient to produce bidirectional movement and that the tail domain is involved in regulating the directionality of Cin8. Consistent with this finding, in cells, Cin8Δtail exhibited a bias in localization to the plus-end of MTs. We also observed that Cin8Δtail failed to cross-link MTs in vitro, which explains the inability of this variant to support viability of cells lacking endogenous copies of Cin8 and Kip1. Thus, our study identifies the tail domain of Cin8 as one of the major regulators of its motor functions in general and directionality in particular.
Experimental Procedures
Yeast Strains, Growth Conditions, and Viability Test
The S. cerevisiae strains (Table 1) used in this work are derivatives of the S288C strain. Rich (YPD) and minimal (synthetic defined) media were described previously (28).
TABLE 1.
Yeast strains used in this study
| Yeast strain | Genotype |
|---|---|
| LGY 620 | MATa, ura3, leu2, his3, ade2, lys2, cyh2r, cin8::HIS3, kip1::HIS3 (pMA1208) |
| LGY 2562 | MATa, ura3, leu2, his3, ade2, lys2, cyh2r, cin8::HIS3, kip1::HIS3 (pVF37) |
| LGY 3412 | MATα, leu2, reg1-501, ura3, pep4-3, prb-1122, gal1, (pAG36) |
| LGY 3707 | MATα, leu2, reg1-501, ura3, pep4-3, prb-1122, gal1, (pVF87) |
| LGY 4013 | MATa, ura3, leu2, his3, lys2, cin8::LEU2, spc42::KanMX-SPC42-tdTomato (pVF37) |
| LGY 4017 | MATa, ura3, leu2, his3, lys2, cin8::LEU2, spc42::KanMX-SPC42-tdTomato (pVF96) |
| LGY 4019 | MATa, ura3, leu2, his3, lys2, cin8::LEU2, spc42::KanMX-SPC42-tdTomato (pVF98) |
| LGY 4021 | MATa, ura3, leu2, his3, lys2, cin8::LEU2, spc42::KanMX-SPC42-tdTomato (pVF100) |
The ability of different Cin8 variants to complement a CIN8 function was tested in a strain with deletions in both CIN8 and KIP1. Because cin8Δ kip1Δ double mutants are not viable, the viability of the tester strain was maintained by a shuffle plasmid (Table 2) containing wt CIN8. On plates containing cycloheximide, the shuffle plasmid is removed and only cells expressing functional Cin8 remain viable (1, 8).
TABLE 2.
Plasmids used in this study
| Plasmid name | Description | Reference |
|---|---|---|
| pAG36 | PGAL1-CIN8-TEV-GFP-6HIS, URA3, CEN | 17 |
| pCIN8KIN | Cin8Kin-Chimera | This study |
| pMA1208 | CIN8, CYH2, LEU2, CEN | 11 |
| pVF37 | CIN8-3GFP, URA3, CEN | 17 |
| pVF87 | PGAL1-cin8-Δtail-NLS(SV40)-TEV-GFP-6HIS, URA3, CEN | This study |
| pVF96 | cin8-Δtail-3GFP, URA3, CEN | This study |
| pVF98 | cin8-Δnls-3GFP, URA3, CEN | This study |
| pVF100 | cin8-Δtail-NLS(SV40)-linker-3GFP, URA3, CEN | This study |
In Vivo Localization of Cin8
To follow Cin8 localization, the different Cin8 variants were fused with three consecutive C-terminal GFPs (Cin8-3GFP) and expressed under Cin8's own promoter from a CEN plasmid in cin8Δ cells expressing Spc42-Tdtomato. In vivo localization was examined by real-time fluorescence microscope, and Z-stacks with 0.5 μm of separation between planes were recorded on a motorized inverted microscope (Axiovert M200 (Zeiss)) on a vibration isolation table (TMC), supplemented with a cooled CCD camera (SensiCAM (PCO)) and supported by acquisition and image processing software (MetaMorph (Universal Imaging)).
DNA Manipulation
Standard techniques were used for DNA manipulation (29). DH5α Escherichia coli bacterial strains were used as the plasmid host. All PCR products and mutagenesis products were sequenced. Plasmids created are listed in Table 2.
Cloning of Cin8Kin Construct
The Cin8Kin chimera was constructed from pPK113 pET5a-FL (DmKHC-His6) and LGB830 pVF18 (Cin8-GFP) (17) using a nested PCR approach to extend the sequence of the Cin8 motor domain (1–534) with the D. melanogaster kinesin-1 (DmKHC) residues (343–426). The DmKHC motor domain in pPK113 was replaced by subcloning a nested PCR fragment using NdeI and AscIII. A GFP-His6 cassette flanked by AscI and XmaI was generated in pT7-7 for insertion downstream of the Cin8Kin construct in pPK113. The motor was expressed in E. coli and purified as described elsewhere (15).
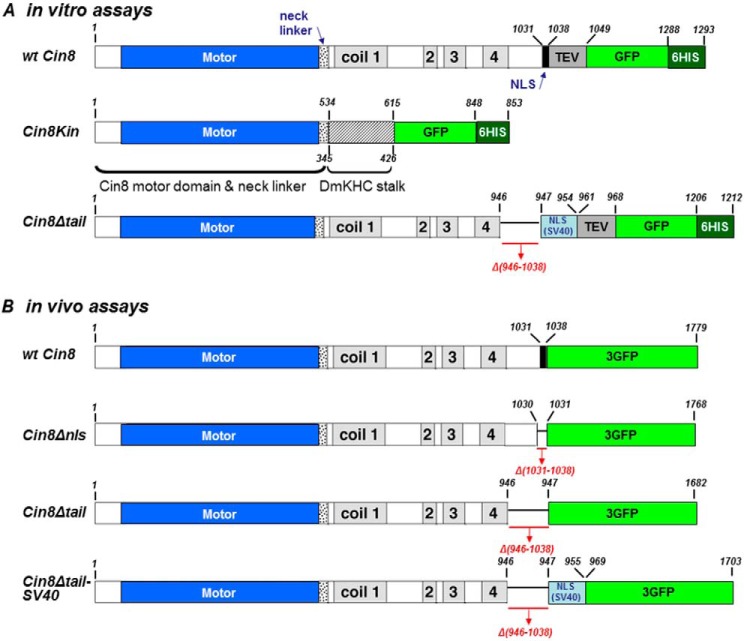
Cloning of Cin8-Δtail-NLS(SV40)-TEV-GFP-His6 and Cin8-3GFP Variants
Cin8-Δtail-NLS(SV40)-TEV-GFP-His6 was created based on the pAG36 plasmid (17) using a PCR-based approach. Because deletion of the tail region (aa 946–1038) eliminates the original NLS sequence, an SV40 NLS (PKKKRKV) sequence was added before TEV to target this protein to the nucleus. The Cin8-3GFP variants (Δnls: deletion of aa 1031–1038; Δtail: deletion of aa 946–1038; and Δtail-NLS(SV40) linker: deletion of aa 946–1038 and addition of SV40 NLS (PKKKRKV) and 12-aa linker (30)) were also created by standard PCR methods based on the pVF37 plasmid (17).
Overexpression and Purification of Cin8 from Yeast Cells
A S. cerevisiae protease-deficient strain was used for protein overexpression (31), and Cin8 overexpression and purification were done as described previously (17). Briefly, S. cerevisiae cells expressing Cin8-TEV-GFP-His6 or Cin8-Δtail-NLS(SV40)-TEV-GFP-His6 under the GAL1 promoter on a CEN plasmid were grown overnight in minimal liquid medium supplemented with 2% raffinose. For overexpression, cells were added to minimal liquid medium containing 2% galactose and grown for an additional 9.5 h. Following overexpression, cells were harvested and pelleted, and 3× volume of lysis buffer was added (30 mm Tris/HCl, 35 mm PIPES/KOH, final pH 7.4, 10% glycerol, 300 mm NaCl, 2 mm EDTA, 1 mm EGTA, 1 mm DTT, 1 mm PMSF, 0.1 mm MgATP, 0.2% Triton X-100, and protease inhibitors). The washed pellet was ground with a mortar and pestle under liquid nitrogen. Following centrifugation, Cin8 was purified on a Ni2+ affinity column as described previously (17). Protein purification was repeated at least three times for each sample, with similar motility results, reducing the likelihood that differences in observed motor behaviors were caused by variability in the quality of purified motors.
Single-molecule Fluorescence Assay
To immobilize MTs, coverslips were silanized with a positively charged silane, 3-[2-(2-aminoethylamino)-ethylamino]propyl-trimethoxysilane (DETA) (Sigma), after plasma cleaning (Harrick Plasma). DETA coverslips were used to assemble assay chambers by attaching them with double-sided tape to a microscope slide. Tetramethylrhodamine-labeled MTs, with Atto 488-labeled seeds, diluted in BRB80 buffer (80 mm PIPES, pH 6.8, 1 mm MgCl2, 1 mm EGTA) containing 10 μm taxol (paclitaxel) were allowed to bind for 5 min to the silanized glass surface of the assay chamber. This was followed by a 5-min incubation with 0.1 mg/ml casein in BRB80. The chamber was then flushed with 7–10 μl of assay buffer BRB80 containing 2 mm ATP, 4 mm MgCl2, and an oxygen-scavenging system based on 10 mm DTT, 0.08 mg/ml catalase, 0.1 mg/ml glucose oxidase, and 10 mm glucose, containing GFP-tagged motor proteins at appropriate single-molecule concentrations.
For relative-sliding assays, the same preparation was used. In addition, short, polarity-marked MTs as well as 4.1 nm Ase1 were added to the motility buffer. Ase1 was purified as described (32).
Fluorescence was observed in a custom-built total internal reflection fluorescence microscope, using a 473-nm laser (Coherent) for excitation of Atto 488, and a 532-nm laser (Viasho) for tetramethylrhodamine, a 100× objective (Nikon, S Fluor, NA 1.49, oil), and an EMCCD camera (iXon Ultra, Andor). Digital videos were recorded with Solis (Andor) with an 800-ms exposure time (1.25 frames/s) and analyzed for motor speeds and run lengths using kymographs generated with open access ImageJ software. Mean-squared displacements (MSDs) for different motor constructs were calculated using the tracked motion of individual molecules performed with MosaicSuite for ImageJ (33). We linearly fitted the parts of the log-log MSD plots that were clearly not affected by localization noise (>4 s). Statistical analyses of the data were performed with OriginPro (OriginLab Corp.). All measurements were performed at 22 °C.
Results and Discussion
Single Molecules of Dimeric Cin8 Are Bidirectional
To examine whether the tail domains of Cin8 regulate its directionality, we first examined the motility of a chimeric Cin8Kin construct lacking most of the stalk and all of the tail domains. We fused motor domain and neck linker of Cin8 (aa 1–534) to the truncated stalk of DmKHC (aa 345–426) and added a C-terminal GFP (Cin8Kin, Fig. 1A). For Eg5 kinesin-5 (14, 15), this strategy had provided a stable dimeric chimera capable of processive motility.
FIGURE 1.
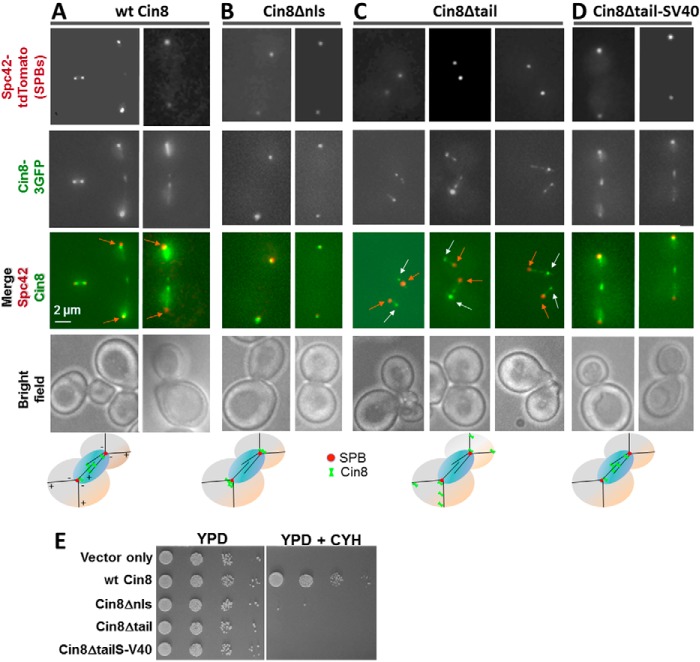
Schematic representation of motor variants used in this study. Structural features of the Cin8 sequence with the corresponding amino acid numbers are shown. Motor domain, high probability coiled-coil domains 1–4, neck linker, and C-terminal NLS are indicated (42). The SV40 NLS sequence (PKKKRKV) (drawn in light blue) was added to Cin8, prior to the sequence of TEV-GFP-His6. Numbers of deleted amino acids are indicated in red below each construct. A, variants used in in vitro assays. Cin8Kin, motor domain and neck linker of Cin8 were fused to DmKHC stalk; numbers below the Cin8Kin scheme refer to the sequence of DmKHC. Cin8Δtail, full-length sequence of Cin8 in which the C-terminal tail was deleted. B, 3GFP-tagged variants used in in vivo assays. Cin8Δnls, aa 1031–1038 that are required for nuclear localization for Cin8 were deleted. Cin8Δtail, C-terminal tail was deleted. Cin8Δtail-SV40, the tail of Cin8 was deleted and the SV40 NLS sequence (PKKKRKV) was added to Cin8, prior to the sequence of 3GFP.
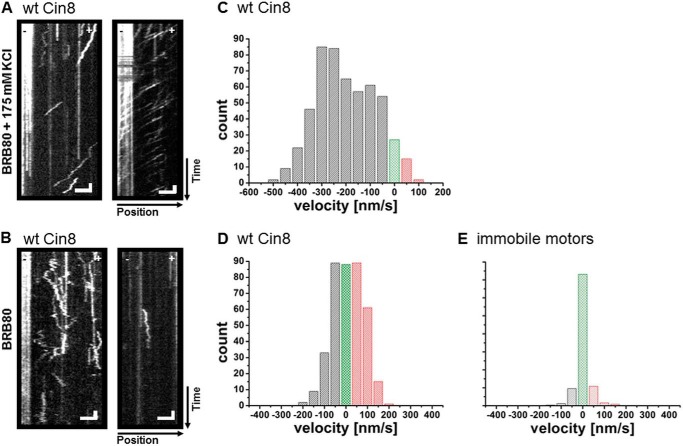
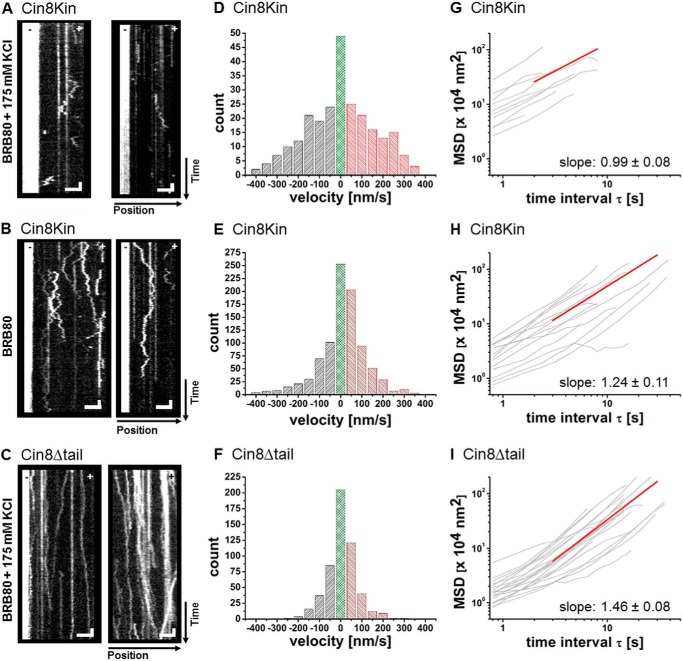
Because our working hypothesis was that the tail regulates switching, we initially expected to see only one directionality, plus or minus, with the dimeric chimeras. We performed single-molecule fluorescence motility experiments with Cin8Kin in low (BRB80) and high-ionic-strength buffer (BRB80 + 175 mm KCl). Ionic strength can determine directionality in the wt motor (17). As a control, we confirmed that wt Cin8 was minus-end-directed at high ionic strength with a velocity of ∼250 nm/s (Fig. 2, A and C) and bidirectional at low ionic strength (Fig. 2, B and D). This behavior is consistent with our previous study (17), although the high velocity of minus-end-directed movement of Cin8 remains largely unexplained. The plus- and minus-end-directed motility of wt Cin8 is clearly ATP-driven and a non-equilibrium phenomenon, rather than mere diffusion, because long directed runs occur in both directions and plus-end motility is evident in antiparallel MT sliding (17). In contrast to wt Cin8, Cin8Kin was bidirectional at both low and high ionic strength (Fig. 3, A and B, and supplemental Movie M1), without any directionality bias. The time the motors remained bound to a MT varied with ionic strength, from seconds at high ionic strength to minutes at low ionic strength (Fig. 3, A and B). This is likely an effect of electrostatic interactions. Because we did not find any clear long-distance motility events at all, it is possible that the Cin8Kin motors merely diffused thermally. Relying on the analogy to wt Cin8, we first assumed actively driven plus- and minus-end motion, however. To quantitate the velocity distribution of Cin8Kin, we segmented the kymographs of the recorded motion. We used segments of 4 frames (resulting in 3.2-s segments using a frame rate of 1.25 frames/s) and fitted the displacement in these segments by straight lines (17). Using the same method to analyze immobile motors on the MT results in a relatively sharp distribution around zero (Fig. 2E), which can be well fitted by a Gaussian with a width of 34 nm/s. This result indicates that the error in velocity in plus and minus direction due to localization errors is about 17 nm/s. The velocity distributions we obtained for moving motors (Fig. 3, D and E) show that in both high-ionic-strength conditions and low-ionic-strength conditions, average speeds for the plus- and minus-end-directed motility of Cin8Kin were about equal.
FIGURE 2.
Single-molecule motility assay with wt Cin8. A and B, kymographs of single-molecule assays of wt Cin8 on surface-immobilized MTs. A, in high-ionic-strength buffer. B, in low-ionic-strength buffer. Representative kymographs are shown. The Atto 488-labeled minus-ends of the MTs are on the left side in all panels. Vertical scale bars: 8 s; horizontal scale bars: 3 μm. C and D, histograms of velocities obtained from segmentation analysis of kymographs for wt Cin8. C, average velocity for high-ionic-strength buffer. vavg = −256 ± 7 nm/s (mean ± S.E.); number of segments, n = 502. D, average velocities for low-ionic-strength buffer. Overall average, vavg = 10 ± 4 nm/s; plus-end average (red), v+ = 80 ± 3 nm/s, n = 166; and minus-end average (gray), v− = −73 ± 3 nm/s, n = 133. Velocities close to zero, in the range between −25 nm/s and 25 nm/s, are indicated in green. E, control, histogram of apparent velocities of immobile Cin8 motors (straight lines in kymographs). The width of the Gaussian fit to this distribution is 34 nm/s.
FIGURE 3.
Single-molecule analysis of Cin8 variants. A–C, kymographs of single-molecule motility assays of Cin8 variants. A, Cin8Kin measured in high-ionic-strength buffer. B, Cin8Kin measured in low-ionic-strength buffer. C, Cin8Δtail measured in high-ionic-strength buffer. Representative kymographs are shown in each case; the Atto 488-labeled minus-ends of the MTs are on the left side in all panels. Vertical scale bars: 8 s; horizontal scale bars: 3 μm. D–F, histograms of the velocity distribution from the segmentation analysis of all corresponding kymographs. D, Cin8Kin, average velocity for high-ionic-strength buffer. vavg = 1 ± 18 nm/s for plus-end-directed motion (red); v+ = 165 ± 10 nm/s, n = 102 and minus-end-directed motion (gray); v− = −172 ± 10 nm/s, n = 106. E, Cin8Kin, average velocity for low-ionic-strength buffer. vavg = 18 ± 5 nm/s for plus-end-directed motion; v+ = 102 ± 3 nm/s, n = 397 and minus-end-directed motion; v− = −126 ± 6 nm/s, n = 256. F, Cin8Δtail, average velocity for high-ionic-strength buffer. vavg = 7 ± 3 nm/s for plus-end-directed motion; v+ = 78 ± 4 nm/s, n = 184 and minus-end-directed motion; v− = −80 ± 3 nm/s, n = 134. Velocities in the range between −25 nm/s and 25 nm/s are indicated in green. G–I, MSD analysis. We linearly fitted the log-log curves above 4 s, beyond localization noise. G, Cin8Kin measured in high-ionic-strength buffer. Exponent = 0.99 ± 0.08 (S.E.), n = 10. H, Cin8Kin measured in low-ionic-strength buffer. Exponent = 1.24 ± 0.11 (S.E.), n = 15. I, Cin8Δtail measured in high-ionic-strength buffer. Exponent = 1.46 ± 0.07 (S.E.), n = 21.
It remains to be confirmed that the bidirectional motility was not merely thermally driven diffusion. Controls with ADP showed only brief interactions, strongly suggesting that the bidirectional motion required ATP hydrolysis and was not thermally driven (Fig. 4A).
FIGURE 4.
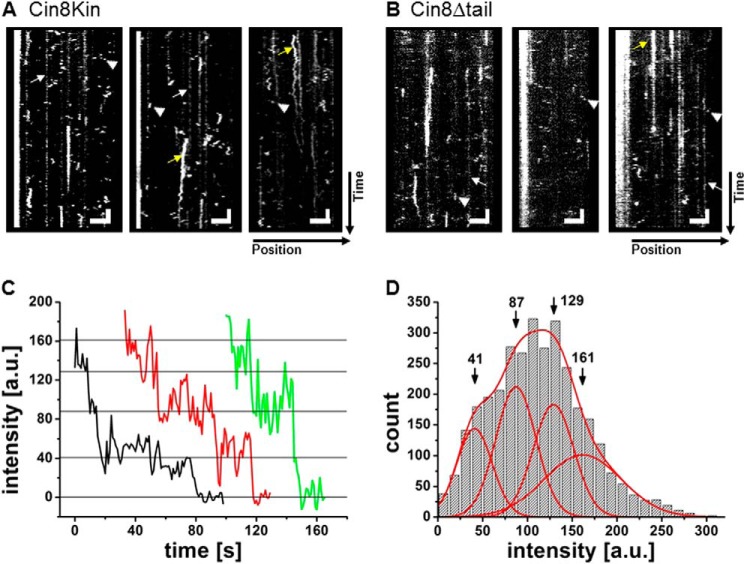
Control experiments: motion in ADP buffer and photobleaching. A and B, kymographs of single-molecule motility assays of Cin8Kin (A) and Cin8Δtail (B) on surface-immobilized MTs in the presence of 2 mm ADP. Cin8Δtail was measured in high-ionic-strength buffer, and Cin8Kin was measured in low-ionic-strength buffer. Brief binding interactions, lasting typically for one frame only, correspond to single motors for which no processive motion is apparent (arrowheads). White arrows point to less intense non-mobile fluorescent spots on the surface that are not Cin8 particles because they largely lack photobleaching. Yellow arrows are high intensity photobleachable spots, present in both Cin8Kin and Cin8Δtail samples, that likely represent higher order Cin8 aggregates that bind for extended times. Vertical scale bars: 8 s; horizontal scale bars: 3 μm. C and D, photobleaching experiments. C, photobleaching-step analysis. Representative photobleaching curves for three Cin8Δtail molecules are shown. a. u., arbitrary units. D, fluorescence intensity distribution for all recorded Cin8Δtail molecules. Indicated are the positions of the intensity peaks corresponding to one to four GFP fluorophores on one protein. The maximum of the distribution is between two and three, which rules out larger aggregates of the tetrameric motor.
The fact that the motor does not bind as strongly in the presence of ADP as in the presence of ATP does not strictly exclude thermal diffusion in the ATP case, however. Therefore, we looked for further proof. A commonly used method to differentiate directed non-equilibrium transport from thermal motion or simple diffusion is the analysis of the ensemble- and or time-averaged MSD of the motor (16). For Cin8Kin, this analysis showed power-law behavior of the MSD, with an exponent 0.99 ± 0.08 (S.E.) in 80 mm buffer with 175 mm added salt and an exponent of 1.24 ± 0.11 in standard 80 mm buffer (Fig. 3, G and H). While an exponent of 1 is no proof for pure thermal driving forces in a viscous medium, but can also result from combinations of particular non-equilibrium driving forces and medium response functions (34), an exponent significantly larger than 1 is a clear indication of non-equilibrium driving forces (34). These results therefore further support the notion that the observed bidirectional motion of Cin8Kin is active and ATP-dependent. Thus, our results indicate that although the stalk and the tail of Cin8 affect its directionality, likely by ensuring the correct geometry of the tetrameric complex, the motor domain and neck linker of Cin8 are intrinsically able to actively move along a single MT in both directions.
The Tail of Cin8 Is One of the Factors That Regulate Its Directionality
We next examined the motility of a tailless tetrameric Cin8 variant in which the stalk was present to test whether the presence of the native stalk and the tetrameric conformation, still in the absence of the tails, affect the motor characteristics. To obtain this construct, we removed aa 946–1038 from the sequence of wt Cin8 and fused it to a C-terminal GFP (Cin8Δtail, Fig. 1A). We chose the cut-out section based on sequence homology to other kinesin-5 motors (24).
In contrast to wt Cin8 (Fig. 2), Cin8Δtail showed no motility in buffers with ionic strength below BRB80 + 175 mm KCl. At this ionic strength, we surprisingly found that Cin8Δtail moved processively and with high directional persistence along MTs in both the plus-end and the minus-end direction (Fig. 3C, supplemental Movie M2). Individual molecules appeared to possess “memory” and moved without changing directionality during individual runs (Fig. 3C). This behavior was distinctly different from both wt Cin8 in low-ionic-strength conditions (Fig. 2) and Cin8Kin under high and low-ionic-strength conditions (Fig. 3, A and B). To rule out aggregation as a cause for this behavior, we performed a photobleaching analysis. In fluorescence-intensity time traces of individual dots, we never observed more than four bleaching steps (Fig. 4C), consistent with the presence of maximally four functional GFP molecules in a single tetrameric Cin8Δtail complex. The distribution of fluorescence intensities of individual dots could be fitted with four Gaussians, with an average of ∼3 GFP fluorophores (Fig. 4D), arguing against aggregates.
To quantitatively compare the different degrees of “directional memory” in the different Cin8 variants exhibited, we used velocity autocorrelation analysis (Fig. 5). A short velocity correlation time would indicate a short memory and a long correlation time would indicate increased velocity persistence. The velocity autocorrelation function of wt Cin8 and Cin8Δtail at high ionic strength decayed with a time constant of τ ≈ 8 s (wt Cin8, τ = 8.9 ± 0.8 s; Cin8Δtail, τ = 7.4 ± 1.0 s) to a plateau, whereas the velocity autocorrelation functions of the dimeric chimera and of wt Cin8 at low ionic strength decayed more rapidly (τ ≈ 2.5 s (wt Cin8, τ = 3.4 ± 0.3 s; Cin8Kin, τ = 2.0 ± 0.2 s)) to zero. This result reflects the persistent directional motility of wt Cin8 and Cin8Δtail in high ionic strength and the frequent directionality changes of Cin8Kin and wt Cin8 in low-ionic-strength buffer.
FIGURE 5.
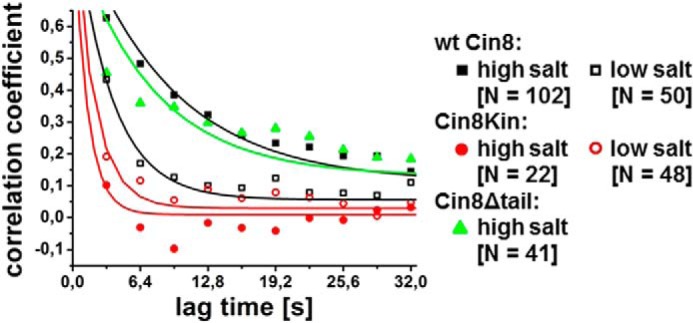
Velocity autocorrelation. Velocity autocorrelation of motility traces of Cin8Δtail, wt Cin8, and Cin8Kin in high and low-ionic-strength buffer is shown. Velocity autocorrelation was calculated from kymographs at a window size of 4 frames (3.2 s) after segmented velocity analysis and fitted with single exponentials. Shown are averaged autocorrelation curves. For Cin8Kin and wt Cin8 in low-ionic-strength buffer, the autocorrelation function decays rapidly to zero, whereas Cin8Δtail and wt Cin8 in high-ionic-strength buffer show a longer velocity memory, indicating processive motility over longer intervals. Values for zero lag-time are omitted. N: number of averaged autocorrelation curves.
In view of the fact that wt Cin8 velocity strongly differs between plus-end motility in high salt conditions and minus- and plus-end motility in low salt conditions (17), we examined whether such differences persist for Cin8Δtail. Velocity analysis of Cin8Δtail motility revealed that the average velocities for plus-end motility (78 ± 4 nm/s, (mean ± S.E.)) and minus-end motility (80 ± 3 nm/s) were indistinguishable (Fig. 3F). Overall, we observed slightly more events of plus-end movement than minus-end movement. In the presence of 2 mm ADP, binding was transient, typically <3 s (Fig. 4B), so that velocities could not be obtained.
Although it is evident from the kymographs that motion of Cin8Δtail was directed and not random, we performed MSD analysis to be able to compare these data with the dimer data. MSD analysis of the motion of Cin8Δtail in the presence of ATP shows a power-law exponent of 1.46 ± 0.08 (S.E.) (Fig. 3I), clearly confirming that movement was not merely thermally driven. Run times of individual Cin8Δtail proteins typically exceeded the recording time of 160 s (Fig. 3C). With an average speed of ∼80 nm/s (Fig. 3F), a lower limit for the run length is therefore ∼14 μm, 5–10 times longer than that of wt Cin8 (17). A comparably long run length was reported for a tetrameric kinesin-5/kinesin-1 chimera (35) and may be caused by both sides of the molecule binding to the same MT, which might also apply here. An interesting speculation is that the lack of the tail domains might facilitate the simultaneous binding of the two opposite ends of the tetramer to a single microtubule. This, in turn, would postulate a role for the tail in the wt motor in preventing the binding of the tetramer to only one MT, which would make immediate physiological sense.
The characteristics of the motile properties of the tailless Cin8 tetramer (Figs. 3, C, F, and I, 4, C and D, and 5) thus imply that the tail domain is one of the factors that regulate directionality in the wt motor. Interestingly, the way in which the tailless tetramer interacts with the microtubule can evidently lock the motor in either plus-end or minus-end motility. It is tempting to speculate that the locking mechanism has to do with how the second set of heads binds with respect to the first.
The Tail of Cin8 Is Required for MT Cross-linking
The mitotic functions of kinesin-5 motors depend on their ability to cross-link and slide apart antiparallel spindle MTs (7, 9, 17). Binding between two antiparallel MTs is the major factor that affects the switching of the direction of movement of Cin8 (17, 18). Because the kinesin-5 tails are known to be able to interact with MTs (24), we next examined whether Cin8Δtail can cross-link and slide apart antiparallel MTs. First, we performed an experiment in which we mixed wt Cin8 or Cin8Δtail with MTs in the presence of AMP-PNP, inducing stable binding of the motors to the MTs. Although we observed extensive bundling of MTs by wt Cin8 (Fig. 6A), almost no bundling of MTs was observed in the presence of Cin8Δtail (Fig. 6B). The tail of Cin8 is thus important even for static cross-linking of MTs.
FIGURE 6.
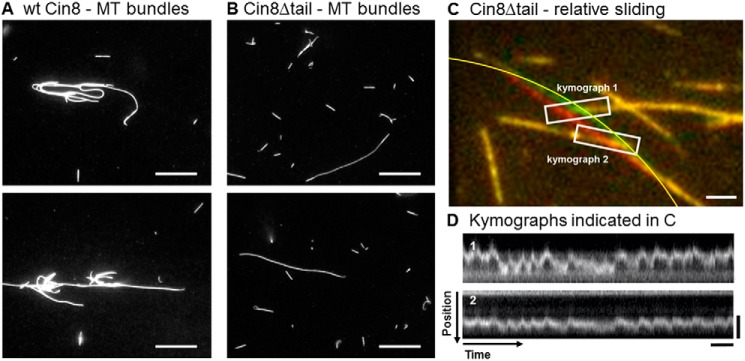
MT cross-linking and relative sliding. A and B, MT cross-linking experiment with wt Cin8 (A) and Cin8Δtail (B). In the presence of AMP-PNP, motors were mixed with MTs in high-ionic-strength buffer. Incubation for 5 min allowed the motors to bind to and cross-link MTs. MT bundles on the DETA surface were imaged with an epifluorescence microscope. Scale bars: 10 μm. C and D, relative-sliding assay with Cin8Δtail. C, overlay of two consecutive frames (one in red, the other in green) from a video recording of a relative-sliding experiment in the presence of Ase1 (4.1 nm) as an MT cross-linker. Surface-attached immobile MT segments appear yellow, whereas a motile segment is visible in two positions (red and green). The long MT containing the motile segment was fixed at both ends and crossed two immobilized, shorter MTs. In the overlap zone, actively driven, approximately periodic lateral motion of the mobile segment over the immobilized MTs was observed. From the contour of the bent MT, approximated by the yellow segment of a circle, we estimated bending energy. Scale bar: 2 μm. D, kymographs of the movement of the motile MT in the two overlap zones as indicated by the white boxes in C. Vertical scale bars: 4 μm; horizontal scale bars: 20 s.
To test dynamic cross-linking, we next performed relative-sliding assays with Cin8Δtail by letting MTs in solution interact with surface-immobilized MTs in the presence of ATP. Because bundling of MTs was not at all possible by Cin8Δtail alone in the presence of ATP (data not shown), we loosely cross-linked the MTs by adding ∼4 nm of the conserved MT-bundling and spindle midzone-organizing protein Ase1 (36–38), which is able to diffuse along the MT lattice while stably cross-linking two MTs (39, 40). Only after the addition of Ase1 could we detect active relative motion of crossing MTs, likely driven by intermittent interactions of Cin8Δtail with the two MTs at the same time (supplemental Movie M3).
In Fig. 6C, an overlay of two video frames (red and green) from a relative-sliding assay is shown. As is evident from the kymographs in Fig. 6D, the central MT that crosses two other MTs moved significantly and in an intermittent, oscillatory fashion. To determine whether this motion was indeed motor-driven and not merely thermally driven, we estimated the elastic energy required to bend the MT to the amplitude observed (Fig. 6C). The bending energy was estimated as
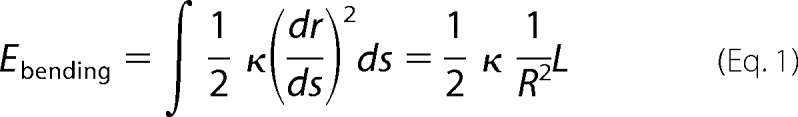 |
where κ denotes the bending stiffness of the MT, R is the radius of curvature of the MT near the crossing point, and L is the length of the bent MT. We estimated a radius of curvature of 20.5 μm over a length of 11.5 μm in this experiment. Assuming a conservative value for the bending stiffness of 10−23 Nm2 (newton meters) (41), we calculated a maximal bending energy of ∼35 kBT. This value is significantly above the available thermal energy. Therefore, thermal fluctuations can be eliminated as an explanation for the observed motions. These relative-sliding assays indicate that, although Cin8Δtail is less efficient in capturing and cross-linking two MTs when compared with the wt Cin8, it is still in principle able to displace two MTs relative to each other, but with short persistence.
The Tail of Cin8 Regulates Its Intracellular Localization and Function
How does the elimination of the tails affect Cin8 in vivo functions in S. cerevisiae? We first imaged the localization of 3GFP-tagged variants of full-length Cin8 (Fig. 1B), co-expressed with Spc42-tdTomato to visualize the SPBs. Consistent with previous studies (1), we found that wt Cin8 localizes to the mitotic spindle within the nucleus between the two SPBs (Fig. 7A). Because the tail of Cin8 (aa 946–1038) contains its nuclear localization sequence (NLS, aa 1031–1038) (42), Cin8Δtail is expected to remain outside the nucleus. We first compared the localization of Cin8Δtail to a variant in which we deleted only the NLS, leaving the rest of the tail intact (Cin8Δnls). The majority of Cin8Δnls localized close to the SPBs (Fig. 7B), likely at the minus-ends of the cytoplasmic MTs, consistent with Cin8 moving toward the minus-ends of the cytoplasmic MTs (17–19). In contrast, Cin8Δtail decorated cytoplasmic MTs along their length as well as SPBs and was concentrated at the distal plus-ends of cytoplasmic MTs (Fig. 7C). A similar localization pattern was found for the S. cerevisiae kinesin-8 homolog, Kip3 (43), which accumulates at plus-ends of MTs in vitro (44) and in vivo (45). Thus, the in vivo localization pattern of Cin8Δtail is consistent with increased movement to the plus-ends of cytoplasmic MTs, suggesting that the C-terminal tail of Cin8 is important for regulating directionality also in vivo.
FIGURE 7.
In vivo localization and viability of Cin8 variants. A–D, deletion of the tail of Cin8 shifts its localization to the plus-ends of the MTs. Representative two-dimensional projections of cells co-expressing GFP-tagged variants of wt Cin8 and the SPB-localizing Spc42-tdTomato are shown. Cin8 variants are indicated on the top of each panel (see Fig. 1B for description). A, wt Cin8. B, Cin8Δnls. C, Cin8Δtail. D, Cin8Δtail-SV40. Orange arrows point toward the SPBs; white arrows point to the accumulation of Cin8 at the plus-ends of cytoplasmic MTs. Bottom row: model for intracellular localization of the different variants of Cin8. E, viability of cin8Δkip1Δ cells expressing vector only or 3GFP-tagged variants of Cin8 from a CEN plasmid. Cells were grown in serial dilutions on either YPD plates (left) or YPD plates containing 7.5 μg/ml cycloheximide (CYH, right).
Because Cin8Δtail localizes outside the nucleus (Fig. 7C), it is in the wrong place to perform any of the normal mitotic functions of wt Cin8 (2, 42). Therefore, we created a nuclear version of tailless Cin8 by adding an SV40 NLS at the C terminus of Cin8Δtail (Fig. 1B, Cin8Δtail-SV40). As expected, this variant localized to the mitotic spindle in the nucleus, similarly to wt Cin8 (Fig. 7, A and D). To test the functionality of Cin8Δtail-SV40 in this location, we examined whether it could support viability of S. cerevisiae cells, being the sole kinesin-5. We used a S. cerevisiae shuffle strain that carries chromosomal deletions of CIN8 and KIP1, covered by a wt Cin8 plasmid that can be shuffled out by growth on cycloheximide (1, 8). GFP-tagged Cin8 variants were transformed into this shuffle strain, and the viability of the transformed strain was observed on cycloheximide and compared with control cells transformed with plasmid expressing wt Cin8 (Fig. 7E). Cells expressing only Cin8Δnls or Cin8Δtail were not viable, consistent with motor localization outside the nucleus (42). Importantly, cells expressing Cin8Δtail-SV40, which localized to the nucleus (Fig. 7, A and D), were also not viable on cycloheximide (Fig. 7E), indicating that this variant is not able to provide the essential functions of Cin8. We conclude that the tail of Cin8 is essential for its intracellular function and that the motile properties of Cin8Δtail do not enable it to perform these functions in vivo.
In this study, we have identified the stalk and the tail domains as one of the crucial structural elements of Cin8 that control directionality switching. Thus far, three kinesin-5 homologs were found to be bidirectional: S. cerevisiae Cin8 (17–19) and Kip1 (46) and Schizosaccharomyces pombe Cut7 (47). At least for Cin8, the switch between minus- to plus-end-directed motility appears to be affected by simultaneous binding between antiparallel MTs (17, 18). The stalk and the tail domains thus appear to be involved in transmitting a signal between the two ends of the tetramer. It is tempting to speculate that this allosteric signaling might use torsional twist in the molecule. This is based on the finding that the two pairs of catalytic heads are rotated by 90° around the motor's long axis to each other in a relaxed state (26). Binding between antiparallel MTs via the tail domains, which appear to be more rigidly connected to the stalk than the heads, must thus cause twist in the stalk that might serve as the switch mechanism (34). Although more pieces in the puzzle are coming together, more dynamic experiments and structural studies are needed to solve the fascinating question of how kinesin-5 motors can sense their binding geometry and adapt their motility in such a complex manner.
Supplementary Material
Acknowledgments
We thank Kerstin von Roden und Charlotte Willms for help with the protein expression and purification and Ofer Shapira and Nurit Siegler for critical reading of the manuscript. Kymograph analysis was performed using ImageJ macros written by J. Rietdorf and A. Seitz.
This work was supported in part by the Lower Saxony Grant 11-76251-99-26/08 (ZN2440) (to L. G. and C. F. S.) and in part by the Deutsche Forschungsgemeinschaft (DFG) through SFB 937 Project A2 as well as by the Center for Nanoscale Microscopy and Molecular Physiology of the Brain (CNMPB) funded by the DFG. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Movies M1–M3.
- MT
- microtubule
- SPB
- spindle pole body
- aa
- amino acid(s)
- TEV
- tobacco etch virus
- NLS
- nuclear localization sequence
- DETA
- 3-[2-(2-aminoethylamino)-ethylamino]propyl-trimethoxysilane
- MSD
- mean-squared displacement
- DmKHC
- D. melanogaster kinesin-1
- AMP-PNP
- 5′-adenylyl-β,γ-imidodiphosphate.
References
- 1. Avunie-Masala R., Movshovich N., Nissenkorn Y., Gerson-Gurwitz A., Fridman V., Kõivomägi M., Loog M., Hoyt M. A., Zaritsky A., Gheber L. (2011) Phospho-regulation of kinesin-5 during anaphase spindle elongation. J. Cell Sci. 124, 873–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gerson-Gurwitz A., Movshovich N., Avunie R., Fridman V., Moyal K., Katz B., Hoyt M. A., Gheber L. (2009) Mid-anaphase arrest in S. cerevisiae cells eliminated for the function of Cin8 and dynein. Cell. Mol. Life Sci. 66, 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fridman V., Gerson-Gurwitz A., Movshovich N., Kupiec M., Gheber L. (2009) Midzone organization restricts interpolar microtubule plus-end dynamics during spindle elongation. EMBO Rep. 10, 387–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Movshovich N., Fridman V., Gerson-Gurwitz A., Shumacher I., Gertsberg I., Fich A., Hoyt M. A., Katz B., Gheber L. (2008) Slk19-dependent mid-anaphase pause in kinesin-5-mutated cells. J. Cell Sci. 121, 2529–2539 [DOI] [PubMed] [Google Scholar]
- 5. Sharp D. J., Rogers G. C., Scholey J. M. (2000) Microtubule motors in mitosis. Nature 407, 41–47 [DOI] [PubMed] [Google Scholar]
- 6. Gordon D. M., Roof D. M. (2001) Degradation of the kinesin Kip1p at anaphase onset is mediated by the anaphase-promoting complex and Cdc20p. Proc. Natl. Acad. Sci. U.S.A. 98, 12515–12520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kashina A. S., Rogers G. C., Scholey J. M. (1997) The bimC family of kinesins: essential bipolar mitotic motors driving centrosome separation. Biochim. Biophys. Acta 1357, 257–271 [DOI] [PubMed] [Google Scholar]
- 8. Gheber L., Kuo S. C., Hoyt M. A. (1999) Motile properties of the kinesin-related Cin8p spindle motor extracted from Saccharomyces cerevisiae cells. J. Biol. Chem. 274, 9564–9572 [DOI] [PubMed] [Google Scholar]
- 9. Kapitein L. C., Peterman E. J. G., Kwok B. H., Kim J. H., Kapoor T. M., Schmidt C. F. (2005) The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature 435, 114–118 [DOI] [PubMed] [Google Scholar]
- 10. Mayer T. U., Kapoor T. M., Haggarty S. J., King R. W., Schreiber S. L., Mitchison T. J. (1999) Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science 286, 971–974 [DOI] [PubMed] [Google Scholar]
- 11. Saunders W. S., Hoyt M. A. (1992) Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell 70, 451–458 [DOI] [PubMed] [Google Scholar]
- 12. Sharp D. J., McDonald K. L., Brown H. M., Matthies H. J., Walczak C., Vale R. D., Mitchison T. J., Scholey J. M. (1999) The bipolar kinesin, KLP61F, cross-links microtubules within interpolar microtubule bundles of Drosophila embryonic mitotic spindles. J. Cell Biol. 144, 125–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Saunders W. S., Koshland D., Eshel D., Gibbons I. R., Hoyt M. A. (1995) Saccharomyces cerevisiae kinesin- and dynein-related proteins required for anaphase chromosome segregation. J. Cell Biol. 128, 617–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lakämper S., Thiede C., Düselder A., Reiter S., Korneev M. J., Kapitein L. C., Peterman E. J., Schmidt C. F. (2010) The effect of monastrol on the processive motility of a dimeric kinesin-5 head/kinesin-1 stalk chimera. J. Mol. Biol. 399, 1–8 [DOI] [PubMed] [Google Scholar]
- 15. Düselder A., Thiede C., Schmidt C. F., Lakämper S. (2012) Neck-linker length dependence of processive Kinesin-5 motility. J. Mol. Biol. 423, 159–168 [DOI] [PubMed] [Google Scholar]
- 16. Kapitein L. C., Kwok B. H., Weinger J. S., Schmidt C. F., Kapoor T. M., Peterman E. J. G. (2008) Microtubule cross-linking triggers the directional motility of kinesin-5. J. Cell Biol. 182, 421–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gerson-Gurwitz A., Thiede C., Movshovich N., Fridman V., Podolskaya M., Danieli T., Lakämper S., Klopfenstein D. R., Schmidt C. F., Gheber L. (2011) Directionality of individual kinesin-5 Cin8 motors is modulated by loop 8, ionic strength and microtubule geometry. EMBO J. 30, 4942–4954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Roostalu J., Hentrich C., Bieling P., Telley I. A., Schiebel E., Surrey T. (2011) Directional switching of the kinesin Cin8 through motor coupling. Science 332, 94–99 [DOI] [PubMed] [Google Scholar]
- 19. Thiede C., Fridman V., Gerson-Gurwitz A., Gheber L., Schmidt C. F. (2012) Regulation of bi-directional movement of single kinesin-5 Cin8 molecules. Bioarchitecture 2, 70–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tytell J. D., Sorger P. K. (2006) Analysis of kinesin motor function at budding yeast kinetochores. J. Cell Biol. 172, 861–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wargacki M. M., Tay J. C., Muller E. G., Asbury C. L., Davis T. N. (2010) Kip3, the yeast kinesin-8, is required for clustering of kinetochores at metaphase. Cell Cycle 9, 2581–2588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hackney D. D. (1995) Highly processive microtubule-stimulated ATP hydrolysis by dimeric kinesin head domains. Nature 377, 448–450 [DOI] [PubMed] [Google Scholar]
- 23. Kaan H. Y., Hackney D. D., Kozielski F. (2011) The structure of the kinesin-1 motor-tail complex reveals the mechanism of autoinhibition. Science 333, 883–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Weinger J. S., Qiu M., Yang G., Kapoor T. M. (2011) A nonmotor microtubule binding site in kinesin-5 is required for filament crosslinking and sliding. Curr. Biol. 21, 154–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Acar S., Carlson D. B., Budamagunta M. S., Yarov-Yarovoy V., Correia J. J., Niñonuevo M. R., Jia W., Tao L., Leary J. A., Voss J. C., Evans J. E., Scholey J. M. (2013) The bipolar assembly domain of the mitotic motor kinesin-5. Nat. Commun. 4, 1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Scholey J. E., Nithianantham S., Scholey J. M., Al-Bassam J. (2014) Structural basis for the assembly of the mitotic motor Kinesin-5 into bipolar tetramers. Elife 3, e02217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fakhri N., Schmidt C. F. (2014) A surprising twist. Elife 3, e02715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sherman F., Fink G. R., Hicks J. B. (1986) Methods in Yeast Genetics, Cold Spring Harbor Laboratory Press, New York [Google Scholar]
- 29. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, New York [Google Scholar]
- 30. Waldo G. S., Standish B. M., Berendzen J., Terwilliger T. C. (1999) Rapid protein-folding assay using green fluorescent protein. Nat. Biotechnol. 17, 691–695 [DOI] [PubMed] [Google Scholar]
- 31. Hovland P., Flick J., Johnston M., Sclafani R. A. (1989) Galactose as a gratuitous inducer of GAL gene expression in yeasts growing on glucose. Gene 83, 57–64 [DOI] [PubMed] [Google Scholar]
- 32. Janson M. E., Loughlin R., Loïodice I., Fu C., Brunner D., Nédélec F. J., Tran P. T. (2007) Crosslinkers and motors organize dynamic microtubules to form stable bipolar arrays in fission yeast. Cell 128, 357–368 [DOI] [PubMed] [Google Scholar]
- 33. Sbalzarini I. F., Koumoutsakos P. (2005) Feature point tracking and trajectory analysis for video imaging in cell biology. J. Struct. Biol 151, 182–195 [DOI] [PubMed] [Google Scholar]
- 34. Fakhri N., Wessel A. D., Willms C., Pasquali M., Klopfenstein D. R., MacKintosh F. C., Schmidt C. F. (2014) High-resolution mapping of intracellular fluctuations using carbon nanotubes. Science 344, 1031–1035 [DOI] [PubMed] [Google Scholar]
- 35. Thiede C., Lakämper S., Wessel A. D., Kramer S., Schmidt C. F. (2013) A chimeric kinesin-1 head/kinesin-5 tail motor switches between diffusive and processive motility. Biophys. J. 104, 432–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pellman D., Bagget M., Tu Y. H., Fink G. R., Tu H. (1995) Two microtubule-associated proteins required for anaphase spindle movement in Saccharomyces cerevisiae. J. Cell Biol. 130, 1373–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. de Gramont A., Barbour L., Ross K. E., Cohen-Fix O. (2007) The spindle midzone microtubule-associated proteins Ase1p and Cin8p affect the number and orientation of astral microtubules in Saccharomyces cerevisiae. Cell Cycle 6, 1231–1241 [DOI] [PubMed] [Google Scholar]
- 38. Khmelinskii A., Schiebel E. (2008) Assembling the spindle midzone in the right place at the right time. Cell Cycle 7, 283–286 [DOI] [PubMed] [Google Scholar]
- 39. Braun M., Lansky Z., Fink G., Ruhnow F., Diez S., Janson M. E. (2011) Adaptive braking by Ase1 prevents overlapping microtubules from sliding completely apart. Nat. Cell Biol. 13, 1259–1264 [DOI] [PubMed] [Google Scholar]
- 40. Kapitein L. C., Janson M. E., van den Wildenberg S. M. J. L., Hoogenraad C. C., Schmidt C. F., Peterman E. J. G. (2008) Microtubule-driven multimerization recruits ase1p onto overlapping microtubules. Curr. Biol. 18, 1713–1717 [DOI] [PubMed] [Google Scholar]
- 41. Mickey B., Howard J. (1995) Rigidity of microtubules is increased by stabilizing agents. J. Cell Biol. 130, 909–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hildebrandt E. R., Gheber L., Kingsbury T., Hoyt M. A. (2006) Homotetrameric form of Cin8p, a Saccharomyces cerevisiae kinesin-5 motor, is essential for its in vivo function. J. Biol. Chem. 281, 26004–26013 [DOI] [PubMed] [Google Scholar]
- 43. Su X., Qiu W., Gupta M. L., Jr., Pereira-Leal J. B., Reck-Peterson S. L., Pellman D. (2011) Mechanisms underlying the dual-mode regulation of microtubule dynamics by Kip3/kinesin-8. Mol. Cell 43, 751–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Varga V., Leduc C., Bormuth V., Diez S., Howard J. (2009) Kinesin-8 motors act cooperatively to mediate length-dependent microtubule depolymerization. Cell 138, 1174–1183 [DOI] [PubMed] [Google Scholar]
- 45. Gupta M. L., Jr., Carvalho P., Roof D. M., Pellman D. (2006) Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat. Cell Biol. 8, 913–923 [DOI] [PubMed] [Google Scholar]
- 46. Fridman V., Gerson-Gurwitz A., Shapira O., Movshovich N., Lakämper S., Schmidt C. F., Gheber L. (2013) Kinesin-5 Kip1 is a bi-directional motor that stabilizes microtubules and tracks their plus-ends in vivo. J. Cell Sci. 126, 4147–4159 [DOI] [PubMed] [Google Scholar]
- 47. Edamatsu M. (2014) Bidirectional motility of the fission yeast kinesin-5, Cut7. Biochem. Biophys. Res. Commun. 446, 231–234 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.